Abstract
The NR4A3 nuclear receptor is implicated in the development of extraskeletal myxoid chondrosarcoma (EMC), primitive sarcoma unrelated to conventional chondrosarcomas, through a specific fusion with EWSR1 resulting in an aberrant fusion protein that is thought to disrupt the transcriptional regulation of specific target genes. We performed an expression microarray analysis of EMC tumors expressing the EWSR1/NR4A3 fusion protein, comparing their expression profiles to those of other sarcoma types. We thereby identified a set of genes significantly over-expressed in EMC relative to other sarcomas, including PPARG and NDRG2. Western blot or immunohistochemical analyses confirm that PPARG and NDRG2 are expressed in tumors positive for EWSR1/NR4A3. Bioinformatic analysis identified a DNA response element for EWSR1/NR4A3 in the PPARG promoter, and band-shift experiments and transient transfections indicate that EWSR1/NR4A3 can activate transcription through this element. Western blots further show that an isoform of the native NR4A3 receptor lacking the C terminal domain is very highly expressed in tumors positive for EWSR1/NR4A3, and co-transfections of this isoform along with EWSR1/NR4A3 indicate that it may negatively regulate the activity of the fusion protein on the PPARG promoter. These results suggest that the overall expression of PPARG in EMC may be regulated in part by the balance between EWSR1/NR4A3 and NR4A3, and that PPARG may play a crucial role in the development of these tumors. The specific up-regulation of PPARG by EWSR1/NR4A3 may also have potential therapeutic implications.
Keywords: EWSR1/NR4A3, PPARG, extraskeletal myxoid chondrosarcoma, nuclear receptors, target genes, fusion proteins, chromosomal translocations
Introduction
NR4A3 (also known as NOR1, CHN, TEC, MINOR, CSMF) is a member of the NR4A subfamily of nuclear receptors originally discovered as an induced mRNA in cultured rat neuronal cells undergoing apoptosis [1]. All three members of the NR4A subfamily have been characterized as immediate-early gene products induced by a variety of mitogenic stimuli such as growth factors and liver regeneration [2–5]. These receptors differ from the other more typical receptors of the family in that they can either homodimerize [6], heterodimerize with each other [7], heterodimerize with RXR [8], or activate transcription constitutively as monomers apparently in the absence of ligand [5, 9–11]. The NR4A subfamily is involved in apoptosis [12, 13], differentiation [14, 15], regulation of the hypothalamo-pituitary-adrenal axis [16], vascular smooth muscle cell proliferation [17], atherogenesis [18], and more recently these receptors have been implicated in the regulation of genes involved in hepatic glucose metabolism [19] and insulin uptake by skeletal muscles and adipose tissues [20].
Given their role as immediate-early gene products in the mitogenic response, it is perhaps not unexpected that these receptors have been implicated in cancer, but their exact role in tumor development remains unclear. For example, all three receptors are down regulated in revertant HeLa cells that have lost anchorage-independent growth and tumorigenicity [21], suggesting that they may act as oncogenes in this model. On the other hand, the double NR4A1/NR4A3 knock-out mouse rapidly develops lethal acute myeloid leukemia [22], suggesting they may act as tumor suppressors in that setting. In extraskeletal myxoid chondrosarcoma (EMC), we and others have shown that the NR4A3 gene is fused most often to EWSR1 [23–25], or less commonly to one of three other gene partners: TAF15 [26–28], TCF12 [29], or TFG [30]. EWSR1 encodes a RNA-binding protein homologous to TFG, TAF15 encodes a TATA binding protein-associated factor, and TCF12 encodes a basic helix-loop-helix transcription factor. The observation that at least 95% of EMC tumors possess a NR4A3 fusion gene strongly suggests that aberrant expression of this nuclear receptor is a necessary step in the development of these tumors. In all cases characterized to date, these fusion proteins consist of the amino-terminal domain of the upstream fusion partner fused to the complete amino acid sequence of NR4A3, and therefore all contain the DNA-binding domain of NR4A3. Transient transfections of mammalian cells have shown that EWSR1/NR4A3 is a very potent transcriptional activator of a minimal promoter containing several copies of the NBRE (NGFIB DNA Response Element; [11]), a DNA response element that all three NR4A receptors can bind to as monomers and through which they constitutively activate transcription [1, 4, 5, 9, 11]. These results suggest that a key role of the NR4A3 fusion proteins in EMC tumorigenesis may be to activate the transcription of specific genes.
One approach to the identification of in vivo targets of EWSR1/NR4A3 relevant to EMC is examine the expression profile of EMC for genes that are differentially overexpressed relative to other sarcomas. However, EMC samples are rare and their histopathologic diagnosis is difficult. So far only two expression profiling studies of EMC have been published, and neither one focused on identifying EWSR1/NR4A3 target genes. One used samples in which the diagnosis of EMC was not independently confirmed by testing for EWS/NR4A3 or other related but less common EMC-specific gene fusions [31] and the other compared the profile of two fusion-positive EMCs to that of a single myxoid liposarcoma [32]. Here, we have performed expression profiling of EWSR1/NR4A3 fusion-positive EMC tumors compared to a very large set of other sarcomas and show that the peroxisome proliferator-activated receptor gamma (PPARG) gene, a significantly differentially overexpressed gene in EMC based on this analysis, is also a putative in vivo direct transcriptional target of EWSR1/NR4A3.
Materials and Methods
Microarray analyses
Three snap-frozen EMC tumor samples were obtained with institutional review board approval from patients operated at Memorial Sloan-Kettering Cancer Center (MSKCC). For all three samples, the presence of the specific EWSR1/NR4A3 fusion transcript was confirmed by reverse-transcription PCR. For expression microarray analysis, RNAs were processed at MSKCC according to procedures recommended by Affymetrix (Santa Clara, CA). Samples were hybridized to Affymetrix U133A microarrays containing 22,215 probes sets representing approximately 18,500 transcripts from approximately 14,500 genes. Array hybridization and scanning were performed at the MSKCC Genomics Core Laboratory. Expression estimates from the Affymetrix probe set data were derived using the robust multi-array average (RMA) [33]. To identify genes significantly differentially expressed in EMC compared to other primitive sarcomas with translocation-derived aberrant transcriptional proteins, we compared the data from these 3 EMC samples to similarly processed data from 137 samples of five other types of sarcomas (see Results for listing). Differentially expressed genes were identified based on two-tailed t-tests with a stringent Bonferroni-adjusted p < 0.05 threshold (i.e. p < 2.25 × 10−6). In addition, we restrict the differential set to those with fold changes of at least 2.
Western blot and immunohistochemistry analyses
Total protein extracts from EMC tumors were separated by SDS-PAGE, transferred on PVDF membranes (Millipore) and reacted with the following antibodies according to the manufacturer's recommendations: SGK1 (Millipore, 07–315), SGK1-pThr256 and NDRG2 (Santa Cruz Biotechnology, sc-16744 and sc-19468 respectively), NDRG2-pThr348 (Kinasource, PB-022), PPARG (Cell Signaling Technology, 2443) and actin (Developmental Studies Hybridoma Bank, University of Iowa, JLA20). EWSR1/NR4A3, NR4A3 and NR4A3ΔC were detected with a polyclonal antibody directed against NR4A3 [34]. Western blots were revealed with a Perkin-Elmer Chemiluminescence kit based on horseradish peroxidase. NDRG2 immunohistochemistry analyses of EMC tumors was performed as previously described [35].
