SUMMARY
The innate immune system provides early defense against infections and also plays a key role in monitoring alterations of homeostasis in the body. DNA is highly immunostimulatory, and recent advances in this field have led to the identification of the innate immune sensors responsible for the recognition of DNA as well as the downstream pathways that are activated. Moreover, information on how cells regulate DNA-driven immune responses to avoid excessive inflammation is now emerging. Finally, several reports have demonstrated how defects in DNA sensing, signaling, and regulation are associated with susceptibility to infections or inflammatory diseases in humans and model organisms. In this review, the current literature on DNA-stimulated innate immune activation is discussed, and important new questions facing this field are proposed.
INTRODUCTION
Innate immune recognition is important for host responses against infections and is also involved in maintaining homeostasis in the organism (1). DNA has been known to stimulate immune reactions for >50 years (2), and the identification of Toll-like receptor 9 (TLR9) as an endosomal sensor for DNA was reported in 2000 (3). In 2006, it was shown that the cytosolic location of DNA also potently stimulates innate immune activities (4, 5), and this discovery sparked a new research line focusing on intracellular DNA sensing (6). This work led to the identification of sensors, signaling pathways, and biological processes stimulated by DNA (6). Most recently, mechanisms of how DNA-activated immune responses are regulated and targeted by microbes have been described, and human inflammatory diseases caused by the constitutive activation of DNA-stimulated pathways have been identified. Given the potent immunostimulatory potential of DNA, mammalian cells seek to achieve full genome function without activating undesired inflammation despite the presence of DNA species such as endogenous retroviral DNA, DNA replication by-products, and damaged genomic DNA in the cellular environment. Recent work described that the DNA sensing machinery interacts closely with basal cellular processes such as the DNA damage response and autophagy to maintain optimal activation and regulation of this immune pathway.
In this article, I review the current understanding of innate DNA sensing and discuss recent discoveries on how this system is activated and regulated. In addition, I discuss how the DNA-stimulated immune responses are modulated by microbes and how this important part of the innate immune system impacts the development and prevention of inflammatory, infectious, and malignant diseases.
RECOGNITION OF DNA
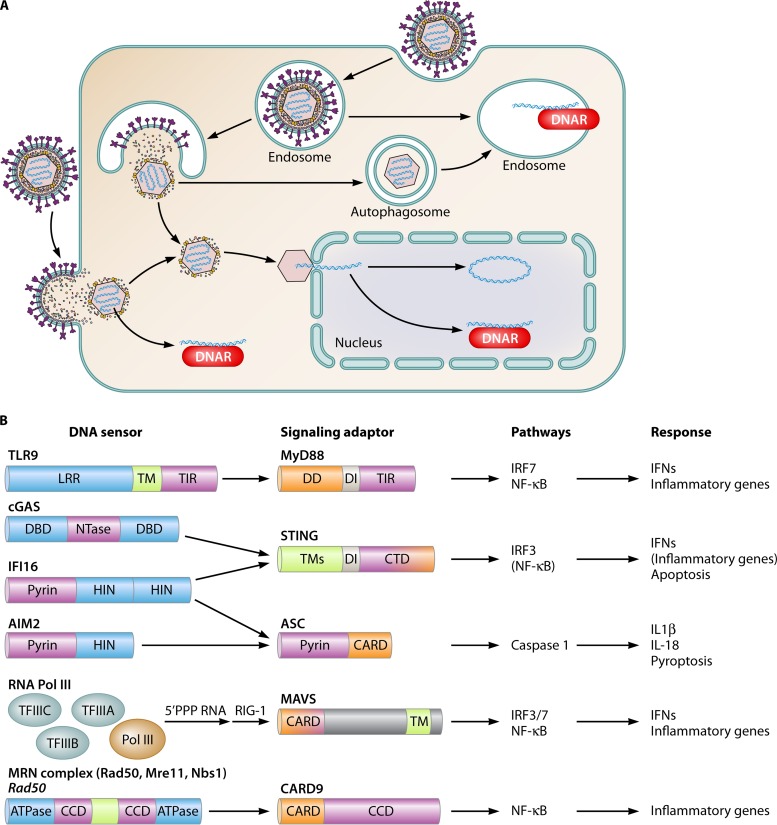
Given the fact that the chemical composition of the core DNA molecule is identical for mammalian cells and microorganisms, the principle of recognition is based mainly on localization as opposed to the pathogen-specific chemistry of the molecule (6). Mammalian DNA is localized in the nucleus, and most DNA sensors are expressed in other cellular compartments (Fig. 1A). Although a dozen DNA sensors have been proposed, only a few have been confirmed through independent studies, hence making these the most likely candidates to actually function as immune sensors of DNA (Fig. 1B). The first identified DNA sensor, TLR9, binds DNA in endosomes, thereby detecting external DNA entering through the endosomal route or perhaps also the cytoplasm through autophagocytic delivery (3, 7, 8). TLR9 preferentially binds unmethylated CpG-rich DNA (3, 9, 10). Recently, it was demonstrated that TLR9, similar to other agonist-bound TLRs, forms an m-shaped 2:2 complex and that each immunostimulatory CpG DNA molecule interacts with both TLR9 molecules in the complex (11). The structure also revealed interactions between TLR9 and the backbone as well as the bases in the DNA, and this could explain the requirement for CpG motifs for high-affinity DNA binding to TLR9.
FIG 1.
Innate DNA sensors and subcellular localization of DNA recognition. (A) DNA sensors are expressed in endosomes, the cytoplasm, and the nucleus. DNA may come into contact with DNA sensors in the endosome through uptake via endosomal or autophagy pathways. Stimulation of the cytoplasmic DNA sensing pathways may occur as a result of (i) bacteriolysis or viral capsid degradation in the cytosol, (ii) accumulation of endogenous retroviral reverse transcripts or DNA replication intermediates, or (iii) leakage of DNA from endosomal compartments. In the nucleus, the features that distinguish stimulatory from nonstimulatory DNAs are not known. Factors that may be involved include epigenetic modifications and interactions with histones. DNAR, DNA receptor. (B) DNA sensors mediate signaling via adaptor proteins to activate specific pathways and biological responses. Blue, DNA-binding domains; purple, domains/proteins relaying signals between the sensors and the adaptor proteins; orange, domains relaying signals from the adaptor proteins and downstream. CARD, caspase activation and recruitment domain; CCD, coiled-coil domain; CTD, C-terminal domain; DD, death domain; DI, dimerization interphase; LLR, leucine-rich repeat; TIR, Toll/interleukin-1 receptor domain; TM, transmembrane region.
A second group of DNA sensors, which includes cyclic GMP-AMP (cGAMP) synthase (cGAS) and absent in melanoma 2 (AIM2), localizes exclusively to the cytoplasm and detects DNA in this location in a sequence-independent manner (12–17). cGAS binds DNA by interactions with the sugar-phosphate backbone of both DNA strands along a positively charged platform of the protein flanked by a helical spine and a zinc thumb (18–20). In addition, cGAS has a second positively charged surface interacting with DNA, and crystal structures have revealed that cGAS forms 2:2 complexes with DNA (21, 22). AIM2 belongs to the family of PYHIN proteins, consisting of a pyrin domain and one or two HIN domains (Fig. 1B) (23). DNA binding is mediated by the HIN domain, where a positively charged surface interacts with the sugar-phosphate backbone through interactions with both DNA strands across both major and minor grooves (24).
Finally, there is a group of DNA sensors that localize in both the nucleus and the cytoplasm. This group includes interferon (IFN)-inducible gene 16 (IFI16), RNA polymerase III (Pol III), and the Mre11-Rad50-Nbs1 (MRN) complex (25–29). Common for these proteins is that, in addition to their proposed role in DNA sensing, they have other well-established functions in DNA biology, including transcription and DNA repair. Like AIM2, IFI16 is a PYHIN protein and binds DNA in a manner similarly to AIM2 (24). In addition, it was recently reported that homotypic interactions between IFI16 pyrin domains allow cooperative binding on double-stranded DNA (dsDNA) and the formation of IFI16 filaments (30). The DNA-binding capacity of IFI16 also includes base-pared single-stranded DNA (ssDNA), hence explaining the immunostimulatory capacity of this DNA species (31). The transcriptional activity of RNA Pol III has long been known to be initiated from TATA boxes or AT-rich sequences (32). This involves the DNA-binding activity of the transcription factors TFIIIA, TFIIIB, and TFIIIC. Similarly, the stimulation of innate immune responses through this pathway was reported to be mediated mainly by AT-rich DNA stimulation, most likely reflecting a mechanism of DNA binding similar to the one known for classical Pol III-mediated transcription (27, 28). The MRN complex recognizes DNA via the N- and C-terminal nucleotide-binding domains of Rad50 and six conserved loops in the N-terminal nuclease domain of Mre11 (29, 33). The interaction of DNA occurs exclusively with the backbone, again repeating the theme of sequence-independent sensing by innate DNA sensors (33). The relative contributions of Mre11 and Rad50 to DNA binding upstream of immune signaling remain to be established. Interestingly, the MRN complex can bind ssDNA, dsDNA ends, dsDNA, and branched DNA, but there is no published data on the relative capacities of these DNA structures to activate immune reactions through the MRN pathway. In addition to the above-mentioned DNA sensors, a group of RNA helicases was also suggested to possess the capacity to bind DNA and promote innate immune signaling. This group includes DHX9, DHX36, DDX41, and DDX60 (34–36). Currently, it is not known how these proteins act in the DNA signaling pathway, and more work is needed to clarify this issue. The findings that DHX9 and DDX60 have the capacity to bind both RNA and DNA (36, 37) and reports on DDX41 acting in both DNA- and cyclic dinucleotide (CDN)-stimulated signaling (35, 38) may suggest a role in downstream signaling rather than DNA sensing.
