Abstract
Background
Fasciola hepatica causes chronic liver disease, fasciolosis, leading to significant losses in the livestock economy and concerns for human health in many countries. The identification of F. hepatica genes involved in the parasite’s virulence through modulation of host immune system is utmost important to comprehend evasion mechanisms of the parasite and develop more effective strategies against fasciolosis. In this study, to identify the parasite’s putative virulence genes which are associated with host immunomodulation, we explored whole transcriptome of an adult F. hepatica using current transcriptome profiling approaches integrated with detailed in silico analyses. In brief, the comparison of the parasite transcripts with the specialised public databases containing sequence data of non-parasitic organisms (Dugesiidae species and Caenorhabditis elegans) or of numerous pathogens and investigation of the sequences in terms of nucleotide evolution (directional selection) and cytokine signaling relation were conducted.
Results
NGS of the whole transcriptome resulted in 19,534,766 sequence reads, yielding a total of 40,260 transcripts (N50 = 522 bp). A number of the parasite transcripts (n = 1,671) were predicted to be virulence-related on the basis of the exclusive homology with the pathogen-associated data, positive selection or relationship with cytokine signaling. Of these, a group of the virulence-related genes (n = 62), not previously described, were found likely to be associated with immunomodulation based on in silico functional categorisation, showing significant sequence similarities with various immune receptors (i.e. MHC I class, TGF-β receptor, toll/interleukin-1 receptor, T-cell receptor, TNF receptor, and IL-18 receptor accessory protein), cytokines (i.e. TGF-β, interleukin-4/interleukin-13 and TNF-α), cluster of differentiations (e.g. CD48 and CD147) or molecules associated with other immunomodulatory mechanisms (such as regulation of macrophage activation). Some of the genes (n = 5) appeared to be under positive selection (Ka/Ks > 1), imitating proteins associated with cytokine signaling (through sequence homologies with thrombospondin type 1, toll/interleukin-1 receptor, TGF-β receptor and CD147).
Conclusions
With a comparative transcriptome profiling approach, we have identified a number of potential immunomodulator genes of F. hepatica (n = 62), which are firstly described here, could be employed for the development of better strategies (including RNAi) in the battle against both zoonotically and economically important disease, fasciolosis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1539-8) contains supplementary material, which is available to authorized users.
Keywords: Fasciola hepatica, Whole transcriptome, Trematode, RNA-Seq, Virulence, Immunomodulation, Evasion
Background
Fasciola hepatica, the liver fluke, is a digenetic trematode helminth, causing highly damaging hepatobiliary disease (fasciolosis) in mammalians including economically important ruminants (such as cattle and sheep) and humans [1]. Fasciolosis results in a significant economic loss in livestock industry worldwide and more cases have been reported in humans in different countries [1-4]. The disease is currently treated with anti-helminthics (such as triclabendazole), but the observed anti-helminthic drug resistance [5] necessitates more effective strategies for the treatment and/or the prevention of fasciolosis.
For the elucidation of infection mechanisms of the liver fluke and the development of better strategies in dealing with fasciolosis, the most critical step is the identification and characterisation of the genes which are important for the establishment of parasitism. The transcriptome of F. hepatica was reported in a previous publication where a previous sequencing platform (454 NGS, Roche) was employed for the purpose of describing general biological characteristics of the parasite [6]. However, the sequence data from that study is likely to be still further from encapturing the entirety of the transcriptome profile of F. hepatica and the virulence factors of the parasite, particularly those related to host immunomodulation, are worthwhile for additional investigation with current NGS platforms (such as HiSeq 2000, Illumina). The current NGS technologies produce greater transcriptomic data in comparison with the previous sequencing systems and increase the chance of detection of parasite transcripts which are expressed at lower levels (relative to house-keeping transcripts) but with significant importance in immune evasion.