Real-time PCR analyses of SGK1 mRNAs
EMC tumor RNA was prepared using the RNeasy Plus Kit and reverse-transcription of the RNA was performed using the Quantitec RT Kit, both from Qiagen. Real-time PCR was performed with intron-spanning primers designed by Simon and colleagues [36] and specific for each mRNA: SGK1+1: forward: 5' gtggtgatgacggtgaaaactg 3', reverse: 5' tgaaagcgatgagaattgcca 3'; SGK1-850: forward: 5' cacaaaaagagggccgagg 3', reverse: 5' atgaaagccagtccgctcag 3'; SGK1-2981: forward: 5' ctctacctccagcctccagaag 3', reverse: 5' ctgcttcatgaaagctttcaaag 3'. The amplification efficiencies of these primers were estimated by generating standard calibration curves with two-fold dilution series. Reactions were carried out using the Power SYBR Green Master Mix and the ABI Prism 7000 Sequence Detector System, both from Applied Biosystems. Amplification conditions were the same for all targets assayed: one cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s and at 60°C for 60 s. Samples were run in triplicate with a final volume of 25 µl per reaction. Formation of the expected PCR product was monitored by melting curve analysis. An 18S ribosomal RNA amplification was used to normalize the data for the difference in starting material and efficiencies of RNA extractions and reverse-transcription reactions for each tumor sample.
Electrophoretic mobility gel-shift assays, cell lines and transient transfections
Band-shift assays were carried out as described previously [11]. Oligonucleotides used were NBRE: 5' gagttttaaaaggtcatgctaatttgg 3', PPARG: 5' caggaaaagaaaaggtcactgtctaccca 3, mutant NBRE: 5' gagttttaagaggtcatgctaatttgg 3', mutant PPARG: 5' caggaaaagaagaggtcactgtctaccca 3'. The cell lines CFK2, pc2, et2, et16 and et19 [37] were maintaind in RPMI-1640 with 10% FBS and antibiotics. G418 was added at 250 µg/ml for pc2, et2, et16 and et19. COS cells were maintaind in DMEM with 10% FBS and antibiotics. Transient transfections were performed with Effectene (Qiagen) following the manufacturer's recommendations. COS cells were transfected with a PPARG isoform 1 expression vector obtained from OriGene Technologies, Inc., and total protein extracts were prepared 48 hours after transfection. CFK2 cell lines were transfected with the indicated vectors and 48 hours afterwards total cell extracts were prepared and luciferase and beta-galactosidase activities were determined using a Luciferase Assay System from Promega, a Beta-Gal Assay Kit from Clontech, and a Berthold MiniLumat LB 9506 Luminometer. Luciferase activities were divided by beta-galactosidase activities to correct for transfection efficiencies.
Results
Expression profiling of EMC tumors
To identify genes significantly differentially expressed in EMC, we compared the expression profile of three EWSR1/NR4A3 fusion-positive EMC tumors to the expression profile of 137 other primitive sarcomas with translocation-derived aberrant transcriptional proteins. These sarcomas consisted of 28 Ewing’s sarcomas (ES), 23 alveolar rhabdomyosarcomas (ARMS), 28 desmoplastic small round cell tumors (DSRCT), 12 alveolar soft part sarcomas (ASPS) and 46 synovial sarcomas (SS), all of which had also been confirmed by RT-PCR to harbor their respective specific fusion transcripts (M. Ladanyi, unpublished data). Using two-tailed t-tests with a stringent Bonferroni correction together with a two-fold cut-off (see Methods), we identified 404 probe sets that were significantly differentially expressed in EMC relative to the 137 samples in the rest of the dataset (Supplementary Tables 1 and 2). Of these 404 probe sets, 383 were higher in EMC and 11 were lower.
The top 25 differentially expressed probes are listed in Table 1. Subramanian and colleagues derived an expression profile of EMC on a different microarray platform and with a different set of sarcomas as a comparison group (cases of SS, gastrointestinal stromal tumour, leiomyosarcoma, malignant fibrous histiocytoma, dermatofibrosarcoma protuberans) [31]. In spite of these differences, there is a substantial overlap in significantly differentially expressed genes between their study and the present one; 20/50 top EMC genes in their study also appear in our 383 significantly differentially overexpressed probe sets (Table 2). We should also note that some genes may not have been represented on both microarray platforms. The overlapping genes are likely to comprise robust components of the EMC expression profile. In particular, DKK1 and NMB appear at or near the very top of both lists, thus validating the data (Tables 1 and 2). Here, we further examine the roles of two other genes identified as EMC-related by both studies, PPARG and NDRG2.
Table 1.
Top 25 probe sets differentially expressed in EMC. All probe sets were more highly expressed in EMC than in the 5 other sarcomas. P-values reflect Bonferonni correction for 22,215 comparisons.
| Probe Set | Fold | P-value | Gene | Gene Description |
|---|---|---|---|---|
| 204602_at | 40 | 4.6E-65 | DKK1 | dickkopf homolog 1 |
| 220115_s_at | 117 | 7.2E-62 | CDH10 | cadherin 10, type 2 |
| 205204_at | 65 | 2.6E-42 | NMB* | neuromedin B |
| 207930_at | 32 | 1.7E-40 | LCN1 | lipocalin 1 |
| 220283_at | 8 | 8.2E-38 | KIAA1822L | KIAA1822-like |
| 220595_at | 15 | 3.8E-36 | PDZRN4 | PDZ domain containing RING finger 4 |
| 220356_at | 15 | 7.8E-36 | CORIN | corin, serine peptidase |
| 207577_at | 5 | 3.4E-35 | HTR4 | 5-hydroxytryptamine (serotonin) receptor 4 |
| 208116_s_at | 8 | 3.6E-35 | MAN1A1 | mannosidase, alpha, class 1A, member 1 |
| 215913_s_at | 19 | 5.1E-33 | GULP1 | GULP, engulfment adaptor PTB domain containing 1 |
| 215972_at | 7 | 4.2E-32 | --- | clone 24820 mRNA sequence |
| 222317_at | 7 | 1.6E-30 | PDE3B | phosphodiesterase 3B, cGMP-inhibited |
| 206730_at | 5 | 4.2E-25 | GRIA3 | glutamate receptor, ionotrophic, AMPA 3 |
| 206869_at | 10 | 3.3E-24 | CHAD | chondroadherin |
| 205694_at | 18 | 3.6E-24 | TYRP1 | tyrosinase-related protein 1 |
| 207012_at | 7 | 1.7E-23 | MMP16 | matrix metallopeptidase 16 |
| 205523_at | 8 | 9.1E-23 | HAPLN1 | hyaluronan and proteoglycan link protein 1 |
| 214390_s_at | 9 | 1.2E-22 | BCAT1 | branched chain aminotransferase 1, cytosolic |
| 207379_at | 12 | 2.8E-22 | EDIL3 | EGF-like repeats and discoidin I-like domains 3 |
| 218197_s_at | 14 | 7.5E-22 | OXR1 | oxidation resistance 1 |
| 206637_at | 10 | 1.5E-21 | P2RY14 | purinergic receptor P2Y, G-protein coupled, 14 |
| 214582_at | 4 | 2.0E-21 | PDE3B | phosphodiesterase 3B, cGMP-inhibited |
| 219564_at | 5 | 6.2E-21 | KCNJ16 | potassium inwardly-rectifying channel J16 |
| 216012_at | 5 | 1.0E-20 | --- | unidentified mRNA, partial sequence |
| 220351_at | 5 | 6.3E-20 | CCRL1 | chemokine (C-C motif) receptor-like 1 |
Table 2.