While the DNA sensing proteins with both cytosolic and nuclear locations are reported to be involved in cytosolic DNA sensing, at least for IFI16, it has been demonstrated that sensing in the nucleus also initiates immune signaling (26, 39). For RNA Pol III, there is indirect evidence to support a role for nuclear DNA sensing in immune activation, with a reported involvement of RNA Pol III in sensing Epstein-Barr virus (EBV) latent genomes, which are localized in the nucleus (27). These findings raise fundamental questions as to what determines whether DNA stimulates innate immune responses or not in the nucleus. This issue remains to be resolved, but factors that are likely to impact immunogenicity include methylation of DNA, binding to histones, and free DNA ends.
Collectively, the innate immune system utilizes a panel of compartmentalized pattern recognition receptors (PRRs) to sense dsDNA, base-pared ssDNA, AT-rich DNA, and CpG DNA and induce antiviral and inflammatory reactions. It is still not fully resolved what distinguishes immunostimulatory from nonstimulatory DNA in the nucleus.
SIGNAL TRANSDUCTION ACTIVATED BY DNA
Signaling for Type I IFN Expression
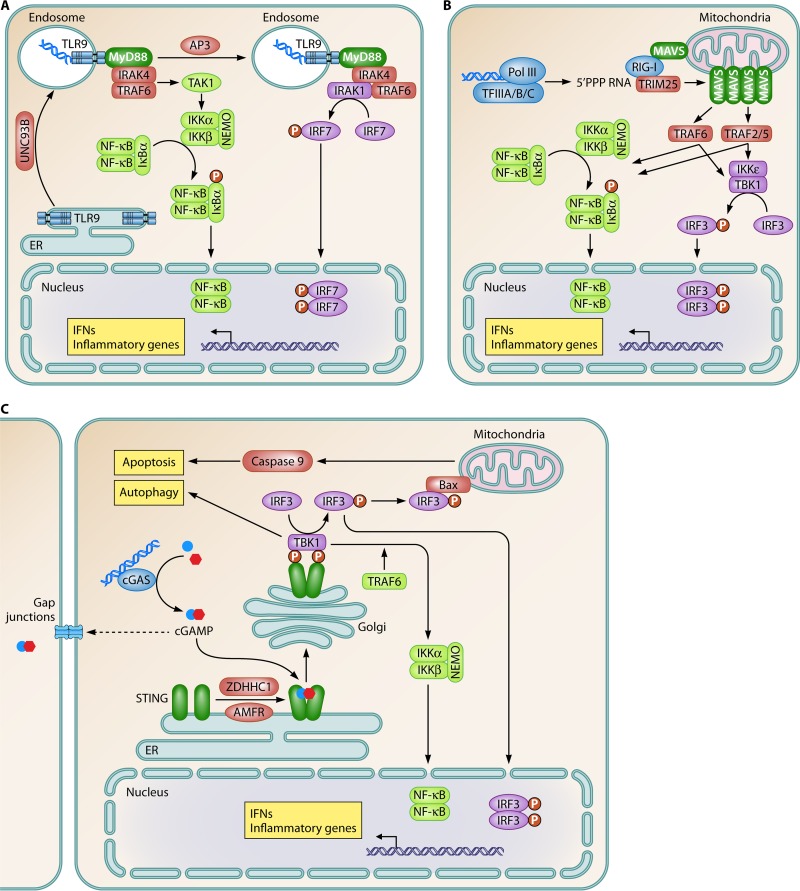
DNA recognition by PRRs initiates intracellular signaling events, eventually activating immune effector functions. These events include the expression of type I IFNs and inflammatory genes, inflammasome activation, and different types of cell death (6). In recent years, knowledge on DNA-stimulated signal transduction has grown immensely, and there is now molecular information on central events in all key DNA-activated signaling pathways. While signaling downstream of TLR9 has been known for some time and is nicely reviewed elsewhere (40, 41) (Fig. 2A), progress has recently been made in the understanding of signaling downstream of intracellular DNA sensors. Following the sensing of AT-rich DNA by RNA Pol III, 5′-PPP RNA is made by the polymerase (27, 28). This RNA species represents the core pathogen-associated molecular pattern (PAMP) for the PRR retinoic acid-inducible gene I (RIG-I), thus leading to the stimulation of this pathway (Fig. 2B). RNA engagement of RIG-I stimulates a conformational change in RIG-I, the ubiquitination of the CARD domain, leading to interactions with the CARD domain on mitochondrial antiviral signaling protein (MAVS) (42). This in turn induces a prion-like conformational switch of MAVS, leading to fibril formation; recruitment of TRAF2, -5, and -6; and propagation of signaling cascades for the IFN regulatory factor 3 (IRF3) and nuclear factor κB (NF-κB) pathways (42, 43). Although it remains unresolved why RNA Pol III-mediated transcription of cellular genes does not activate this pathway, a likely explanation is that cellular genes are transcribed by RNA Pol III in the nucleus and that the resulting RNAs are 5′ modified before being exported to the cytoplasm. Poly(dA·dT) is transcribed in the cytoplasm by cytosolic RNA Pol III, and therefore, the resulting RNA contains 5′-PPP, which activates RIG-I. However, this model does not explain the finding that IFN-α induction by the EBV-positive BL Mutu III cell line (in which the EBV genome is expected to be in the nucleus) is inhibited by an inhibitor of RNA Pol III (27).
FIG 2.
Signal transduction pathways activated by DNA to stimulate type I IFN expression. Innate DNA sensing induces the activation of three signaling pathways to stimulate type I IFN expression. (A) TLR9; (B) RNA polymerase III; (C) cGAS. ER, endoplasmic reticulum. Blue, sensor; green, adaptor; purple, IRF pathway; light green, NF-κB pathway; claret, other signaling molecules; yellow boxes, biological effector functions.
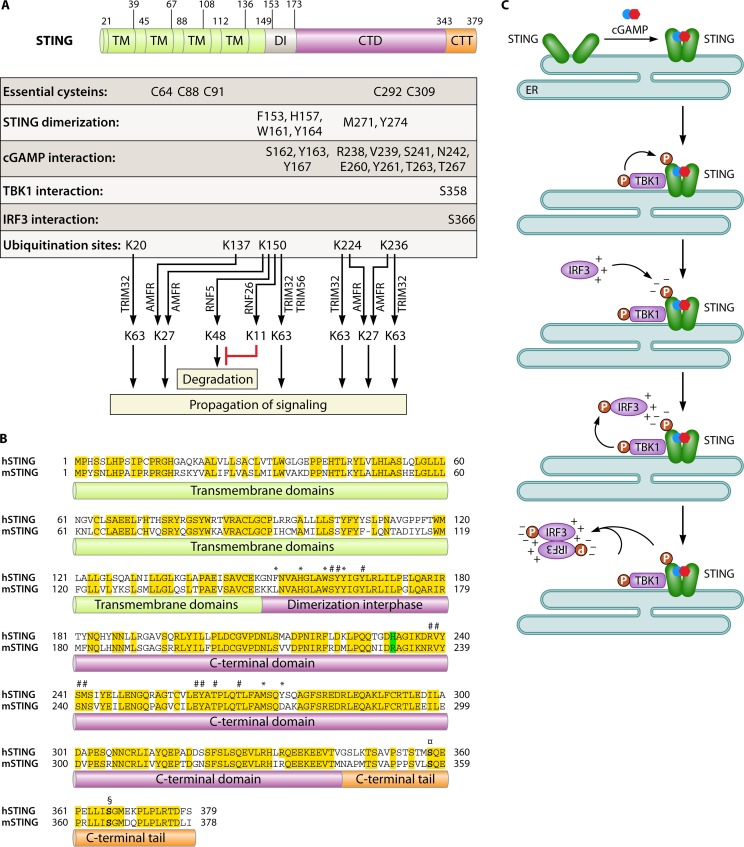
A third pathway for the induction of IFNs is the cGAS-STING pathway (Fig. 2C). Upon DNA binding by cGAS, synthesis of the CDN 2′,3′-cGAMP is initiated (12, 44). cGAMP binds with high affinity to the STING dimer, hence inducing a conformational change in the C-terminal region of STING, which creates an optimal platform for interaction with TBK1 and the phosphorylation of IRF3 (45, 46). Interestingly, DNA-induced cGAMP can be delivered to neighboring cells through gap junctions, thus allowing lateral propagation of the signal and activation of antiviral responses prior to infection (47). With the recent intense interest in the STING pathway, there is now significant knowledge on the molecular biology of this protein (Fig. 3A). STING is a tetra-spanning membrane protein (i.e., having 4 transmembrane regions) with the transmembrane regions being localized in the N-terminal part (48–50). The protein is dimeric in the active form, and a dimerization region is localized between residues 152 and 173 in human STING. The C-terminal region of the protein faces toward the cytoplasmic side of organelles and vesicles where STING is localized, and it is this part of the molecule that serves as the platform for interactions with TBK1 (51). The binding of cGAMP to STING involves residues in the dimerization region as well as in the C-terminal domain (Fig. 3A). Interestingly, human STING and murine STING, although exhibiting high overall sequence homology (63%), differ at key positions in the loops that cover the CDN-binding pocket (Fig. 3B). Most notably, while reference human STING has a histidine at position 232, murine STING has an arginine at the corresponding position (residue 231 in murine STING) (52). Consequently, murine STING is activated by both 2′,3′- and 3′,5′-linked CDNs, while human STING selectively responds to 2′,3′-linked CDNs (52).