One of the most promising approaches for determining the virulence factors of parasitic organisms is in silico comparison of parasites’ transcripts with publicly available data [7-10]. A recent data resource, the helminth secretome database (HSD) [11,12], contains a broad repertoire of excretory/secretory (ES) protein sequences (n > 18,000) of various parasitic helminths including trematodes, cestodes and nematodes. ES proteins of endoparasites are, in general, thought to play vital roles in the establishment of infection [11-14]. Additionally, Vaccine Investigation and Online Information Network (Violin; http://www.violinet.org), provides gene and protein sequences that are affiliated with infection mechanisms of various micro (virus, bacteria, protozoon) and macro (helminth) infectious organisms [15]. Compared to other resources providing whole genome data of numerous pathogens, both HSD and Violin contain filtered data better suiting to glean pathogen-related molecules of infectious organisms. However, the major caveat of the specialised databases is the presence of insufficiently refined data (such as house keeping genes/proteins in particularly HSD database) that are related with regular physiological events in both parasitic and non-parasitic organisms, but not indeed linked to virulence.
Parasitism genes which are not directly involved in virulence, but rather associated with regular physiological mechanisms, could be uncovered by sequence homology with taxonomically similar, free-living (non-parasitic) organisms [9]. Very recently, a large nucleotide sequence collection from free-living/non-infectious trematodes (taxonomically close to F. hepatica) of Dugesiidae family (including Dugesia sp. and Schmidtea sp.) has become publicly available at DNA Data Bank of Japan (DDBJ; www.ddbj.nig.ac.jp). Additionally, a comprehensive data for a well studied free-living model nematode, Caenorhabditis elegans, is freely accessible from a regularly updated resource, WormBase (http://www.wormbase.org).
The data of non-parasitic organisms from current resources have been useful tool to investigate the genes that are under directional selection through the comparative analysis of nucleotide diversity by assessing nonsynonymous/synonymous (Ka/Ks) substitution rates [16-18]. It is a well known fact that parasitism related genes important for the evasion of defensive systems of host and the corresponding genes in the host are under constant selective pressure favoring nucleotide subsitutions [18]. For example, the strong influence of directional selection in the evolution of avirulence genes to gain immunomodulatory properties has been clearly reported in multiple studies [19-21]. Furthermore, lineage-specific sub- or neo-functionalisation of genes which are vital for the establishment and maintenance of parasitism could be identified through comparative genomics [21-23].
Apart from the analysis of sequence homology with infectious and non-infectious organisms, the virulence genes associated with modulation of host immune responses (such as cytokine signaling) can be identified by the manual inspection of encoded protein motifs in public databases [24,25].
To date, in vivo and in vitro studies have shown that the liver fluke modulates host immune responses for enhancing its virulence [17,18,26]; however, which genes of the parasite imitate the components of host immune system have not yet been elucidated in detail.
The main purpose of this study was to glean virulence and immunomodulatory F. hepatica genes through comparative transcriptome profiling with the transcriptomes of non-parasitic related organisms by focusing on the genes which are evolved in lineage-specific manner, under positive selection and show similar motifs of host immune system genes involved in cytokine signaling.
Results
Transcriptome profile of F. hepatica
Workflow illustrating the experimental steps of the study is demonstrated in Figure 1. From a total of 19,534,766 sequence reads, generated by the sequencing instrument (HiSeq 2000, Illumina) with paired-end 2X 100 bp reading, 81,090 contigs (contig N50 = 377) were de novo assembled, of which, 40,260 transcripts were annotated with blast searches (blastx and blastn/tblastx) as described in Additional file 1. The obtained base number in this study was approximately 12.5 times higher than that reported in the previous transcriptome study of F. hepatica [6]. The transcript N50 was 522 bp and the length of a total of 7,861 transcripts was equal or greater than the observed N50 length in this work. In the present study, F. hepatica G + C content was 48.01%, which was similar to that reported in the previous related studies of F. hepatica (44,5%, 47.0 ± 14.1%) [6,27]. The identified transcript sequences in our study corresponded to a total of 28,142 unique accession numbers [n = 24,243 (NCBI related), n = 3,899 (GeneDB, CHGC, and SchistoDB related)].
Figure 1.
