Abstract
Although the oncogenic functions of activating BRAF mutations have been clearly demonstrated in human cancer, their roles in nontransformed epithelial cells remain largely unclear. Investigating the cellular response to the expression of mutant BRAF in nontransformed epithelial cells is fundamental to the understanding of the roles of BRAF in cancer pathogenesis. In this study, we used two nontransformed cyst108 and RK3E epithelial cell lines as models in which to compare the phenotypes of cells expressing BRAFWT and BRAFV600E. We found that transfection of the BRAFV600E, but not the BRAFWT, expression vector suppressed cellular proliferation and induced apoptosis in both cell types. BRAFV600E generated reactive oxygen species, induced DNA double-strand breaks, and caused subsequent DNA damage response as evidenced by an increased number of pCHK2 and γH2AX nuclear foci as well as the up-regulation of pCHK2, p53, and p21. Because BRAF and KRAS (alias Ki-ras) mutations have been correlated with GLUT1 up-regulation, which encodes glucose transporter-1, we demonstrated here that expression of BRAFV600E, but not BRAFWT, was sufficient to up-regulate GLUT1. Taken together, our findings provide new insights into mutant BRAF-induced oncogenic stress that is manifested by DNA damage and growth arrest by activating the pCHK2-p53-p21 pathway in nontransformed cells, while it also confers tumor-promoting phenotypes such as the up-regulation of GLUT1 that contributes to enhanced glucose metabolism that characterizes tumor cells.
BRAF probably represents the most frequently mutated oncogene within the kinase family and activating point mutation at the hot spot V600E of BRAF has been found in several types of human neoplasms, most frequently in melanoma,1 papillary thyroid carcinoma,2,3 high-grade malignant astrocytoma4 and ovarian low-grade serous neoplasms.5 BRAF protein is a downstream effector of KRAS and participates in the signal transduction of the mitogen activated protein kinase (MAPK) pathway that controls cellular growth, differentiation, and survival.6,7 Dimerization of the BRAF kinase domain with KSR or with other RAF molecules has been recently shown to be central to its activation mechanism.8 Activating mutations in BRAF and KRAS appear to exert equivalent tumor-promoting effects as based on the mutual exclusive mutation in both genes.5,9 Constitutive activation of BRAF due to V600E mutation activates the MAPK pathway and results in up-regulation of several genes with tumor-promoting functions including cyclin D1,10,11 and targeting BRAF and its downstream effectors has emerged as a new therapeutic strategy for those tumors harboring the BRAF mutation.12–16
Ovarian low-grade serous tumor represents a unique type of ovarian epithelial neoplasm and is distinct from ovarian high-grade serous carcinoma, the conventional type of ovarian cancer, based on their clinical, pathological, and molecular features.17,18 Ovarian low-grade serous tumors include a benign form, serous borderline tumor, and the malignant counterpart, low-grade serous carcinoma. Low-grade serous carcinoma develops from serous borderline tumor, which in turn may arise from an ovarian serous cystadenoma. Both ovarian low-grade serous carcinoma and serous borderline tumor harbor BRAF, KRAS, or ERBB2 sequence mutation in more than 50% of cases.5,19–21 Expression of active MAPK was more frequently observed in low-grade serous tumors than in high-grade ovarian serous carcinomas that have rare mutations in either BRAF or KRAS.22 Moreover, BRAF and KRAS mutation status is a useful predictor of sensitivity to MEK inhibition in ovarian cancer.11,23 Interestingly, BRAF or KRAS mutations can be detected in morphologically normal-appearing cyst epithelium that is adjacent to a serous borderline tumor but not in the cystadenomas without concurrent borderline tumors, suggesting the mutations may occur early during tumor progression of ovarian low-grade serous tumors.24 Although the oncogenic roles of BRAF mutations have been established in mouse models,25 it remains largely unclear what are the biological effects of BRAF mutations in the very beginning of tumor formation such as in nontransformed epithelial cells. Thus, in this study, we ectopically expressed either BRAFV600E or BRAFWT in nontransformed epithelial cells isolated from ovarian cystadenoma and RK3E cells, an epithelial cell model frequently used to test the oncogenic effects, to determine the phenotypes in both cell lines. Furthermore, a recent study has demonstrated that BRAF expression is required for the expression of GLUT1, which encodes glucose transporter-1, and glucose deprivation is associated with the development of KRAS pathway mutations in tumor cells.26 Thus, in this study, we also tried to determine whether mutant BRAF plays a causal role in up-regulating GLUT1 expression in our cellular model.
Materials and Methods
Cell Growth Assay
Expression vectors including the empty vector, wild-type BRAF (BRAFWT), and mutant BRAF (BRAFV600E) were kind gifts from Dr. Raquel Seruca (Institute of Molecular Pathology and Immunology of the University of Porto, Portugal). To determine the effects of BRAFV600E on nontransformed epithelial cells, we established the cyst108 cell line, which was derived from a benign ovarian serous cystadenoma. The reason to use the epithelial cells from a cystadenoma was because cystadenoma represents the immediate precursor lesion of serous borderline tumor. To establish cyst108, we scraped the epithelial cells directly from a benign serous cystadenoma after incubating a fragment of cystadenoma with 0.5% trypsin and EDTA at 37°C for 15 minutes. The epithelial cells were then rigorously suspended to obtain a single cell population. After overnight culture, the cells were immortalized with SV40 large T antigen, and the epithelial cells were enriched using cold trypsin treatments to eliminate stromal cells afterward. Cyst108 cells were maintained in RPMI1640 medium supplemented with 10% fetal bovine serum and have been passed for at least 30 passages; these cells exhibited epithelioid morphology under phase-contrast microscopy and expressed epithelial cell markers, including cytokeratin 18 and Epi-CAM in >98% of cells. They showed contact inhibition in vitro and were not tumorigenic in nu/nu mice for more than 3 months. In this study, we also included the RK3E cell line because it has been widely used to assess transformation ability of potential oncogenes.27–33 Cells transfected with BRAFV600E were grown in 96-well plates at a density of 3000 cells per well. As controls, the empty vector and BRAFWT vector were also transfected into the cells. Cell number was measured daily for 4 consecutive days using the SYBR green I staining method (Molecular Probes, Eugene, OR). The data were expressed as mean ± SD from five replicates.