FIG 3.
STING, the adaptor protein in DNA-activated signaling for IFN expression. (A) Summary of the known molecular details on the residues and regions of STING involved in the biological functions of the molecule. CTT, C-terminal tail. (B) Alignment of human STING (hSTING) and murine STING (mSTING), which have 63% sequence homology. Position 232 (position 231 in murine STING), which contributes critically to the differential responsiveness of human and murine STINGs to 3′,5′-linked CDNs, is highlighted in green. Conserved residues are highlighted in yellow. Residues involved in STING dimerization are marked with *. Residues involved in cGAMP binding are marked with #. Serine residues in the C-terminal tail involved in the recruitment of TBK1 and IRF3 are shown in boldface type and marked with ¤ and §, respectively. (C) Model for IRF3 activation through the cGAS-STING pathway. In the first step, cGAMP binding induces a conformational change in the C-terminal domain of STING. The second step involves the recruitment and activation of TBK1, followed by TBK1-mediated phosphorylation of STING at serine 366. The third step is the recruitment of monomeric IRF3 to STING at phosphoserine 366, through an interaction with a positively charged surface on IRF3. The fourth step is TBK1-mediated phosphorylation of IRF3 at C-terminal serines, including positions 385 and 386. The fifth step involves the homodimerization of phosphorylated IRF3 molecules and the activation of transcriptional functions.
Upon cGAMP binding, STING undergoes a ligand-induced conformational change in the C-terminal domain, which may underlie the downstream activation of TBK1 (45, 46) (Fig. 3C). The binding of TBK1 to STING is dependent on the phosphorylation of STING at serine 358, which is mediated by TBK1 (49). Once recruited to STING, TBK1 in turn phosphorylates STING at serine 366 and thus creates a negatively charged surface, which attracts IRF3 monomers through a conserved positive surface of the molecule (53). Once in proximity, TBK1 phosphorylates IRF3, which triggers IRF3 dimerization and activation. In addition to phosphorylation, ubiquitination represents another posttranslational modification of importance for the control of the activity of STING. Tsuchida and associates first reported that the E3 ubiquitin ligase TRIM56 interacts with STING to catalyze K63-linked ubiquitination at K150 (54) (Fig. 3A). This modification promotes STING dimer formation and interactions with TBK1. Subsequently, TRIM32 was also demonstrated to mediate K63-linked ubiquitination of STING at several lysine residues, including K150 (55), which was associated with elevated IFN-β expression. Most recently, the endoplasmic reticulum (ER)-associated E3 ubiquitin ligase AMFR was reported to interact with STING and catalyze K27-linked polyubiquitination, in a manner dependent on insulin-induced gene 1 (56). This modification of STING facilitated the recruitment of TBK1. Together, these studies suggest an important role for the ubiquitin system in STING activation, although the mechanistic details remain to be described, including the interaction with the cGAS-cGAMP pathway.
Other important aspects of STING biology in relation to IFN induction include the role of the local protein-protein networks and interactions with the organelles and membrane structures in which the protein resides. One study mapped the STING interactome and its dependence on DNA stimulation (57). This resource is now available for further studies. Another study demonstrated that STING constitutively interacts with the ER-associated protein ZDHHC1 and that this interaction is essential for STING dimerization and the recruitment of TBK1 (58). Finally, it has been reported that a series of cysteines in the membrane-spanning regions of STING are essential for IFN-β induction (59). Given the covalent binding between the two STING monomers in the STING dimer and its sensitivity to reducing agents, the cysteine residues in STING likely form disulfide bridges (59). At present, it is not known how this redox-sensitive step in STING activation is regulated, nor is it known where in the cell this takes place. However, given the reported involvement of STING in the cellular ER stress response (60), it is tempting to speculate that this organelle is where the covalently linked STING dimer is formed via disulfide bridges.
Interactions between the cGAS-cGAMP-STING Pathway and Other DNA Sensing Systems
Prior to the identification of cGAS, several DNA sensors stimulating IFN expression were proposed (6). Although it is now generally agreed by most investigators in the field that several of these proteins are not actual DNA sensors, there are published data of high quality on other proposed DNA sensors where the relation to the cGAS-cGAMP-STING pathway remains to be clarified (25, 35, 61). Here, IFI16, DDX41, and DNA-dependent protein kinase are discussed.
IFI16 was identified as a DNA sensor in a mass spectrometry analysis of cytosolic proteins coprecipitated with immunostimulatory DNA (25). Since then, data supporting a role for IFI16 in DNA sensing, STING activation, inflammasome activation, genome silencing, and gene expression have been reported (25, 26, 31, 62, 63; for a recent review and discussion of the literature on IFI16 in DNA sensing and genome regulation, see reference 23). The focus here is on IFI16 in the STING pathway. IFI16 was demonstrated to associate with microbial DNA during different viral and bacterial infections and to interact with STING (25, 31, 64, 65). In human THP1 cells differentiated with phorbol myristate acetate into a macrophage-like phenotype, it was demonstrated that IFI16, cGAS, and STING were all required for the stimulation of type I IFN expression by either human immunodeficiency virus type 1 (HIV-1) or Listeria monocytogenes (31, 65). This indicates an interaction between the IFI16- and cGAS-dependent pathways. Differentiation with phorbol myristate acetate leads to a reduction of cGAS expression, while IFI16 levels are upregulated (65). It is possible that the cGAS-cGAMP-STING pathway is amplified by signals from other DNA-binding factors under conditions of low levels of cGAS expression. Recently, IFI16 was demonstrated to bind DNA in a cooperative manner to form IFI16 filaments on dsDNA (30). It will be interesting to learn whether this function of IFI16 acts upstream of cGAS in the initial process of DNA sensing, and this could be of particular biological importance under conditions of low levels of cytosolic DNA. Alternatively, the reported interaction between IFI16 and STING could stabilize the STING dimer in cells with only low-level cGAMP production. Given the dual nuclear-cytoplasmic localization of IFI16, as opposed to the strictly cytoplasmic localization of cGAS, insight into the mechanism of IFI16-dependent induction of expression is of central importance for the understanding of innate immune responses stimulated by both cytoplasmic and nuclear DNA (23).
DDX41 was first suggested to be a sensor for microbial DNA and was reported to associate with DNA and to colocalize with STING (35). Subsequently, DDX41 was found to sense bacterial CDNs and stimulate the STING pathway (38). It has now also been demonstrated that short hairpin RNA (shRNA)-mediated knockdown of DDX41 reduces the responsiveness to both dsDNA and cGAMP in human cells (65). Collectively, the data available suggest that DDX41 acts in the DNA-activated signaling STING pathway but downstream of DNA sensing, most likely as a cofactor for cGAMP in mediating the activation of the STING pathway.
As a third example, DNA-dependent protein kinase, which is well established to play a central role in the nonhomologous end joining pathway of DNA repair (66), is also reported to interact with DNA in the cytoplasm and to be essential for the full induction of innate immune responses to DNA and DNA viruses in fibroblasts (61, 67, 68). Ferguson et al. found that the DNA-dependent protein kinase subunit Ku70 is in a complex with STING in resting cells, but this interaction is abrogated following DNA stimulation (61). These authors did not formally demonstrate that the responses evoked were dependent on STING, but the data nevertheless suggest a role for the DNA-dependent protein kinase in the initial steps of DNA-stimulated signaling. Further mechanistic evidence supporting a link between DNA damage and STING-activated IFN pathways was provided by a study demonstrating that DNA damage or defects in the DNA repair system stimulate type I IFN expression in a STING-dependent manner (69). It was also revealed that DNA damage or a failure to repair DNA leads to the accumulation of cellular DNA in the cytoplasm, thus raising the question as to whether the proposed nuclear DNA sensing is in fact the sensing of DNA leaking into the cytoplasm (69–71). This issue needs further attention and will help clarify the involvement of DNA repair proteins in DNA-driven innate immunity.
Collectively, although the emerging picture is that cGAS is the essential DNA sensor, the extent to which the initially proposed DNA sensors fit into the cGAS-cGAMP-STING pathway still remains unresolved. It is also worth investigating whether there are mechanisms through which intracellular DNA can stimulate IFN expression independently from cGAS. The Pol III–RIG-I pathway represents an example of this, but its physiological importance remains unclear.