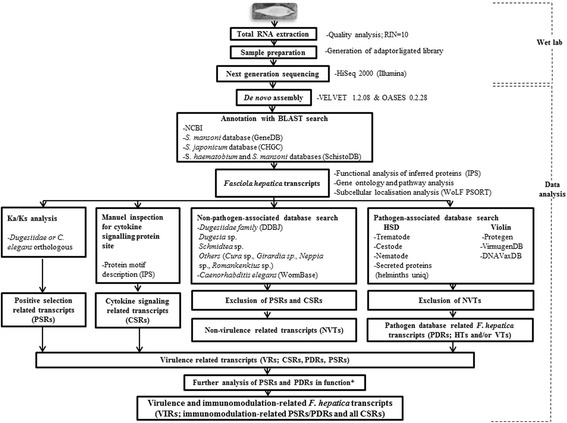
Experimental design. The transcriptome study is consisted of wet lab procedures and data analysis processes. The parasite transcript sequences were analysed in terms of positive selection, cytokine signaling and pathogen database homology. The parasite sequences showing the signs of positive selection (Ka/Ks > 1) were termed PSRs (positive selection related transcripts). The cytokine signaling relation for the inferred parasite proteins were detected by the protein motif information and the related transcripts were termed CSRs (cytokine signaling related transcripts). Non-virulence-related transcripts (NVTs) were determined by the sequence homology of the parasite transcripts with the data of the non-pathogen associated databases (DDBJ, WormBase; E value < 10−5) by excluding the non-parasite organism homologous PSRs and CSRs. The parasite transcripts showing the exclusive sequence homology with the pathogen-associated databases (HSD and Violin) (E value < 10−5), but not being NVTs, were termed PRDs (pathogen database related transcripts). The parasite transcripts including PSRs, CSRs and PDRs were considered virulence-related transcripts (VRs). Immunomodulation categorisable VRs (PSRs/PDRs and all CSRs) were termed virulence and immunomodulation-related transcripts (VIRs). Asterisk (*) indicates that data from other resources (NCBI, UniProt, GeneDB, Gene Ontology and referred publications) were used for functional categorisation of some PSRs and PDRs.
The identity of species was genetically confirmed by the presence of a F. hepatica transcript (showing significant similarity with previously known species-specific heat shock protein 70 of F. hepatica [28], #ABS52704.1; E value = 8.00−21), in addition to the morphological identification of the isolated parasite. The amino acid sequence for this protein at the correct frame of the transcript sequence was valine (V-599) as in F. hepatica, but not leucine (L-599) as in F. gigantica [28], confirming the species specificity of the isolated parasite in this study. In terms of drug resistance, the isolated parasite in the present study was found to be susceptible to albendazole (an anthelmintic benzimidazole drug). This was extrapolated by the comparative analysis of the translated aminoacid sequences of a F. hepatica transcript (annotated with tubulin beta-2 of F. hepatica; #CAP72050.1; E value = 0) with the drug susceptibility associated amino acid residues (N-165; F-167; E-198; F-200; R-241) of tubulin beta-2 of F. hepatica [29].
Nonsynonymous/synonymous substitution rate of F. hepatica transcripts
A total of 16,832 orthologous pairs (E value < 10−3) could be subjected to nonsynonymous/synonymous substitution rate analysis (13,288 F. hepatica transcripts showing homology with the sequences of Dugesiidae species and the remaining parasite transcripts showing homology with the sequences of C. elegans). Ka/Ks ratio was calculable for a total of 12,394 transcript pairs (Additional file 2). Ka/Ks analysis revealed that majority of the analysed F. hepatica transcripts (89.63%) have Ka/Ks < 0.5, hinting a purifiying selection against nonsynonymous changes as expected, a minority of the transcripts (3.37%) have Ka/Ks > 1, and a tiny portion of the transcripts (0.24%) have Ka/Ks = 1 (Figure 2). F. hepatica transcripts with Ka/Ks > 1 were hereafter called transcripts under positive selection (termed PSRs within PSR subgroup). Only the small percentage of the orthologous transcripts were under positive selection, which confirms the hypothesis that some of the genes are diversing from the former versions to possess virulence capabilities as previously suggested [18].
Figure 2.