Genes differentially overexpressed in EMC in present study overlapping with top 50 EMC-overexpressed genes in Subramanian and colleagues [31].
| Probe Set | Fold | P-value | Gene | Gene Description |
|---|---|---|---|---|
| 204602_at | 40 | 4.6E-65 | DKK1 | dickkopf homolog 1 |
| 205204_at | 65 | 2.6E-42 | NMB | neuromedin B |
| 208116_s_at | 8 | 3.6E-35 | MAN1A1 | mannosidase, alpha, class 1A, member 1 |
| 201732_s_at | 14 | 2.0E-16 | CLCN3 | chloride channel 3 |
| 205236_x_at | 5 | 3.5E-13 | SOD3 | superoxide dismutase 3, extracellular |
| 217997_at | 34 | 8.5E-12 | PHLDA1 | pleckstrin homology-like domain, member A1 |
| 214279_s_at | 9 | 3.4E-10 | NDRG2 | NDRG family member 2 |
| 209468_at | 4 | 3.6E-08 | LRP5 | low density lipoprotein receptor-related protein 5 |
| 209618_at | 3 | 1.5E-07 | CTNND2 | catenin (cadherin-associated protein), delta 2 |
| 204467_s_at | 3 | 9.6E-07 | SNCA | synuclein, alpha |
| 204284_at | 15 | 3.9E-05 | PPP1R3C | protein phosphatase 1, regulatory subunit 3C |
| 211571_s_at | 25 | 2.2E-04 | CSPG2 | chondroitin sulfate proteoglycan 2 (versican) |
| 217865_at | 6 | 4.2E-04 | RNF130 | ring finger protein 130 |
| 202922_at | 5 | 8.0E-04 | GCLC | glutamate-cysteine ligase, catalytic subunit |
| 202218_s_at | 8 | 1.1E-03 | FADS2 | fatty acid desaturase 2 |
| 214620_x_at | 11 | 1.6E-03 | PAM | peptidylglycine alpha-amidating monooxygenase |
| 205736_at | 4 | 1.7E-03 | PGAM2 | phosphoglycerate mutase 2 (muscle) |
| 201941_at | 4 | 1.0E-02 | CPD | carboxypeptidase D |
| 203571_s_at | 19 | 1.1E-02 | C10orf116 | chromosome 10 open reading frame 116 |
| 208510_s_at | 2 | 2.3E-02 | PPARG | peroxisome proliferative activated receptor, gamma |
Note: for genes represented in Supplementary Table 2 by more than one probe set, only the most significant is shown. P-values reflect Bonferonni correction for 22,215 comparisons.
Western blot and immunohistochemistry analyses of EMC tumors
To validate the microarray results, we performed western blot analyses on three EMC tumor samples, all three with evidence of the EWSR1/NR4A3 fusion (Fig. 1A). Tumors 2 and 3 express the EWSR1/NR4A3 type 2 fusion mRNA [25] as determined by RT-PCR, while tumor 1 was cytogenetically positive for the typical t(9;22) chromosome translocation characteristic of an EWSR1/NR4A3 gene fusion (data not shown). Only one of these 3 EMCs was among the 3 studied above by expression profiling, and therefore two of the samples were also independent of the above mRNA-level expression data. The fusion protein was detected using a polyclonal antibody made in our laboratory and directed against NR4A3 [34], and the results were confirmed using a commercial monoclonal antibody to NR4A3 (Perseus Proteomics Inc., data not shown). We then analyzed the expression of PPARG because its promoter contains a perfect predicted NBRE binding site (5' AAAGGTCA 3') located at −675 bp upstream of the transcriptional start site, making it a potential target gene for EWSR1/NR4A3. As shown in Figure 1B, tumors 2 and 3 are clearly positive for PPARG, while a faint band can be detected in tumor 1. In four additional EMC tumors analyzed, three showed prominent PPARG expression (data not shown) indicating that, in aggregate, 5/7 (71%) EMCs show clear expression of PPARG by western blotting.
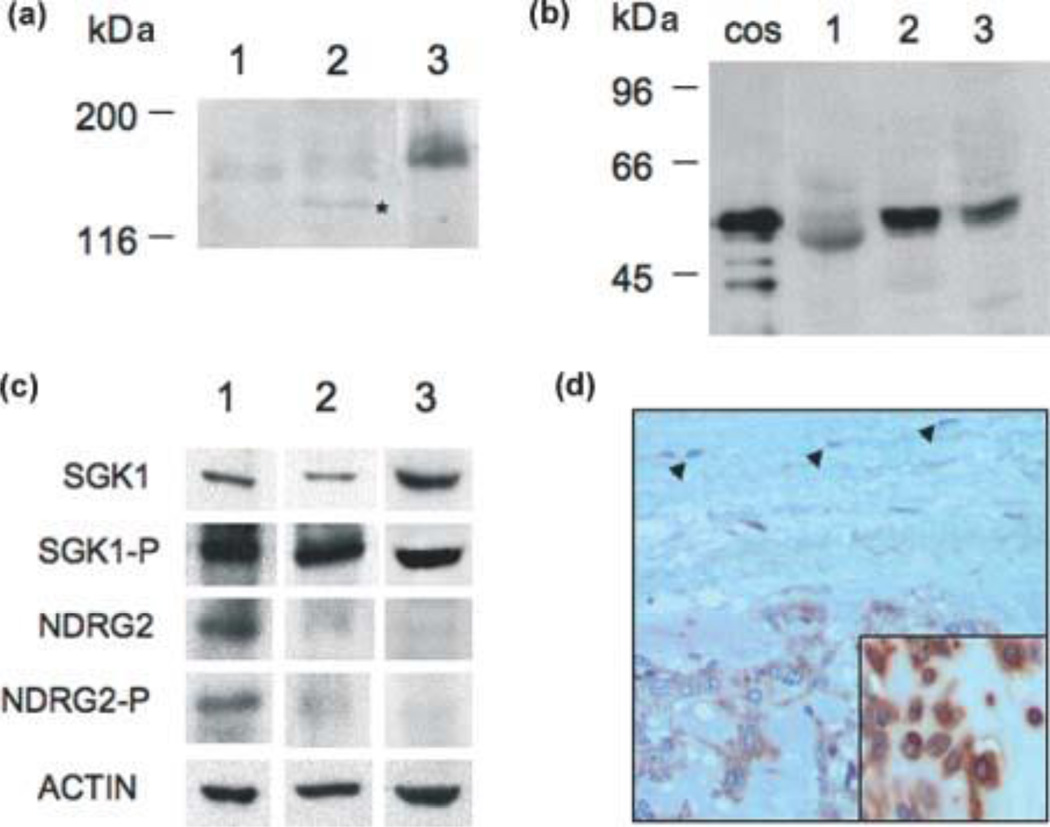
Figure 1.