DNA-Stimulated Signaling to Inflammasomes, NF-κB, and Apoptosis
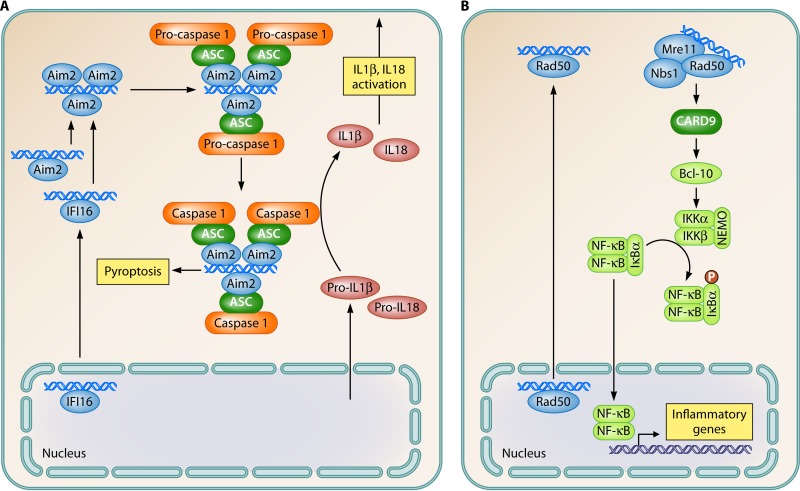
In addition to the induction of IFNs, DNA also stimulates inflammasome activation, the expression of inflammatory genes, and apoptosis (Fig. 2C and 4A and B). In particular, the activation of inflammasomes by DNA has been well described (14–17). Upon DNA binding by AIM2 or IFI16, the proteins take a more open conformation, allowing the N-terminal pyrin domain to engage in interactions with the pyrin domain of ASC, the essential inflammasome adaptor protein (Fig. 4A). This interaction in turn leads to the nucleation of ASC and caspase 1 recruitment through interactions between the CARD domains in the two proteins, eventually leading to proximity-induced cleavage and the activation of caspase 1 (72). The best-described inflammasome substrates are pro-interleukin-1β (IL-1β) and pro-IL-18 (73), but several others have also been reported. Therefore, the inflammasome has functions beyond cytokine maturation, most notably stimulation of pyroptotic cell death (68, 74). Since IFI16 has been ascribed functions in both the STING and inflammasome pathways, a key question is what determines whether DNA binding by IFI16 stimulates IFN or inflammasome pathways. The answer to this question could contribute to answering the more fundamental questions of what determines which of the potential DNA-stimulated pathways are activated by cytosolic DNA in cells and what the interactions between the pathways may be.
FIG 4.
Signal transduction pathways activated by DNA to stimulate activation of inflammasomes and the NF-κB pathway. (A) AIM2-like receptors; (B) Rad50. Blue, sensor; green, adaptor; light green, NF-κB pathway; orange, inflammasome pathway; claret, other signaling molecules; yellow boxes, biological effector functions.
Among the signaling pathways activated by DNA, most attention has been drawn toward IRF3 (6). However, it has long been known that the NF-κB pathway is also stimulated upon DNA recognition. At present, there are data to support that this occurs through at least 2 pathways. First, in murine fibroblasts and myeloid cells lacking STING, the activation of NF-κB and the induction of NF-κB-stimulated genes are impaired (75, 76). The STING-dependent pathway for the activation of NF-κB was proposed to involve TRAF6 acting upstream of TBK1 to trigger the classical IκB kinase α/β (IKKα/β) pathway (75) (Fig. 2C). In addition to the STING-dependent pathway for NF-κB activation, it was recently reported that Rad50 (most likely through the MRN complex together with Mre11 and Nbs1) selectively stimulates NF-κB activation and that this proceeds through a pathway dependent on CARD9 and Bcl-10 (29) (Fig. 4B). Rad50 localized to the nucleus in untreated cells but was recruited to the cytoplasm along with Mre11 and Nbs1 after DNA stimulation, thus leaving open the questions as to where the initial DNA sensing takes place and what triggers the relocation of the MRN complex to the cytoplasm. Downstream signaling was mediated through the interaction between Rad50 and CARD9, which recruits Bcl-10 for signaling to the NF-κB pathway (29). With the selective activation of the NF-κB pathway and, hence, the induction of genes, including pro-IL-1β, the Rad50–CARD–Bcl-10 pathway is predicted to work together with the AIM2/IFI16 inflammasome pathways to enable DNA to provide both signals required for the production of bioactive IL-1β. This is in contrast to most other PAMPs, which stimulate only one step in the biogenesis of bioactive IL-1β, and may contribute significantly to the pronounced immunostimulatory activity of DNA.
Finally, there are now also data available on how DNA can induce apoptosis. The phenomenon of DNA-driven apoptosis has been known for many years to researchers performing transfection experiments, and it was previously proposed that DNA-induced cell death may have a physiological role in antiviral defense (77). In a study using human monocytes, Sze et al. reported that the accumulation of DNA in SAMHD1-deficient cells infected with the retrovirus human T cell leukemia virus type 1 led to STING-dependent IRF3 phosphorylation. This stimulated apoptosis through a previously reported mechanism where IRF3 associates with Bax on mitochondria to activate the mitochondrial apoptosis pathway dependent on caspases 9 and 3 (78, 79). A similar mechanism of DNA-driven apoptosis has been reported by others (60). This mechanism of apoptosis, which is independent of IFNs and the transcriptional activity of IRF3, may be of importance in a range of pathological conditions. Therefore, it will be interesting to learn which cofactors are involved in determining whether IRF3 is translocated to the nucleus to act as a transcription factor or remains in the cytoplasm to stimulate apoptosis.
Collectively, intracellular DNA stimulates a series of signaling cascades with particularly the STING pathway being of central importance. This leads to the expression of IFN and inflammatory genes and also the activation of inflammasomes and different types of death.
CELL TYPE DIFFERENCES
The innate DNA sensing system is operative in many cell types. These cell types include both leukocytes, such as macrophages and dendritic cells (DCs), as well as nonprofessional immune cells, such as fibroblasts (25, 80, 81). However, there is now emerging evidence that some cell types do not respond to cytosolic DNA with the production of type I IFNs. For instance, activated T cells do not produce IFN-β upon DNA stimulation but do so after stimulation with RNA (82). However, this cell type expresses the proteins known to be important for the STING pathway and in fact activates the pathway to the level of phosphorylation of TBK1 (82). This could indicate that an unknown factor of central importance for the execution of the STING pathway downstream of TBK1 is not expressed in activated T cells. Interestingly, other studies have revealed DNA-driven stimulation of pyroptosis in T cells during HIV-1 infection, further highlighting the cell type-specific nature of the innate immune responses to intracellular DNA (68).
A second cell type now known to exhibit intrinsic tolerance to stimulation through the DNA sensing pathway is keratinocytes (13). Interestingly, upon stimulation with inflammatory cytokines such as IL-1β and tumor necrosis factor alpha, these cells gain the capacity to induce innate immune responses. In resting keratinocytes, IFI16, cGAS, and STING are expressed, but STING is not mobilized after DNA transfection, and IFI16 is largely restricted to the nucleus. Cytokine treatment redistributes a pool of IFI16 to the cytoplasm, which associates with DNA and STING to allow the induction of cytokine expression in an IFI16-dependent manner (13).
One common theme for the two cell types discussed in this section is that they both have high proliferation potential. It has been demonstrated that DNA replication and cell division lead to the accumulation of DNA-containing replication by-products in the cytoplasm (83). Therefore, it is possible that the presence of a fully operative DNA sensing system in the cytoplasm that stimulates type I IFN responses in these cell types would lead to constitutive IFN expression and therefore associated pathological conditions. Why the two cell types discussed have different mechanisms of unresponsiveness to DNA remains an open question. One possibility is that they have different requirements regarding adaption to environmental changes. For instance, for keratinocytes, it may be advantageous to be able to respond to DNA when the challenge is sustained (13), and for activated T cells, DNA sensing may preferentially stimulate death pathways as opposed to IFN responses (82, 84).
NEGATIVE REGULATION OF DNA-DRIVEN IMMUNE RESPONSES
Given the strong immunostimulatory function of DNA, it is important for cells to be able to control the responses stimulated by this PAMP. This is important for curbing the response after clearance of an infection and also for preventing the stimulation of inflammation by endogenous DNA sources. With the constitutive production of reverse transcription products from endogenous retroviruses, the accumulation of DNA replication by-products in the cytosol upon cell division, and the release of mitochondrial DNA into the cytoplasm during mitochondrion-driven apoptosis, cells need a certain DNA load threshold, which has to be passed to stimulate immune responses (85, 86).
