Abstract
Overexpression of CD30 and JunB is a hallmark of tumor cells in Hodgkin's lymphoma (HL) and anaplastic large-cell lymphoma (ALCL). We reported that CD30–extracellular signal–regulated kinase (ERK)1/2 mitogen-activated protein kinase (MAPK) signaling induces JunB, which maintains constitutive activation of the CD30 promoter. Herein, we localize a cis-acting enhancer in the JunB promoter that is regulated by Ets-1. We show that E26 transformation-specific-1 (Ets-1) (−146 to −137) enhances JunB promoter activation in a manner that is dependent on CD30 or the nucleophosmin-anaplastic lymphoma kinase (NPM-ALK)–ERK1/2 MAPK pathway. Ets-1 knockdown reduces the expression of both JunB and CD30, and CD30 knockdown significantly reduces JunB expression in HL and ALCL cell lines. NPM-ALK knockdown also reduces JunB expression in ALCL cell lines expressing NPM-ALK. Collectively, these results indicate that CD30 and NPM-ALK cooperate to activate the ERK1/2 MAPK–Ets-1 pathway. Ets-1, constitutively activated by ERK1/2-MAPK, plays a central role in the overexpression of JunB and CD30, which are both involved in the pathogenesis of HL and ALCL.
CD30 is a member of the tumor necrosis factor receptor superfamily that was initially identified on the surface of Hodgkin and Reed-Sternberg cells of Hodgkin's lymphoma (HL). Immunohistochemical (IHC) analysis of a large range of human tumors has shown that CD30 is overexpressed by Hodgkin and Reed-Sternberg cells and by a subset of large-cell neoplasms with anaplastic features, called anaplastic large-cell lymphoma (ALCL).1 Approximately 50% of systemic ALCLs express the nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) chimeric protein, which exerts a distinct role in CD30- overexpressing lymphoma.2,3
JunB, a member of the activator protein-1 (AP-1) transcription family, is composed of homodimers or heterodimers of the related Jun (c-Jun, JunB, and JunD), Fos (Fos, FosB, Fra1, and Fra2), activating transcription factor (ATF), and cAMP response element-binding (CREB) families.4 AP-1 functions by binding to 12-O-tetra-decanoylphorbol-13-acetate (TPA)-responsive elements within the promoter of numerous genes. AP-1 is involved in multiple biological processes, including cell differentiation, proliferation, and apoptosis. Transcription of AP-1 family members is rapidly and transiently stimulated by multiple extracellular signals.5 c-Jun and JunB have antagonistic functions in biological processes, such as oncogenic transformation and cell proliferation.6 However, JunB also regulates distinct target genes in a c-Jun–independent manner and exerts specific functions.7,8 Overexpression of JunB has been associated with neoplastic transformation.9–12
Overexpression of CD30 and JunB is characteristic of HL and ALCL.13–16 Ligand-independent signals, triggered by highly expressed CD30, induce activation of NF-κB and the extracellular signal–regulated kinase (ERK) 1/2 mitogen-activated protein kinase (MAPK) pathway.15,17 These pathways contribute to the tumorigenesis and maintenance of survival of HL and ALCL cells.2,17–21 We also showed that ligand-independent CD30-ERK-MAPK signals induce enhanced JunB protein expression, which acts on the unmethylated CD30 promoter to maintain overexpression of CD30 in HL and ALCL cells.15,22
In this report, we localized a cis-acting enhancer in the JunB promoter of HL and ALCL and investigated the mechanisms that maintain the overexpression of JunB and CD30 in these lymphomas.
Materials and Methods
Cell Lines and Cell Cultures
Jurkat and K562 cell lines were obtained from Fujisaki Cell Biology Center (Okayama, Japan). ALCL cell lines (SUDHL1 and Karpas299) and HL cell lines (L428, KMH2, HDLM2, and L540) were obtained from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). Cells were cultured in RPMI 1640 medium supplemented with fetal bovine serum and antibiotics.
Inhibitors
The UO126 inhibitor of MAPK kinase (MEK) 1/2 was obtained from Cell Signaling Technology (Beverly, MA), dissolved in dimethyl sulfoxide, and used for experiments at the specified concentrations.
EMSA Data
Electrophoretic mobility shift analyses (EMSAs) were performed according to methods described by Andrews and Faller.23 The double-stranded oligonucleotide probe (−151 to −134) spanning the Ets-1 consensus motif of the JunB promoter was used for this analysis. Anti-Ets-1 (H150) X rabbit polyclonal antibody and the Ets-1/polyomavirus enhancer activator protein 3 (PEA3) gel shift oligonucleotide (both from Santa Cruz Biotechnology, Santa Cruz, CA) were used for supershift and competition assays, respectively.
ChIP Data
Chromatin immunoprecipitation (ChIP) was performed using the Epi Quick Chromatin Immunoprecipitation Kit (Epigentek, Brooklyn, NY). Briefly, HL and ALCL cell lines (2 × 106) were treated with 1% formaldehyde and washed with PBS without calcium and magnesium, and the remainder of the protocol was performed according to the manufacturer's instructions. Anti-Ets-1 (C-4) X mouse antibody (Santa Cruz Biotechnology) and anti-RNA polymerase II antibody (positive control) or isotype-matched antibody (negative control) were used for immunoprecipitation. Input DNA and immunoprecipitated DNA were assayed by PCR using two sets of primer pairs for amplification of the JunB promoter. The primers used were as follows: forward primer for pair A, 5′-CCCTGAAACCCCTCACTCATGTG-3′; forward primer for pair B, 5′-CCGTGGCCGCTGTTTACAAGG-3′; and the common reverse primer for A and B, 5′-GAAGTGCGCTCCGATTGGCGG-3′.
