Abstract
Dysbindin is a schizophrenia susceptibility factor and subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) required for lysosome-related organelle biogenesis, and in neurons, synaptic vesicle assembly, neurotransmission, and plasticity. Protein networks, or interactomes, downstream of dysbindin/BLOC-1 remain partially explored despite their potential to illuminate neurodevelopmental disorder mechanisms. Here, we conducted a proteome-wide search for polypeptides whose cellular content is sensitive to dysbindin/BLOC-1 loss of function. We identified components of the vesicle fusion machinery as factors downregulated in dysbindin/BLOC-1 deficiency in neuroectodermal cells and iPSC-derived human neurons, among them the N-ethylmaleimide-sensitive factor (NSF). Human dysbindin/BLOC-1 coprecipitates with NSF and vice versa, and both proteins colocalized in a Drosophila model synapse. To test the hypothesis that NSF and dysbindin/BLOC-1 participate in a pathway-regulating synaptic function, we examined the role for NSF in dysbindin/BLOC-1-dependent synaptic homeostatic plasticity in Drosophila. As previously described, we found that mutations in dysbindin precluded homeostatic synaptic plasticity elicited by acute blockage of postsynaptic receptors. This dysbindin mutant phenotype is fully rescued by presynaptic expression of either dysbindin or Drosophila NSF. However, neither reduction of NSF alone or in combination with dysbindin haploinsufficiency impaired homeostatic synaptic plasticity. Our results demonstrate that dysbindin/BLOC-1 expression defects result in altered cellular content of proteins of the vesicle fusion apparatus and therefore influence synaptic plasticity.
Keywords: BLOC-1, DTNBP1, dysbindin, NSF, schizophrenia, SILAC
Introduction
Genetic polymorphisms associated with neurodevelopmental disorders frequently reside in noncoding regions, arguing that most genetic variants modify gene and/or protein levels rather than protein primary structure (Nicolae et al., 2010; Maurano et al., 2012; Richards et al., 2012; Schizophrenia Working Group of the Psychiatric Genomics, 2014). This is the case with genetic polymorphisms in the gene encoding dysbindin, DTNBP1, which are risk factors for schizophrenia onset (Straub et al., 2002; Van Den Bogaert et al., 2003; Bray et al., 2005; Mullin et al., 2011; Ghiani and Dell'Angelica, 2011; Ayalew et al., 2012). DTNBP1 polymorphisms reside in noncoding regions of the dysbindin gene and are thought to produce moderate effects in polypeptide expression (Talbot et al., 2009). This observation is in rapport with postmortem studies of adult schizophrenia brains, which reveal a partial reduction in dysbindin transcripts and protein (Talbot et al., 2004; Weickert et al., 2008). Nearly 80% of schizophrenia subjects possess 30–50% reduced dysbindin protein content in neuroanatomical regions affected by this disorder (Talbot et al., 2004, 2011). The cellular consequences of DTNBP1 polymorphisms remain unknown, yet vertebrate and invertebrate animal models show that reductions of 50% in the content of dysbindin are sufficient to generate molecular and functional phenotypes in neurons and the synapse (Jentsch et al., 2009; Karlsgodt et al., 2011; Larimore et al., 2014). These findings argue for partial loss of function in dysbindin as a penetrant link connected to the chain of events associated with schizophrenia development.
Dysbindin associates with seven other polypeptides to form the biogenesis of lysosome-related organelles complex 1 (BLOC-1; Starcevic and Dell'Angelica, 2004; Mullin et al., 2011; Ghiani and Dell'Angelica, 2011). Null mutations in mouse dysbindin reduce the expression of other BLOC-1 subunit mRNAs and polypeptides (Mullin et al., 2011; Ghiani and Dell'Angelica, 2011; Larimore et al., 2014). This suggests that dysbindin genetic downregulation could elicit multiple alterations of protein content in cells (Foss et al., 2007; Albert et al., 2014). We identified 224 proteins whose content was modified by dysbindin/BLOC-1 partial loss of function using unbiased quantitative mass spectrometry. Our screen prominently identified components of the N-ethylmaleimide-sensitive factor (NSF)-dependent vesicle fusion machinery. We focused on NSF, a catalytic component of the fusion machinery, and asked whether NSF participates in dysbindin/BLOC-1-dependent synaptic mechanisms. We used as an assay Drosophila presynaptic plasticity produced by the inhibition of postsynaptic receptors. As previously reported by Dickman and Davis (2009) and Dickman et al., (2012), we observed that mutations in fly dysbindin precluded the establishment of homeostatic synaptic plasticity, a phenotype that we rescued by presynaptic expression of dysbindin (Dickman and Davis, 2009; Dickman et al., 2012). Neuron-specific expression of dNSF1, the gene encoding Drosophila NSF, by itself does not modulate this form of plasticity, yet NSF1 expression at the synapse of dysbindin mutants rescued homeostatic synaptic plasticity defects to the same extent as dysbindin re-expression in the presynaptic compartment. Our results demonstrate that partial dysbindin/BLOC-1 loss of function alters the cellular content of proteins that specifically have roles in synaptic mechanisms.
Materials and Methods
Antibodies cell culture.
Antibodies are listed in Table 1. SH-SY5Y (ATCC) cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin and streptomycin (Hyclone) at 37°C in 10% CO2. The SH-SY5Y cell line carrying 3x-FLAG Dysbindin (catalog #EX-Mm12550-M12) was previously described (Gokhale et al., 2012). MNT-1 cells were a gift of Dr. Vincent Hearing (National Cancer Institute, NIH, Bethesda, MD) (Kushimoto et al., 2001). Cells were maintained in DMEM supplemented with 20% AIM-V medium (Life Technologies), 10% FBS (heat inactivated at 65°C for 60 min), and 100 μg/ml penicillin/streptomycin at 37°C and 5% CO2. Bloc1s5mu/mu, Bloc1s6pa/pa, and rescued melanocytes were a gift of Dr. Michael Marks (Department of Pathology & Laboratory Medicine, University of Pennsylvania, Philadelphia, PA) (Setty et al., 2007, 2008).
Table 1.
Antibodies used
| Antibody | Catalog number | Source | Dilutions |
|
|---|---|---|---|---|
| Blot | Immunofluorescence | |||
| Polyclonal anti-muted | Dell'Angelica laboratory | 1:1000 | ||
| Polyclonal anti-pallidin | 10891-AP | Proteintech | 1:1000 | |
| Monoclonal anti-pallidin (2G6) | Dell'Angelica laboratory | 1:500 | ||
| Polyclonal anti-dysbindin | HPA029616 | Sigma-Aldrich | 1:125 | |
| Polyclonal anti-NSF (D31C7) | 3924 | Cell Signaling Technology | 1:1000 | |
| Monoclonal anti-Actin (AC-15) | A5451 | Sigma-Aldrich | 1:1000 | |
| Polyclonal anti-GFP | 132002 | Synaptic Systems | 1:2000 | |
| Monoclonal anti-FLAG (M2) | F3165 | Sigma-Aldrich | 1:1000 | |
| Polyclonal anti-FLAG | A190-102A | Bethyl Laboratories | 1:1000 | |
| Monoclonal VAMP7 | A.A. Peden | |||
| Monoclonal anti-TrFr (H84) | 12-6800 | Zymed | 1:1000 | |
| Monoclonal anti-tomosyn | 611296 | BD Biosciences | 1:500 | |
| Monoclonal anti-Munc18 | 610336 | BD Biosciences | 1:2000 | |
| Polyclonal anti-Syntaxin 7 | A.A. Peden | 1:1000 | ||
| Monoclonal MAP2 | M1406 | Sigma-Aldrich | 1:1000 | |
| Polyclonal B3 tubulin | 5568 | Cell Signaling Technology | 1:200 | |
Amniotic epithelial (AE) cells-induced pluripotent stem cells (iPSCs) were obtained and cultured on human embryonic stem cell-qualified Matrigel (BD Biosciences) with mTeSR-1 medium (STEMCELL Technologies), as described previously (Easley et al., 2012). The AE cells-iPSCs were grown for a minimum of 5 d before being infected by the control and pallidin lentiviral particles for 7 d in selection media, as described above. After the 7 d treatment, the iPSCs were lifted and placed in a neural stem cell medium (DMEM F-12 +N2 supplement) containing high concentrations of EGF and FGF-2 (100 ng/ml; Peprotech), and heparin (5 μg/ml; Sigma-Aldrich) to produce cell aggregates termed EZ spheres (Ebert et al., 2013). To induce neuronal differentiation EZ spheres were removed from the N2/EGF/FGF/heparin media and differentiated as follows: EZ spheres were cultured on poly-d-lysine/laminin-coated, acid-etched glass coverslips for immunofluorescence, or 10 cm culture dishes for biochemical analyses in Neural Differentiation Medium containing Neurobasal, B27, and Glutamax (all from Life Technologies), with 20 ng/ml GDNF (Peprotech) and 10 ng/ml BDNF (Peprotech). Medium changes on differentiating cells occurred weekly. Neuronal differentiation was confirmed by immunofluorescence microscopy using MAP2 and β-III-tubulin as neuronal markers.