Cellular DNases play a central role in eliminating undesired DNA from different cell compartments. For instance, the lysosomal endonuclease DNase II digests endogenous and exogenous DNA through a lysosomal pathway (Fig. 5A). Correspondingly, the lack of DNase II leads to the STING-dependent activation of DNA-stimulated responses induced by apoptotic cells as well self DNA (87). In the cytoplasm, Trex1, which possesses 3′-to-5′ exonuclease activity, is essential for keeping the level of DNA below the threshold for stimulating immune responses (85, 88). Trex1 has a substrate preference for ssDNA, and accordingly, the responsiveness to ssDNA is increased to the greatest extent compared to other DNA species in Trex1-deficient cells (89). The absence of Trex1 stimulates the cGAS-STING pathway (90), but there are so far no data on other DNA-driven pathways being activated in Trex1-deficient cells. Interestingly, since Trex1 cannot degrade DNA with modifications commonly introduced by oxidation (e.g., 8-hydroxyguanosine), different types of oxidative stress generate DNA species with elevated immunostimulatory function (91). With respect to the cellular mechanism of Trex1 function, it is interesting that this DNase, like STING, is associated with the ER. One study reported that Trex1 and other components of the SET complex associate with factors of the DNA sensing machinery, including the murine PYHIN protein p204, which has the same domain organization as that of IFI16 (25, 92). This suggests that the process of Trex1-mediated degradation of DNA is spatially linked to immunostimulatory DNA sensing (57).
FIG 5.
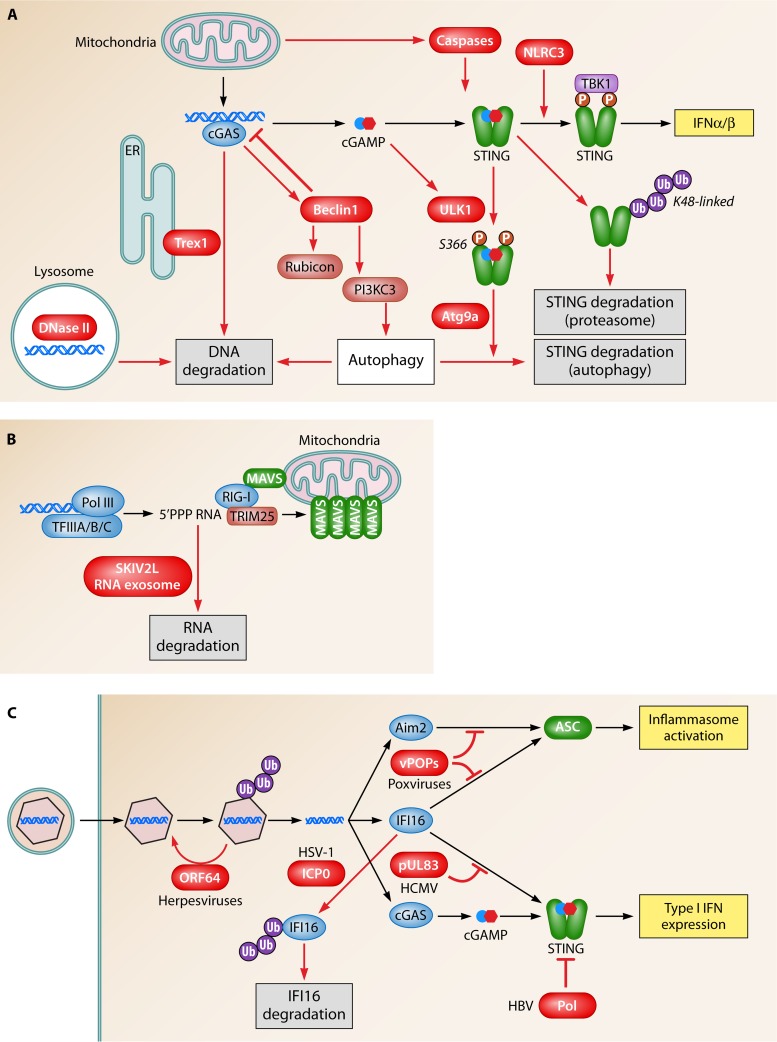
Endogenous and microbial mechanisms to downmodulate and evade DNA-driven innate immune responses. (A) DNA is degraded by DNase II in lysosomal compartments and by Trex1 in the cytoplasm. DNA is also targeted for degradation by an autophagy pathway stimulated upon DNA binding by cGAS, leading to the release of Rubicon from Beclin-1. In addition to DNA turnover, the pathway is also regulated at the level of STING. After activation, this protein can be degraded through either proteasomal or autophagocytic pathways. Moreover, caspases 3, 7, and 9, activated during mitochondrial apoptosis, suppress DNA-stimulated IFN production by targeting a step upstream of TBK1 phosphorylation. Finally, the ULK1 kinase is activated by DNA and cGAMP to catalyze specific serine phosphorylation of STING in the C-terminal domain, leading to the downmodulation of the IRF3 pathway. (B) The SKIV2L exosome pathway degrades RNA that stimulates the RIG-I pathway and is thus likely to also be involved in the control of the Pol III–RIG-I pathway. (C) Viruses evade and counteract DNA-driven immune responses at several levels, including prevention of the release of DNA into the cytoplasm, degradation of specific DNA sensors, and inhibition of signaling proteins via protein-protein interactions.
Regarding the RNA Pol III pathway, the RIG-I agonist produced from AT-rich DNA may also be subject to negative regulation. Eckard and associates recently reported that the SKIV2L RNA exosome, which possesses 3′-to-5′ exoribonuclease activity, limits the activation RIG-I by degrading the PAMP (93) (Fig. 5B). A role for the SKIV2L RNA exosome in the control of the RNA Pol III pathway remains to be formally demonstrated.
In addition to limiting the availability of the PAMPs, innate immune responses can be controlled at the level of signaling. For instance, the NOD-like receptor family member NLRC3 reduces STING-dependent signaling, and mice deficient in NLRC3 respond to herpes simplex virus 1 (HSV-1) infection with elevated IFN expression (94). Mechanistically, NLRC3 interacts directly with STING to reduce the TBK1-STING association, thus inhibiting downstream signaling (94). As a second mechanism of negative control of the STING pathway, the E3 ubiquitin ligase RNF5 was identified to interact with STING and mediate K48-linked ubiquitination and degradation of STING (95). This study was based on work with RNA viruses and also used an experimental setup with overexpression of the proteins of interest. Although data on RNF5 from primary cells or in vivo models are still lacking, this study delivers a proof-of-principle that K48-linked ubiquitination of STING at K150 can provide a negative regulatory signal for the DNA sensing pathway by targeting the protein for degradation. A subsequent publication from the same group most recently reported that K150 of STING is also targeted for K11-linked ubiquitination by RNF26 and that this protects the protein from RNF5-mediated K48-linked polyubiquitination and degradation (96). These results seem to indicate a complex interplay between ubiquitin ligases in the control of STING stability and activation (Fig. 3A).
Recently, it was reported that the endogenous control of DNA sensing also represents a mechanism through which cells render mitochondrial apoptosis immunologically silent. The stimulation of the mitochondrial Bak/Bax-mediated apoptosis pathway leads to the release of mitochondrial DNA with the potential to stimulate the cGAS-STING pathway (97, 98). However, this pathway was inhibited by a mechanism dependent on caspases 3, 7, and 9. At the mechanistic level, the specific target of caspases in the cGAS-STING pathway was not identified, but the caspases were demonstrated to act upstream of TBK1 phosphorylation (97). It will be interesting to learn whether caspase-dependent regulation of the cGAS-STING pathway is also involved in the regulation of signaling from exogenous DNA sources.
Finally, it was recently demonstrated that autophagy represents a central mechanism through which the STING pathway is regulated (99, 100). This is in contrast to the TLR9 pathway, where autophagy appears to facilitate the delivery of the DNA PAMP to the TLR9-containing endosomal compartment (8). It was previously described that cytosolic DNA is a trigger of autophagy (101, 102) (Fig. 2C), and the first demonstration of the involvement of this basic catabolic process in the regulation of the DNA-activated pathway was provided by Saitoh and associates (103). These investigators reported that cells lacking Atg9a, a protein involved in supplying membranes for autophagosomes, induced elevated expression of type I IFN and inflammatory cytokines, and this correlated with a sustained activation of the STING signalosome. Moreover, electron microscopy revealed that the DNA-activated autophagy-related membranous compartments did not contain the classical double membrane, thus indicating that a noncanonical type of autophagy is involved in the regulation of DNA sensing. More recently, it was demonstrated that STING, but not TBK1, is degraded in this process (99). Upon DNA stimulation, STING is phosphorylated at serine 366 by the kinase ULK1, which is centrally involved in the initiation of autophagy (99). This process also includes two other autophagy-related kinases, namely, AMPK and LKB1. The downstream effect of S366 phosphorylation on STING was found to be an impairment of IRF3 activation. In contrast, a link between S366 phosphorylation and autophagocytic STING degradation was not conclusively established. While these authors demonstrated a very pronounced effect of DNA on STING degradation, this effect was more modest after stimulation with cGAMP (99). Despite this, cGAMP modulated the phosphorylation of ULK1, AMPK, and LKB1 to the same extent as did dsDNA, suggesting that different processes involving components of the autophagy machinery are operating in parallel in DNA.