Expression of EWSR1/NR4A3, PPARG, NDRG2 and SGK1 proteins in EMC tumors. (A) Western blot analyses of three EMC tumors demonstrating the expression of the EWSR1/NR4A3 fusion protein using a polyclonal antibody directed against NR4A3 [34]. The lower band detected in tumor sample 2 and marked by a star may correspond to either a degradation product of the full-length fusion protein or a truncated isoform generated by alternative splicing of the fusion mRNA [23]. (B) Western blot analysis of PPARG expression in the same three tumor samples. The lane labeled COS contains a sample of COS cells transfected with a PPARG isoform 1 expression vector. (C) Western blot analysis of NDRG2 and SGK1 proteins in the same three tumor samples. (D) Immunohistochemistry staining for NDRG2 showing diffuse cytoplasmic staining of EMC tumor cells and lack of staining in fibroblasts in fibrotic capsule (arrows) (200×). Inset shows higher magnification (400×) of cytoplasmic staining in another EMC case.
We analyzed the expression of the serum- and glucocorticoid-regulated kinase 1 (SGK1) because although our tumor expression microarray data suggest a lower level of this mRNA in EMC than in other sarcomas (Supplementary Table 1, also consistent with the data of Subramanian and colleagues [31]), we have previously shown by immunohistochemistry that EMC tumors express significant levels of this kinase [38]. Figure 1C confirms the immunohistochemistry observations and furthermore we show that SGK1 is activated in EMC tumors using an antibody specific to a phosphorylated form of the protein. Finally we analyzed the expression of N-myc downstream-regulated gene 2 (NDRG2) because it is a known phosphorylation target of SGK1 [39] and its mRNA is up-regulated in our microarray analyses and those of Subramanian et al [31](Table 2). Figure 1C shows that tumor sample 1 is clearly positive for both total and phosphorylated NDRG2, while this is not clear for samples 2 and 3. To further document NDRG2 expression in EMC, we performed immunohistochemistry analyses which confirmed that NDRG2 is moderately to strongly expressed in the cytoplasm of EMC tumor cells in 9/9 EWSR1/NR4A3-positive EMCs (Fig. 1D). These data suggest that NDRG2 may be a pathophysiological target of SGK1 in EMC.
SGK1 mRNAs expression in EMC tumors
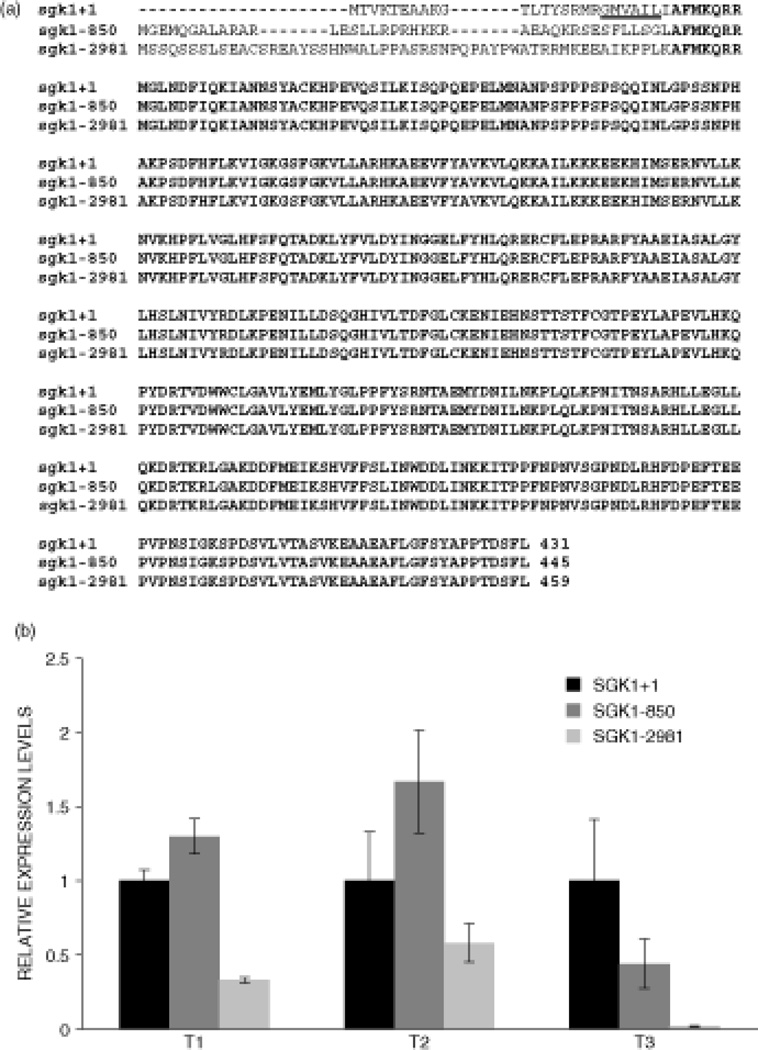
The expression data indicate that the SGK1 mRNA appears to be downregulated in EMC compared to other sarcomas, while the protein data shows that the protein is actually over-expressed in EMC. The reason for this discrepancy is unknown, however it may be related to the fact that there are three different mRNAs encoding three SGK1 isoforms [36], two of which lack the proteasomal degradation signal located in the amino-terminal domain of the protein [40] (Fig. 2A). Simon and colleagues have compared the expression of the three SGK1 mRNAs (named SGK1+1, SGK1-850 and SGK1-2981, with SGK1+1 encoding the isoform containing the degradation signal) in various tissues and cell lines, and they found that SGK1+1 was the most highly expressed, about 20-fold more than SGK1-850 and 500-fold more than SGK1-2981 [36]. Using the same primers for real-time PCR as Simon and colleagues, we compared the expression levels of these SGK1 isoforms in three EMC tumors, and the results show that for two tumors, SGK1-850 appears to be more highly expressed than SGK1+1, while in the third it is approximately 2-fold less expressed than SGK1+1 (Fig. 2B). Thus, the SGK1 protein isoform expressed in most EMC tumors may lack the proteasomal degradation signal, leading to the accumulation of SGK1 protein observed by immunohistochemistry, in spite of relatively low transcript levels.
Figure 2.
Real-time PCR analyses of SGK1 mRNAs in EMC tumors. (A) Schematic diagram of the three SGK1 protein isoforms encoded by the SGK1+1, SGK1-850 and SGK1-2981 mRNAs. Common amino acids are in bold characters and the proteasomal degradation signal present in the SGK1+1 isoform is underlined. (B) Real-time PCR of the three SGK1 mRNAs in three EMC tumors. For each tumor, a total RNA extract was prepared from which three different reverse-transcription reactions were performed followed by a real-time PCR for each SGK1 mRNA and the 18S ribosomal RNA. The data were normalized with respect to the 18S RNA and the results are expressed as relative expression levels with the SGK1+1 mRNA set at one. The standard deviations are shown as vertical bars.