DNA Constructs
The 5′ region of the JunB gene was cloned into the pGL3–basic luciferase reporter vector (Promega, Madison, WI) by PCR using the promoter sequence of the JunB gene in the L540 cell line as a template and was mutated using the mutagenesis method of Kunkel et al.24 The nucleotide sequence of various deletion constructs was confirmed using the 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). The gene regions contained in the resultant pGL3-JunBpro reporters were as follows: −998 to 344, −278 to 344, −184 to 344, and −47 to 344. The expression vectors for CD30 and NPM-ALK are described elsewhere.2
Reporter Gene Assays
Activities of the promoter were measured by transient reporter gene assays using the Dual Luciferase assay kit (Promega), according to the manufacturer's instructions. The Renilla luciferase expression vector driven by the herpes simplex virus thymidine kinase promoter, pRL-TK (Promega), was cotransfected to standardize the transfection efficiency in each experiment. If not indicated otherwise, 0.1 μg of the reporter construct and 0.05 μg of pRL-TK, with or without 0.5 μg of each expression vector, were used per transfection. Cells (2 × 105) were transfected using the Lipofectamine 2000 reagent (Invitrogen, Groningen, the Netherlands), according to the manufacturer's instructions. Cells were harvested after 16 hours, and luciferase activities were measured.
Immunoblot Analysis
Immunoblotting experiments were performed as previously described.25 Antibodies (in parentheses) against the following proteins were used: Ets-1 (C-20, rabbit polyclonal), JunB (C-11, mouse monoclonal), α-tubulin (TU-02, mouse IgM), glyceraldehyde-3-phosphate dehydrogenase (FL-335, rabbit polyclonal), ALK (C-19, goat polyclonal) (all from Santa Cruz Biotechnology), and CD30 (Ber-H2, mouse monoclonal) (Dako, Kyoto, Japan). Alkaline phosphatase–conjugated secondary antibodies were as follows: anti-mouse IgG, anti-rabbit IgG, and anti-goat IgG (all obtained from Chemicon International, Temecula, CA) and anti-mouse IgM (Santa Cruz Biotechnology).
IHC Data
Immunostaining of Ets-1 was performed on cell lines and paraffin-embedded specimens of primary samples, which had been obtained after informed consent. After deparaffinization, samples were subjected to autoclave antigen retrieval in 10 mmol/L citrate buffer (pH 6.0) for 1 minute. Sections were then treated with 3% H2O2 for 15 minutes, blocked using 5% skim milk for 1 hour, and then incubated with the anti-Ets-1 antibody (C-20, rabbit polyclonal) (Santa Cruz Biotechnology) at 4°C overnight. Sections were then washed, and bound antibodies were detected using the Histofine Simple Stain MAX PO (MULTI) and diaminobenzidine substrate kit (both from Nichirei Biosciences, Tokyo, Japan).
Knockdown Experiments Using RNA Interference
HL and ALCL cells were transfected with 5 μg of control [AllStars Negative Control small-interefering RNA (siRNA), which is the most thoroughly tested and validated negative control nonsilencing siRNA available] or with HP GenomeWide siRNA (Qiagen, Hilden, Germany) by nucleofection, as described later. The sequence targeted in each molecule was as follows: Hs_ETS_2, 5′-CTCGGATTACTTCATTAGCTA-3′; Hs_ALK_1, 5′-CTCGACCATCATGACCGACTA-3′; Hs_ALK_6, 5′-CTGGGCCTGTATACCGGATAA-3′; CD30 no. 1, 5′-ACCAATAACAAGATTGAGAAA-3′; and CD30 no. 2, 5′-ATGCAAATGAGTGATGGATAA-3′. ALK and CD30 siRNAs were mixed in a 1:1 ratio before transfection. The effect of siRNA transfection on protein expression was monitored by immunoblot analysis. Cells were transfected using nucleofector technology (Amaxa, Cologne, Germany) with the solution and programs indicated in parentheses: KMH2 (V, A-24), HDLM2 (T, T-01), Karpas299 (V, A-30), and SUDHL1 (V, G-16). Transfection conditions were optimized using AllStars Negative Control siRNA fluorescein (Qiagen). Transfection of siRNAs for Jurkat cells was performed using the Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions.
Cell Proliferation Assays
Effects of knockdown by Ets-1 siRNA and CD30 siRNA on cell proliferation were assayed using the methyl thiazolyl tetrazolium method, as previously described.17 After incubation of cells transduced with CD30 siRNA, Ets-1 siRNA, or negative control siRNA, cells were harvested, treated with methyl thiazolyl tetrazolium solution, and measured with a microplate reader (Bio-Rad Laboratories, Hercules, CA) at a reference wavelength of 570 nm and a test wavelength of 450 nm.
Results
Identification of a cis-Acting Enhancer for the JunB Promoter in HL and ALCL Cell Lines
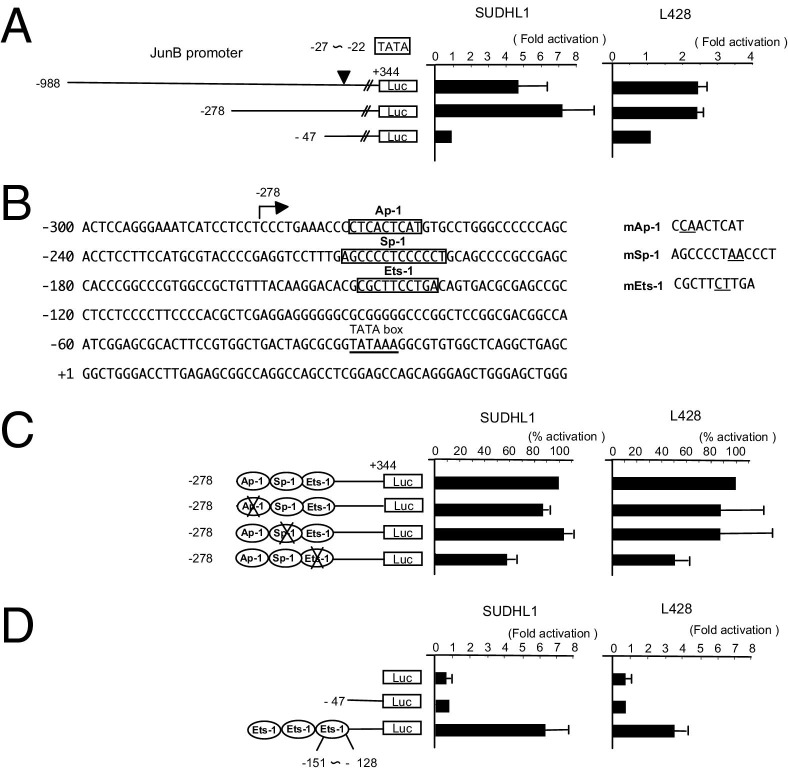
We first examined the activity of deletion constructs of the JunB promoter in the HL cell line L428 and the ALCL cell line SUDHL1 using promoter reporter assays. As shown in Figure 1A, a significant decrease in luciferase activity was detected when the construct with a deletion in the region between −278 and −47 was tested, indicating that a cis-acting enhancer is located within this region. A computer search using TFSEARCH version 1.3 (Kyoto University, Kyoto, Japan) indicated the presence of binding sites for AP-1 (−267 to −259), Sp-1 (−208 to −196), and Ets-1 (−146 to −137) in this region (Figure 1B). The introduction of a mutation into the Ets-1 site significantly decreased JunB promoter activity, whereas mutation of the AP-1 or the Sp-1 site did not decrease this activity (Figure 1C). These results show that the Ets-1 binding site (−146 to −137) is responsible for the constitutive JunB promoter activity in these cell lines. A construct that contained only three Ets-1 sites just upstream of the core JunB promoter was sufficient to enhance core JunB promoter activity in L428 and SUDHL1 cells (Figure 1D). Collectively, these results indicate that the Ets-1 site (−146 to −137) is responsible for JunB promoter induction in these HL and ALCL cells. However, because mutation of the Ets-1 binding site only resulted in a 50% decrease in the activity of the JunB promoter, there may also be additional factors responsible for this promoter activation.