For shRNA-mediated Bloc1s5 muted and Bloc1s6 pallidin knockdowns, shRNA in a pLKO.1 vector for lentiviral infection was obtained from Open Biosystems (Pallidin, Clone ID: TRCN0000122781; Muted, Clone ID: TRCN0000128812). Control shRNA in pLKO.1 was obtained from Addgene (vector 1864). For shRNA-mediated dysbindin knockdowns, shRNA in a psiHIV-U6 vector for lentiviral infection was obtained from Genecopoeia (Dysbindin, catalog #HSH020444-1HIVU6). Control shRNA in a psiHIV-U6 vector was also obtained from Genecopoeia (Control, catalog #CSCHCTR001-HIVU6). SH-SY5Y cells were treated with lentiviral particles for 7 d to obtain efficient knockdown. After day 3 of lentiviral infection, cells were maintained in DMEM supplemented with 10% FBS and selected with puromycin (2 μg/ml; Invitrogen).
Stable isotope labeling by amino acids in cell culture labeling and mass spectrometry analysis.
SH-SY5Y cells were labeled using the protocol described (Gokhale et al., 2012; Ryder et al., 2013). Briefly, cells were grown in DMEM with either “light” unlabeled arginine and lysine amino acids (R0K0) or “heavy” 13C- and 15N-labeled arginine, and 13C- and 15N-labeled lysine amino acids (R10K8) supplemented with 10% FBS and 100 μg/ml penicillin and streptomycin, and in some cases 2 μg/ml neomycin. Cells were grown for a minimum of seven passages ensuring maximum incorporation (97.5%) of the amino acids in the cellular proteins. All reagents for stable isotope labeling by amino acids in cell culture (SILAC) labeling were obtained from Dundee Cell Products. Cell lysates were prepared, as described below, and analyzed by mass spectrometry, as described previously (Gokhale et al., 2012; Ryder et al., 2013) using the services of MS Bioworks. Briefly, SILAC-labeled samples were separated on a 4–12% Bis-Tris Novex mini-gel (Invitrogen) using the MOPS [3-(N-morpholino)propanesulfonic acid] buffer system. The gel was stained with Coomassie blue, and the lane was excised into 20 equal segments using a grid. Gel pieces were processed using a robot (ProGest, DigiLab) with the following protocol. First, slices were washed with 25 mm ammonium bicarbonate followed by acetonitrile; then they were reduced with 10 mm dithiothreitol at 60°C followed by alkylation with 50 mm iodoacetamide at room temperature. Samples were digested with trypsin at 37°C for 4 h and quenched with formic acid, and the supernatant was analyzed directly without further processing. Each gel digest was analyzed by nano liquid chromatography-tandem mass spectrometry (MS/MS) with a Waters nanoAcquity HPLC system interfaced to a Thermo Fisher Scientific LTQ Orbitrap Velos. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350 nl/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with MS performed in the Orbitrap at 60,000 FWHM resolution and MS/MS performed in the LTQ. The 15 most abundant ions were selected for MS/MS. Data were processed through MaxQuant software version 1.0.13.13, which served the data recalibration of MS, filtering of database search results at the 1% protein and peptide false discovery rate, and calculation of SILAC heavy/light ratios. Data were searched using a local copy of Mascot.
Immunoprecipitation.
To confirm interactions between BLOC-1 subunits and NSF, we performed cross-linking in intact cells with dithiobis succinimidylpropionate (DSP) followed by immunoprecipitation as previously described (Zlatic et al., 2010; Gokhale et al., 2012; Ryder et al., 2013). Briefly, untransfected SH-SY5Y cells or SH-SY5Y cells stably transfected with FLAG-dysbindin were placed on ice, rinsed twice with PBS, and incubated either with 10 mm DSP (Pierce), or as a vehicle control DMSO, diluted in PBS for 2 h on ice. Tris, pH 7.4, was added to the cells for 15 min to quench the DSP reaction. The cells were then rinsed twice with PBS and lysed in buffer A (150 mm NaCl, 10 mm HEPES, 1 mm EGTA, and 0.1 mm MgCl2, pH 7.4) with 0.5% Triton X-100 and Complete Anti-Protease (catalog #11245200, Roche), followed by incubation for 30 min on ice. Cells were scraped from the dish, and cell homogenates were centrifuged at 16,100 × g for 10 min. The clarified supernatant was recovered, and at least 500 μg of protein extract was applied to 30 μl of DYNAL Magnetic Beads (catalog #110.31, Life Technologies) coated with antibody, and incubated for 2 h at 4°C. In some cases, immunoprecipitation was performed in the presence of the antigenic 3x-FLAG peptide (340 μm; F4799, Sigma) or antigenic GST-NSF (66 nm; H00004905-P01, Novus Biologicals) as a control. The beads were then washed four to six times with buffer A with 0.1% Triton X-100. Proteins were eluted from the beads with Laemmli sample buffer. Samples were resolved by SDS-PAGE, and contents were analyzed by immunoblot.
Sucrose density sedimentation.
Control or BLOC-1 knock-down SH-SY5Y cells were treated with DSP, as described previously (Zlatic et al., 2010; Gokhale et al., 2012). Cells were rinsed twice with PBS and lysed in buffer A with 0.5% Triton X-100 supplemented with Complete Anti-Protease, followed by incubation for 30 min on ice. Cells were scraped from the dish, and cell homogenates were centrifuged at 16,100 × g for 10 min. The clarified supernatant was recovered and measured for total protein content. Samples were then analyzed by immunoblot, or cell lysates were resolved by sucrose sedimentation in 5–30% sucrose gradients, as previously described (Gokhale et al., 2012).
Immunofluorescence.
Glass coverslips were coated with poly-d-lysine. The next day, coverslips were washed two times with cell culture water and air dried. The coverslips were then coated with laminin diluted in HBSS and placed in a 37°C tissue culture incubator for 2 h. The coverslips were washed twice with cell culture water and air dried, and the cells (EZ spheres resuspended in conditioned media) were seeded onto the coverslips. The EZ spheres were grown on the coverslips for 2 weeks. At the end of the 2 weeks, the cells are fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After fixation, all coverslips were washed two times in PBS, and then blocked and permeabilized for 30 min at room temperature in a solution of 5% BSA and 0.1% Triton X-100 in PBS. Primary antibodies (Table 1) were diluted in blocking solution and applied overnight at 4°C. After primary antibody incubation, coverslips were washed three times in a solution containing 0.1% Triton X-100 in PBS. Coverslips were incubated for 1 h at room temperature with fluorophore-conjugated secondary antibodies (Alexa Fluor mouse 488, Alexa Fluor rabbit 568) diluted in blocking solution. Coverslips were washed two times in blocking solution, one time in PBS, and one time in ultrapure water; and then mounted on glass slides in ProLong antifade mounting medium. Coverslips were imaged by confocal microscopy, as described previously (Zlatic et al., 2013).
S2 Drosophila and HeLa cell secretion assay.
We used S2 or HeLa cells stably expressing a secretory reporter carrying a mutant FKBP protein (F36M) EGFP. Secretion was induced by AP21998 to resolve EGFP protein aggregates in a synchronous pulse of reporter secretion. Secretion was estimated from the amount of EGFP fluorescence remaining in cells after siRNA downregulation using flow cytometry or in 96-well fluorescence plate reader (Synergy HT, Biotek). S2 cells were incubated with 20 μg of double-stranded RNA (http://genomernai.dkfz.de/GenomeRNAi/) targeting syntaxin 5 as a positive control to inhibit constitutive secretion, misfire as a negative control (reagents DRSC10543 and AMB34062), and the Drosophila BLOC-1 subunits dysbindin (reagents DRSC10730 and DRSC35459), and muted (reagents DRSC36270 and DRSC36270). S2 cells were maintained for 4 d in the presence of double-stranded RNA. Secretion pulse lasted 80 min. Detailed procedures were already described (Gordon et al., 2010).
Drosophila stocks, rearing, genetics, and biochemical procedures.