Detection of Reactive Oxygen Species
Cyst108 and RK3E cells were seeded on chamber slides (Nunc, Roskilde, Denmark), and subsequently transfected with empty vector, BRAFWT, or BRAFV600E vectors on the second day. Seventy-two hours after transfection, cells were stained with 5 μmol/L CellROX Deep Red reagent (Invitrogen, Carlsbad, CA) in complete medium for 30 minutes at 37°C, followed by three washes with PBS and fixation with 4% formaldehyde. Cell nuclei were counterstained with DAPI (Sigma, St. Louis, MO). Reactive oxygen species (ROS)-positive cells were detected and counted under a fluorescent microscope (excitation wave: 644 nm and emission wave: 665 nm). The data were expressed as mean ± SD from triplicates.
DNA Strand Break Assay
DNA strand breaks were quantified using a Comet assay kit (Trevigen, Gaithersburg, MD) as previously described.33,34 Briefly, transfected cells were harvested in ice-cold PBS, and the cell number was adjusted at a density of 1 × 105 cells/mL. Cells were mixed with LMAgarose at 1:10 ratio (v/v) and spread onto the CometSlide immediately. After gel solidification, cells on slides were lysed, and DNA in the cells was denatured by using the buffers provided by the kit. Fragmented DNA strands were separated from nuclei by electrophoresis and detected by SYBR Green staining. The percentage of comet-like nuclei (with DNA strand breaks) was counted under a fluorescent microscope from five randomly selected high-power fields (×40) with each approximately containing 100 nuclei. UVC-treated cells at sublethal dose were used as the positive control in this assay.
Immunofluorescence Staining
To determine whether BRAFV600E expression resulted in a DNA damage response, transfected cells were seeded in chamber slides at a density of 5000 cells per well. At different time points, cells were fixed with para-formaldehyde and incubated with anti–phospho-CHK2 (pCHK2) antibody (clone ab38461; Abcam, Cambridge, MA) or anti-γH2AX antibody (clone ab11174; Abcam) for 2 hours followed by rhodamine-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) and nuclei were counterstained with DAPI (Sigma, St. Louis, MO). Cells transfected with an empty vector or BRAFWT vector were used as controls.
Western Blot Analysis
Protein lysates from different groups were collected at different time points after gene transfection. Proteins were then separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. To determine whether BRAFV600E caused activation of the DNA damage response pathway, we performed Western blot analysis by hybridizing membranes with antibodies against γH2AX (clone ab11174; Abcam), phosphor-CHK2 (clone ab38461; Abcam), p53 (clone sc-6243; Santa Cruz Biotechnology, Santa Cruz, CA), and p21 (clone sc-6246; Santa Cruz Biotechnology), all of which are involved in the DNA damage response pathway35,36 for 2 hours at room temperature. Antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as the loading control. To detect GLUT1, the membranes were hybridized with an affinity-purified rabbit anti-GLUT1 polyclonal antibody (Millipore, Bedford, MA). After three washes with 0.01% Tween 20 in Tris-buffered saline, the membranes were blotted with horseradish peroxidase-conjugated anti-mouse (Pierce, Rockford, IL) or anti-rabbit (Cell Signaling Technology, Danvers, MA) antibodies for 1 hour at room temperature. Protein bands were revealed by chemiluminescence (Amersham Biosciences, Arlington Heights, IL).
GLUT1 Immunohistochemistry
Paraffin-embedded tissues from 33 cases of ovarian low-grade serous tumors (serous borderline tumors and low-grade serous carcinomas) were obtained from the Department of Pathology at the Johns Hopkins Hospital, Baltimore, MD. Acquisition of tissue specimens and clinical information were approved under the regulations of the institutional review board. There were 10 cases of low-grade serous carcinomas that metastasized or disseminated to intraperitoneal soft tissues. For immunohistochemistry, the unstained slides were subjected to antigen retrieval by boiling the slides in citrate buffer (pH 6.0) (Zymed, South San Francisco, CA) for 20 minutes. After blocking, samples were then stained with an affinity-purified rabbit anti-GLUT1 polyclonal antibody (Millipore) at a 1:600 dilution at room temperature for 1 hour. An EnVision+System peroxidase kit (DAKO, Carpentaria, CA) was used for chromogen development. Immunointensity was independently scored by two investigators based on membrane immunoreactivity and labeled as negative (0), weakly positive (1+), moderately positive (2+), and strongly positive (3+) groups. For discordant cases, a third investigator scored, and the final intensity score was determined by the majority scores.