Figure 1.
Identification of a cis-acting enhancer for the JunB promoter in HL and ALCL cell lines. A: The activity of deletion constructs of the JunB promoter was examined using a reporter gene assay in the L428 HL and the SUDHL1 ALCL cell lines. The constructs used for reporter gene assays are indicated on the left. The relative luciferase activity of each construct compared with the core promoter activity (fold activation) is shown. The cell lines used are indicated on the top. Data are given as the mean ± SD of more than three triplicate experiments. B: Nucleic acid sequence of the JunB promoter between −300 and 60. Potential AP-1 (−267 to −259), Sp-1 (−208 to −196), or Ets-1 (−146 to −137) binding sites are indicated by open boxes. Mutations introduced into each potential binding sequence are indicated on the right, and nucleic acids changed are underlined. C: Activity of a JunB promoter construct (−278) with a strong cis-acting enhancer region in which AP-1 (−267 to −259), Sp-1 (−208 to −196), or Ets-1 (−146 to −137) binding sites were mutated. The relative activity of each construct compared with the construct without mutation (percentage activation) is shown. Data are given as the mean ± SD of more than three triplicate experiments. D: Effect of the Ets-1 binding site on core promoter activity. Three Ets-1 binding sequences (5′-ACACGCGCTTCCTGACAGTGACGC-3′) were directly linked to the core JunB promoter (−47). The activity (fold activation) of this three times Ets-1 construct or the reporter vector without an insert (Luc) relative to that of the core promoter (−47) is shown. Data are given as the mean ± SD of more than three triplicate experiments.
Ets-1 Binds a cis-Acting Enhancer of the JunB Promoter in HL and ALCL Cell Lines
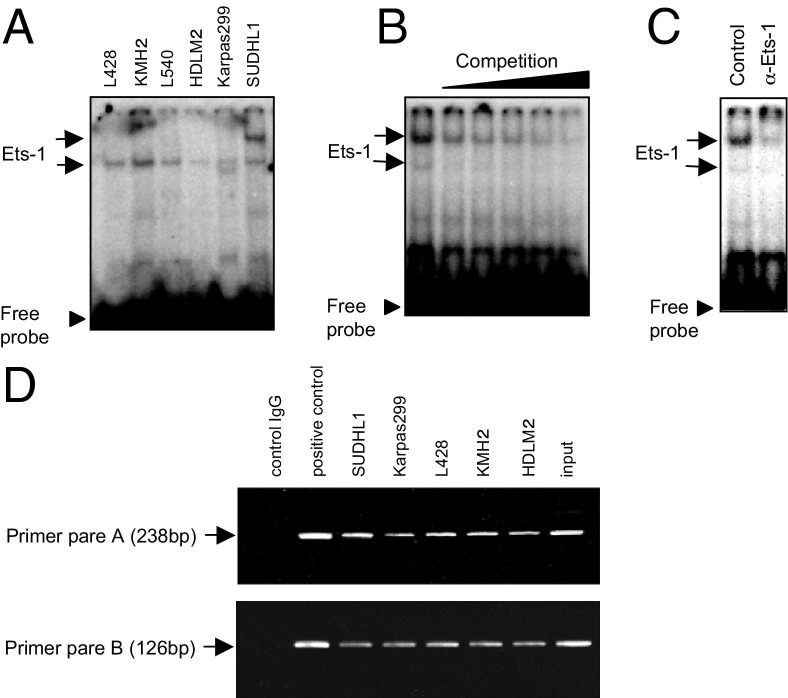
We next determined whether Ets-1 binds the enhancer sequence of the JunB promoter (−151 to −134) in HL and ALCL cell lines. EMSA analyses revealed two probe-reactive bands (Figure 2A). The faster-migrating band was observed in both HL and ALCL cell lines, whereas the slower-migrating band was of strong intensity in the SUDHL1 ALCL cell line (Figure 2A). Both signals were decreased in a competition assay and after supershift analysis with an anti-Ets-1 antibody, suggesting that both signals contain Ets-1 (Figure 2, B and C). To confirm these results, we performed ChIP assays. The results using different primer pairs for JunB promoter clearly show association of Ets-1 with the enhancer sequence (−146 to −137) in HL and ALCL cell lines (Figure 2D). Collectively, these results indicate that Ets-1 binds the enhancer motif (−146 to −137) in the JunB promoter and contributes to the constitutive strong JunB expression in HL and ALCL cells.
Figure 2.