All fly stocks were raised throughout life at 25°C and maintained on normal food. Appropriate second and third chromosome balancers were used for all crosses. dysb1A and UAS-Dysb were from Graeme Davis (University of California, San Francisco, San Francisco, CA), and UAS-NSF::FLAG was from Richard Ordway (Penn State University, State College, PA). w1118, Elav-GAL4C155, and other fly strains such as balancer chromosome containing stocks are part of the Sanyal laboratory collection. dNSF1 RNAi lines were obtained from the Vienna Drosophila Resource Center (catalog #31470 and #31666). Antibodies to dNSF1 and α-SNAP were a gift from Leo Pallanck (University of Washington, Seattle, WA). Drosophila neuromuscular junctions were stained as described previously (Franciscovich et al., 2008).
Intracellular recordings from muscle 6 of abdominal segment 2 or 3 of female, wandering third instar larvae were performed in modified HL3 saline solution containing the following (in mm): NaCl 70, KCl 5, CaCl2 0.3, MgCl2 10, NaHCO3 10, sucrose 115, Trehalose 5, BES [2 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid, pH 7.25. For all physiological recordings, severed motor neurons were taken up into a stimulating electrode and stimulated at 1 Hz for 50 s. Only recordings where the resting membrane potential was between −60 and −90 mV, and the muscle input resistance was >5 MΩ were used. For acute pharmacological homeostatic challenge, semi-intact preparations were maintained with the CNS, fat bodies, and gut intact, and perfused with Phillanthatoxin-433 (PhTx; Sigma-Aldrich), as previously described (Dickman and Davis, 2009; Dickman et al., 2012). PhTx was prepared as a stock solution (4 mm in DMSO) and diluted in modified HL3 to 4 μm. After 10 min in PhTx, preparations were rinsed in modified HL3, and dissections were completed. Only recordings where the observed miniature excitatory junction potentials of untreated synapses (mEJP) amplitude following PhTx incubation was ≤60% of baseline were used, indicating that the PhTx had gained access to the muscle. Microelectrodes were prepared on a magnetic glass microelectrode horizontal puller (PN-30, Narishige) to 30–70 MΩ resistance and filled with 3 m KCl. Signals were amplified using an Axoclamp 900A, digitized using a Digidata 1440A, and recorded in Clampfit version 10.1. Signals were analyzed in MiniAnalysis (Synaptosoft Inc.) and Microsoft Excel. Quantal content was calculated by dividing the average EJP by the average mEJP. Correction for nonlinear summation was applied. Quantal content for each recording was calculated and then averaged across all animals for the given genotype.
Fly heads were prepared as described previously (van de Goor et al., 1995) Briefly, ∼100 flies per genotype were flash frozen in liquid nitrogen and decapitated. Frozen heads were collected by passing the tissue through a microsieve in liquid nitrogen. The tissue was then ground into a powder in a mortar and pestle. The powder was combined with 100 μl of lysis buffer [Buffer A (150 mm NaCl, 10 mm HEPES, 1 mm EGTA, and 0.1 mm MgCl2, pH 7.4) plus 0.5% Triton X-100 with Complete Anti-Protease (catalog #11245200, Roche), also was frozen and ground into a powder, and stored at −80°C. After thawing, samples were sonicated, tissue debris was removed by centrifugation, and protein concentrations were determined by Bradford assay (Bio-Rad). Proteins were resolved by SDS-PAGE on a 4–20% gel (Invitrogen), and immunoblot analysis was performed as previously described (Gokhale et al., 2012). dNSF1 and dSNAP antibodies were gifts from Dr. Leo Pallanck (Babcock et al., 2004).
Drosophila immunolabeling.
Immunohistochemical staining of third instar larval neuromuscular junctions was as described previously (Freeman et al., 2011). Briefly, wandering third instar larvae of specific genotypes reared at 25°C were dissected in Ca2+-free HL3 saline solution (Stewart et al., 1994) and fixed in 4% paraformaldehyde for 1 h at room temperature. Preparations were then incubated in primary antibody overnight in PBS containing 0.1% Triton X-100 (PBST). Following washes in PBST, preparations were incubated in Alexa Fluor dye-conjugated secondary antibodies for 2 h at room temperature, washed, and mounted in SlowFade mounting medium (Life Technologies). Primary antibodies to GFP (mouse) and dNSF (rabbit) were used at a 1:1000 dilution (Yu et al., 2011), and FITC-conjugated anti-HRP was used at 1:500 as a control counterstain for quantitative immunofluorescence. Samples were imaged using the Zeiss 710 laser-scanning confocal microscope at the Biogen imaging core. Identical imaging conditions were used for all preparations, taking care to not saturate images in any sample. UAS-NSF::FLAG animals were used to calibrate maximum intensity settings. Large Ib boutons at the muscle 6/7 synapse in abdominal segment A2 were imaged in all cases, and the terminal 3 boutons in a branch were used for analysis. For these three boutons, average intensity was measured using ImageJ for both HRP and NSF, and the NSF staining intensity was normalized to the HRP staining intensity in the same sample to account for sample-to-sample variability. Twelve animals were imaged for each genotype. The anti-dNSF antibody was a gift from Leo Pallanck.
Statistical analysis.
Experimental conditions were compared using Synergy Kaleida-Graph, version 4.1.3, or StatPlus Mac Built5.6.0pre/Universal (AnalystSoft).
Results
Reduction in the abundance of dysbindin/BLOC-1 complex could result in altered protein content in other polypeptides, providing insight into dysbindin/BLOC-1 complex functions relevant to neurodevelopmental disorders. We tested this hypothesis by screening for proteome-wide modifications associated with dysbindin/BLOC-1 partial loss of function in mammalian neuronal cells. We reasoned that studying the whole cellular proteome would identify proteins involved in the genesis or compensation of dysbindin/BLOC-1 partial loss-of-function phenotypes, regardless of whether proteins downstream of dysbindin/BLOC-1 either directly or indirectly interact with this complex.
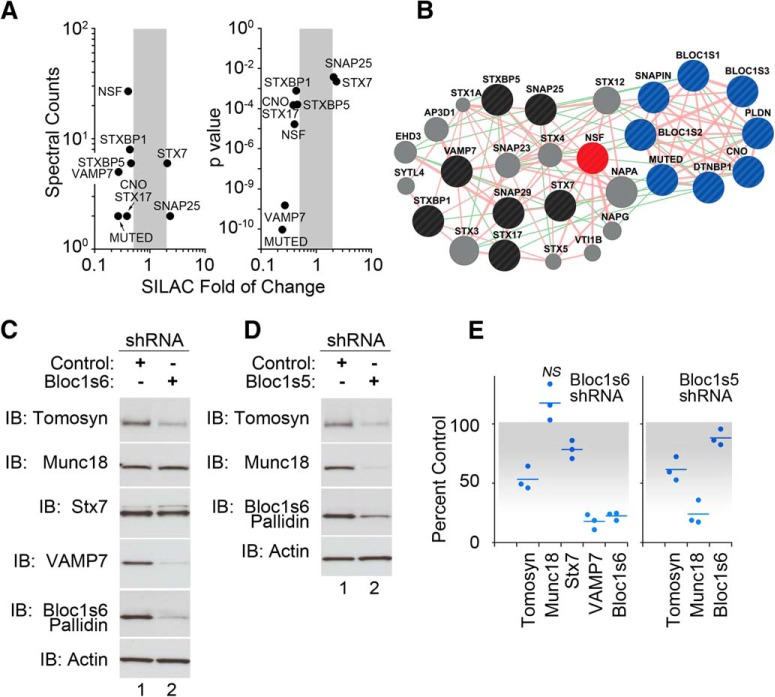
We profiled the whole cellular proteome from control and shRNA BLOC-1 downregulated neuroblastoma cells using shRNAs directed against Bloc1s5 muted or Bloc1s6 pallidin. These hairpins partially reduce the levels of dysbindin by 50–75%, emulating the reduction of dysbindin observed in schizophrenia brains (Talbot et al., 2004, 2011; Ryder et al., 2013). Quantitative mass spectrometry with SILAC identified 224 proteins sensitive to BLOC-1 downregulation defined as proteins upregulated or downregulated at least twofold after BLOC-1 knockdown (our unpublished observations; Ong et al., 2002; Gokhale et al., 2012; Ryder et al., 2013). These proteins included subunits of the BLOC-1 complex (Bloc1s5 muted and Bloc1s4 cappuccino; Fig. 1A). In this study, we confirmed and focused on vesicle fusion machinery components whose content increased or decreased by at least twofold compared with control shRNA-treated cells. These dysbindin/BLOC-1-sensitive proteins include the SNAREs syntaxin 7 and 17, SNAP25, VAMP7; and the SNARE-binding proteins munc18 (STXBP1), tomosyn (STXBP5), and NSF (Fig. 1A; Rizo and Sudhof, 2012). We and others have documented phenotypes of SNAP25, SNAP29, and various syntaxins in BLOC-1/dysbindin-null animals and diverse cells; thus, these proteins were not further studied here (Numakawa et al., 2004; Kumamoto et al., 2006; Salazar et al., 2006a; Ghiani et al., 2010; Newell-Litwa et al., 2010; Larimore et al., 2013; Saggu et al., 2013).