Results
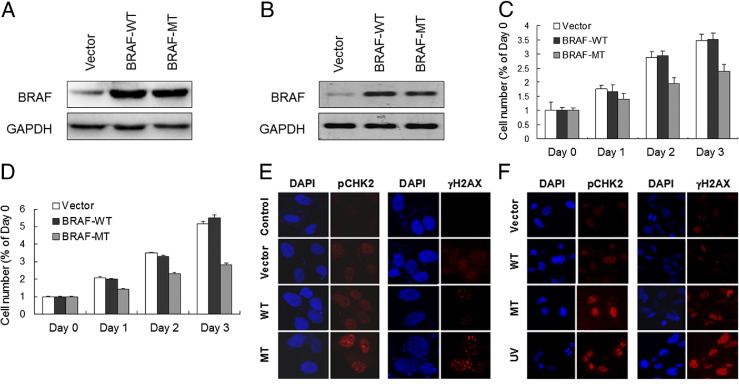
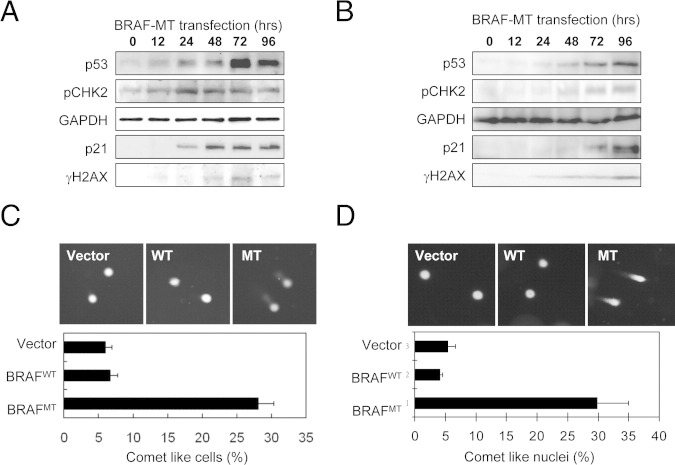
To determine the effect of mutant BRAF on cyst108 and RK3E cells, we transfected both cell lines with constructs that expressed mutant (V600E) and wild-type BRAF and compared the proliferative activity of cells in vitro (Figure 1, A and B). As shown in Figure 1, C and D, both cyst108 and RK3E cells expressing wild-type BRAF continued growing as in the vector control groups. By contrast, the proliferative activity significantly decreased in cyst108 and RK3E cells when they expressed BRAFV600E. One of the explanations for the growth-inhibitory effects by mutant BRAF is oncogenic stress that describes growth arrest and cellular senescence as a result of expression of oncogenes in otherwise normal cells.37,38 Because oncogenic stress is a poorly defined process, we sought to determine whether it was directly related to DNA damage response. First, we performed immunofluoresence staining for two representative markers of DNA damage, including phosphorylated checkpoint kinase 2 (pCHK2) and phosphorylated histone 2AX (γH2AX) in cyst108 and RK3E cells. We found that BRAFV600E-expressing cells demonstrated an increased number of nuclear foci of pCHK2 and γH2AX as compared to the cells in the control groups, including BRAFWT-expressing, vector control, and parental cells (Figure 1, E and F). UV-irradiated cells served as the positive control which induced numerous pCHK2 and γH2AX foci in the nuclei. Western blot analysis further demonstrated a time-dependent increase in protein levels of pCHK2, γH2AX, p53, and p21 in BRAFV600E, but not in BRAFWT, transfected cyst108 and RK3E cells (Figure 2, A and B).
Figure 1.
Expression of mutant BRAF (V600E) suppresses cellular proliferation and induces pCHK2 and γH2AX nuclear foci. Forty-eight hours after transfecting epithelial cells, robust expression levels of wild-type and mutant BRAF can be detected in cyst108 cells (A) and in RK3E cells (B). A reduced cellular proliferation was recorded in cyst108 cells (C) and in RK3E cells (D) that expressed mutant BRAF. Forty-eight hours after transfection, cells were stained for pCHK2 and γH2AX. Nuclear foci for pCHK2 and γH2AX immunofluoresence were observed in mutant BRAF-expressing cyst108 cells (E) and RK3E cells (F). UV light-treated cells serve as the positive control for immunofluoresence staining. MT, mutant; WT, wild type.
Figure 2.
Mutant BRAF activates DNA damage response pathway and induces DNA strand breaks. Western blot analysis demonstrates a time-dependent increase in protein expression of p53, pCHK2, γH2AX, and p21 in cyst108 cells (A) and in RK3E cells (B). GAPDH serves as the protein loading control. Cells with DNA strand breaks were analyzed by the Comet assay. For both cyst108 cells (C) and RK3E cells (D), a significantly higher percentage of comet-like nuclei are found in the BRAFV600E expressing group than in control group transfected with BRAFWT or vector only 48 hours after transfection. MT, mutant; WT, wild type.
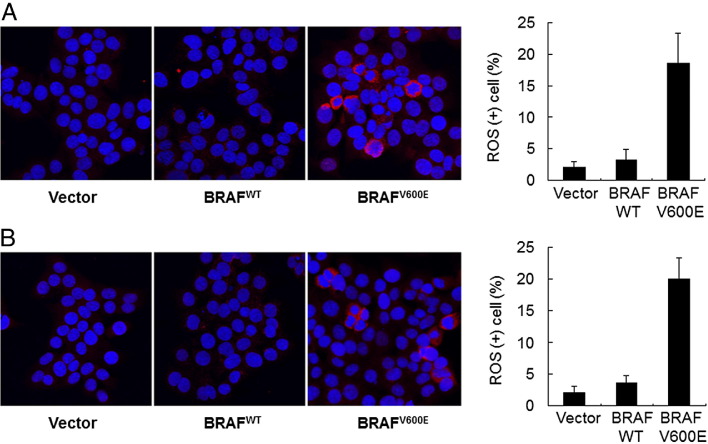
The above findings indicated that expression of mutant BRAF activated the DNA damage pathway and subsequently induced growth arrest because of up-regulation of p53 and p21 proteins. This observation also suggests that DNA strand breaks occur due to mutant BRAF, but not wild-type BRAF, expression. To determine whether this was the case, we directly visualized the individual cells with DNA strand breaks in cyst108 and RK3E cells. On electrophoresis, DNA with double-strand breaks migrated out of the nuclei, forming a comet tail–like structure, whereas the undamaged DNA remained within the nuclei. Figure 2, C and D, showed a higher percentage of comet-like cells in the BRAFV600E-expressing group than in control group transfected with BRAFWT or vector only as early as 48 hours after transfection. These findings suggest that ectopic expression of mutant BRAF proteins caused DNA strand breaks, initiated DNA damage response and subsequently up-regulated p53 and p21, leading to growth arrest in nontransformed epithelial cells. Although DNA double-strand breaks have several causes, we asked in this study whether expression of mutant BRAFV600E was associated with generation of reactive oxygen species, which were detected by CellROX Deep Red reagent. Both cyst108 and RK3E cells were analyzed, and their percentage of positive cells was determined under a fluorescent microscope. As shown in Figure 3, the percentage of positive cells was significantly higher in cells expressing BRAFV600E than cells expressing BRAFWT or in vector control cells.