Binding of Ets-1 to an enhancer sequence for the JunB promoter in HL and ALCL cell lines. EMSA analysis of Ets-1 binding to an enhancer sequence for the JunB promoter using 2 μg of nuclear extracts of HL cell lines (L428, KMH2, L540, and HDLM2) and ALCL cell lines (Karpas299 and SUDHL1) (A). Competition analysis using a maximum 200-fold molar excess of the unlabeled probe containing the Ets-1–PEA3 consensus sequence (B), and supershift analysis using the anti-Ets-1 antibody (2 μg) (C), was performed using the SUDHL1 ALCL cell line. D: ChIP analysis. DNA immunoprecipitated with the anti-Ets-1 antibody was assayed by PCR using two sets of primer pairs for the JunB promoter (A and B) and was analyzed by agarose gel electrophoresis. Immunoprecipitation of SUDHL1 DNA with an anti-RNA polymerase II antibody and an isotype-matched IgG antibody served as positive and negative controls, respectively. Direct amplification of SUDHL1 DNA also served as a positive control and is indicated as input.
JunB Induction by Ets-1 Is Mediated by CD30 and NPM-ALK via the ERK1/2-MAPK Pathway
CD30 and NPM-ALK also are involved in constitutive ERK1/2-MAPK activation.15,21,26 Ets-1, a member of the Ets family of transcription factors, which includes an evolutionarily conserved C-terminal Ets DNA binding domain and an N-terminal pointed domain, is a target for Ras-mediated phosphorylation.27 We, therefore, focused on the role of the ERK1/2-MAPK pathway in Ets-1–mediated JunB induction in HL and ALCL cells. The MEK1/2 inhibitor UO126 decreased Ets-1–mediated induction of the JunB promoter in an ALCL cell line that expresses NPM-ALK (SUDHL1) in a dose-dependent manner, indicating that CD30 and/or the NPM-ALK-ERK1/2 MAPK pathway is involved in Ets-1–mediated JunB promoter induction (Figure 3A). We next examined the effect of overexpressed CD30 or NPM-ALK on ERK1/2 MAPK–Ets-1–mediated JunB promoter induction. The MEK1/2 inhibitor UO126 reduced both CD30- and NPM-ALK–mediated induction of the JunB promoter (Figure 3B). CD30- and NPM-ALK–mediated JunB promoter induction was also inhibited by Ets-1 knockdown (Figure 3C). The protein expression of each construct was confirmed by immunoblot analysis (data not shown), which also confirmed reduction in Ets-1 expression by knockdown, as shown in Figure 3C. These results indicate that JunB induction by Ets-1 is mediated by CD30 and NPM-ALK via the ERK1/2-MAPK pathway.
Figure 3.
JunB induction by Ets-1 is mediated by CD30 and NPM-ALK via ERK1/2-MAPK. A: Dose-dependent inhibition of Ets-1 binding site–mediated induction of the JunB promoter by the MEK1/2 inhibitor UO126 in SUDHL1 cells. Cotransfected pGL3-JunBpro (−184 to 344; 0.2 μg) was used for the reporter gene assay. Luciferase activities are expressed as a percentage of the luciferase activity without UO126, which was set at 100%. Data are given as the mean ± SD of more than three triplicate experiments. B: Inhibition of CD30- and NPM-ALK–mediated JunB promoter induction by the MEK1/2 inhibitor, UO126. Jurkat cells were transfected with an expression vector for CD30 or NPM-ALK, with or without pretreatment with 10 μmol/L of UO126, which was started 30 minutes before transfection. The promoter activity of cotransfected pGL3-JunBpro (−184 to 344) was then assayed using a dual-luciferase assay, as described. Luciferase activities are expressed as a percentage of the luciferase activity of the empty vector without UO126, which was set at 100%. Data are given as the mean ± SD of more than three triplicate experiments. C: Induction of JunB promoter activity by CD30 and NPM-ALK is mediated by Ets-1. At 24 hours after transduction of 50 pmol of Ets-1 or control siRNA using the Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions, Jurkat cells were transfected with a CD30 or an NPM-ALK expression vector and cultured for 24 hours. Luciferase activities of cotransfected pGL3-JunBpro (−184 to 344) were measured using a dual-luciferase assay, as described. Activities are expressed as a percentage of the luciferase activity of the sample treated with control siRNA and empty vector, which was set at 100%. Data are given as the mean ± SD of more than three triplicate experiments (left panel). The protein expression of Ets-1 was analyzed at 24 and 48 hours by immunoblotting 30 μg of whole cell lysates (right panel).
Expression of Ets-1 in HL and ALCL Cells
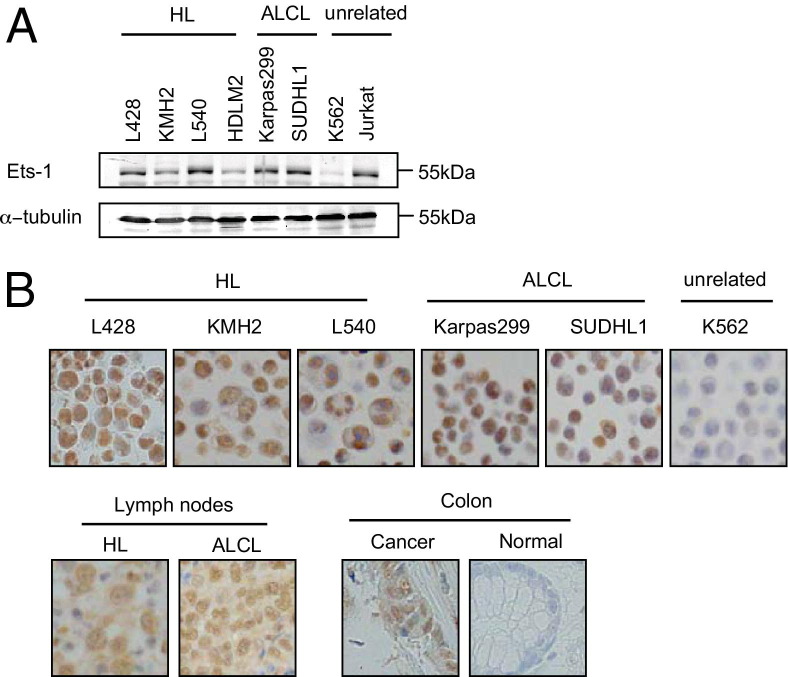
The expression of Ets-1 in HL and ALCL cells has been poorly characterized. We, therefore, examined Ets-1 expression in cell lines and clinical samples of HL and ALCL. Immunoblot analysis of HL and ALCL cell lines indicated that Ets-1 is constitutively expressed in these cells, although its expression level differs between the cell lines tested (Figure 4A). Immunostaining of these cell lines showed predominant nuclear localization of Ets-1. The same Ets-1 staining pattern was observed in biopsy samples of HL and ALCL (n = 5 for each). Based on previous reports,28,29 we used normal and neoplastic colon samples as negative and positive controls, respectively. Representative results are shown in Figure 4B. These results indicate that Ets-1 is constitutively expressed mainly in the nucleus of HL and ALCL cells.