Figure 1.
Fusion apparatus content is altered in BLOC-1 deficiency. A, Plots depict fusion machinery components whose content was modified by targeting the BLOC-1 subunits Bloc1s6 pallidin or Bloc1s5 muted. The x-axis depicts the SILAC fold change; y-axes indicate spectral counts and the p value of the change. B, Interactome map of proteins modified after BLOC-1 downregulation. Pink and green lines depict predicted protein–protein and genetic interactions, respectively, as per GeneMANIA. BLOC-1 complex subunits are in blue, NSF is in red, BLOC-1-sensitive proteins are in black, and proteins are not identified here but are predicted to be part of the interactome, which are shown in gray. C, D, Downregulation of BLOC-1 subunits Bloc1s6 pallidin or Bloc1s5 muted decreases the content of other fusion machinery components, as follows: tomosyn, munc18, syntaxin 7 (Stx7), and VAMP7 in neuroblastoma cells. E, Quantification of data in C and D. All p values <0.05, unless otherwise indicated, are not significant (NS), Wilcoxon–Mann–Whitney rank sum test. Dots represent independent biological replicates.
The BLOC-1 complex subunits along with the fusion machinery form a predicted network of protein–protein/genetic interactions (Fig. 1B), where NSF is a central hub of this interactome and a catalytic activity that is required for the resolution of membrane fusion reaction products (Rizo and Sudhof, 2012).
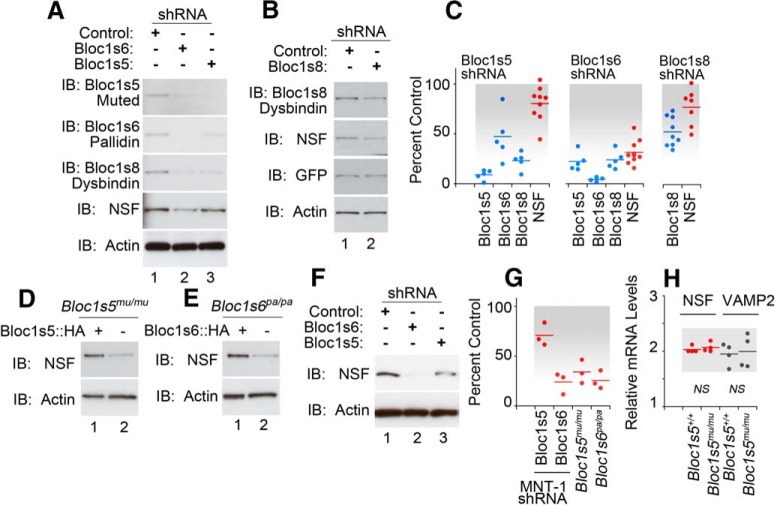
We first confirmed that the total cellular levels of tomosyn, munc18, syntaxin 7, and VAMP7 were significantly reduced in BLOC-1 downregulated SH-SY5Y neuroblastoma cells (Fig. 1C–E). Of these proteins, only munc18 was sensitive to Bloc1s5 muted, but not to Bloc1s6 pallidin, downregulation. This observation is reminiscent of differences in phenotypes among loss-of-function alleles affecting different BLOC-1 subunits in mice and Drosophila (Larimore et al., 2014; Mullin et al., 2015). Similarly, shRNAs against the BLOC-1 subunits Bloc1s5 muted, Bloc1s6 pallidin, or Bloc1s8 dysbindin all significantly reduced the total cellular levels of Bloc1s8 dysbindin and NSF in neuroblastoma cells (Fig. 2A–C). Exogenous expression of VAMP7 did not rescue the NSF phenotype in neuroblastoma cells, indicating that the NSF phenotype is parallel to the VAMP7 content reduction (data not shown). The effects of BLOC-1 downregulation were also observed in two neuroectodermal cell lines of the melanocytic lineage. We chose this lineage because it is prominently affected in mouse and human genetic defects in Bloc1s8 dysbindin and Bloc1s6 pallidin (Fig. 2D–G; Raposo and Marks, 2007). Reduced NSF content in mouse melanocytes carrying BLOC-1-null alleles Bloc1s5mu/mu or Bloc1s6pa/pa is increased by re-expression of the missing BLOC-1 subunit (Fig. 2D,E,G; Setty et al., 2008). Similarly, MNT-1 human melanoma cells treated with shRNAs against either Bloc1s5 muted or Bloc1s6 pallidin downregulated NSF expression (Fig. 2F,G). Reduced expression of NSF polypeptides in mouse melanocytes was not due to decreased NSF transcripts (Fig. 2H). NSF sensitivity to dysbindin/BLOC-1 downregulation with an shRNA against Bloc1s6 pallidin was also observed in iPSC-derived human neurons (Fig. 3A–C). NSF content was proportionately reduced with the expression of dysbindin/BLOC-1 in human neurons (Fig. 3D). These results establish NSF as a BLOC-1 sensitive factor.
Figure 2.
NSF content is reduced in BLOC-1 deficiency. A, B, Downregulation of BLOC-1 subunits decreases NSF content in neuroblastoma cells. C, Quantification of data in A and B. D–H, We used two neuroectodermal-derived cells, MNT1 melanoma cells, and immortalized melanocytes from BLOC-1-null mice. Loss of BLOC-1 reduced the content of NSF in both cells. Controls were the mutant melanocytes rescued by expression of the missing BLOC-1 gene (lane 1). G presents the quantification of D–F. H presents quantitative RT-PCR data measuring transcript levels of NSF and VAMP2 from immortalized melanocytes from BLOC-1-null mice and the above-described controls. All p values <0.05, unless otherwise indicated, as not significant in H (NS), Wilcoxon–Mann–Whitney rank sum test. Dots represent independent biological replicates.
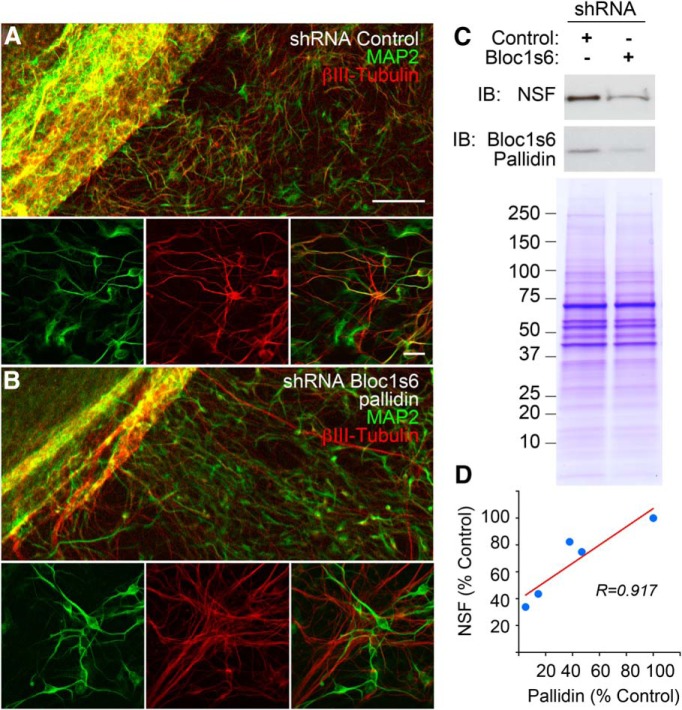
Figure 3.
Downregulation of Bloc1s6 pallidin in iPSC-derived human neurons. Human iPSCs carrying shRNA controls or targeting Bloc1s6 pallidin were differentiated into neuronal cells for 14 d in vitro. A, B, Cell differentiation was assessed by confocal microscopy with antibodies against the neuronal markers MAP2 and β-III-tubulin. C, Cellular content of NSF was determined by immunoblot. Loading controls were performed in parallel samples resolved by SDS-PAGE and stained with Coomassie blue. D, Graph depicts NSF quantifications in four independent experiments.