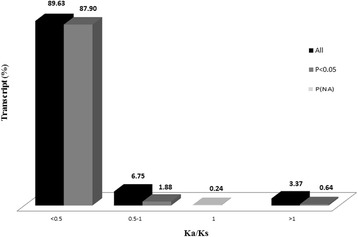
Nonsynonymous/synonymous substitution rate of the liver fluke transcripts. Ka/Ks ratio distribution (%) of a total of 12,394 Dugesiidae or C. elegans orthologous F. hepatica transcripts is shown. The majority of the transcripts [89.63% (87.90% at P < 0.05)] were found to be associated with negative selection (Ka/Ks < 1). A group of transcripts [6.75% (1.88% at P < 0.05)] could be under directional selection (0.5 < Ka/Ks < 1). Another group of transcripts (0.24%) were related to neutral selection. Only a small portion of the transcripts (3.37%, n = 418) showed the signs of positive selection (Ka/Ks > 1) and, of this, a minority (0.64%, n = 79) reached statistical significance (P < 0.05).
The level of homology of F. hepatica transcripts in cytokine signaling
The detailed analysis of InterProScan descriptions for all functionally categorised transcripts revealed that some of the protein motifs (family, domain or functional site), inferred from a group of transcript sequences (n = 35), suggesting possible involvement of them in cytokine signaling (named CSRs under CSR subgroup).
Sequence homology of F. hepatica transcripts with the specialised secondary databases and virulence-related transcripts of the parasite
Approximately half of the total liver fluke transcript number (i.e. 51.87%) showed sequence homology with nucleotide or protein sequences of non-parasitic organisms including Dugesiidae species and C. elegans (E value < 10−5) (Table 1a). The level of sequential homology of F. hepatica transcripts with Dugesia sp. and Schmidtea sp. was slightly higher (~1.2%) than that with the transcripts of C. elegans (37.77 %), but this increased to 46.36% when considering the parasite transcripts sequentially homologous to any of Dugesiidae species (E value < 10−5). The parasite transcripts which were similar to the sequences of Dugesiidae species or C. elegans (E value < 10−5) but not showed the signs of positive selection (Ka/Ks > 1) and/or cytokine signaling relation, a total of 20,483 liver fluke sequences (50.88%), were named non-virulence-related transcripts (NVTs). Based on the transcript number, approximately a quarter of F. hepatica transcriptome (26.56%) showed homology with the sequences from helminth secretome database (HSD; Table 1a), but only a minor part of these transcripts (3.1%; n = 1,251) were out of the category of NVTs that these transcripts were named HTs (E value < 10−5; Table 1b). A smaller percentage of F. hepatica transcriptome showed homology with the data of Vaccine Investigation and Online Information Network (Violin), but only a minority of these transcripts (0.29%; n = 117) were not observed in the category of NVTs and these transcripts were termed VTs (E value < 10−5; Table 1b). A number of non-NVTs (n = 23) were found sequentially homologous to both HSD and Violin databases (HTs/VTs; E value < 10−5). All HTs, VTs and HTs/VTs were called PDR(s) [pathogen database related transcript(s)] under PDR subgroup.
Table 1.
Sequence homology of the F. hepatica transcripts with the secondary databases and virulence-related transcripts of the parasite
| a) F. hepatica transcripts predicted to be similar to: | Number | Percentage (%) | Data type | Analysis/Database |
| Dugesia sp. | 15,784 | 39.21 | Non-parasite-associated | Blast/DDBJ |
| Schmidtea sp. | 15,546 | 38.61 | Non-parasite-associated | Blast/DDBJ |
| other trematodes (Cura sp., Girardia sp., Neppia sp., Romankenkius sp.) | 524 | 1.30 | Non-parasite-associated | Blast/DDBJ |
| Dugesiidae family (in total, excluding common transcripts) | 18,663 | 46.36 | Non-parasite-associated | Blast/DDBJ |
| C. elegans | 15,208 | 37.77 | Non-parasite-associated | Blast/WormBase |
| C. elegans specific (not associated with Dugesiidae family) | 2,218 | 5.51 | Non-parasite-associated | Blast/WormBase |
| Free-living worms (in total) | 20,881 | 51.87 | Non-parasite-associated | Blast/WormBase |
| Positive selection related transcripts (Dugesiidae or C. elegans orthologous) | 377 | 1.04 | Virulence-associated | Ka/Ks//DDBJ/WormBase |
| Cytokine signaling related transcripts (Dugesiidae or C. elegans orthologous) | 21 | 0.05 | Virulence-associated | InterProScan/EBI |
| NVTs (excluding the positive selection and cytokine signaling related transcripts) | 20,483 | 50.88 | Non-parasite-associated | - |