Figure 4.
Constitutive expression of Ets-1 in HL and ALCL cells. A: Immunoblot analysis of Ets-1. Whole cell lysates, 30 μg, of four HL cell lines, two ALCL cell lines, and two unrelated cell lines were examined. The migration of Ets-1 and α-tubulin relative to a molecular weight marker is indicated. B: IHC analysis of Ets-1. Staining of Ets-1 in HL, ALCL, and K562 cell lines (top panel) and representative Ets-1 staining of clinical examples of classic HL and ALCL (n = 5 each; bottom panel) and of healthy and neoplastic colon samples that served as negative and positive controls, respectively (n = 5 each; bottom panel), are shown. Original magnification, ×400.
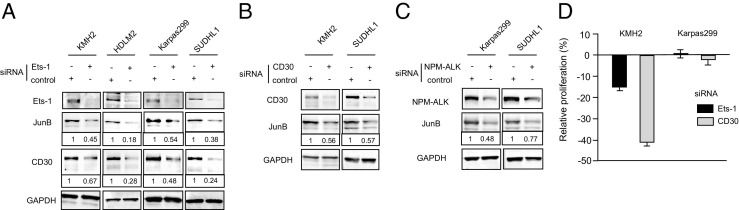
Ets-1 Knockdown Reduces the Expression of JunB and CD30 in HL and ALCL Cell Lines
To further evaluate the involvement of Ets-1 in JunB induction in HL and ALCL cells, we examined the effect of Ets-1 knockdown on the expression of JunB and CD30. Ets-1 knockdown reduced the expression of JunB and CD30, suggesting that knockdown of Ets-1 triggers down-regulation of JunB, which, in turn, decreases CD30 expression in these cells (Figure 5A). In addition to CD30, NPM-ALK has been involved in JunB induction via ERK1/2-MAPK.15,21,30 To elucidate the effect of CD30 and NPM-ALK on JunB induction in HL and ALCL, we next examined the effect of CD30 and NPM-ALK knockdown on JunB expression. Knockdown of CD30 in HL and ALCL cell lines (KMH2 and SUDHL1, respectively) and of NPM-ALK in ALCL cell lines expressing NPM-ALK (Karpas299 and SUDHL1) significantly reduced JunB expression (Figure 5, B and C). Immunoblot analysis showed that the expression of Ets-1, JunB, CD30, and glyceraldehyde-3-phosphate dehydrogenase was almost the same in both types of cells transfected with the negative control siRNA or with vehicle alone, indicating that the negative control siRNA did not cause nonspecific effects on the expression of these proteins (data not shown). Combined with the results shown in Figure 3, these experiments indicate that Ets-1–mediated JunB induction is triggered by CD30 in HL and ALCL, and also by NPM-ALK in ALCL, via the ERK1/2-MAPK pathway.
Figure 5.
Interrelationship among Ets-1, JunB, CD30, and NPM-ALK in HL and ALCL cell lines. A: Ets-1 knockdown reduced the expression of JunB and CD30 in HL and ALCL cell lines. Immunoblot analyses of Ets-1, JunB, and CD30 after 24 hours of Ets-1 knockdown were performed as described in Materials and Methods. Whole cell lysate, 30 μg, was used for each analysis. The level of expression of JunB and CD30 in cells treated with Ets-1 or control siRNA was measured using densitometry. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Protein expression levels in cells treated with Ets-1 siRNA are expressed relative to those treated with control siRNA, which were set at 1. B and C: CD30 or NPM-ALK knockdown reduced the expression of JunB in HL and ALCL cell lines. Immunoblot analyses of JunB, CD30, and NPM-ALK 24 hours after CD30 or NPM-ALK knockdown was performed as described in Materials and Methods. In the KMH2 HL cell line, only CD30 was knocked down (B), whereas in the ALCL cell lines, CD30 and NPM-ALK were knocked down by their respective siRNA (B and C, respectively). Whole cell lysate, 30 μg, was used for each analysis. The expression levels of JunB in cells treated with CD30 or NPM-ALK siRNA relative to controls were calculated as described as in A. D: Cell proliferation analysis after Ets-1 and CD30 knockdown. The KMH2 HL and the Karpas299 ALCL cell lines were transduced with Ets-1, CD30, or control siRNAs, and their proliferations were determined by methyl thiazolyl tetrazolium assays. Proliferation levels in cells treated with Ets-1 or CD30 siRNA are expressed relative to those treated with control siRNA, which were set at 100%. The results are presented as reduction rates. Data are given as the mean ± SD of more than three triplicate experiments.
We next examined the effects of Ets-1 knockdown in the proliferation of HL and ALCL cell lines. We recently reported that knockdown of CD30 reduces the proliferation in HL cell lines.31 Knockdown of Ets-1 and CD30 caused a maximum and significant reduction (15.1%±1.5% and 40.8%±1.7%, respectively) in the proliferation of the KMH2 HL cell line 4 days after knockdown (Figure 5D). The relatively low reduction in the proliferation by Ets-1 knockdown compared with the reduction by CD30 knockdown may be due to indirect and partial repression of CD30 via JunB repression. In contrast, knockdown of Ets-1 and CD30 did not cause a significant reduction in the proliferation of the Karpas299 ALCL cell line (Figure 5D). Immunoblot analysis confirmed successful reduction in Ets-1 and CD30 expression by knockdown in these cell lines (data not shown).