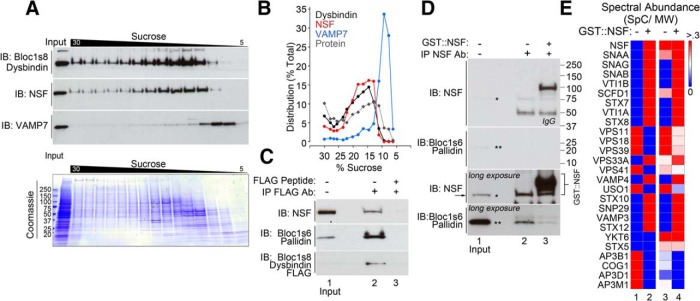
We next asked whether changes in NSF and SNARE levels detected in BLOC-1-deficient cells reflected molecular associations of BLOC-1 with components of the fusion apparatus. To determine whether BLOC-1, SNAREs, and NSF might exist in a complex together, we analyzed their sedimentation in sucrose gradients. We used the cross-linker DSP to stabilize transient interactions. We previously used this procedure to detect interactions between the SNAREs VAMP7 and SNAP29 with BLOC-1 complexes (Gokhale et al., 2012; Ryder et al., 2013). The BLOC-1 complex, detected with Bloc1s8 dysbindin antibodies, mostly cosediments with NSF, with minimal overlap with the SNARE VAMP7 even though cell lysates were treated with the cross-linker DSP (Fig. 4A,B). Endogenous NSF associates with BLOC-1 complexes containing FLAG-tagged dysbindin (Fig. 4C). FLAG antibodies precipitated the BLOC-1 subunit Bloc1s6 pallidin together with NSF (Fig. 4C, lane 2). This coprecipitation was prevented by the addition of an excess of FLAG peptide (Fig. 4C, lane 3). Conversely, antibodies against NSF precipitated endogenous NSF together with Bloc1s6 pallidin (Fig. 4D, lane 2). Controls with an excess of recombinant full-length GST::NSF prevented endogenous NSF-BLOC-1 coprecipitation (Fig. 4D, lane 3).
Figure 4.
BLOC-1 interacts with NSF or SNAREs. A, Sucrose sedimentation of detergent-soluble cross-linked complexes from neuroblastoma cells. BLOC-1 was detected with antibodies against dysbindin. SNARE sedimentation was determined with VAMP7 antibodies. B, Relative distribution plot of data in A. C, DSP-treated neuroblastoma cells expressing FLAG-dysbindin immunoprecipitate NSF with FLAG antibodies (lane 2). D, NSF antibodies immunoprecipitate Bloc1s6 pallidin (lane 2). Short and long exposures are presented. Lane 3 in C and D shows controls with an excess of antigen FLAG peptide or GST-NSF. E, Mass spectrometry analysis of the replicated experiment, as in D. Heat map depicts spectral abundance, which is defined as the ratio of spectral counts to the molecular weight of a polypeptide. Columns 1 and 2 depict the spectral abundance where rows 1 and 2 have a mean of 0 and a variance of 1; thus, the red in column 2 represents an increase in spectral abundance. Columns 3 and 4 depict absolute spectral abundance values per row.
To address whether this association between NSF and BLOC-1 detected by coimmunoprecipitation (Fig. 4C,D) occurs in the context of SNAREs, we profiled by quantitative mass spectrometry proteins present in anti-NSF antibody immunoprecipitates such as those depicted in Figure 4, D (lane 2) and E (heat maps). In the absence of competing GST::NSF, an experimental condition where we can detect BLOC-1 subunits associated with NSF (Fig. 4D, lane 2), we found molecules known to interact with BLOC-1, such as subunits of the adaptor complex AP-3 and components of the HOPS complex (homotypic fusion and vacuole protein sorting complex) (Salazar et al., 2009; Fig. 4E). α-SNAP and low levels of two SNARES, YKT6 and syntaxin 5, were detected coprecipitating with endogenous NSF. In contrast, the addition of GST::NSF increased the content of recombinant NSF bound to anti-NSF beads 17-fold, which in turn recruited three SNAP isoforms from cell lysates (24-fold increase), and increased the number of SNAREs identified in content and number (11 SNAREs detected; Fig. 4E). Among these SNAREs were SNAP29 and syntaxin 7, both of which are BLOC-1-sensitive SNAREs. The increase in these fusion machinery components bound by GST::NSF occurred even though there was no detectable increase in BLOC-1 binding determined by immunoblot (Fig. 4D,E). These results and the cosedimentation profile of SNAREs, NSF, and BLOC-1 indicate that BLOC-1 indirectly forms complexes preferentially with either NSF or SNAREs, but not both at the same time.
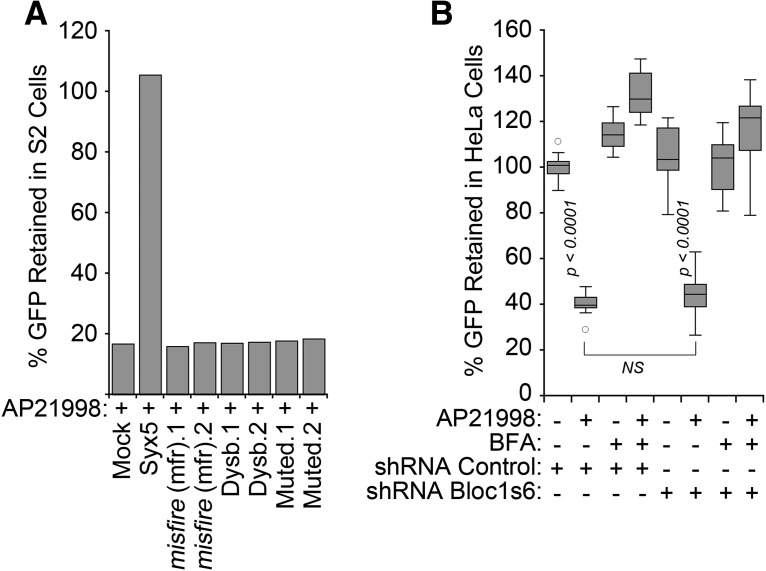
The functional consequences of reduced NSF cellular levels in BLOC-1 deficiencies were assessed measuring the constitutive secretion of a pulse of GFP released from the endoplasmic reticulum by the addition of the AP21998 compound to either Drosophila S2 or HeLa cells (Gordon et al., 2010; Fig. 5A,B). We used as positive controls RNAi-mediated depletion of Drosophila syntaxin 5 (Fig. 5A) or brefeldin A treatment of mammalian cells (Fig. 4B), both of which stopped GFP secretion (Gordon et al., 2010). These controls impaired secretion, yet downregulation of either Drosophila or human BLOC-1 subunits dysbindin, Bloc1s5 muted, or Bloc1s6 pallidin did not affect constitutive GFP secretion along the exocytic route in either cell type (Fig. 5A,B). Thus, we conclude that BLOC-1 deficiency does not globally impair NSF-dependent processes.
Figure 5.
A pulse of a constitutive secretion cargo is not impaired by BLOC-1 deficiency. A, S2 Drosophila cells expressing a signal-peptide EGFP-FKBP chimera retained in the endoplasmic reticulum were treated with mock transfection, one or two siRNA duplexes against syntaxin 5 (positive control), misfire (negative control), and the Drosophila BLOC-1 subunits dysbindin and muted. EGFP secretion was induced by disaggregation of chimeras by incubation with AP21998, and the amount of EGFP retained in cells after 80 min of secretion was measured by flow cytometry. B, HeLa cells were treated with shRNA control or against Bloc1s6 pallidin for 7 d. EGFP secretion was induced by incubation with AP21998. We measured EGFP retained in cells after 80 min of secretion using a fluorescence plate reader in 96-well plates. Cells were incubated with brefeldin A 10 μg/ml as a control to inhibit secretion. Data are depicted as box plots and compared by one-way ANOVA followed by Bonferroni's all-pair comparisons (n = 16 from two biological replicates).
The Drosophila neuromuscular junction undergoes acute homeostatic synaptic plasticity in response to the blockage of postsynaptic glutamatergic receptors with PhTx-433. This plasticity mechanism localizes to the presynaptic terminal and requires the BLOC-1 subunits dysbindin, SNAPIN and SNAP25 and RIM, which are components of the vesicle fusion machinery and (Dickman and Davis, 2009; Dickman et al., 2012; Müller et al., 2012). Thus, we asked whether NSF function is required for this dysbindin-dependent plasticity at the Drosophila neuromuscular junction. We focused on this presynaptic homeostatic process since a requirement for dysbindin/BLOC-1 in postsynaptic homeostatic mechanisms has been excluded in Drosophila (Dickman and Davis, 2009; Dickman et al., 2012). Moreover, there have not been studies yet in mammals addressing whether dysbindin/BLOC-1 participates in presynaptic or postsynaptic homeostatic mechanisms, even though these subunits localize to the mammalian presynapse and postsynapse (Talbot et al., 2006; Larimore et al., 2013; Jia et al., 2014).