| HSD data | 10,694 | 26.56 | Pathogen-associated | Blast/HSD |
| Violin data | 1,731 | 4.30 | Pathogen-associated | Blast/Violin |
| b) Virulence-related F. hepatica transcripts (VRs) | Number | Percentage (%) | ||
| HTs (HSD related transcripts, not identified as NVTs) | 1,251 | 3.11 | ||
| VTs (Violin related transcripts, not identified as NVTs) | 117 | 0.29 | ||
| HTs/VTs (transcripts homologous to both HSD and Violin data, not identified as NVTs) | 23 | 0.06 | ||
| PDRs (pathogen database related transcripts, including HTs and VTs or HTs/VTs) | 1,391 | 3.46 | ||
| PSRs (positive selection related transcripts; PSR subgroup) | 246 | 0.61 | ||
| CSRs (cytokine signaling related transcripts; CSR subgroup) | 31 | 0.08 | ||
| PSRs and PDRs (common for PSR and PDR subgroups) | 168 | 0.42 | ||
| PSRs and CSRs (common for PSR and CSR subgroups) | 3 | 0.007 | ||
| PDR, PSR and CSR (common for all subgroups) | 1 | 0.002 | ||
| Virulence-related transcripts (VRs) identified as PDRs, PSRs or CSRs | 1,671 | 4.15 |
Numerical values for the F. hepatica transcripts which show similarity with nucleotide/protein sequences of the pathogen-associated and non-pathogen-associated databases (a) and the numbers of the virulence-related F. hepatica transcripts (VRs) (b) are listed. A total of 18,663 liver fluke transcripts (46.36%) and an additional 2,218 transcripts (5.51%) showed homology with sequences of Dugesiidae species and C. elegans, respectively, yielding a total of 20,881 transcripts (51.87%) (E value < 10−5) (a). Overall, 20,483 liver fluke transcripts (50.88%) (orthologous to the non-parasitic organisms, E value < 10−5) were determined to be non-virulence-related (NVTs) after excluding the transcripts showing the signs of positive selection (Ka/Ks > 1) (n = 377) and the relationship with cytokine signaling (n = 21). Approximately a quarter of the total liver fluke transcripts (n = 10,694) showed sequence homology with the HSD data and 4.30% of the total liver fluke transcripts showed sequential similarity with the Violin data. A total of 1,251 transcripts (3.11%) and another set of 117 transcripts (0.29%) were exclusively homologous to the HSD (HTs) and Violin data (VTs), respectively but not identified in the category of NVTs (b). A small number of VRs (n = 23) were common for both HTs and VTs (HTs/VTs). In total, 1,391 VRs (HTs, VTs and HTs/VTs) were included in PDR subgroup (containing pathogen database related transcripts). A total of 246 VRs showing the signs of positive selection (Ka/Ks > 1) were observed in PSR group alone. Some of VRs (n = 31) were only identified by CSR subgroup. A number of PSRs (n = 168) were common for PDR subgroup. Three of VRs were detected by both PSR and CSR subgroups. Only one of VRs was determined by all the subgroups (PDR, PSR and CSR). Overall, a total of 1,671 transcripts, identified at least by one of the subgroups, were predicted to be virulence-related. The percentage values indicate the ratio of transcript number to total transcript number (n = 40,260). NVTs: Non-virulence-related transcripts.
Overall, 4.15 % of all identified F. hepatica transcripts was assumed to be virulence-related transcripts (ascribed as VRs under VR group) on the basis of the degree of homology with sequences from publicly available databases; i) pathogen-associated data (PDR subgroup; n = 1,391; E value < 10−5), ii) observation of the signs of positive selection (PSR subgroup; n = 418; Ka/Ks > 1) and iii) cytokine signaling relation (CSR subgroup; n = 35; manual inspection), yielding a total of 1,671 transcripts (excluding transcripts commonly determined by the different analyses) (Additional file 3). A number of the transcripts with Ka/Ks > 1 (n = 169) were also observed in PDR subgroup (n = 141 for HTs, n = 5 for VTs, and n = 23 for HTs/VTs). Of the cytokine signaling related transcripts (CSRs), one transcript showed the signs of positive selection and sequence homology with the HSD database (#20661), three transcripts (#6733, #23314 and #64440) showed the signs of positive selection without sequential similarity of any pathogen related databases, and the others showed sequence homology with the non-parasitic databases (n = 21). A number of CSRs (n = 10) showed non-similarity with any specialised databases used in this study.