Transcriptional activation of PPARG by EWSR1/NR4A3
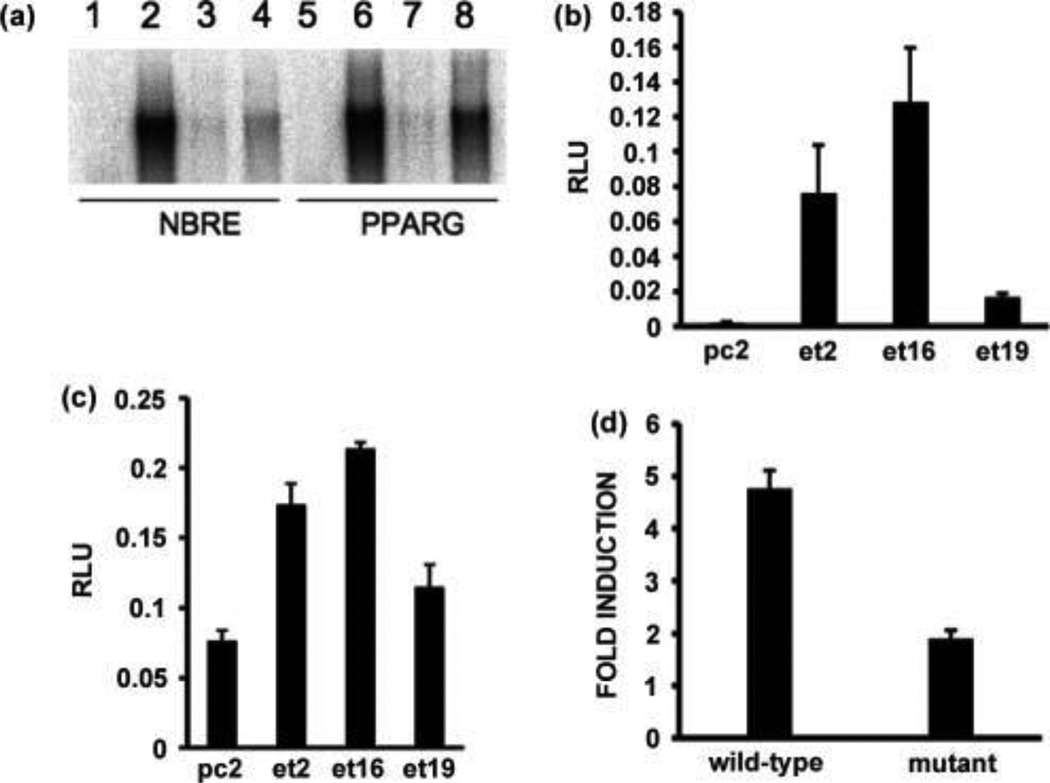
To examine the possibility that PPARG may be directly transcriptionally up-regulated by EWSR1/NR4A3, we performed band-shift experiments with the fusion protein and oligonucleotides containing the NBRE of the PPARG promoter. As a control we used the NBRE that we have previously characterized as a binding site for EWSR1/NR4A3 [11]. The results indicate that the fusion protein binds these two NBRE sites with similar affinity, and mutant oligonucleotide controls confirmed the specificity of the binding (Fig. 3A). To determine if EWSR1/NR4A3 could activate transcription from this element, we used a pGL3 luciferase reporter vector containing a 2.8 kb insert corresponding to the human PPARG isoform 1 promoter including the NBRE site [41]. We transfected this reporter vector in CFK2 cell lines that stably express EWSR1/NR4A3 [37]. As a control we transfected the cell lines with a p(B1a)8luc reporter vector that is strongly activated by EWSR1/NR4A3 [11]. As shown in Figure 3B, the three cell lines expressing EWSR1/NR4A3 all significantly activate p(B1a)8luc compared to the control pc2 cell line, and the same activation pattern is observed in the cell lines when the PPARG reporter is used (Fig. 3C). The activation ratio is smaller for the PPARG promoter, however this can be explained by the fact that it contains only one NBRE site located −675 bp from the transcriptional start site, whereas the control reporter vector contains 8 NBRE sites adjacent to the transcriptional start site. To extend these observations, we subcloned into pGL3 a 1 kb fragment of the PPARG promoter and mutated the NBRE from 5' AAAGGTCA 3' to 5' AGAGGTCA 3' (the same mutation used for the band-shift assays in Fig. 3A). Transient transfections of CFK2 cells with EWSR1/NR4A3 and the wild-type or mutated promoter clearly show that this single nucleotide change drastically reduces the ability of the fusion protein to activate transcription of PPARG (Fig. 3D). Thus, the PPARG promoter is a target of aberrant transactivation by the EWSR1/NR4A3 fusion protein in EMC tumors.
Figure 3.
Activation of the PPARG promoter by EWSR1/NR4A3 through the NBRE site. (A) Band-shift analyses of in vitro translated EWSR1/NR4A3 (lanes 2–4 and 6–8) with NBRE (lanes 1–4) and PPARG (lanes 5–8) oligonucleotides. Lanes 3 and 7 reactions contained a 25-fold excess of cold wild-type oligonucleotides, and lanes 4 and 8 reactions contained a 25 fold excess of mutant oligonucleotides. The reactions in lanes 1 and 5 used a control rabbit reticulocyte lysate sample. (B) and (C) Transient transfections of CFK2 cell lines stably expressing EWSR1/NR4A3 (et2, et16, et19) with a NBRE luciferase reporter vector (B) or a PPARG promoter luciferase reporter vector (C). The pc2 cell line is a negative control cell line stably transfected with an empty expression vector. The results are expressed as relative light units (RLU) and the standard deviation is shown (vertical bars). (D) Transient transfections of wild-type CFK2 cells with an EWSR1/NR4A3 expression vector, a wild-type or mutated PPARG promoter luciferase reporter vector, and a pBIND (Promega) normalization vector. Transfections were carried out in triplicate, the results are expressed as fold induction relative to the empty expression vector, and the standard deviation is shown (vertical bars).