Further evidence for the role of autophagy in the control of DNA-driven immune responses was provided by the interaction between cGAS and Beclin-1 and the elevated responsiveness to DNA in cells not expressing Beclin-1 (Fig. 5A) (100). This cGAS-dependent, STING-independent autophagy pathway was reported to be dependent on the DNA-dependent interaction between cGAS and Beclin-1, which in turn releases Rubicon from Beclin-1 to initiate the autophagy pathway for the degradation of DNA (100). In addition, data were provided to support the idea that cGAMP induces autophagy exclusively through a STING-dependent pathway, whereas DNA-induced autophagy is critically dependent on cGAS but proceeds through both STING-dependent and -independent pathways (104).
Collectively, the data available on DNA sensing and autophagy suggest that both DNA and STING are targeted for degradation by this pathway. However, whether the degradations of DNA and STING are triggered by the same signal and also whether there is selectivity in the delivery of cargo from the DNA sensing pathway to autophagosomes remain unknown. The finding that STING but not TBK1 is degraded in response to DNA stimulation suggests that there is selectivity in the system.
DNA-ACTIVATED INNATE IMMUNE RESPONSES IN DEFENSE AND DISEASE
In parallel to the progress made on the molecular mechanisms in DNA sensing and signaling, there is now also ample evidence that this system plays central roles in protective immunity, autoinflammation, and, most recently, cancer development. The first demonstration of the role of the intracellular DNA sensing pathway in host defense was a report of STING-deficient mice being highly susceptible to intravenous HSV-1 infection, with accelerated mortality and elevated viral loads in the brain (80). With the same infection model, a very similar picture was observed for cGAS-deficient mice (105). Following these data, the question arises as to which cell types in the brain sense HSV-1 DNA through the cGAS-STING pathway. It will also be important to reveal how the cGAS-STING pathway interacts with the TLR3 pathway, known to be central for the control of HSV infection in the central nervous system (106). cGAS−/− mice also exhibit elevated susceptibility to intraperitoneal infection with murine gammaherpesvirus 68 and intranasal infection with vaccinia virus (107). While viral DNA is indeed detected directly by DNA sensors (64), a recent article now demonstrates that mitochondrial DNA stress occurs during infection with herpesviruses and that this leads to a leakage of mitochondrial DNA into the cytoplasm for the activation of the cGAS-STING pathway (108). This innate antiviral response triggered by mitochondrial DNA was demonstrated to contribute significantly to the control of infection in vitro and in vivo. Such data highlight that innate immune activation is mediated not only directly by microbes but also through a disturbance of homeostasis as it occurs during infection.
In addition to a role in the defense against pathogens carrying DNA genomes, STING-deficient and cGAS-deficient mice were also demonstrated to be more susceptible to infection with the RNA viruses vesicular stomatitis virus and West Nile virus, respectively (80, 107). This has been suggested to be due to a lower antiviral tone (i.e., lower constitutive antiviral activity) in mice with a defective DNA sensing pathway, since lower constitutive levels of IFN-stimulating genes (ISGs) were observed in cGAS-deficient cells (107). It is possible that low constitutive levels of cytoplasmic DNA derived, e.g., from endogenous retroelements or mitochondrial DNA could stimulate this response and hence be beneficial for the host. Alternatively, the observed phenotypes may be explained by the stimulation of protective immune responses by host DNA released from cells as a consequence of lytic viral replication. These issues remain to be addressed in detail. Studies on bacterial infections in the murine system have revealed that Mycobacterium tuberculosis DNA stimulates autophagy in a manner dependent on STING and TBK1 and that this pathway is essential for the optimal control of infection in vivo (109). At the same time, M. tuberculosis exploits the DNA sensing pathway, as evidenced by the DNA-dependent activation of IRF3 and the impaired establishment of chronic infection in IRF3-deficient mice (110). With respect to parasitic infections, there are also data on a role for DNA sensing in the promotion of infection. It has been demonstrated that Plasmodium falciparum DNA as well as Plasmodium berghei-infected red blood cells induce type I IFN expression in a STING-dependent manner and that mice lacking IRF3/7 or TBK1 were protected from lethal infection with P. berghei (111). Such data highlight the complexity in the interplay between microbes and the immune system, at least for pathogens establishing persistent infections.
Studies with primary cells have now also documented the importance of the DNA sensing system in the recognition of pathogens in humans. For instance, small interfering RNA (siRNA)-mediated knockdown of IFI16 in human primary macrophages impairs the expression of IFN-β during infection with HSV-1 and human cytomegalovirus (HCMV) (64). By using a similar system, the type I IFN response to wild-type and mutant HIV-1 was found to be dependent on both IFI16 and cGAS (31, 112). In the case of HIV, reverse transcription DNA intermediates were also found to induce bystander cell death in abortively infected CD4+ T cells, through a mechanism involving caspases 1 and 3 (84). This work was subsequently extended by the demonstration of IFI16 as the DNA sensor in CD4+ T cells mediating pyroptotic cell death (68). At present, it is not known how the sensing of HIV DNA by IFI16 in macrophages promotes signaling through the IFN-inducing STING pathway while it stimulates inflammasome activation in CD4+ T cells. This represents a major unresolved issue of importance for HIV-1 immunology and DNA immunobiology in general. During bacterial infections, the DNA sensing system is also of importance in human cells. Although initial studies with murine cells proposed that bacterium-derived CDNs were the main PAMP inducing IFN expression (113, 114), subsequent work with human macrophages established that the induction of IFN-β expression during L. monocytogenes infection was driven by DNA, as evidenced by the dependence on IFI16, cGAS, and STING (65). This difference between humans and rodents is most likely explained by human STING binding cGAMP, which has a 2′,3′ linkage between the two nucleotide molecules (19, 115), with a much higher affinity than that of the bacterial CDN cyclic di-GMP, which has a 3′,5′ linkage between the two nucleotide molecules (45). In contrast, murine STING binds efficiently to both 2′,3′- and 3′,5′-linked CDNs (115).
For humans, it is now well documented that defects in the cytosolic DNA metabolism pathway can lead to chronic systemic activation of the IFN system and the development of Aicardi-Goutières syndrome (AGS), which is an early-onset encephalopathy (116). Mutations in Trex1 leading to impaired clearance of cytosolic DNA, most likely derived from retroelements and DNA replication by-products, are frequently associated with severe cases of AGS. In addition, mutations in SAMHD1 and RNase H have been identified in AGS patients (117). While SAMHD1 controls nucleotide synthesis by exhibiting phosphohydrolase activity on nucleotide triphosphates, RNase H has endonuclease activity on RNA in RNA-DNA duplexes. Collectively, these data provide compelling evidence for inappropriate DNA-driven stimulation of inflammation playing a key role in the pathogenesis of AGS. Similarly to humans, mice lacking Trex1 also develop fatal systemic inflammatory disease in a type I IFN-dependent manner (85). In contrast to humans, where encephalitis is the most severe clinical manifestation, Trex1-deficient mice exhibit severe inflammatory myocarditis and succumb to circulatory failure (88). STING-deficient mice are protected from Trex1-regulated pathological inflammatory responses, and it was demonstrated that nonhematopoietic cells initiate autoimmune disease in Trex1-deficient mice in a manner dependent on type I IFN signaling in αβ T cells (118). A subsequent study expanded on this initial work by reporting that macrophages also play an important role in driving the pathology in the heart in Trex1−/− mice (119). Although it remains to be demonstrated whether cGAS-Trex1-double-deficient mice are protected from disease, one study demonstrated that deletion of cGAS from Trex1-deficient murine fibroblasts prevents the spontaneous expression of IFN-β and IFN-stimulated genes (90).
Dysregulation of the DNA sensing pathway is also associated with other inflammatory diseases beyond AGS, including systemic lupus erythematosus, and in mice, it has been demonstrated that a lack of DNase II leads to embryonic lethality, which is alleviated in DNase II-STING-double-deficient mice (120). Recently, it was reported that six patients with early-onset systemic inflammation, cutaneous vasculopathy, and pulmonary inflammation were all heterozygous for mutations in exon 5 of the gene encoding STING (121). The authors of this study classified this disease as STING-associated vasculopathy with onset in infancy (SAVI). These mutations lead to an amino acid shift of one of the conserved residues V147, N154, and V155, all of which are localized in or around the STING dimerization interphase. The expression of the mutant STING proteins, either in patient cells or in a HEK293T cell system, led to constitutive IFN-β expression with a limited additional elevation of levels upon cGAMP stimulation. Together, knowledge on these inflammatory diseases driven by the constitutive activation of DNA-driven inflammation, and the STING pathway, underscores the importance of well-controlled mechanisms in the negative regulation of the pathway. At the mechanistic level, data on the SAVI-associated STING mutants indicate that under certain conditions, STING can be activated to stimulate IFN-β expression in the absence of cGAMP. This could be mediated, e.g., by STING-interacting proteins inducing the same conformational change in the C-terminal domain as the one induced by cGAMP. Although the noncanonical mechanisms of STING activation remain to be identified, data from reports on STING-dependent signaling pathways activated by ER stress and virus-cell fusion support the idea that STING can be activated by more than the cGAS-cGAMP axis (60, 122).
