Abstract
miR-31 is well known as an anti-metastatic microRNA (miRNA) in the context of breast cancer. However, to date the mechanism of regulation of this miRNA has yet to be elucidated. The recent publication by Viré et al. in Molecular Cell [1] demonstrates for the first time that one mechanism of regulation of miR-31 is through the putative oncogene EMSY, whose amplification in breast cancer patients correlates with reduced expression of the miRNA. This regulation is dependent on the DNA-binding transcription factor ETS-1 which recruits EMSY and the histone demethylase KDM5B to the miR-31 promoter, thus repressing its transcription.
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-014-0467-x) contains supplementary material, which is available to authorized users.
Background
miRNAs are small non-coding RNA species which primarily negatively regulate gene expression, thus affecting a plethora of cancer-associated phenotypes. miR-31, a well-known anti-metastatic miRNA in breast cancer, was first shown to inhibit breast cancer metastasis both in vitro and in vivo[1] through the targeting of integrin-α5 (ITGA5), radixin (RDX) and RhoA [2] and later through targeting of WAVE3 [3]. Further investigation reported that suppression of ITGA5, RDX and RhoA recapitulated the phenotype produced by ectopic expression of miR-31 in vitro and in vivo[4]. Moreover, re-introduction of miR-31 expression in established xenograft MDA-MB-231 lung metastases was shown to result in metastatic regression, indicating that re-expression of miR-31 may be of therapeutic benefit in clinically advanced patients [5].
EMSY, a putative oncogene involved in gene regulation, is known to be amplified in approximately 13% of sporadic breast cancers [6]. However, the locus in which this gene is contained (11q13-14) includes a number of other genes implicated in breast cancer including CCND1, PAK1, CTTN, and FGF3, making it difficult to decipher the exact contribution of EMSY to the poor prognosis associated with amplification of this region. A recent article in Molecular Cell has gone some way to elucidating the function of this gene in breast cancer, describing a role for EMSY in oncogenic transformation as well as invasion/migration in breast cancer through the regulation of miR-31 [7].
Article
The publication from the Kouzarides group reports that the oncogene EMSY targets miR-31 through co-operation with the transcription factor ETS-1 and the histone lysine demethylase KDM5B, resulting in transcriptional repression of the miRNA and numerous phenotypic effects (Figure 1).
Figure 1.
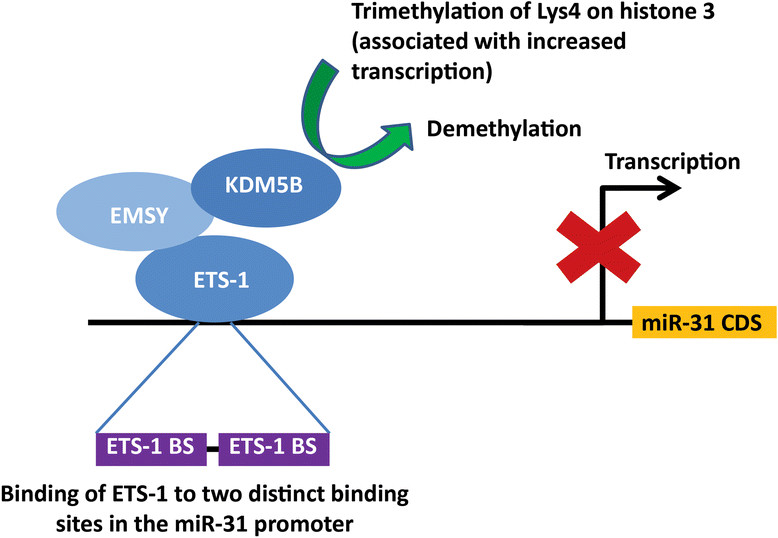
Proposed mechanism of regulation of miR-31 in breast cancer (adapted from [[7]]). ETS-1 recruits oncogene EMSY and H3K4me3 demethylase KDM5B to the promoter of miR-31, repressing transcription of the pri-miRNA.
Firstly, the group investigated the effect of modified expression of EMSY in cell lines and xenograft models. Ectopic expression of the oncogene in MCF7 cells resulted in increased growth in an orthotopic mammary fat pad xenograft model while an experimental metastasis tail vein injection model of the same cells demonstated increased lung micrometastases relative to the control line. Given the potential role for this gene in transcriptional regulation, a qRT-PCR-based screen was employed to profile 88 miRNAs in MCF7 cells in which EMSY had been depleted revealing 38 to be altered in this context. A second approach was then utilised whereby re-analysis of the METABRIC cohort [8],[9] for miRNAs altered in EMSY-amplified (cis-acting aberrations of the 11q13-14 locus) versus non-amplified cases revealed dysregulation of 12 miRNAs. As miR-31 was the only miRNA to be identified using both approaches, it was chosen for further studies.
miR-31 expression levels inversely correlated with EMSY levels in patient samples from the METABRIC dataset and in cell lines with amplification or ectopic expression of EMSY. Finding that expression of EMSY increased both oncogenic transformation and migration in vitro, the Kouzarides group then demonstrated that re-expression of miR-31 had the ability to abrogate both phenotypic effects of EMSY oncogenicity, while modification of the expression of the miRNA alone altered invasion and migration and phenocopied the effects of EMSY depletion.