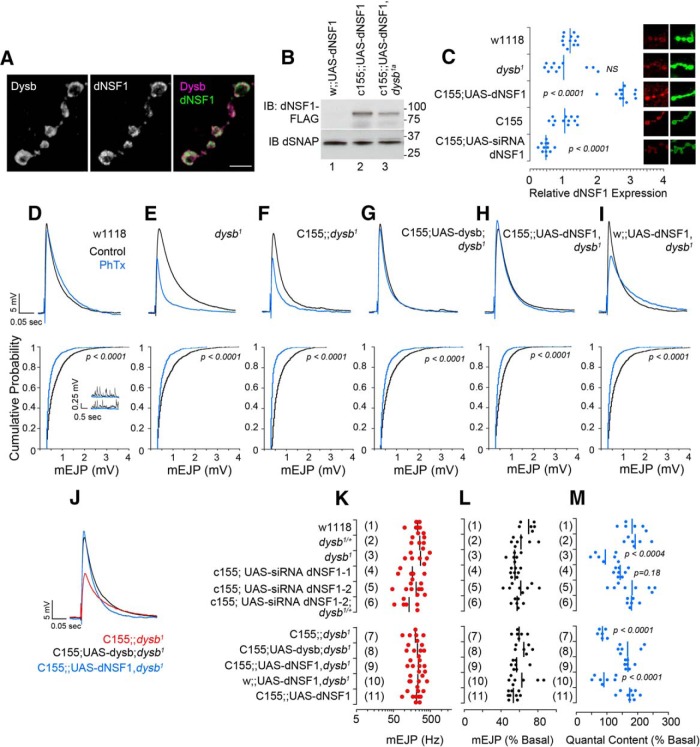
First, we colocalized endogenous dNSF1 and Venus-Dysbindin in the presynaptic compartment of the Drosophila neuromuscular junction (Fig. 6A). dNSF1 confocal microscopy detected either a twofold increase or reduction of dNSF1 expression after neuronal expression of a dNSF1-FLAG transgene (Fig. 6B,C) or dNSF1 RNAi, respectively (Fig. 6C). However, we did not observe a significant reduction of dNSF1 by quantitative confocal microscopy in the neuromuscular junction of animals carrying mutations in dysbindin (dysb1; Fig. 6C), although recombinant dNSF1-FLAG content was reduced by the dysb1 allele in neurons (Fig. 6B, compare lanes 2 and 3).
Figure 6.
NSF presynaptically rescues dysbindin synaptic homeostasis defect. A, Confocal microscopy of Drosophila neuromuscular junction stained with antibodies against dNSF1 and GFP to detect functional recombinant neuronal expressed Venus-Dysbindin. Scale bar, 5 μm. B, immunoblot demonstrating the expression of the UAS-dNSF1 transgene in animals with or without the dysb1a mutation. dSNAP was used as a loading control. C, Confocal microscopy of boutons stained with HRP as a control and dNSF1. Fluorescence was normalized per bouton to HRP and to wild-type genotype. D–J, Representative EJP and mEJP traces without PhTx-433 (black) and following PhTx incubation (blue). Representative mEJP traces are shown only for wild-type w1118 animals. J, Overlay of EJP traces after PhTx presented in F–H. K, No differences across genotypes in mEJP frequency in the absence of PhTx. L, No differences across genotypes in mEJP amplitude after PhTx. M, Quantal content across different genotypes. Dots represent individual animals; n > 6 for all genotypes and all conditions. The p values were obtained with one-way ANOVA followed by Bonferroni's all-pair comparisons.
Homozygotic mutations in BLOC-1 subunits dysbindin and snapin each block philanthotoxin-induced homeostatic increase in quantal content, while heterozygotic mutations have no effect (Dickman and Davis, 2009; Dickman et al., 2012). However, double heterozygotic mutations in dysbindin and snapin also reduce homeostatic compensation compared with wild type (Dickman and Davis, 2009; Dickman et al., 2012). This suggested to us that this paradigm would be ideal for testing the genetic interactions between dysbindin/BLOC-1 and NSF predicted from our coimmunoprecipitation experiments. We genetically modified the expression of NSF and dysbindin in the presynaptic compartment of the fly neuromuscular junction. These genetic manipulations affected neither the amplitude of evoked EJPs (Fig. 6D–I) nor the amplitude (Fig. 6D–I) and frequency of mEJPs (Fig. 6K). However, our results show that wild-type synapses increase their quantal content by 182 ± 14.8% following PhTx treatment (Fig. 6M, row 1). In contrast, and consistent with previous reports (Dickman and Davis, 2009; Dickman et al., 2012), dysb1 animals did not display a homeostatic increase in quantal content (94.9 ± 13.5%; Fig. 6E,M, row 3). Importantly, neuron-specific expression of dysbindin rescued the dysb1 synaptic homeostasis defect (c155-GAL4;UAS-dysbindin;dysb1; Fig. 6G,J,M, row 8). We next tested the effects of neuronal dNSF1 upregulation or downregulation on homeostatic synaptic plasticity. Neuron-specific downregulation of dNSF1 by RNAi or neuronal expression of dNSF1-FLAG (Fig. 6C) did not affect quantal content increase after PhTx treatment (Fig. 6M, compare rows 1, 4–6 and 11). Similarly, downregulation of dNSF1 in dysb1/+ heterozygotes animals did not differ from wild-type or dysb1/+ heterozygote responses to toxin (Fig. 6M, compare rows 1, 2, and 6). Strikingly, presynaptic expression of dNSF1-FLAG rescued the dysb1 synaptic homeostasis defect (c155-GAL4;UAS-dNSF1-FLAG, dysb1) to the same extent as presynaptic dysbindin rescue (Fig. 6, compare J and F, G and H; compare row 7 with M, rows 8 and 9). Presynaptic addition of either dysbindin or dNSF1 restored the homeostatic quantal content increase to wild-type levels, 182.9 ± 17.5% and 171.3 ± 10.4%, respectively (Fig. 6M, rows 8 and 9). These results demonstrate that dysbindin and NSF genetically interact in a molecular pathway necessary for synaptic homeostasis at the presynapse.
Discussion
Genetic polymorphisms associated with schizophrenia mostly reside in noncoding regions modifying gene and/or protein levels rather protein sequence (Nicolae et al., 2010; Maurano et al., 2012; Richards et al., 2012; Schizophrenia Working Group of the Psychiatric Genomics, 2014). The question of how widespread the effects are of a single mutation or polymorphism across the proteome has been poorly explored. Here we addressed this question by modeling a partial reduction in the cellular content of dysbindin/BLOC-1 using shRNAs against BLOC-1 complex subunits. We identified 224 proteins (our unpublished observations) whose content is affected by a partial loss of function of dysbindin/BLOC-1 and focused on an interactome centered around a schizophrenia susceptibility gene, dysbindin, and NSF (Fig. 1B), a component of the membrane fusion machinery that localizes to the synapse and was previously implicated in schizophrenia mechanisms (Mirnics et al., 2000). We confirmed functional outcomes of the dysbindin/BLOC-1 and NSF association using a Drosophila synaptic adaptive response. Our results demonstrate that dysbindin/BLOC-1 expression defects induce multiple downstream quantitative protein expression traits associated with the vesicle fusion apparatus, which influence synaptic plasticity in an invertebrate model synapse.
Our proteomic search prominently highlights the following components of the vesicle fusion apparatus: munc18, tomosyn, NSF; and the SNAREs syntaxin 7, syntaxin 17, SNAP23, SNAP25, SNAP 29, and VAMP7. Importantly, most of the aforementioned vesicle fusion machinery components have been implicated by genomic and postmortem studies in several neurodevelopmental disorders, including schizophrenia (Thompson et al., 1998; Mirnics et al., 2000; Saito et al., 2001; Halim et al., 2003; Behan et al., 2009; Gil-Pisa et al., 2012), intellectual disability (Hamdan et al., 2009), and autism spectrum disorder (Matsunami et al., 2013; Cukier et al., 2014). Our strategy is validated by the identification of proteins previously known to be downregulated in null alleles of BLOC-1 subunits and/or known to interact with BLOC-1. These proteins include subunits of the BLOC-1 complex (Bloc1s5 muted and Bloc1s4 cappuccino) and the SNARE VAMP7 (Li et al., 2003; Starcevic and Dell'Angelica, 2004; Salazar et al., 2006; Newell-Litwa et al., 2010; Mullin et al., 2011; Ghiani and Dell'Angelica, 2011; Yang et al., 2012). We further authenticated these fusion machinery components as part of a dysbindin/BLOC-1 network by (1) coimmunoprecipitation of a fusion machinery component with dysbindin/BLOC-1 subunits and/or (2) downregulation of a fusion machinery component after genetic or shRNA-mediated reduction of dysbindin/BLOC-1 subunits. We centered on NSF since it is a hub of protein–protein interactions with components of the fusion machinery, and is a catalytic activity that is required for the resolution of fusion reaction products and other protein–protein complexes (Rizo and Sudhof, 2012). We found that NSF associates with dysbindin and BLOC-1 subunits in neuroblastoma cells in culture. However, our efforts to document the association of NSF and dysbindin-BLOC-1 by immunoprecipitation with NSF antibodies were unsuccessful in brain. This outcome occurred regardless of whether NSF was immunoprecipitated from brain homogenates or cross-linked synaptosomal lysates from adult mouse brain. We attribute this negative result to the high abundance of NSF in brain compared with dysbindin/BLOC-1. Reverse immunoprecipitations with dysbindin/BLOC-1 antibodies were not possible, as none of the antibodies available to us are suitable for immunoprecipitation. Since most of the associations between NSF and dysbindin/BLOC-1 are detected in the presence of the cross-linker DSP in cell lines, it is likely that the biochemical interactions between NSF and dysbindin/BLOC-1 are indirect. However, NSF cellular levels are decreased following shRNA-mediated or genomic reduction of BLOC-1 complex members, arguing in favor of a functional outcome of this association. We could not detect a NSF downregulation phenotype in hippocampal extracts of Bloc1s8sdy/sdy mice at days 7 or 50 of postnatal development (data not shown). This suggests that NSF phenotypes may be anatomically restricted either to a region of the hippocampus or to an earlier and transient developmental stage. However, this reduced NSF trait is robustly and reversibly induced by genetic disruption of the dysbindin/BLOC-1 complex or by downregulation of dysbindin/BLOC-1 subunits in neuroblastoma and human embryonic kidney cells, neuroectodermal cells, and iPSC-derived human neurons.