Figure 3.
Measurement of reactive oxygen species in cyst108 (A) and RK3E cells (B). CellROX Deep Red reagent was used to detect the reactive oxygen species and percentage of positive cells was recorded under a fluorescent microscope. Cell nuclei were counter stained with DAPI (blue fluorescence). The percentage of positive cells (red fluorescence in cytoplasm) is significantly higher in cells expressing BRAFV600E than cells expressing BRAFWT or vector control cells.
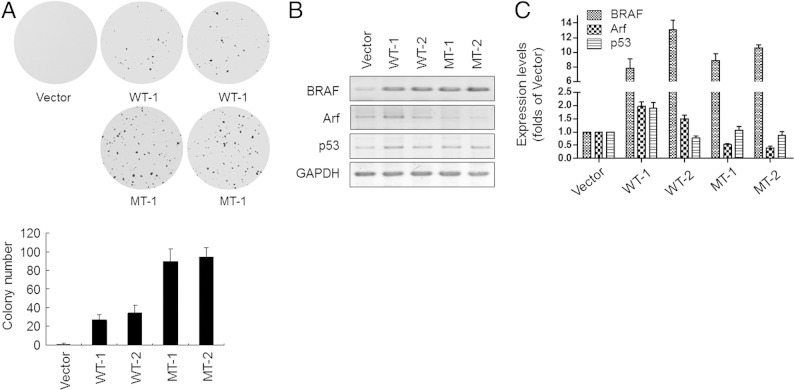
Several studies have shown that oncogene-induced DNA damage response serves as a molecular pressure to select tumorigenic clones during cancer development.39–41 To study whether BRAFV600E can also promote tumor progression, we selected two cyst108 cell clones, MT-1 and MT-2, that were refractory to BRAFV600E-induced cell death at a low cell density (1000 cells/25 cm2). Two BRAFWT cell clones, WT-1 and WT-2, were also analyzed as controls. Anchorage-independent assay showed that cell clones that overexpress either BRAFWT or BRAFV600E formed colonies in soft-agar (Figure 4A). Constitutive expression of BRAFV600E confirmed MT-1 and MT-2 clones to be highly transformed as evidenced by more colonies (Figure 4A). Consistent with our previous study,42 Western blot analysis (Figure 4B) and quantitative PCR (Figure 4C) confirmed lower expression levels of Arf in BRAFV600E-treated, but not in BRAFWT-treated, clones that accounted for the failure of p53 up-regulation.
Figure 4.
Long-term expression of BRAFV600E promotes a more transforming phenotype in cyst108 cells. A soft-agar assay was performed to detect the tumorigenic activity of cell clones that overexpress BRAFV600E or BRAFWT under G418 selection (A). Western blot analysis (B) and quantitative PCR (C) were performed to detect expression levels of BRAF, Arf, and p53 in selected cell clones. GAPDH was used as the protein loading control. Vector-treated cells serve as controls in both assays. MT, mutant; WT, wild type.
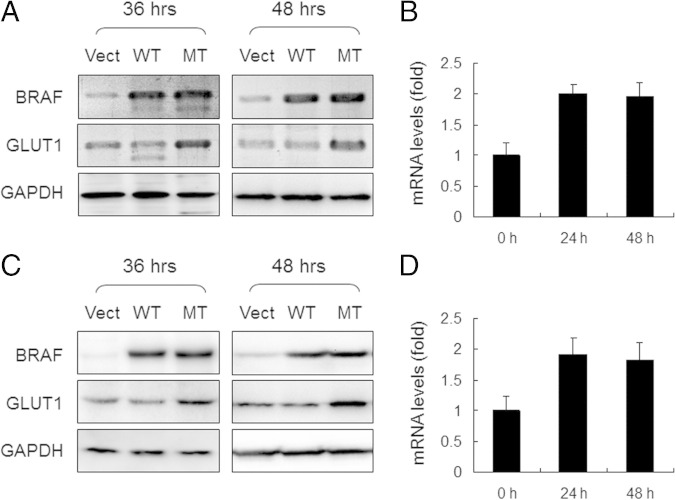
Previous study demonstrated that BRAF mutation was associated with up-regulation of GLUT1, which was responsible for an increased glucose uptake and promotion of cellular survival and growth in cancer cells.26 However, it is not known whether mutant BRAF is sufficient to up-regulate GLUT1 expression. Therefore, we applied Western blot analysis to demonstrate that expression of GLUT1 significantly increased in cyst108 and RK3E cells expressing BRAFV600E as compared to those cells expressing BRAFWT 36 hours and 48 hours after transfection (Figure 5, A and C). Similarly, real-time quantitative PCR also demonstrated a significant increase at the mRNA levels of GLUT1 in cells expressing BRAFV600E (Figure 5, B and D). To extrapolate the in vitro finding to human specimens, we performed immunohistochemistry of GLUT1 on a panel of 33 cases of ovarian low-grade serous tumors, including 23 serous borderline tumors and 10 low-grade serous carcinomas, and correlated their GLUT1 immunoreactivity and the mutation status of BRAF and KRAS. Consistent with our previous study,5 we found that mutations in BRAF and KRAS were mutually exclusive, and mutations in either one of the genes were detected in 18 (55%) of 33 specimens. Specifically, mutations of BRAF or KRAS were found in 6 (60%) of 10 metastatic/disseminated low-grade serous carcinomas, whereas the mutations were recorded in 12 (52%) of 23 serous borderline tumors.