Finally, there is now also emerging evidence that the DNA sensing pathway plays a role in cancer. First, it was reported that mutagens induce nuclear DNA leakage into the cytosol and that the resulting STING-dependent inflammatory response mediated by infiltrating leukocytes drives carcinogenesis. Consequently, STING-deficient mice were largely resistant to mutagen-induced cancer (70). This was followed by papers reporting that STING deficiency renders mice more prone to the development of different cancers (123–125). Mechanistically, the observed phenomenon was explained by tumor-derived DNA activating DCs through the cGAS-STING pathway to induce IFN-dependent cross-priming of tumor antigens and the activation of tumor-specific T cells (123, 126, 127). Together, this series of papers suggests that the DNA sensing system plays both positive and negative roles in cancer by impacting both inflammation-stimulated carcinogenic processes and antitumor T cell responses.
Altogether, the activation and control of the innate DNA sensing machinery are of importance for host defense and the avoidance of inflammatory diseases, respectively, and are also centrally involved in tumor immunology.
VIRAL EVASION OF THE DNA SENSING MACHINERY
Perhaps the best evidence for a given immunological pathway being of physiological importance is the demonstration of the pathway being targeted by a microbe. Several viral mechanisms to evade the DNA sensing machinery have now been reported. The HCMV pUL83 protein was first shown to play a role in the downmodulation of innate immune responses during infection (128, 129). This was followed by the demonstration that IFI16 acts as a nuclear DNA sensor for HCMV infection in fibroblasts and that pUL83 interacts directly with IFI16 through the pyrin domain, hence blocking oligomerization upon DNA sensing and subsequent immune signals (130) (Fig. 5C). HCMV utilizes the pUL83-IFI16 interaction to further redirect IFI16 to the major HCMV immediate early promoter, thus augmenting viral replication at the early stages of the replication cycle (131).
HSV-1 encodes a ring finger domain-containing protein, ICP0, with E3 ubiquitin ligase activity. ICP0 catalyzes the K48-linked polyubiquitination of target proteins, thereby promoting proteasomal degradation (132). Viruses deficient in ICP0 induce elevated expression levels of type I IFN compared to those induced by the wild-type virus (133). Orzalli and colleagues reported that cellular sensing of viral DNA in the nucleus induced IFN responses in human fibroblasts after HSV-1 infection in a manner dependent on IFI16 and STING (39). Moreover, they reported that ICP0 counteracts this by promoting the degradation of IFI16 in a proteasome- and ring finger domain-dependent manner (39). The HSV-1-mediated degradation of IFI16 has been confirmed by others (134). However, this study, which used a different strain of HSV-1, found that the degradation of IFI16 was independent of ICP0 and that IFI16 is not efficiently degraded in all cell types productively infected, hence also suggesting the contribution of cellular factors to the effectuation of IFI16 degradation. Therefore, the molecular mechanisms of HSV-1-mediated degradation of IFI16, including ICP0-dependent and -independent pathways, remain unresolved.
Hepatitis B virus (HBV) is a small DNA virus with a replication cycle including at least two distinct forms of DNA, namely, relaxed circular DNA and covalently closed circular DNA. A recent study demonstrated that the HBV polymerase inhibits STING-dependent signaling in a manner independent of the catalytic activity of the enzyme (135). This was mediated by a direct interaction between HBV Pol and STING, which led to the inhibition of K63-linked ubiquitination of STING (135). At this stage, there are no published data on viral targeting of cGAS. Other viral proteins affecting DNA-driven IFN expression have also been reported, such as the vaccinia virus C16 protein, which binds DNA-dependent protein kinase (136). Knowledge on how DNA-dependent protein kinase interacts with the cGAS-cGAMP-STING pathway will allow further insight into how viruses evade immune responses.
Inflammasome activation is now increasingly being appreciated to contribute to optimal innate and adaptive immune defenses against viral infections, as evidenced by data from in vivo models (74, 137). Further support for this comes from the fact that poxviruses encode pyrin-only proteins (POPs), which inhibit inflammasome activation. Mammalian POPs were recently reported to inhibit inflammasome binding to the pyrin domains of either ASC (POP1 and -2) or AIM2 and IFI16 (POP3) (138–140), thus inhibiting the activation of these inflammasomes by DNA viruses (138). Many poxviruses encode POPs that share a high degree of similarity with mammalian POPs. Poxvirus POPs act similarly to POP1 and -2 via pyrin-pyrin interactions with ASC to prevent the proper assembly of the inflammasome (141). Importantly, a myxomavirus mutant lacking the M13L POP was not pathogenic in vivo in a rabbit model, thus emphasizing the importance of the inflammasome in the pathogenesis of viral infections (141).
The above-described mechanisms all target a specific DNA-stimulated pathway. Herpesviruses were recently demonstrated to also have evolved a mechanism to counteract the exposure of viral DNA to cytosolic DNA sensors (76). It was first reported that the HSV-1 capsid is ubiquitinated in the cytosol in macrophages and that this stimulates a degradation pathway leading to the release of DNA into the cytosol (64). All herpesviruses encode a deubiquitinase in the large tegument protein, and it has now been reported that this enzymatic activity is essential for the efficient delivery of viral DNA to the nucleus versus the cytoplasm and that alpha-, beta-, and gammaherpesviruses lacking the deubiquitinase activity stimulate elevated expression of type I IFN through the STING pathway (76).
Collectively, viruses have evolved strategies to hide their DNA from the immune system and also to inactivate specific DNA-activated pathways. It should be noted that some microorganisms do not inhibit but rather exploit the DNA sensing system to promote infection. For example, M. tuberculosis stimulates the DNA-driven STING-dependent IRF3 pathway, and this is essential for long-term infection (110). Interestingly, these bacteria stimulate this pathway via limited perforation of the phagosome membrane mediated by the ESX-1 secretion system, most likely allowing low-grade leakage of bacterial DNA into the cytoplasm.
CONCLUSION
DNA is a potent inducer of innate immune responses, and the past couple of years have revealed mechanistic insight into how the molecule carrying the genetic code also functions as an alarm molecule stimulating the production of type I IFNs and inflammatory molecules, the activation of inflammasomes, as well as different types of cell death. We are now also starting to understand how cell types differ in their responses to DNA sensing and how cells are equipped with systems to downmodulate DNA-driven immune responses. The importance of this is highlighted by the recent identification of STING gain-of-function mutations in patients with SAVI (121). A key challenge for the field is now to obtain mechanistic insight into the function of the DNA sensing machinery in vivo. Studies on immune stimulation by DNA have additionally revealed that proteins previously demonstrated to function in the DNA damage response also work in this immune pathway. Therefore, in addition to generating fundamental new knowledge on an important part of the host defense system against infections, the field of DNA immunology research continues to provide further insight into the mechanistic link between infectious diseases, autoimmunity, and cancer.
ACKNOWLEDGMENTS
The work in my laboratory related to the topic discussed in the present review is supported by grants from the Danish Medical Research Council (grant no. 12-124330), the Novo Nordisk Foundation, the Lundbeck Foundation (grant no. R34-3855), the Kathrine og Vigo Skovgaards Fond, the Aarhus University Research Foundation, and Fonden til Lægevidenskabens Fremme.
I have no conflicts of interest.
Biography
Søren R. Paludan has an educational background in molecular biology and biochemistry obtained through studies at Aarhus University and the University of Oxford. He received his Ph.D. and D.M.Sc. from Aarhus University based on research in the fields of herpesvirology and innate immunology. He is now professor in virology and immunology at Aarhus University, with research interest in how microbes are detected by the innate immune system and how this leads to protective and pathological immune responses.