Discussion
In this report, we localized an Ets-1–modulated cis-acting enhancer to the region between −146 and −137 of the JunB promoter in HL and ALCL cells. Previous reports indicated the existence of various cis-acting enhancer elements in the murine JunB promoter (eg, a CRE-like site located between −135 and −128, NF-κB sites located further downstream, and Ets sites located between −848 and −574 or between −196 and −91).32–35 Because different cell lines and stimuli were used in these studies, cis-acting enhancer sites in the JunB promoter in the murine system may differ, depending on cell types and stimuli. Although the induction of c-fos and JunB by activation of the human μ-opioid receptor has been dependent on the MAPK pathway,36 little is known about the molecule(s) responsible for JunB induction in the human system. Our results indicate that Ets-1 is critical for JunB overexpression in HL and ALCL. We did not investigate if a downstream JunB gene fragment, which includes sequences that bind NF-κB, might act as a cis-acting enhancer of the JunB promoter. Our previous study15 showed that NF-κB is not involved in JunB induction in HL and ALCL. Therefore, involvement of NF-κB in JunB induction in these cells is unlikely. The Ets-1 sequence identified as a cis-acting enhancer of the JunB promoter in our study is highly conserved between humans and mice, supporting the importance of this Ets-1 site for JunB promoter induction.
Previous reports37,38 suggested that the Ets protein functions cooperatively with AP-1 transcription factors to regulate the expression of a wide variety of genes. An AP-1 binding site has been identified adjacent to Ets binding sites in many of these genes, and in some cases, mutation of either the AP-1 site or the Ets site has eliminated the transcriptional activity mediated by these elements. In addition to this functional evidence of cooperative activity, direct physical interaction between Ets and AP-1 transcription factors has been demonstrated. The Ets–AP-1 association is mediated by binding of the basic domain of AP-1 proteins to the Ets domain of Ets proteins.39 In the JunB promoter, we found an AP-1 binding site adjacent to an Ets binding site and hypothesized that the JunB promoter might be self-activated by an Ets-1–JunB complex. Although immunoprecipitation analyses using HL and ALCL cell lines showed interaction of Ets-1 and JunB, and Ets-1 and JunB partially colocalized in the nucleus of these cells, functional evidence of cooperative activity was not obtained using luciferase assays (data not shown). This result is supported by experiments that show that the Ets-1 site alone is critical for JunB promoter induction (Figure 1, B and C) and that Ets-1 knockdown is sufficient for inhibition of JunB expression (Figure 5A). Collectively, these results suggest that the observed Ets-1 transcriptional enhancement occurs without the cooperation of JunB.
CD30 knockdown significantly reduced JunB expression in HL and ALCL cell lines. NPM-ALK knockdown also reduced JunB expression in ALCL cell lines that express NPM-ALK. We previously showed that ligand-independent CD30-ERK1/2-MAPK signals induce enhanced JunB protein expression, which acts on the unmethylated CD30 promoter to maintain a high expression of CD30 in HL and ALCL cells.13,15,22 Another report30 indicated that NPM-ALK-ERK1/2-MAPK is responsible for JunB induction. Therefore, in ALCL cells that express chimeric ALK, both CD30 and chimeric ALK appear to be responsible for Ets-1–mediated induction of JunB, which, in turn, activates the CD30 promoter and contributes to high expression of CD30. In HL and ALCL without chimeric ALK, CD30 appears to be a major factor for Ets-1–mediated induction of JunB and CD30.
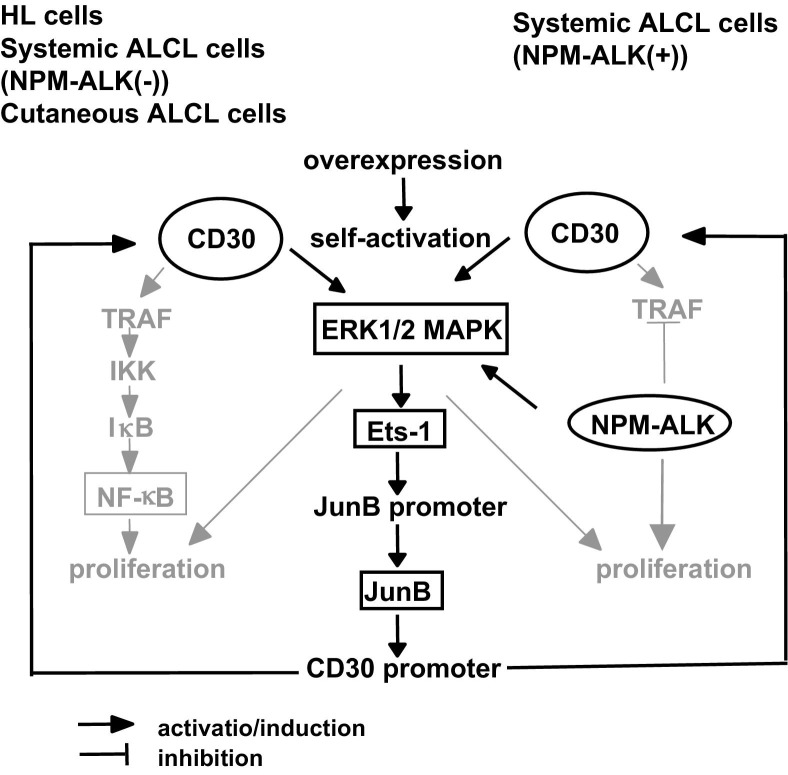
We showed that knockdown of Ets-1 and CD30 caused a significant decrease in the proliferation of the KMH2 HL cell line. In contrast, knockdown of Ets-1 and CD30 did not cause a significant decrease in the proliferation of the Karpas299 ALCL cell line. This result may be because of the expression of NPM-ALK in Karpas299, which also supports proliferation of these cells via ERK1/2-MAPK, phosphoinositide 3-kinase–AKT, Janus kinase–signal transducer and activator of transcription 3, and phospholipase C γ pathways.40 These results indicate that the Ets-1–mediated JunB-CD30 pathway affects the proliferation of HL and ALCL cells without NPM-ALK, whereas this pathway can be compensated by NPM-ALK in the proliferation of ALCL cells expressing NPM-ALK. A schematic outline of the findings in this study, combined with those of our previous study,15 is indicated in Figure 6.