Figure 5.
Increased GLUT1 protein expression in cells that express mutant BRAF. Western blot analysis was performed 36 hours and 48 hours after transfection. Expression of GLUT1 is significantly increased in cyst108 (A) and RK3E cells (C) expressing BRAFV600E as compared to those cells expressing BRAFWT at both time points. GAPDH serves as the protein loading control. Quantitative PCR was performed to detect mRNA levels of GLUT1 in cyst108 (B) and RK3E cells (D) 48 hours after gene transfection. MT, mutant.
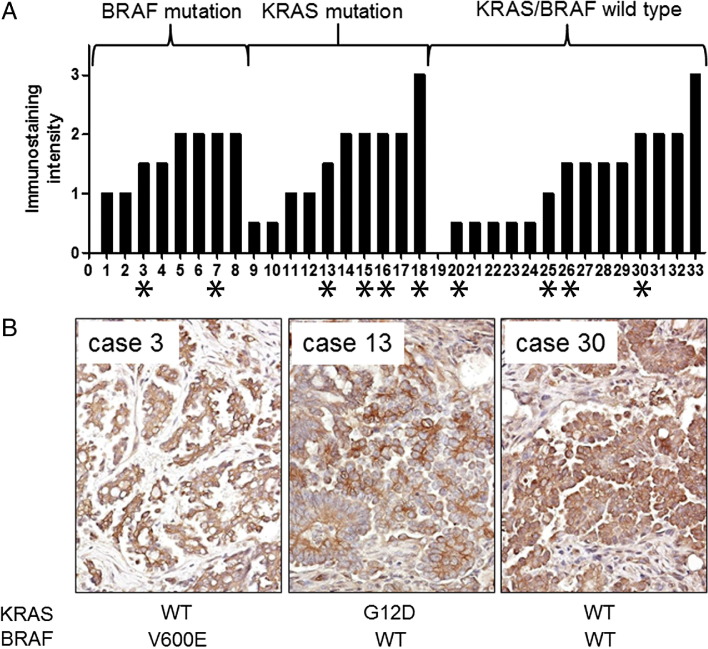
Immunohistochemically, we found that all 33 tumor samples were positive for GLUT1 staining except one serous borderline tumor (case 19) (Figure 6A). GLUT1 immunoreactivity was only detected in the cell membrane and cytoplasm of tumor cells, but not in stromal cells. All of the tumor specimens harboring either BRAF or KRAS mutations showed GLUT1 immunostaining; however, there was no significance in staining intensity among groups with BRAF mutation, KRAS mutation, and wild-type (P > 0.1, Mann-Whitney test). All 10 cases of low-grade serous carcinomas expressed GLUT1 protein, of which the immunostaining intensity score ranged from 1 to 3. Representative photomicrographs of GLUT1 staining in three advanced-stage low-grade serous carcinomas with different mutation status of BRAF and KRAS are illustrated in Figure 6B.
Figure 6.
Expression of GLUT1 in low-grade ovarian serous tumors and the correlation of GLUT1 immunoreactivity with mutation status of BRAF and KRAS. Immunohistochemistry was performed in 33 cases of low-grade ovarian serous tumors including 23 serous borderline tumors and 10 advanced stage low-grade serous carcinomas. A: The GLUT1 immunostaining intensity is shown for all cases, which are grouped into BRAF mutation, KRAS mutation, and wild-type groups. Mutations of KRAS and BRAF are mutually exclusive. Low-grade serous carcinomas are labeled with asterisks, otherwise they are serous borderline tumors. B: Representative photomicrographs of low-grade serous carcinomas from each group are illustrated, and their mutation status in KRAS and BRAF is indicated below. The case numbers correspond to those shown above.