REFERENCES
- 1.Medzhitov R. 2008. Origin and physiological roles of inflammation. Nature 454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 2.Rotem Z, Cox RA, Isaacs A. 1963. Inhibition of virus multiplication by foreign nucleic acid. Nature 197:564–566. doi: 10.1038/197564a0. [DOI] [PubMed] [Google Scholar]
- 3.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 4.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 5.Stetson DB, Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. 2002. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol 32:1958–1968. doi:. [DOI] [PubMed] [Google Scholar]
- 8.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 9.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol 5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda K, Richez C, Uccellini MB, Richards RJ, Bonegio RG, Akira S, Monestier M, Corley RB, Viglianti GA, Marshak-Rothstein A, Rifkin IR. 2009. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J Immunol 183:3109–3117. doi: 10.4049/jimmunol.0900399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, Miyake K, Shimizu T. 9 February 2015 Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiliveru S, Rahbek SH, Jensen SK, Jorgensen SE, Nissen SK, Christiansen SH, Mogensen TH, Jakobsen MR, Iversen L, Johansen C, Paludan SR. 2014. Inflammatory cytokines break down intrinsic immunological tolerance of human primary keratinocytes to cytosolic DNA. J Immunol 192:2395–2404. doi: 10.4049/jimmunol.1302120. [DOI] [PubMed] [Google Scholar]
- 14.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 17.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 18.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. 2013. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. 2013. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. 2013. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep 3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, Zhang X, Chen ZJ. 2014. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep 6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. 2013. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsen MR, Paludan SR. 2014. IFI16: at the interphase between innate DNA sensing and genome regulation. Cytokine Growth Factor Rev 25:649–655. doi: 10.1016/j.cytogfr.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Jin TC, Perry A, Jiang JS, Smith P, Curry JA, Unterholzner L, Jiang ZZ, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois C, Jin T, Xiao T, Fitzgerald P, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth S, Rottach A, Lotz-Havla AS, Laux V, Muschaweckh A, Gersting SW, Muntau AC, Hopfner KP, Jin L, Vanness K, Petrini JH, Drexler I, Leonhardt H, Ruland J. 2014. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1beta production. Nat Immunol 15:538–545. doi: 10.1038/ni.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. 2014. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A 111:E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Jin T, Laustsen A, Hansen K, Ostergaard LJ, Fitzgerald KA, Xiao T, Mikkelsen JG, Mogensen TH, Paludan SR. 2013. IFI16 senses DNA forms of the retroviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A 110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannini A, Cramer P. 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Kanaar R, Wyman C. 2008. DNA repair by the MRN complex: break it to make it. Cell 135:14–16. doi: 10.1016/j.cell.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 34.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A 107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZQ, Yuan B, Bao MS, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyashita M, Oshiumi H, Matsumoto M, Seya T. 2011. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol 31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang ZQ, Yuan B, Lu N, Facchinetti V, Liu YJ. 2011. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol 187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu Z, Modlin RL, Liu YJ, Cheng G. 2012. The helicase DDX41 recognizes the bacterial secondary messenger cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orzalli MH, DeLuca NA, Knipe DM. 2012. HSV-1 ICP0 redistributes the nuclear IFI16 pathogen sensor and promotes its degradation. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 41.Gilliet M, Cao W, Liu YJ. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 42.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. 2013. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. 2013. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu Y, Grishin N, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 54.Tsuchida T, Zou JA, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C. 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41:919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 57.Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, Dotiwala F, Sen J, Doench JG, Orzalli MH, Kramnik I, Knipe DM, Lieberman J, Carr SA, Hacohen N. 2013. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol 14:179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y, Ye W, Xiong X, Zhong B, Shu HB, Wang YY. 2014. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe 16:450–461. doi: 10.1016/j.chom.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Jin L, Lenz LL, Cambier JC. 2010. Cellular reactive oxygen species inhibit MPYS induction of IFN beta. PLoS One 5:e15142. doi: 10.1371/journal.pone.0015142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, Fitzgerald KA, Szabo G. 2013. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A 110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferguson B, Mansur D, Peters N, Ren H, Smith GL. 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110:E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA. 2014. Interferon gamma-inducible protein (IFI) 16 transcriptionally regulates type I interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem 289:23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horan KA, Hansen K, Jakobsen MR, Holm CK, Waggoner L, West JA, Unterholzner L, Iversen MB, Soby S, Thomson M, Jensen SB, Rasmussen SB, Ellermann-Eriksen S, Kurt-Jones EA, Landolfo S, Melchjorsen J, Bowie AG, Damania B, Fitzgerald KA, Paludan SR. 2013. Proteasomal degradation of herpes simplex virus capsids in macrophage releases DNA to the cytosol for recognition by DNA sensors. J Immunol 190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbek SH, Jensen SB, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. 2014. Listeria monocytogenes induces IFNb expression through an IFI16, cGAS, and STING dependent pathway. EMBO J 33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis AJ, Chen BP, Chen DJ. 2014. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. 2011. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. 2014. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kroger A, Nilsson JA, Ek T, Weiss S, Gekara NO. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 70.Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. 2014. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun 5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lan YY, Londono D, Bouley R, Rooney MS, Hacohen N. 2014. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep 9:180–192. doi: 10.1016/j.celrep.2014.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrilli V, Dostert C, Muruve DA, Tschopp J. 2007. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol 19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abe T, Barber GN. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O'Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stacey KJ, Ross IL, Hume DA. 1993. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol 71(Part 2):75–85. [DOI] [PubMed] [Google Scholar]
- 78.Sze A, Belgnaoui SM, Olagnier D, Lin R, Hiscott J, van Grevenynghe J. 2013. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14:422–434. doi: 10.1016/j.chom.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 79.Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, Williams BR, Sen GC. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 82.Berg RK, Rahbek SH, Kofod-Olsen E, Holm CK, Melchjorsen J, Jensen DG, Hansen AL, Jørgensen LB, Ostergaard L, Tolstrup M, Larsen CS, Paludan SR, Jakobsen MR, Mogensen TH. 2014. T cells detect intracellular DNA but fail to induce potent type I IFN responses—implications for restriction of HIV replication. PLoS One 9:e84513. doi: 10.1371/journal.pone.0084513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang YG, Lindahl T, Barnes DE. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 84.Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang ZY, Santiago ML, Hebbeler AM, Greene WC. 2010. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stetson DB, Ko JS, Heidmann T, Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. 2005. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med 202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. 2014. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol 192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 91.Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, Tuting T, Hartmann G, Barchet W. 2013. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Hasan M, Koch J, Rakheja D, Pattnaik AK, Brugarolas J, Dozmorov I, Levine B, Wakeland EK, Lee-Kirsch MA, Yan N. 2013. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat Immunol 14:61–71. doi: 10.1038/nri3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. 2014. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, Ouyang S, Liu ZJ, Damania B, Shu HB, Duncan JA, Ting JP. 2014. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 96.Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, Zhong B, Shu HB. 2014. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, Kile BT. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Konno H, Konno K, Barber GN. 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, Jung JU. 2014. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. 2011. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J Virol 85:4212–4221. doi: 10.1128/JVI.02435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. 2011. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol 187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomas PG, Dash P, Aldridge JR Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC, Kanneganti TD. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reinert LS, Harder L, Holm CK, Iversen MB, Horan KA, Dagnaes-Hansen F, Ulhoi B, Holm T, Mogensen TH, Owens T, Nyengaard JR, Thomsen AR, Paludan SR. 2012. TLR3-deficiency renders astrocytes permissive to HSV infection and facilitates establishment of CNS infection in mice. J Clin Invest 122:1368–1376. doi: 10.1172/JCI60893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, Macduff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. 2 February 2015 Mitochondrial DNA stress primes the antiviral innate immune response. Nature doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watson RO, Manzanillo PS, Cox JS. 2012. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang ZZ, Chan JN, Bartholomeu DLC, Lauw F, Hall JP, Barber GN, Gazzinelli RT, Fitzgerald KA, Golenbock DT. 2011. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity 35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. 2012. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun 80:1537–1545. doi: 10.1128/IAI.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. 2006. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 117.Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U, Barnerias C, Bernard G, Bodemer C, Botella MP, Cereda C, Chandler KE, Dabydeen L, Dale RC, De Laet C, De Goede CG, Del Toro M, Effat L, Enamorado NN, Fazzi E, Gener B, Haldre M, Lin JP, Livingston JH, Lourenco CM, Marques W Jr, Oades P, Peterson P, Rasmussen M, Roubertie A, Schmidt JL, Shalev SA, Simon R, Spiegel R, Swoboda KJ, Temtamy SA, Vassallo G, Vilain CN, Vogt J, Wermenbol V, Whitehouse WP, Soler D, Olivieri I, Orcesi S, Aglan MS, Zaki MS, et al. 2013. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol 12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Barber GN, Stetson DB. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ahn J, Ruiz P, Barber GN. 2014. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol 193:4634–4642. doi: 10.4049/jimmunol.1401337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ahn J, Gutman D, Saijo S, Barber GN. 2012. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A 109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CC, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R. 2014. Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, Rixon FJ, Herold BC, Fitzgerald KA, Paludan SR. 2012. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol 13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. 2014. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF. 2014. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. 2014. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol 193:4779–4782. doi: 10.4049/jimmunol.1402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Campanella GS, Tager AM, El Khoury JK, Thomas SY, Abrazinski TA, Manice LA, Colvin RA, Luster AD. 2008. Chemokine receptor CXCR3 and its ligands CXCL9 and CXCL10 are required for the development of murine cerebral malaria. Proc Natl Acad Sci U S A 105:4814–4819. doi: 10.1073/pnas.0801544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klarquist J, Hennies CM, Lehn MA, Reboulet RA, Feau S, Janssen EM. 2014. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J Immunol 193:6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Abate DA, Watanabe S, Mocarski ES. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol 78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Browne EP, Shenk T. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A 100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol 84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Everett RD, Orr A, Preston CM. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J 17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol 78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cuchet-Lourenco D, Anderson G, Sloan E, Orr A, Everett RD. 2013. The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J Virol 87:13422–13432. doi: 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Y, Li J, Chen J, Li Y, Wang W, Du X, Song W, Zhang W, Lin L, Yuan Z. 2015. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol 89:2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, Arribas-Bosacoma R, Pearl LH, Ren H, Smith GL. 2013. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog 9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, Wallin MC, Gangopadhyay A, Forte E, Gottwein E, Perlman H, Reed JC, Greaves DR, Dorfleutner A, Stehlik C. 2014. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol 15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. 2003. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J 373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dorfleutner A, Bryan NB, Talbott SJ, Funya KN, Rellick SL, Reed JC, Shi X, Rojanasakul Y, Flynn DC, Stehlik C. 2007. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun 75:1484–1492. doi: 10.1128/IAI.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricciuto D, Wang G, McFadden G. 2005. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]