Expression of NR4A3∆C in EMC tumors
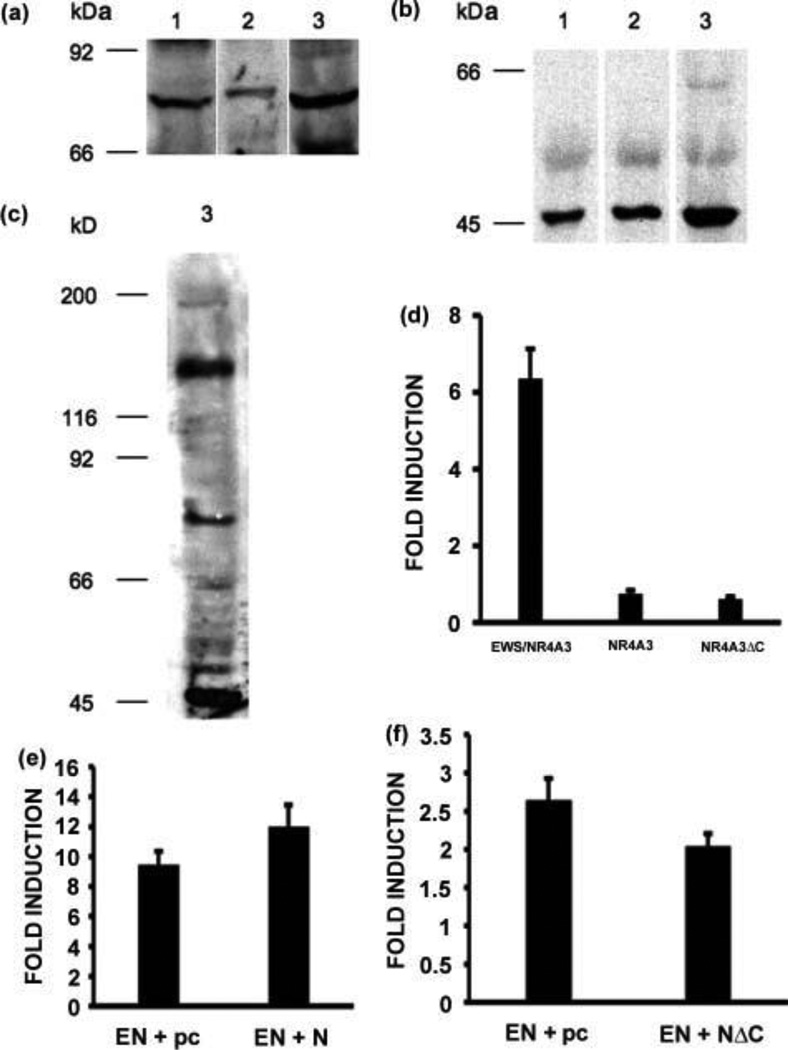
As mentioned previously, the NBRE is recognized by all three members of the NR4A subfamily, therefore it was of interest to analyze the expression of these receptors in the tumors. Western blot analyses were performed and we could not detect either NR4A1 or NR4A2 (data not shown). We could however detect a moderate expression level of NR4A3 (Fig. 4A), and a very high expression level of a 48 kD isoform of NR4A3 (Fig. 4B), called NR4A3ΔC, lacking the entire carboxy-terminal domain but retaining the amino-terminal and DNA-binding domains [11]. These results were confirmed using two different antibodies made in our laboratory and directed against the amino-terminal domain of NR4A3. To have an idea of the relative expression levels of the fusion protein versus the native and truncated receptors, the three proteins were detected on the same western blot strip from tumor sample 3 (Fig. 4C). The results show that for this tumor sample, and compared to the fusion protein, the native receptor appears to be expressed at lower levels and the truncated receptor at higher levels. Transient transfections of CFK2 cells were performed to compare the ability of these proteins to activate transcription from the PPARG promoter, and the results show that both the native and truncated receptors do not activate PPARG transcription under the same conditions in which it is readily activated by the fusion protein (Fig. 4D).
Figure 4.
Expression of NR4A3 and NR4A3∆C in EMC tumors. (A), (B) and (C) Western blot analyses of the same three EMC tumors presented in Fig. 1 showing expression of the full-length NR4A3 nuclear receptor (A) and of the truncated NR4A3ΔC isoform (B), and co-expression of EWSR1/NR4A3, NR4A3, and NR4A3ΔC in tumor sample 3 (C). (D), (E) and (F) Transient transfections of CFK2 cells with an EWSR1/NR4A3, NR4A3, or NR4A3∆C expression vector and the PPARG promoter luciferase reporter vector (D), with a five-fold excess of the EWSR1/NR4A3 expression vector compared to the NR4A3 expression vector (E), or with a five-fold excess of the NR4A3∆C expression vector compared to the EWSR1/NR4A3 expression vector (F). Transfections were carried out in triplicate, pBIND (Promega) was used for normalization, results are expressed as fold induction with respect to the empty expression vector, and the standard deviation is shown (vertical bars).
To investigate the possibility that the two native forms of NR4A3 may interfere with the activity of the EWSR1/NR4A3 fusion protein on the PPARG promoter, we performed transfections of CFK2 cells combining various expression vectors at ratios based on the western blot data shown in Figure 4C. When 100 nanograms of NR4A3 expression vector was co-transfected with 500 nanograms of EWSR1/NR4A3 expression vector, the activity of the fusion protein on the PPARG promoter showed a moderate but significant increase (Fig. 4E, P = 0.021). Conversely, when 500 nanograms of NR4A3ΔC expression vector was co-transfected with 100 nanograms of EWSR1/NR4A3 expression vector, the activity of the fusion protein on the PPARG promoter decreased (Fig. 4F, P = 0.049). This latter finding is consistent with results obtained by Ohkura and colleagues showing that an isoform of the NR4A2 receptor, lacking the carboxy-terminal domain but retaining the DNA-binding and amino-terminal domains, can act as a negative regulator of the NR4A receptors on NBRE sites [42]. Taken together, these results suggest that NR4A3∆C may negatively regulate the activity of EWSR1/NR4A3 on the PPARG promoter in EMC tumors.
Discussion
The EWS/NR4A3 fusion oncoprotein was identified in EMC in 1995 [24, 25] but little progress has been made in identifying its critical downstream targets. PPARG is the first direct transcriptional target of the EWS/NR4A3 fusion protein identified to date. PPARG is known as a major regulator of adipogenesis and glucose homeostasis [43]. Numerous studies have also implicated this receptor in a wide range of biological processes including inflammation, angiogenesis, bone morphogenesis, vascularization, atherosclerosis, proliferation, differentiation, and apoptosis (reviewed in [44]). PPARG is also expressed in many cancers including the colon, breast, lung, and prostate [45], where it is generally believed to act as a tumor suppressor through induction of differentiation or apoptosis, or inhibition of proliferation or angiogenesis [46]. However some studies are not entirely consistent with this view: murine models of transgenic mice over-expressing PPARG and mice lacking PPARG in various organs suggest that the receptor may be cancer-permissive [47]; an extensive survey of various cancer samples and cell lines has not identified any mutations in PPARG [48]; and families with germline mutations in PPARG, although presenting abnormalities of either fat metabolism or glucose homeostasis, do not show a higher incidence of cancer to date [49]. Our results indicating that the overall expression level of the PPARG promoter in EMC may be determined in part by the balance between EWS/NR4A3, NR4A3 and NR4A3∆C suggest that the amount of PPARG may be an important factor with regards to its tumoral function. The high expression level of NR4A3∆C in EMC may very well reflect the need to tightly control the transcription of this gene and the ensuing accumulation of the receptor.
We also examined here two other genes implicated in EMC. SGK1 was found to be induced by EWSR1/NR4A3 in a cellular model and immunohistochemistry has shown that this kinase is prominently expressed in EMC tumors [38]. Our western blot results confirm our previous immunohistochemistry data, and furthermore we show that SGK1 is phosphorylated in EMC. SGK1 has prosurvival properties [50–52], and therefore its over-expression and activation is likely to be an important feature of EMC tumorigenesis. We also analyzed the expression of NDRG2 because in our microarray analyses the gene was over-expressed in EMC and previous reports had indicated that NDRG2 was a phosphorylation target of SGK1 [39]. We have shown that NDRG2 is expressed in EMC, and since both SGK1 and NDRG2 are found in the cytoplasm of EMC tumor cells, there is a possibility that SGK1 may phosphorylate NDRG2. The impact of NDRG2 phosphorylation on its function is presently unknown, however NDRG2 is generally considered a tumor suppressor as its expression is down-regulated in several cancer cell lines [53]. The possibility that SGK1 plays a prosurvival role by inhibiting the tumor suppressor properties of NDRG2 through phosphorylation warrants further study.