Discussion
Using both ovarian cystadenoma epithelial cells and RK3E cells as the models, we were able to demonstrate that mutant BRAF induced growth arrest in both cell types and such “oncogenic stress” is attributed, at least in part, by the DNA damage response pathway that subsequently activates p53 and p21. We provide cogent evidence in this report that expression of mutant, but not wild-type, BRAF directly causes DNA strand breaks and accounts for the activation of the DNA damage response. Transcription-induced DNA double-strand breaks have been proposed to occur when novel transcription is induced during tumor development.43 It is likely that expression of mutant BRAF, like deregulated expression of Myc,44 induces oxidative stress that is responsible for topoisomerase TOP2B-dependent DNA double-strand breaks in epithelial cells. In fact, we have also observed that expression of mutant BRAF was associated with generation of reactive oxygen species in epithelial cells. As occurs in oncogenic stress, increased p53 levels due to the ATM-pCHK2-p53-p21 pathway activation lead to cell growth arrest at G1 or G2/M and/or in apoptosis.45 Consistent with this view, it has also been reported that ovarian serous borderline tumors have a much lower proliferative activity46,47 and a significantly lower TP53 mutation frequency than ovarian high-grade serous carcinoma, the conventional type of ovarian cancer, that harbor neither BRAF nor KRAS mutations.48 Although this is our preferred view, other mechanisms for mutant BRAF-induced p53 activation should be also pointed out. For example, a recent study demonstrates that the Jnk pathway signaling is involved in the activation of p53 in response to both KRAS and Neu oncogene expression.49
The results from this study may help further understanding of the molecular pathogenesis of ovarian low-grade serous carcinoma. It can be speculated that epithelial cells from a serous borderline tumor may evolve a mechanism to restrain tumor progression.39–41 In response to BRAF mutations, activation of ATM/pCHK2/p53/p21 is thus important to suppress tumor cell proliferation. Although not frequently occurring, serous borderline tumors may progress to low-grade serous carcinomas, which are frankly malignant neoplasm and are often associated with high morbidity and mortality. How do tumor cells in low-grade serous carcinoma overcome the growth inhibitory effect due to activating mutations of BRAF? We propose that additional molecular genetic alterations occur during progression from a serous borderline tumor to a low-grade serous carcinoma, and such molecular alterations abolish the checkpoint controlled by the ATM-p53 pathway, allowing cells to proliferate despite the presence of mutant BRAF-induced DNA damage and up-regulation of p53 and p21. To this end, a recent study based on whole exome sequencing reveals rare somatic mutations in ovarian low-grade serous carcinomas except activating mutations in KRAS and BRAF, suggesting that molecular genetic changes other than sequence mutations may be responsible for tumor progression.21 In fact, a previous study that analyzed the genome-wide copy number alterations in ovarian serous neoplasms has reported that hemizygous ch1p36 deletion and ch9p21 homozygous or hemizygous deletions were much more common in ovarian low-grade serous carcinomas than in serous borderline tumor.42 The ch1p36 region contains several candidate tumor suppressors, including miR-34a, which is required for DNA damage response and is the direct p53 target that mediates its tumor suppressor functions.50,51 Similarly, the ch9p21 region corresponding to the CDKN2A/B locus encodes three well-known tumor suppressor proteins, p14 (Arf), p16, and p15. CDKN2A and CDKN2B share similar function in inhibiting cyclin-dependent kinase. Arf is a potent tumor suppressor that blocks cell cycle progression by interfering with the p53-negative regulator, MDM2, thereby stabilizing p53 protein expression. Besides, the expression level of CDKN2A was enhanced in response to oncogene-induced stress such as by the activation of the RAS/RAF/MEK signaling pathway. Thus, deletions or silencing of miR-34a and CDKN2A/B loci may uplift the p53 checkpoint on BRAF mutations and permit tumor cells to escape from cell-cycle arrest and become more aggressive, as shown in Figure 4. The above view is supported by the fact that expression of BRAFV600E in the lung epithelium or in melanocytes fails to result in frankly malignancy unless tumor suppressor genes such as Pten are inactivated.52,53
If BRAF mutations result in growth arrest in serous borderline tumor, why is this genotype clonally selected and can be detected in low-grade serous carcinomas even when they are at advanced stages (Figure 6A)? We reasoned that once tumor cells bypass the oncogene-induced growth arrest, they may benefit from tumor-promoting phenotypes conferred by BRAF mutations, such as metabolic switches among several tumor-promoting functions. To explore this possibility, we focused on GLUT1, a gene that encodes glucose transporter-1, which has been implicated to play a critical role in regulating glucose metabolism and energy consumption in cancer cells.54 The reason to focus on GLUT1 stems from a recent report showing that either BRAF or KRAS mutation is required for GLUT1 overexpression and glucose deprivation contributes to the development of mutations in BRAF and KRAS in colorectal cancer cells.26 In that report, the glycolysis inhibitor, 3-bromopyruvate, preferentially inhibited the growth of cells with either BRAF or KRAS mutations, suggesting that cancer cells may develop dependency on increased glucose metabolism due to GLUT1 overexpression. The observation in the current study demonstrating that BRAF mutation induced up-regulation of GLUT1 expression, thus, provides direct evidence that mutant BRAF is not only required, but also sufficient, to up-regulate GLUT1 expression.
We also report in this study that the great majority of low-grade ovarian serous tumors express GLUT1. More specifically, although all borderline tumors harboring either BRAF or KRAS mutations express GLUT1, those cases with wild-type BRAF and KRAS also show GLUT1 immunoreactivity except in one case. This observation indicates that some low-grade ovarian serous tumors with wild-type BRAF and KRAS up-regulate GLUT1 using different mechanism not directly related to mutations of BRAF or KRAS, such as those mediated by hypoxia.55 Our data demonstrate that mutation of BRAF and KRAS represents one of the mechanisms to up-regulate GLUT1. It can be speculated that for those tumors with either BRAF or KRAS mutation, blocking the pathway by MEK or BRAF inhibitors may be responsible for tumor suppression due to down-regulation of GLUT1 expression. For those tumors with wild-type BRAF and KRAS, MEK or BRAF inhibitor may not work well because alternative mechanisms are used by those tumor cells to up-regulate GLUT1. Because expression of GLUT1 has been shown as a reliable marker to predict positive fluorodeoxyglucose uptake by positron emission tomography in ovarian cancer,56 our data suggest the potential to apply fluorodeoxyglucose uptake imaging to detect low-grade ovarian serous carcinomas.
In conclusion, our findings provide new insights into further defining oncogenic stress induced by mutant BRAF in nontransformed epithelial cells. We demonstrate for the first time that expression of mutant, but not wild-type, BRAF leads to DNA double-strand breaks, followed by activation of pCHK2-p53 DNA damage response pathway that is responsible for growth inhibition and tumor suppression. On the other hand, similar to other oncogenes that regulate cellular metabolism in favor of tumor growth,57 mutant BRAF also confers oncogenic phenotypes by up-regulating GLUT1 of which abundant GLUT1 proteins contribute to enhanced glucose uptake and metabolism that characterize cancer cells.