Figure 6.
Schematic outline of CD30 signaling and CD30 promoter induction by Ets-1–JunB in ALCL and HL. Although the contribution of the tumor necrosis factor receptor-associated factor (TRAF)–IκB kinase (IKK)–IκB–NF-κB pathway differs in HL and ALCL cells, the CD30-ERK1/2 MAPK–Ets-1–JunB pathway is a common pathway that maintains overexpression of CD30 and supports cell survival. NPM-ALK supports overexpression of CD30 and survival of lymphoma cells by activating the NPM-ALK-ERK1/2 MAPK–Ets-1–JunB pathway in systemic ALCL NPM-ALK(+).
In conclusion, the results of this study indicate that CD30 and NPM-ALK collaborate to activate the ERK1/2 MAPK–Ets-1 pathway. Ets-1 is constitutively activated by ERK1/2-MAPK and plays a central role in the high expression of JunB and CD30 in both HL and ALCL.
Footnotes
Supported by a Grant-in-Aid from the Ministry of Education, Science, and Culture of Japan (20590374 to R.H.) and an NIH grant (5P20RR018757-09 to M.E.K.).
References
- 1.Horie R., Watanabe T. CD30: expression and function in health and disease. Semin Immunol. 1998;10:457–470. doi: 10.1006/smim.1998.0156. [DOI] [PubMed] [Google Scholar]
- 2.Horie R., Watanabe M., Ishida T., Koiwa T., Aizawa S., Itoh K., Higashihara M., Kadin M.E., Watanabe T. The NPM-ALK oncoprotein abrogates CD30 signaling and constitutive NF-kappaB activation in anaplastic large cell lymphoma. Cancer Cell. 2004;5:353–364. doi: 10.1016/s1535-6108(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 3.Wasik M.A., Zhang Q., Marzec M., Kasprzycka M., Wang H.Y., Liu X. Anaplastic lymphoma kinase (ALK)-induced malignancies: novel mechanisms of cell transformation and potential therapeutic approaches. Semin Oncol. 2009;36:S27–S35. doi: 10.1053/j.seminoncol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Wagner E.F. AP-1: introductory remarks. Oncogene. 2001;20:2334–2335. doi: 10.1038/sj.onc.1204416. [DOI] [PubMed] [Google Scholar]
- 5.Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 6.Jochum W., Passegue E., Wagner E.F. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 7.Li B., Tournier C., Davis R.J., Flavell R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvamurugan N., Kwok S., Partridge N.C. Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells. J Biol Chem. 2004;279:27764–27773. doi: 10.1074/jbc.M312870200. [DOI] [PubMed] [Google Scholar]
- 9.Robinson C.M., Prime S.S., Huntley S., Stone A.M., Davies M., Eveson J.W., Paterson I.C. Overexpression of JunB in undifferentiated malignant rat oral keratinocytes enhances the malignant phenotype in vitro without altering cellular differentiation. Int J Cancer. 2001;91:625–630. [PubMed] [Google Scholar]
- 10.Mao X., Orchard G., Lillington D.M., Russell-Jones R., Young B.D., Whittaker S.J. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphomas. Blood. 2003;101:1513–1519. doi: 10.1182/blood-2002-08-2434. [DOI] [PubMed] [Google Scholar]
- 11.Leaner V.D., Kinoshita I., Birrer M.J. AP-1 complexes containing cJun and JunB cause cellular transformation of Rat1a fibroblasts and share transcriptional targets. Oncogene. 2003;22:5619–5629. doi: 10.1038/sj.onc.1206644. [DOI] [PubMed] [Google Scholar]
- 12.Ohyama M., Hirayama Y., Tanuma J., Hirano M., Semba I., Shisa H., Hiai H., Sugihara K., Kitano M. Expressions of junB and c-fos are enhanced in 4-nitroquinoline 1-oxide-induced rat tongue cancers. Pathol Int. 2004;54:35–40. doi: 10.1046/j.1440-1827.2003.01587.x. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe M., Ogawa Y., Ito K., Higashihara M., Kadin M.E., Abraham L.J., Watanabe T., Horie R. AP-1 mediated relief of repressive activity of the CD30 promoter microsatellite in Hodgkin and Reed-Sternberg cells. Am J Pathol. 2003;163:633–641. doi: 10.1016/S0002-9440(10)63690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathas S., Hinz M., Anagnostopoulos I., Krappmann D., Lietz A., Jundt F., Bommert K., Mechta-Grigoriou F., Stein H., Dorken B., Scheidereit C. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002;21:4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M., Sasaki M., Itoh K., Higashihara M., Umezawa K., Kadin M.E., Abraham L.J., Watanabe T., Horie R. JunB induced by constitutive CD30-extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase signaling activates the CD30 promoter in anaplastic large cell lymphoma and Reed-Sternberg cells of Hodgkin lymphoma. Cancer Res. 2005;65:7628–7634. doi: 10.1158/0008-5472.CAN-05-0925. [DOI] [PubMed] [Google Scholar]
- 16.Rassidakis G.Z., Thomaides A., Atwell C., Ford R., Jones D., Claret F.X., Medeiros L.J. JunB expression is a common feature of CD30+ lymphomas and lymphomatoid papulosis. Mod Pathol. 2005;18:1365–1370. doi: 10.1038/modpathol.3800419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie R., Watanabe T., Morishita Y., Ito K., Ishida T., Kanegae Y., Saito I., Higashihara M., Mori S., Kadin M.E. Ligand-independent signaling by overexpressed CD30 drives NF-kappaB activation in Hodgkin-Reed-Sternberg cells. Oncogene. 2002;21:2493–2503. doi: 10.1038/sj.onc.1205337. [DOI] [PubMed] [Google Scholar]