Finally, we should note that because transcription factors are generally difficult to target pharmacologically, the identification of their critical target genes can be of therapeutic interest. For instance, PPARG agonists have been reported to have anti-neoplastic effects in a variety of PPARG-expressing cancers [54, 55]. Thus, the direct up-regulation of PPARG by EWSR1/NR4A3 demonstrated here provides a rationale for studies of these agents (several of which are already clinically available) in EMC, either in vitro if genuine EMC cell lines become available, or in vivo in the context of Phase I trials.
Supplementary Material
Acknowledgments
We thank Dr. Lluis Fajas (Université Montpelier 1, France) for the PPARG promoter reporter plasmid and Dr. Agnes Viale and the staff of the MSKCC Genomics Core Laboratory for microarray hybridizations. This work was supported by grants from The Cancer Research Society Inc. to YL and from the National Institutes of Health to M. Ladanyi (P01 CA47179; P01 CA106450; R01 CA95785).
Footnotes
There are no conflicts of interest for any of the authors
The raw microarray data files for this study are available at: http://cbio.mskcc.org/Public/sarcoma_array_data/
References
- 1.Ohkura N, Hijikuro M, Yamamoto A, Miki K. Molecular cloning of a novel thyroid/steroid receptor superfamily gene from cultured rat neuronal cells. Biochemical and biophysical research communications. 1994;205:1959–1965. doi: 10.1006/bbrc.1994.2900. [DOI] [PubMed] [Google Scholar]
- 2.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 3.Hazel TG, Misra R, Davis IJ, Greenberg ME, Lau LF. Nur77 is differentially modified in PC12 cells upon membrane depolarization and growth factor treatment. Molecular and cellular biology. 1991;11:3239–3246. doi: 10.1128/mcb.11.6.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scearce LM, Laz TM, Hazel TG, Lau LF, Taub R. RNR-1, a nuclear receptor in the NGFI-B/Nur77 family that is rapidly induced in regenerating liver. The Journal of biological chemistry. 1993;268:8855–8861. [PubMed] [Google Scholar]
- 5.Hedvat CV, Irving SG. The isolation and characterization of MINOR, a novel mitogen-inducible nuclear orphan receptor. Molecular endocrinology (Baltimore, Md. 1995;9:1692–1700. doi: 10.1210/mend.9.12.8614405. [DOI] [PubMed] [Google Scholar]
- 6.Philips A, Lesage S, Gingras R, Maira MH, Gauthier Y, Hugo P, et al. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Molecular and cellular biology. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Molecular and cellular biology. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes & development. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 9.Wilson TE, Fahrner TJ, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science (New York, NY. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 10.Paulsen RF, Granas K, Johnsen H, Rolseth V, Sterri S. Three related brain nuclear receptors, NGFI-B, Nurr1, and NOR-1, as transcriptional activators. J Mol Neurosci. 1995;6:249–255. doi: 10.1007/BF02736784. [DOI] [PubMed] [Google Scholar]
- 11.Labelle Y, Bussieres J, Courjal F, Goldring MB. The EWS/TEC fusion protein encoded by the t(9;22) chromosomal translocation in human chondrosarcomas is a highly potent transcriptional activator. Oncogene. 1999;18:3303–3308. doi: 10.1038/sj.onc.1202675. [DOI] [PubMed] [Google Scholar]
- 12.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. The EMBO journal. 1997;16:1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohkubo T, Ohkura N, Maruyama K, Sasaki K, Nagasaki K, Hanzawa H, et al. Early induction of the orphan nuclear receptor NOR-1 during cell death of the human breast cancer cell line MCF-7. Molecular and cellular endocrinology. 2000;162:151–156. doi: 10.1016/s0303-7207(00)00222-7. [DOI] [PubMed] [Google Scholar]
- 14.Castro DS, Hermanson E, Joseph B, Wallen A, Aarnisalo P, Heller A, et al. Induction of cell cycle arrest and morphological differentiation by Nurr1 and retinoids in dopamine MN9D cells. The Journal of biological chemistry. 2001;276:43277–43284. doi: 10.1074/jbc.M107013200. [DOI] [PubMed] [Google Scholar]
- 15.Pirih FQ, Aghaloo TL, Bezouglaia O, Nervina JM, Tetradis S. Parathyroid hormone induces the NR4A family of nuclear orphan receptors in vivo. Biochemical and biophysical research communications. 2005;332:494–503. doi: 10.1016/j.bbrc.2005.04.132. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez PM, Brunel F, Jimenez MA, Saez JM, Cereghini S, Zakin MM. Nuclear receptors Nor1 and NGFI-B/Nur77 play similar, albeit distinct, roles in the hypothalamo-pituitary-adrenal axis. Endocrinology. 2000;141:2392–2400. doi: 10.1210/endo.141.7.7562. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Gonzalez J, Rius J, Castello A, Cases-Langhoff C, Badimon L. Neuron-derived orphan receptor-1 (NOR-1) modulates vascular smooth muscle cell proliferation. Circulation research. 2003;92:96–103. doi: 10.1161/01.es.0000050921.53008.47. [DOI] [PubMed] [Google Scholar]
- 18.Arkenbout EK, de Waard V, van Bragt M, van Achterberg TA, Grimbergen JM, Pichon B, et al. Protective function of transcription factor TR3 orphan receptor in atherogenesis: decreased lesion formation in carotid artery ligation model in TR3 transgenic mice. Circulation. 2002;106:1530–1535. doi: 10.1161/01.cir.0000028811.03056.bf. [DOI] [PubMed] [Google Scholar]
- 19.Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nature medicine. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y, Luo L, Luo N, Zhu X, Garvey WT. NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: Potential role in insulin resistance. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M701132200. [DOI] [PubMed] [Google Scholar]
- 21.Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer research. 2004;64:8208–8212. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- 22.Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nature medicine. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 23.Brody RI, Ueda T, Hamelin A, Jhanwar SC, Bridge JA, Healey JH, et al. Molecular analysis of the fusion of EWS to an orphan nuclear receptor gene in extraskeletal myxoid chondrosarcoma. The American journal of pathology. 1997;150:1049–1058. [PMC free article] [PubMed] [Google Scholar]
- 24.Clark J, Benjamin H, Gill S, Sidhar S, Goodwin G, Crew J, et al. Fusion of the EWS gene to CHN, a member of the steroid/thyroid receptor gene superfamily, in a human myxoid chondrosarcoma. Oncogene. 1996;12:229–235. [PubMed] [Google Scholar]
- 25.Labelle Y, Zucman J, Stenman G, Kindblom LG, Knight J, Turc-Carel C, et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Human molecular genetics. 1995;4:2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 26.Sjogren H, Meis-Kindblom J, Kindblom LG, Aman P, Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer research. 1999;59:5064–5067. [PubMed] [Google Scholar]