Protein function profile for whole transcriptome and VR group
The InterProScan search for whole transcriptome of the parasite revealed a total of 5,089 unique accession number indicating protein families, domains or functional sites. Protein sequences inferred from a total of 20,160 transcripts (50.07% of all the annotated transcripts) were categorised in various functional groups by considering the biological functions of the parasite in previous publications [24,25,27]. The majority of the protein sequences (93.23%) were functionally categorisable with InterProScan information and the rest (particularly virulence-related transcripts) were classifiable on the basis of the information from the other resources such as Gene Ontology, UniProt, NCBI or referred publications (Additional file 4). The analysis indicated that the abundant transcripts were mostly involved in nucleic acid binding/transcription (20.91%) and signaling (16.66%), and only 2.08% of all the categorised transcripts (i.e. 419 transcripts) was associated with immunomodulation (Figure 3a). Biological functions of the virulence-related transcripts under VR group, and PDR and PSR subgroups were found to be mostly related to nucleic acid binding/transcription, signaling and unknown mechanisms, respectively (Figure 3b). Interestingly, the relative abundance of the transcripts with unknown mechanisms in PSR subgroup (26.79 %) was higher than those in PDR subgroup (19.12%), suggesting possible novel functions enhancing the virulence of the parasite (Figure 3b). The relative quantity of immunomodulation related transcripts within PDR subgroup (2.66 %) was higher more than twice, compared to those within PSR subgroup (1.20 %) (Figure 3b), likely hinting the importance of high diversity at lineage/genus level for gaining immunomodulation capability. The protein function of all the transcripts under CSR group was inherently related to immunomodulation.
Figure 3.
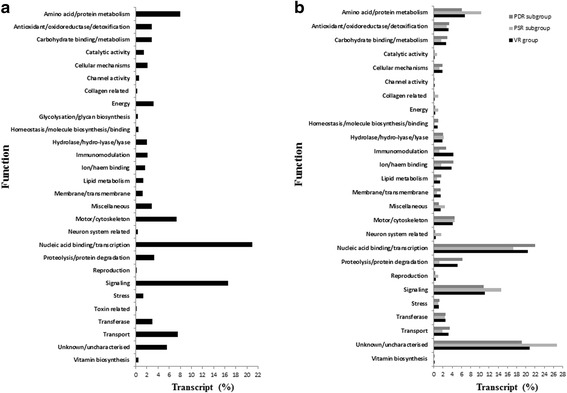
Relative abundances of the liver fluke transcripts in various biological functions based on manual inspection of protein motifs. The relative abundances (%) of all the functionally categorised transcripts within whole transcriptome (a), VR group and its PSR and PDR subgroups (b) in various functions are demonstrated. Nucleic acid binding/transcription and signaling mechanisms were highly pronounced functions of the whole transcriptome, with relative transcript abundances of 20.91% and 16.66%, respectively (Figure 3a). This pattern was similar within VR group except the relative transcript abundance of unknown mechanisms was higher within this group (20.47%), PSR subgroup (26.79%) and PDR subgroup (19.12 %). The transcript proportions of amino acid/protein metabolism, catalytic activity, energy, neuron system and signaling within PSR subgroup were higher (around 1.5-5 times) than those within PDR subgroup. The relative transcript ratios for immunomodulation, ion/haem binding and proteolysis/protein degradation functions were higher (about 2–5 times) in PDR subgroup in comparison with PSR subgroup.
Subcellular localisation profile for whole transcriptome and VR group
Subcellular localisation signals of the parasite protein sequences were mostly related to cytosol (21.68%), extracellular space (20.57%) and nucleus (19.15%) (Figure 4a). Similarly, most of the detected subcellular localisation signals of VRs were associated with the cytosol, extracellular and nucleus parts, in order (Figure 4b). The extracellular localisation signal was more oftenly observed within VR group (20.42%) and PDR subgroup (20.80%) and CSR subgroup (21.80%), in comparison to PSR subgroup (17.81%). Further details about the subcellular localisation for the transcripts are shown individually in Additional file 5.