- 18.Bargou R.C., Emmerich F., Krappmann D., Bommert K., Mapara M.Y., Arnold W., Royer H.D., Grinstein E., Greiner A., Scheidereit C., Dorken B. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J Clin Invest. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe M., Dewan M.Z., Taira M., Shoda M., Honda M., Sata T., Higashihara M., Kadin M.E., Watanabe T., Yamamoto N., Umezawa K., Horie R. IkappaBalpha independent induction of NF-kappaB and its inhibition by DHMEQ in Hodgkin/Reed-Sternberg cells. Lab Invest. 2007;87:372–382. doi: 10.1038/labinvest.3700528. [DOI] [PubMed] [Google Scholar]
- 20.Zheng B., Fiumara P., Li Y.V., Georgakis G., Snell V., Younes M., Vauthey J.N., Carbone A., Younes A. MEK/ERK pathway is aberrantly active in Hodgkin disease: a signaling pathway shared by CD30, CD40, and RANK that regulates cell proliferation and survival. Blood. 2003;102:1019–1027. doi: 10.1182/blood-2002-11-3507. [DOI] [PubMed] [Google Scholar]
- 21.Marzec M., Kasprzycka M., Liu X., Raghunath P.N., Wlodarski P., Wasik M.A. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene. 2007;26:813–821. doi: 10.1038/sj.onc.1209843. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe M., Ogawa Y., Itoh K., Koiwa T., Kadin M.E., Watanabe T., Okayasu I., Higashihara M., Horie R. Hypomethylation of CD30 CpG islands with aberrant JunB expression drives CD30 induction in Hodgkin lymphoma and anaplastic large cell lymphoma. Lab Invest. 2008;88:48–57. doi: 10.1038/labinvest.3700696. [DOI] [PubMed] [Google Scholar]
- 23.Andrews N.C., Faller D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel T.A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 25.Horie R., Ito K., Tatewaki M., Nagai M., Aizawa S., Higashihara M., Ishida T., Inoue J., Takizawa H., Watanabe T. A variant CD30 protein lacking extracellular and transmembrane domains is induced in HL-60 by tetradecanoylphorbol acetate and is expressed in alveolar macrophages. Blood. 1996;88:2422–2432. [PubMed] [Google Scholar]
- 26.Staber P.B., Vesely P., Haq N., Ott R.G., Funato K., Bambach I., Fuchs C., Schauer S., Linkesch W., Hrzenjak A., Dirks W.G., Sexl V., Bergler H., Kadin M.E., Sternberg D.W., Kenner L., Hoefler G. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- 27.Yordy J.S., Muise-Helmericks R.C. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama T., Ito M., Ohtsuru A., Naito S., Sekine I. Expression of the ets-1 proto-oncogene in human colorectal carcinoma. Mod Pathol. 2001;14:415–422. doi: 10.1038/modpathol.3880328. [DOI] [PubMed] [Google Scholar]
- 29.Ito Y., Takeda T., Okada M., Matsuura N. Expression of ets-1 and ets-2 in colonic neoplasms. Anticancer Res. 2002;22:1581–1584. [PubMed] [Google Scholar]
- 30.Hsu F.Y., Johnston P.B., Burke K.A., Zhao Y. The expression of CD30 in anaplastic large cell lymphoma is regulated by nucleophosmin-anaplastic lymphoma kinase-mediated JunB level in a cell type-specific manner. Cancer Res. 2006;66:9002–9008. doi: 10.1158/0008-5472.CAN-05-4101. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe M., Kinji I., Togano T., Nakashima M., Higashihara M., Kadin M.E., Watanabe T., Horie R. Targeted repression of overexpressed CD30 downregulates NF-κB and ERK1/2 pathway in Hodgkin lymphoma cell lines. Oncol Res. 2012 doi: 10.3727/096504012x13285365944292. (in press) [DOI] [PubMed] [Google Scholar]
- 32.Amato S.F., Nakajima K., Hirano T., Chiles T.C. Transcriptional regulation of the junB gene in B lymphocytes: role of protein kinase A and a membrane Ig-regulated protein phosphatase. J Immunol. 1997;159:4676–4685. [PubMed] [Google Scholar]
- 33.Brown R.T., Ades I.Z., Nordan R.P. An acute phase response factor/NF-kappa B site downstream of the junB gene that mediates responsiveness to interleukin-6 in a murine plasmacytoma. J Biol Chem. 1995;270:31129–31135. doi: 10.1074/jbc.270.52.31129. [DOI] [PubMed] [Google Scholar]
- 34.Frazier-Jessen M.R., Thompson C.D., Brown R., Rawat R., Nordan R.P., Feldman G.M. NF-kappaB elements contribute to junB inducibility by lipopolysaccharide in the murine macrophage cell line RAW264.7. FEBS Lett. 2002;513:203–207. doi: 10.1016/s0014-5793(02)02295-0. [DOI] [PubMed] [Google Scholar]
- 35.Coffer P., de Jonge M., Mettouchi A., Binetruy B., Ghysdael J., Kruijer W. junB promoter regulation: ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene. 1994;9:911–921. [PubMed] [Google Scholar]
- 36.Shoda T., Fukuda K., Uga H., Mima H., Morikawa H. Activation of mu-opioid receptor induces expression of c-fos and junB via mitogen-activated protein kinase cascade. Anesthesiology. 2001;95:983–989. doi: 10.1097/00000542-200110000-00030. [DOI] [PubMed] [Google Scholar]
- 37.Wasylyk B., Wasylyk C., Flores P., Begue A., Leprince D., Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature. 1990;346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 38.Gottschalk L.R., Giannola D.M., Emerson S.G. Molecular regulation of the human IL-3 gene: inducible T cell-restricted expression requires intact AP-1 and Elf-1 nuclear protein binding sites. J Exp Med. 1993;178:1681–1692. doi: 10.1084/jem.178.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassuk A.G., Leiden J.M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 40.Palmer R.H., Vernersson E., Grabbe C., Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]