Acknowledgments
We are grateful for the technical support from I-Wen Chiu and Carmen Chan (China Medical University Hospital) and for the kind gift of BRAF expression vectors from Dr. Raquel Seruca (University of Porto, Portugal).
Footnotes
Supported by China Medical University (CMU97-001), the National Science Council (NSC98-2320-B-039-033-MY3), Taiwan, and by NIH/NCIRO1CA103937, RO1CA129080, RO1CA116184, RO1CA148826 and U24CA160036.
Contributor Information
Jim Jinn-Chyuan Sheu, Email: jimsheu@mail.cmu.edu.tw.
Ie-Ming Shih, Email: ishih@jhmi.edu.
References
- 1.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J.W., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Cohen Y., Xing M., Mambo E., Guo Z., Wu G., Trink B., Beller U., Westra W.H., Ladenson P.W., Sidransky D. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 3.Kimura E.T., Nikiforova M.N., Zhu Z., Knauf J.A., Nikiforov Y.E., Fagin J.A. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTCRAS- BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 4.Schiffman J.D., Hodgson J.G., VandenBerg S.R., Flaherty P., Polley M.Y., Yu M., Fisher P.G., Rowitch D.H., Ford J.M., Berger M.S., Ji H., Gutmann D.H., James C.D. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer G., Oldt R., 3rd, Cohen Y., Wang B.G., Sidransky D., Kurman R.J., Shih Ie M. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 6.Ji H., Wang Z., Perera S.A., Li D., Liang M.C., Zaghlul S., McNamara K., Chen L., Albert M., Sun Y., Al-Hashem R., Chirieac L.R., Padera R., Bronson R.T., Thomas R.K., Garraway L.A., Janne P.A., Johnson B.E., Chin L., Wong K.K. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 8.Rajakulendran T., Sahmi M., Lefrancois M., Sicheri F., Therrien M. A dimerizationdependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan H., Bardelli A., Lengauer C., Kinzler K.W., Vogelstein B., Velculescu V.E. Tumorigenesis: rAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 10.Solit D.B., Garraway L.A., Pratilas C.A., Sawai A., Getz G., Basso A., Ye Q., Lobo J.M., She Y., Osman I., Golub T.R., Sebolt-Leopold J., Sellers W.R., Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2005;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pohl G., Ho C.L., Kurman R.J., Bristow R., Wang T.L., Shih Ie M. Inactivation of the mitogenactivated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with KRAS or BRAF mutations. Cancer Res. 2005;65:1994–2000. doi: 10.1158/0008-5472.CAN-04-3625. [DOI] [PubMed] [Google Scholar]
- 12.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur A.G. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vultur A., Villanueva J., Herlyn M. BRAF inhibitor unveils its potential against advanced melanoma. Cancer Cell. 2010;18:301–302. doi: 10.1016/j.ccr.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Vultur A., Villanueva J., Herlyn M. Targeting BRAF in advanced melanoma: a first step toward manageable disease. Clin Cancer Res. 2011;17:1658–1663. doi: 10.1158/1078-0432.CCR-10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K., Wubbenhorst B., Xu X., Gimotty P.A., Kee D., Santiago-Walker A.E., Letrero R., D'Andrea K., Pushparajan A., Hayden J.E., Brown K.D., Laquerre S., McArthur G.A., Sosman J.A., Nathanson K.L., Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuveson D.A., Weber B.L., Herlyn M. BRAF as a potential therapeutic target in melanoma and other malignancies. Cancer Cell. 2003;4:95–98. doi: 10.1016/s1535-6108(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 17.Shih Ie M., Kurman R.J. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho K.R., Shih Ie M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayama K., Nakayama N., Kurman R.J., Cope L., Pohl G., Samuels Y., Velculescu V.E., Wang T.L., Shih Ie M. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–785. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 20.Sieben N.L., Macropoulos P., Roemen G.M., Kolkman-Uljee S.M., Jan Fleuren G., Houmadi R., Diss T., Warren B., Al Adnani M., De Goeij A.P., Krausz T., Flanagan A.M. In ovarian neoplasms, BRAF, but not KRAS, mutations are restricted to low-grade serous tumours. J Pathol. 2004;202:336–340. doi: 10.1002/path.1521. [DOI] [PubMed] [Google Scholar]
- 21.Jones S., Wang T.L., Kurman R.J., Nakayama K., Velculescu V.E., Vogelstein B., Kinzler K.W., Papadopoulos N., Shih I.M. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu C.-Y., Bristow R., Cha M., Wang B.G., Ho C.-L., Kurman R.J., Wang T.-L., Shih I- M. Characterization of active mitogen-activated protein kinase in ovarian serous carcinomas. Clin Cancer Res. 2004;10:6432–6436. doi: 10.1158/1078-0432.CCR-04-0893. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama N., Nakayama K., Yeasmin S., Ishibashi M., Katagiri A., Iida K., Fukumoto M., Miyazaki K. KRAS or BRAF mutation status is a useful predictor of sensitivity to MEK inhibition in ovarian cancer. Br J Cancer. 2008;99:2020–2028. doi: 10.1038/sj.bjc.6604783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho C.-L., Kurman R.J., Dehari R., Wang T.-L., Shih I.-M. Mutations of BRAF and KRAS precede the development of ovarian serous borderline tumors. Cancer Res. 2004;64:6915–6918. doi: 10.1158/0008-5472.CAN-04-2067. [DOI] [PubMed] [Google Scholar]