The authors then investigated the mechanism of miR-31 repression by EMSY demonstrating recruitment of EMSY to the miR-31 promoter, thus repressing transcription of the primary miRNA (pri-miRNA). However, as EMSY contains no DNA-binding domain another factor was necessary for promoter recruitment. Bioinformatic analysis of the miR-31 promoter revealed binding sites for ETS-1, ETV4/PEA3 and GATA1 with site-directed mutagenesis of any of the ETS-1 binding sites in the promoter reducing promoter activation. ETS-1 was subsequently confirmed to bind to the miR-31 promoter. Furthermore, this transcription factor was then shown to recruit the histone H3K4me3 demethylase KDM5B to the miR-31 promotor, thus inhibiting transcription of miR-31 through the demethylation of Lys4 on histone 3.
Viewpoint
This article is the first to mechanistically anchor an oncogenic role for EMSY and demonstrate a direct mechanism of regulation of miR-31, an important metastasis-associated miRNA, in breast cancer. Numerous mechanisms of miRNA regulation exist, including epigenetic silencing via DNA methylation or histone modification [10], direct regulation by nuclear receptors [11] or indirect regulation through alteration of the miRNA biogenesis pathway [12]. miR-31 has been seen to be hypermethylated in some cell lines [13] and a recent study demonstrated that the miR-31 promoter also undergoes aberrant methylation in breast cancer patients [14]. However until now, no direct regulatory mechanisms have been described.
The evidence presented from both in vitro and in vivo studies provides a convincing argument for the classification as EMSY as an oncogene, as well as the direct regulation of miR-31 by this oncogene. However, the story is undoubtedly much more complex, as EMSY may regulate a number of different miRNAs/genes. Nor is this the only potential mechanism of direct regulation of miR-31. Notably, as EMSY amplification is only seen in approximately 13% of sporadic breast cancers, it is imperative to also study the regulation of this miRNA in an EMSY non-amplified context. Moreover, given that there were only 45 samples with EMSY amplification and corresponding miRNA data in the cohort studied, expanding the analysis into larger cohorts would provide additional evidence as to the relevance of this discovery in breast cancer patients.
Viré and colleagues have elegantly documented a novel role for EMSY in breast cancer. However, they have also presented the case for a second scenario, that in which the oncogenic properties of this gene may be functionally propagated by potential oncomiRs. Indeed, EMSY depletion in cell lines led to the upregulation of 29 miRNAs and 6 miRNAs were found to be overexpressed in breast cancer patients with amplification of the gene. The mechanism of targeting of these miRNAs by EMSY, either directly or indirectly, has yet to be investigated.
To date, the focus of the majority of miRNA publications has been to evaluate gene targets observed to affect a functional response. This study highlights a need to fully elucidate the mechanisms of regulation of the miRNAs themselves, not just their targets. This may lead to the discovery of a myriad of feedback loops similar to that between the miR-200 family of miRNAs and the transcriptional repressors ZEB1/SIP1 which functions to regulate epithelial-mesenchymal transition (EMT) [15]. As miRNA inhibitors/mimics have only recently begun to enter clinical trials, ascertaining the mechanism of regulation of key miRNAs may greatly improve the chances of finding a surrogate gene candidate which could be successfully targeted in the clinic.
Acknowledgements
Funding is acknowledged from the Irish Research Council for Science, Engineering and Technology (IRCSET), Science Foundation Ireland through the Molecular Therapeutics for Cancer, Ireland Strategic Research Cluster (award 08/SRC/B1410; http://www.mtci.ie) and the Irish Cancer Society Collaborative Cancer Research Centre BREAST-PREDICT grant, CCRC13GAL (http://www.breastpredict.com).
Abbreviations
- EMT
epithelial-mesenchymal transition
- miRNA
microRNA
- pri-miRNA
primary microRNA
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-014-0467-x) contains supplementary material, which is available to authorized users.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Laoighse Mulrane, Email: laoighse.mulrane@ucd.ie.
William M Gallagher, Email: william.gallagher@ucd.ie.
Darran P O’Connor, Email: darran.oconnor@ucd.ie.
References
- 1.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–2597. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129:1331–1343. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147–5154. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev. 2011;25:646–659. doi: 10.1101/gad.2004211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/S0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 7.Vire E, Curtis C, Davalos V, Git A, Robson S, Villanueva A, Vidal A, Barbieri I, Aparicio S, Esteller M, Caldas C, Kouzarides T. The Breast Cancer Oncogene EMSY Represses Transcription of Antimetastatic microRNA miR-31. Mol Cell. 2014;53:806–818. doi: 10.1016/j.molcel.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, METABRIC Group, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, Green A, Ellis I, Aparicio S, Caldas C. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 10.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1:31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulrane L, McGee SF, Gallagher WM, O'Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 13.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrba L, Munoz-Rodriguez JL, Stampfer MR, Futscher BW. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8:e54398. doi: 10.1371/journal.pone.0054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]