- 27.Panagopoulos I, Mencinger M, Dietrich CU, Bjerkehagen B, Saeter G, Mertens F, et al. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11) Oncogene. 1999;18:7594–7598. doi: 10.1038/sj.onc.1203155. [DOI] [PubMed] [Google Scholar]
- 28.Attwooll C, Tariq M, Harris M, Coyne JD, Telford N, Varley JM. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. doi: 10.1038/sj.onc.1203156. [DOI] [PubMed] [Google Scholar]
- 29.Sjogren H, Wedell B, Meis-Kindblom JM, Kindblom LG, Stenman G. Fusion of the NH2-terminal domain of the basic helix-loop-helix protein TCF12 to TEC in extraskeletal myxoid chondrosarcoma with translocation t(9;15)(q22;q21) Cancer research. 2000;60:6832–6835. [PubMed] [Google Scholar]
- 30.Hisaoka M, Ishida T, Imamura T, Hashimoto H. TFG is a novel fusion partner of NOR1 in extraskeletal myxoid chondrosarcoma. Genes, chromosomes & cancer. 2004;40:325–328. doi: 10.1002/gcc.20044. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian S, West RB, Marinelli RJ, Nielsen TO, Rubin BP, Goldblum JR, et al. The gene expression profile of extraskeletal myxoid chondrosarcoma. The Journal of pathology. 2005;206:433–444. doi: 10.1002/path.1792. [DOI] [PubMed] [Google Scholar]
- 32.Sjogren H, Meis-Kindblom JM, Orndal C, Bergh P, Ptaszynski K, Aman P, et al. Studies on the molecular pathogenesis of extraskeletal myxoid chondrosarcoma-cytogenetic, molecular genetic, and cDNA microarray analyses. The American journal of pathology. 2003;162:781–792. doi: 10.1016/S0002-9440(10)63875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 34.Laflamme C, Filion C, Bridge JA, Ladanyi M, Goldring MB, Labelle Y. The homeotic protein Six3 is a coactivator of the nuclear receptor NOR-1 and a corepressor of the fusion protein EWS/NOR-1 in human extraskeletal myxoid chondrosarcomas. Cancer research. 2003;63:449–454. [PubMed] [Google Scholar]
- 35.Lusis EA, Watson MA, Chicoine MR, Lyman M, Roerig P, Reifenberger G, et al. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer research. 2005;65:7121–7126. doi: 10.1158/0008-5472.CAN-05-0043. [DOI] [PubMed] [Google Scholar]
- 36.Simon P, Schneck M, Hochstetter T, Koutsouki E, Mittelbronn M, Merseburger A, et al. Differential regulation of serum- and glucocorticoid-inducible kinase 1 (SGK1) splice variants based on alternative initiation of transcription. Cell Physiol Biochem. 2007;20:715–728. doi: 10.1159/000110432. [DOI] [PubMed] [Google Scholar]
- 37.Filion C, Labelle Y. The oncogenic fusion protein EWS/NOR-1 induces transformation of CFK2 chondrogenic cells. Experimental cell research. 2004;297:585–592. doi: 10.1016/j.yexcr.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 38.Poulin H, Filion C, Ladanyi M, Labelle Y. Serum- and glucocorticoid-regulated kinase 1 (SGK1) induction by the EWS/NOR1(NR4A3) fusion protein. Biochemical and biophysical research communications. 2006;346:306–313. doi: 10.1016/j.bbrc.2006.05.134. [DOI] [PubMed] [Google Scholar]
- 39.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. The Biochemical journal. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogusz AM, Brickley DR, Pew T, Conzen SD. A novel N-terminal hydrophobic motif mediates constitutive degradation of serum- and glucocorticoid-induced kinase-1 by the ubiquitin-proteasome pathway. The FEBS journal. 2006;273:2913–2928. doi: 10.1111/j.1742-4658.2006.05304.x. [DOI] [PubMed] [Google Scholar]
- 41.Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. The Journal of biological chemistry. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 42.Ohkura N, Hosono T, Maruyama K, Tsukada T, Yamaguchi K. An isoform of Nurr1 functions as a negative inhibitor of the NGFI-B family signaling. Biochimica et biophysica acta. 1999;1444:69–79. doi: 10.1016/s0167-4781(98)00247-4. [DOI] [PubMed] [Google Scholar]
- 43.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 44.Heikkinen S, Auwerx J, Argmann CA. PPARgamma in human and mouse physiology. Biochimica et biophysica acta. 2007;1771:999–1013. doi: 10.1016/j.bbalip.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han S, Roman J. Peroxisome proliferator-activated receptor gamma: a novel target for cancer therapeutics? Anti-cancer drugs. 2007;18:237–244. doi: 10.1097/CAD.0b013e328011e67d. [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Xu J, Yu X, Yang R, Han ZC. Peroxisome proliferator-activated receptor gamma in malignant diseases. Critical reviews in oncology/hematology. 2006;58:1–14. doi: 10.1016/j.critrevonc.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Koeffler HP. Peroxisome proliferator-activated receptor gamma and cancers. Clin Cancer Res. 2003;9:1–9. [PubMed] [Google Scholar]
- 48.Ikezoe T, Miller CW, Kawano S, Heaney A, Williamson EA, Hisatake J, et al. Mutational analysis of the peroxisome proliferator-activated receptor gamma gene in human malignancies. Cancer research. 2001;61:5307–5310. [PubMed] [Google Scholar]
- 49.Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 50.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Molecular and cellular biology. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikosz CA, Brickley DR, Sharkey MS, Moran TW, Conzen SD. Glucocorticoid receptor-mediated protection from apoptosis is associated with induction of the serine/threonine survival kinase gene, sgk-1. The Journal of biological chemistry. 2001;276:16649–16654. doi: 10.1074/jbc.M010842200. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Cui R, Cheng X, Du J. Antiapoptotic effect of serum and glucocorticoid-inducible protein kinase is mediated by novel mechanism activating I{kappa}B kinase. Cancer research. 2005;65:457–464. [PubMed] [Google Scholar]
- 53.Liu N, Wang L, Liu X, Yang Q, Zhang J, Zhang W, et al. Promoter methylation, mutation, and genomic deletion are involved in the decreased NDRG2 expression levels in several cancer cell lines. Biochemical and biophysical research communications. 2007;358:164–169. doi: 10.1016/j.bbrc.2007.04.089. [DOI] [PubMed] [Google Scholar]
- 54.Cellai I, Benvenuti S, Luciani P, Galli A, Ceni E, Simi L, et al. Antineoplastic effects of rosiglitazone and PPARgamma transactivation in neuroblastoma cells. British journal of cancer. 2006;95:879–888. doi: 10.1038/sj.bjc.6603344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grommes C, Landreth GE, Sastre M, Beck M, Feinstein DL, Jacobs AH, et al. Inhibition of in vivo glioma growth and invasion by peroxisome proliferator-activated receptor gamma agonist treatment. Molecular pharmacology. 2006;70:1524–1533. doi: 10.1124/mol.106.022194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.