- 25.Charles R.P., Lezza G., Amendola E., Dankort D., McMahon M. Mutationally activated BRAFV600E elicits papillary thyroid cancer in the adult mouse. Cancer Res. 2011;71:3863–3871. doi: 10.1158/0008-5472.CAN-10-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J.K., Markowitz S., Zhou S., Diaz L.A., Jr, Velculescu V.E., Lengauer C., Kinzler K.W., Vogelstein B., Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolligs F.T., Hu G., Dang C.V., Fearon E.R. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolligs F.T., Nieman M.T., Winer I., Hu G., Van Mater D., Feng Y., Smith I.M., Wu R., Zhai Y., Cho K.R., Fearon E.R. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 29.Komiya T., Park Y., Modi S., Coxon A.B., Oh H., Kaye F.J. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25:6128–6132. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- 30.Bommer G.T., Jager C., Durr E.M., Baehs S., Eichhorst S.T., Brabletz T., Hu G., Frohlich T., Arnold G., Kress D.C., Goke B., Fearon E.R., Kolligs F.T. DRO1, a gene down-regulated by oncogenes, mediates growth inhibition in colon and pancreatic cancer cells. J Biol Chem. 2005;280:7962–7975. doi: 10.1074/jbc.M412593200. [DOI] [PubMed] [Google Scholar]
- 31.Foster K.W., Ren S., Louro I.D., Lobo-Ruppert S.M., McKie-Bell P., Grizzle W., Hayes M.R., Broker T.R., Chow L.T., Ruppert J.M. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 32.Hendrix N.D., Wu R., Kuick R., Schwartz D.R., Fearon E.R., Cho K.R. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 33.Sheu J.J., Guan B., Choi J.H., Lin A., Lee C.H., Hsiao Y.T., Wang T.L., Tsai F.J., Shih I.M. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285:38260–38269. doi: 10.1074/jbc.M110.138735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 35.Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 36.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 37.Haigis K.M., Wistuba I.I., Kurie J.M. Lung premalignancy induced by mutant B-Raf, what is thy fate?: To senesce or not to senesce, that is the question. Genes Dev. 2007;21:361–366. doi: 10.1101/gad.1532107. [DOI] [PubMed] [Google Scholar]
- 38.Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 39.Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Orntoft T., Lukas J., Bartek J. DNA damage response as a candidate anticancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 40.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C.L., Dyrskjot L., Orntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T.D., Bartek J., Gorgoulis V.G. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 41.Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr, Kastrinakis N.G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T.D. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 42.Kuo K.T., Guan B., Feng Y., Mao T.L., Chen X., Jinawath N., Wang Y., Kurman R.J., Shih I.M., Wang T.L. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haffner M.C., De Marzo A.M., Meeker A.K., Nelson W.G., Yegnasubramanian S. Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target? Clin Cancer Res. 2011;17:3858–3864. doi: 10.1158/1078-0432.CCR-10-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sagun K.C., Carcamo J.M., Golde D.W. Antioxidants prevent oxidative DNA damage and cellular transformation elicited by the over-expression of c-MYC. Mutat Res. 2006;593:64–79. doi: 10.1016/j.mrfmmm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 46.Garzetti G.G., Ciavattini A., Goteri G., De Nictolis M., Stramazzotti D., Lucarini G., Biagini G. Ki67 antigen immunostaining (MIB 1 monoclonal antibody) in serous ovarian tumors: index of proliferative activity with prognostic significance. Gynecol Oncol. 1995;56:169–174. doi: 10.1006/gyno.1995.1026. [DOI] [PubMed] [Google Scholar]
- 47.Huettner P.C., Weinberg D.S., Lage J.M. Assessment of proliferative activity in ovarian neoplasms by flow and static cytometry: Correlation with prognostic features. Am J Pathol. 1992;141:699–706. [PMC free article] [PubMed] [Google Scholar]
- 48.Singer G., Stöhr R., Cope L., Dehari R., Hartmann A., Cao D.F., Wang T.L., Kurman R.J., Shih IeM. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29:218–224. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 49.Haigis K.M., Sweet-Cordero A. New insights into oncogenic stress. Nat Genet. 2011;43:177–178. doi: 10.1038/ng0311-177. [DOI] [PubMed] [Google Scholar]
- 50.Kato M., Paranjape T., Muller R.U., Nallur S., Gillespie E., Keane K., Esquela-Kerscher A., Weidhaas J.B., Slack F.J. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A.L., Linsley P.S., Chen C., Lowe S.W., Cleary M.A., Hannon G.J. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dankort D., Filenova E., Collado M., Serrano M., Jones K., McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dankort D., Curley D.P., Cartlidge R.A., Nelson B., Karnezis A.N., Damsky W.E., Jr, You M.J., DePinho R.A., McMahon M., Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amann T., Hellerbrand C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets. 2009;13:1411–1427. doi: 10.1517/14728220903307509. [DOI] [PubMed] [Google Scholar]
- 55.Marotta D., Karar J., Jenkins W.T., Kumanova M., Jenkins K.W., Tobias J.W., Baldwin D., Hatzigeorgiou A., Alexiou P., Evans S.M., Alarcon R., Maity A., Koch C., Koumenis C. In vivo profiling of hypoxic gene expression in gliomas using the hypoxia marker EF5 and laser-capture microdissection. Cancer Res. 2011;71:779–789. doi: 10.1158/0008-5472.CAN-10-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurokawa T., Yoshida Y., Kawahara K., Tsuchida T., Okazawa H., Fujibayashi Y., Yonekura Y., Kotsuji F. Expression of GLUT-1 glucose transfer, cellular proliferation activity and grade of tumor correlate with [F-18]-fluorodeoxyglucose uptake by positron emission tomography in epithelial tumors of the ovary. Int J Cancer. 2004;109:926–932. doi: 10.1002/ijc.20057. [DOI] [PubMed] [Google Scholar]
- 57.Levine A.J., Puzio-Kuter A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]