Our studies indicate that the functional outcome of NSF reduction in BLOC-1 loss of function become evident only when the synapse is challenged. Constitutive secretion in Drosophila or mammalian non-neuronal cells is unaffected, as are spontaneous and evoked neurotransmission at the Drosophila neuromuscular junction. However, a requirement for NSF in BLOC-1 loss-of-function phenotypes can be localized to a presynaptic homeostatic mechanism, which is engaged when postsynaptic receptors are blocked with philanthotoxin. After a brief incubation with philanthotoxin, the resultant reduction in postsynaptic signal transduction rapidly induces a compensatory increase in quantal content, a response known as homeostatic synaptic plasticity (Davis, 2013; Frank, 2014). This adaptive compensatory mechanism is precluded by dysbindin mutations, and can be rescued by presynaptic expression of dysbindin (Dickman and Davis, 2009). However, we were able to rescue this phenotype in the dysbindin mutants to the exact same extent through presynaptic expression NSF. The observation that RNAi downregulation or overexpression of NSF in the neuromuscular junction does not interfere with homeostatic synaptic plasticity argues that the NSF is not an obligate component downstream of dysbindin/BLOC-1 in a linear pathway, but rather is an adaptive response to network perturbation induced by a dysbindin mutant allele. This hypothesis predicts that transheterozygotic reduction of NSF and Dysbindin should impair plasticity, a result that is at odds with our finding that plasticity is normal in dysb1−/+;UAS-NSF RNAi flies (Fig. 6M, row 6). We believe that this may be a consequence of a modest reduction of dysbindin polypeptide in dysb1−/+ animals, which we predict is ∼25% (Shao et al., 2011).
How does the BLOC-1–NSF interaction affect synaptic mechanisms? We believe a model integrating our findings has to consider three key elements. First, BLOC-1 subunits reside at endosomes as well as on synaptic vesicles in presynaptic terminals in neurons (Di Pietro et al., 2006; Setty et al., 2007; Ryder and Faundez, 2009; Mullin et al., 2011). Second, BLOC-1 binds monomeric SNAREs rather than tetrahelical SNARE bundles in vitro (Ghiani et al., 2010). Finally, NSF and SNAREs bind to dysbindin/BLOC-1, yet they do not seem to form a ternary complex. Thus, we propose that BLOC-1 bound to a single SNARE (perhaps for SNARE sorting into vesicles) is resolved by NSF, making SNAREs permissive for vesicle fusion. Therefore, when dysbindin and NSF levels are reduced by hypomorphic mutations in the fly or as a quantitative expression trait in humans, SNARE-dependent mechanisms might be impaired due to defective SNARE sorting, a consequence of the reduced levels of BLOC-1 complex and, additionally, by decreased NSF content that would impair the resolution of remaining SNARE–BLOC-1 complexes. Thus, noncoding polymorphisms in several genes and their quantitative expression traits may converge to impair synaptic mechanisms. We propose that unbiased identification of quantitative traits across the proteome of neurodevelopmental deficiency models is a simple approach to unravel mechanisms of complex neurodevelopmental disorders.
Footnotes
This work was supported by grants from the National Institutes of Health (Grant GM077569) and the Neuroscience Center of Children's Healthcare of Atlanta to V.F.; S.A.Z. was supported by Graduate and Postdoctoral Training in Toxicology Training Grant T32–1P50NS071669 from the National Institute of Environmental Health Sciences. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core and Viral Cores of the Emory Neuroscience NINDS Core Facilities Grant, P30NS055077. We thank the Faundez laboratory members for their comments.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript. The authors declare no competing financial interests.
References
- Albert FW, Treusch S, Shockley AH, Bloom JS, Kruglyak L. Genetics of single-cell protein abundance variation in large yeast populations. Nature. 2014;506:494–497. doi: 10.1038/nature12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O'Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M, Macleod GT, Leither J, Pallanck L. Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles. J Neurosci. 2004;24:3964–3973. doi: 10.1523/JNEUROSCI.5259-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan AT, Byrne C, Dunn MJ, Cagney G, Cotter DR. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry. 2009;14:601–613. doi: 10.1038/mp.2008.7. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, Owen MJ, O'Donovan MC. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Dueker ND, Slifer SH, Lee JM, Whitehead PL, Lalanne E, Leyva N, Konidari I, Gentry RC, Hulme WF, Booven DV, Mayo V, Hofmann NK, Schmidt MA, Martin ER, Haines JL, Cuccaro ML, Gilbert JR, Pericak-Vance MA. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 2014;5:1. doi: 10.1186/2040-2392-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GW. Homeostatic signaling and the stabilization of neural function. Neuron. 2013;80:718–728. doi: 10.1016/j.neuron.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman DK, Davis GW. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science. 2009;326:1127–1130. doi: 10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman DK, Tong A, Davis GW. Snapin is critical for presynaptic homeostatic plasticity. J Neurosci. 2012;32:8716–8724. doi: 10.1523/JNEUROSCI.5465-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro SM, Falcón-Pérez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell'Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley CA, 4th, Miki T, Castro CA, Ozolek JA, Minervini CF, Ben-Yehudah A, Schatten GP. Human amniotic epithelial cells are reprogrammed more efficiently by induced pluripotency than adult fibroblasts. Cell Reprogram. 2012;14:193–203. doi: 10.1089/cell.2011.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Shelley BC, Hurley AM, Onorati M, Castiglioni V, Patitucci TN, Svendsen SP, Mattis VB, McGivern JV, Schwab AJ, Sareen D, Kim HW, Cattaneo E, Svendsen CN. EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res. 2013;10:417–427. doi: 10.1016/j.scr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, Kruglyak L. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- Franciscovich AL, Mortimer AD, Freeman AA, Gu J, Sanyal S. Overexpression screen in Drosophila identifies neuronal roles of GSK-3 beta/shaggy as a regulator of AP-1-dependent developmental plasticity. Genetics. 2008;180:2057–2071. doi: 10.1534/genetics.107.085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CA. Homeostatic plasticity at the Drosophila neuromuscular junction. Neuropharmacology. 2014;78:63–74. doi: 10.1016/j.neuropharm.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Franciscovich A, Bowers M, Sandstrom DJ, Sanyal S. NFAT regulates pre-synaptic development and activity-dependent plasticity in Drosophila. Mol Cell Neurosci. 2011;46:535–547. doi: 10.1016/j.mcn.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani CA, Dell'Angelica EC. Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade. ASN Neuro. 2011;3:e00058. doi: 10.1042/AN20110010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, Malvar JS, de Vellis J, Sabatti C, Dell'Angelica EC. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol Psychiatry. 2010;15:115, 204–215. doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Pisa I, Munarriz-Cuezva E, Ramos-Miguel A, Urigüen L, Meana JJ, García-Sevilla JA. Regulation of munc18–1 and syntaxin-1A interactive partners in schizophrenia prefrontal cortex: down-regulation of munc18–1a isoform and 75 kDa SNARE complex after antipsychotic treatment. Int J Neuropsychopharmacol. 2012;15:573–588. doi: 10.1017/S1461145711000861. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, Lupashin VV, Smith Y, Faundez V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012;32:3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Bond LM, Sahlender DA, Peden AA. A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic. 2010;11:1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- Halim ND, Weickert CS, McClintock BW, Hyde TM, Weinberger DR, Kleinman JE, Lipska BK. Presynaptic proteins in the prefrontal cortex of patients with schizophrenia and rats with abnormal prefrontal development. Mol Psychiatry. 2003;8:797–810. doi: 10.1038/sj.mp.4001319. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Piton A, Gauthier J, Lortie A, Dubeau F, Dobrzeniecka S, Spiegelman D, Noreau A, Pellerin S, Côté M, Henrion E, Fombonne E, Mottron L, Marineau C, Drapeau P, Lafreniere RG, Lacaille JC, Rouleau GA, Michaud JL. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009;65:748–753. doi: 10.1002/ana.21625. [DOI] [PubMed] [Google Scholar]