Figure 4.
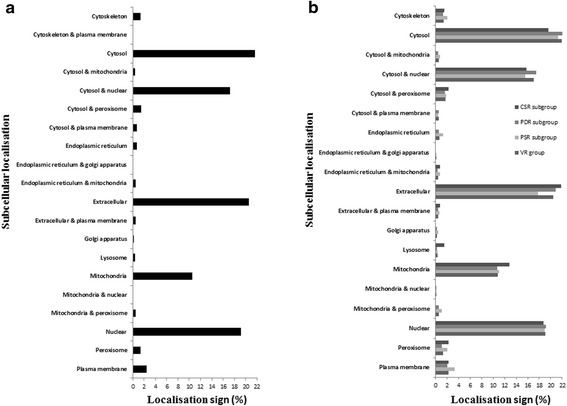
Subcellular localisation signals of the liver fluke transcripts. The relative amount (%) of the detected subcellular localisation signals within whole transcriptome (a), VR group and its subgroups including CSR subgroup, PDR subgroup and PSR subgroup (b) are demonstrated. The relative abundances for the cytosol, extracellular part and nucleus signals were found high levels within the total transcriptome, 21.68%, 20.57%, 19.15%, respectively. This profile was similarly found within VR. The relative percentages of the extracellular signal in CSR subgroup, PDR subgroup and VR group are higher (~2%) than those in PSR subgroup. The abundances of the signals of mitochondria&peroxisome, endoplasmic reticulum and plasma membrane were proportionally slightly higher (about 0.5-1.5%) in PSR subgroup than those within VR group and other subgroups (CSR and PDR subgroups).
Virulence and immunomodulation-related transcripts and genes of F. hepatica
Of VRs (n = 1,671), the immunomodulation categorised PDRs, PSRs, and all CSRs, a total of seventy-one transcripts, corresponding to 64 putative genes, were named virulence and immunomodulation-related transcripts (VIRs) under VIR set (Table 2). Further details about the sequence characteristics including available InterProScan accesion number for VIRs are shown in Additional file 6. The majority of VIRs were specifically detectable through the level of homology with the pathogen related databases (PDR subgroup) (49.3%) and cytokine signaling relation (CSR subgroup) (43.7%) and the minor part of VIRs, observable in PDR subgroup (1.4%), CSR subgroup (4.7%) or both (1.4%), showed the signs of positive selection (PSR subgroup) (Figure 5).
Table 2.
Virulence and immunomodulation-related transcripts of F.hepatica
| Transcript no | Accession no | Description | Subgroup | HPI | Immunological property | Resource |
|---|---|---|---|---|---|---|
| 1584 | XP_002572744.1 | Sarm1 [Schistosoma mansoni] | CSR | * | Toll/interleukin-1 receptor homology | IPS |
| 3866 | CAX70351.1 | LIN1-like protein [Schistosoma japonicum] | PDR (HT) | Proline binding | NCBI | |
| 5195 | BC055836.1 | Neutrophil cytosolic factor 1, mRNA (cDNA clone MGC:67831 IMAGE:3983481), complete cds [Mus musculus] | PDR (VT) | * | Leukocyte mediated cytotoxicity | UniProt |
| 6556 | GAA27790.2 | Hypothetical protein CLF_107202 [Clonorchis sinensis] | CSR | * | Toll/interleukin-1 receptor homology | IPS |
| 6733 | GAA34190.1 | Hypothetical protein CLF_100679 [Clonorchis sinensis] | PSR/CSR | Toll/interleukin-1 receptor homology | IPS | |
| 7694 | XP_002573383.1 | Hypothetical protein [Schistosoma mansoni] | PDR (HT) | * | Complement associated | NCBI/GeneDB |
| 7920 | GAA36580.2 | T-cell immunomodulatory protein [Clonorchis sinensis] | PDR (HT) | * | Modulator of T-cell function | UniProt |
| 8653 | NP_001137161.1 | Histocompatibility 2, Q region locus 4 precursor [Mus musculus] | PDR (HT) | * | MHC class I homology | IPS |