- Jentsch JD, Trantham-Davidson H, Jairl C, Tinsley M, Cannon TD, Lavin A. Dysbindin modulates prefrontal cortical glutamatergic circuits and working memory function in mice. Neuropsychopharmacology. 2009;34:2601–2608. doi: 10.1038/npp.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JM, Hu Z, Nordman J, Li Z. The schizophrenia susceptibility gene dysbindin regulates dendritic spine dynamics. J Neurosci. 2014;34:13725–13736. doi: 10.1523/JNEUROSCI.0184-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Robleto K, Trantham-Davidson H, Jairl C, Cannon TD, Lavin A, Jentsch JD. Reduced dysbindin expression mediates N-methyl-D-aspartate receptor hypofunction and impaired working memory performance. Biol Psychiatry. 2011;69:28–34. doi: 10.1016/j.biopsych.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto N, Matsuzaki S, Inoue K, Hattori T, Shimizu S, Hashimoto R, Yamatodani A, Katayama T, Tohyama M. Hyperactivation of midbrain dopaminergic system in schizophrenia could be attributed to the down-regulation of dysbindin. Biochem Biophys Res Commun. 2006;345:904–909. doi: 10.1016/j.bbrc.2006.04.163. [DOI] [PubMed] [Google Scholar]
- Kushimoto T, Basrur V, Valencia J, Matsunaga J, Vieira WD, Ferrans VJ, Muller J, Appella E, Hearing VJ. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc Natl Acad Sci U S A. 2001;98:10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J, Ryder PV, Kim KY, Ambrose LA, Chapleau C, Calfa G, Gross C, Bassell GJ, Pozzo-Miller L, Smith Y, Talbot K, Park IH, Faundez V. MeCP2 regulates the synaptic expression of a Dysbindin-BLOC-1 network component in mouse brain and human induced pluripotent stem cell-derived neurons. PLoS One. 2013;8:e65069. doi: 10.1371/journal.pone.0065069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J, Zlatic SA, Gokhale A, Tornieri K, Singleton KS, Mullin AP, Tang J, Talbot K, Faundez V. Mutations in the BLOC-1 subunits dysbindin and muted generate divergent and dosage-dependent phenotypes. J Biol Chem. 2014;289:14291–14300. doi: 10.1074/jbc.M114.553750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell'Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami N, Hadley D, Hensel CH, Christensen GB, Kim C, Frackelton E, Thomas K, da Silva RP, Stevens J, Baird L, Otterud B, Ho K, Varvil T, Leppert T, Lambert CG, Leppert M, Hakonarson H. Identification of rare recurrent copy number variants in high-risk autism families and their prevalence in a large ASD population. PLoS One. 2013;8:e52239. doi: 10.1371/journal.pone.0052239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/S0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Müller M, Liu KS, Sigrist SJ, Davis GW. RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J Neurosci. 2012;32:16574–16585. doi: 10.1523/JNEUROSCI.0981-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin AP, Gokhale A, Larimore J, Faundez V. Cell biology of the BLOC-1 complex subunit dysbindin, a schizophrenia susceptibility gene. Mol Neurobiol. 2011;44:53–64. doi: 10.1007/s12035-011-8183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin AP, Sadanandappa MK, Ma W, Dickman DK, VijayRaghavan K, Ramaswami M, Sanyal S, Faundez V. Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. J Neurosci. 2015;35:325–338. doi: 10.1523/JNEUROSCI.3542-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, Faundez V. Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci. 2010;30:820–831. doi: 10.1523/JNEUROSCI.3400-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, Ozaki N, Taguchi T, Tatsumi M, Kamijima K, Straub RE, Weinberger DR, Kunugi H, Hashimoto R. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13:2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, Owen MJ, Holmans P, O'Donovan MC. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- Ryder PV, Faundez V. Schizophrenia: the “BLOC” may be in the endosomes. Sci Signal. 2009;2:pe66. doi: 10.1126/scisignal.293pe66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Vistein R, Gokhale A, Seaman MN, Puthenveedu MA, Faundez V. The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type II alpha. Mol Biol Cell. 2013;24:2269–2284. doi: 10.1091/mbc.E13-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggu S, Cannon TD, Jentsch JD, Lavin A. Potential molecular mechanisms for decreased synaptic glutamate release in dysbindin-1 mutant mice. Schizophr Res. 2013;146:254–263. doi: 10.1016/j.schres.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Guan F, Papolos DF, Rajouria N, Fann CS, Lachman HM. Polymorphism in SNAP29 gene promoter region associated with schizophrenia. Mol Psychiatry. 2001;6:193–201. doi: 10.1038/sj.mp.4000825. [DOI] [PubMed] [Google Scholar]
- Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, Wainer BH, Falcon-Perez JM, Dell'Angelica EC, Peden AA, Werner E, Faundez V. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem. 2009;284:1790–1802. doi: 10.1074/jbc.M805991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, Dell'Angelica EC, Raposo G, Marks MS. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–1146. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Shuai Y, Wang J, Feng S, Lu B, Li Z, Zhao Y, Wang L, Zhong Y. Schizophrenia susceptibility gene dysbindin regulates glutamatergic and dopaminergic functions via distinctive mechanisms in Drosophila. Proc Natl Acad Sci U S A. 2011;108:18831–18836. doi: 10.1073/pnas.1114569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O'Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, Kamins J, Hahn CG, Blake DJ, Arnold SE. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum Mol Genet. 2006;15:3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- Talbot K, Ong WY, Blake DJ, Tang D, Louneva N, Carlson GC, Arnold SE. Dysbindin-1 and its protein family, with special attention to the potential role of dysbindin-1 in neuronal functions and the pathophysiology of schizophrenia. In: Kantrowitz J, editor. Handbook of Neurochemistry and Molecular Neurobiology. New York: Springer Science; 2009. pp. 107–241. [Google Scholar]
- Talbot K, Louneva N, Cohen JW, Kazi H, Blake DJ, Arnold SE. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One. 2011;6:e16886. doi: 10.1371/journal.pone.0016886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Sower AC, Perrone-Bizzozero NI. Altered levels of the synaptosomal associated protein SNAP-25 in schizophrenia. Biol Psychiatry. 1998;43:239–243. doi: 10.1016/S0006-3223(97)00204-7. [DOI] [PubMed] [Google Scholar]
- van de Goor J, Ramaswami M, Kelly R. Redistribution of synaptic vesicles and their proteins in temperature-sensitive shibire(ts1) mutant Drosophila. Proc Natl Acad Sci U S A. 1995;92:5739–5743. doi: 10.1073/pnas.92.12.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Bogaert A, Schumacher J, Schulze TG, Otte AC, Ohlraun S, Kovalenko S, Becker T, Freudenberg J, Jönsson EG, Mattila-Evenden M, Sedvall GC, Czerski PM, Kapelski P, Hauser J, Maier W, Rietschel M, Propping P, Nöthen MM, Cichon S. The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am J Hum Genet. 2003;73:1438–1443. doi: 10.1086/379928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, He X, Yang L, Zhou Z, Cullinane AR, Wei A, Zhang Z, Hao Z, Zhang A, He M, Feng Y, Gao X, Gahl WA, Huizing M, Li W. The BLOS1-interacting protein KXD1 is involved in the biogenesis of lysosome-related organelles. Traffic. 2012;13:1160–1169. doi: 10.1111/j.1600-0854.2012.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Kawasaki F, Ordway RW. Activity-dependent interactions of NSF and SNAP at living synapses. Mol Cell Neurosci. 2011;47:19–27. doi: 10.1016/j.mcn.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Ryder PV, Salazar G, Faundez V. Isolation of labile multi-protein complexes by in vivo controlled cellular cross-linking and immuno-magnetic affinity chromatography. J Vis Exp. 2010;9:1855. doi: 10.3791/1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Grossniklaus EJ, Ryder PV, Salazar G, Mattheyses AL, Peden AA, Faundez V. Chemical-genetic disruption of clathrin function spares adaptor complex 3-dependent endosome vesicle biogenesis. Mol Biol Cell. 2013;24:2378–2388. doi: 10.1091/mbc.E12-12-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]