| 10264 | CCD82398.1 | Putative tgf-beta family [Schistosoma mansoni] | CSR | * | TGF-β homology | IPS |
| 10702 | NP_112313.1 | Macrophage migration inhibitory factor [Rattus norvegicus] | CSR | * | Macrophage migration inhibition | IPS |
| 11419 | GAA47989.1 | Suppressor of cytokine signaling 7 [Clonorchis sinensis] | CSR | Suppressor of cytokine signalling | IPS | |
| 11610/45296//64440 | GAA27846.2 | TGF-beta receptor type-1 [Clonorchis sinensis] | CSR/CSR//PSR/CSR | * | TGF-β receptor homology | IPS/IPS//IPS |
| 12300 | AAW24666.1 | SJCHGC04616 protein [Schistosoma japonicum] | CSR | * | Interleukin-4/interleukin-13 homology | IPS |
| 13771 | 1401243A | Major histocompatibility complex HLA I [Mus musculus] | PDR (VT) | * | MHC class I homology | IPS |
| 16002 | CCD77150.1 | Hypothetical protein Smp_194540 [Schistosoma mansoni] | CSR | * | Toll/interleukin-1 receptor homology | IPS |
| 19626/20908 | GAA37431.2 | TGF-beta receptor type-1 [Clonorchis sinensis] | CSR/CSR | * | TGF-β receptor homology | IPS/IPS |
| 19810/53747/68185 | GAA28730.2 | Thrombospondin-2 [Clonorchis sinensis] | PDR (HT)/PDR (HT)/PDR(HT) | * | TGF-β stimulation | NCBI/NCBI/NCBI |
| 20661 | GAA53897.1 | Activin receptor type-2B [Clonorchis sinensis] | PDR (HT)/PDR (VT)/PSR/CSR | * | TGF-β receptor homology | IPS |
| 22528 | AAA39677.1 | MHC K-bm6 transplantation antigen, partial [Mus musculus] | PDR (VT) | * | MHC class I homology | IPS |
| 23314 | GAA55000.1 | Hypothetical protein CLF_106334 [Clonorchis sinensis] | PSR/CSR | ** | CD147 homology | NCBI/IPS |
| 26180 | XP_005669379.1 | PREDICTED: TGF-beta receptor type-2 [Sus scrofa] | CSR | * | TGF-β receptor homology | IPS |
| 26955 | GAA56301.1 | Bone morphogenetic protein receptor type-1 invertebrate [Clonorchis sinensis] | CSR | * | TGF-β receptor homology | IPS |
| 27939 | NP_001153015.1 | Dipeptidyl peptidase 4 isoform 2 [Mus musculus] | PDR (VT) | * | T-cell receptor homology | NCBI |
| 31945 | CAA36183.1 | Unnamed protein product [Mus musculus] | CSR | * | IL-10 stimulation | IPS |
| 32835 | NP_033865.2 | beta-2-microglobulin precursor [Mus musculus] | PDR (HT) | * | MHC class I homology | IPS |
| 32989 | AIE76460.1 | CD59-like protein [Fasciola hepatica] | PDR (HT) | * | Complement associated | IPS |
| 34645 | CCD61018.1 | Hypothetical protein Smp_194470 [Schistosoma mansoni] | PDR (HT) | * | TGF-β stimulation | NCBI |
| 34729 | BAB30997.1 | Unnamed protein product [Mus musculus] | CSR | T-cell receptor homology | IPS | |
| 37409 | GAA49741.1 | Tumor necrosis factor receptor superfamily member 16 [Clonorchis sinensis] | PDR (HT) | * | TNF receptor homology | NCBI |
| 37746 | GAA51051.1 | Activin receptor type-2B [Clonorchis sinensis] | CSR | * | TGF-β receptor homology | IPS |
| 38312 | XP_003945784.1 | PREDICTED: h-2 class I histocompatibility antigen, K-D alpha chain-like isoform 6 [Mus musculus] | PDR (VT) | * | MHC class I homology | IPS |
| 38341 | XP_002570313.1 | Hypothetical protein [Schistosoma mansoni] | CSR | * | Toll/interleukin-1 receptor homology | IPS |
| 38525 | CCD58880.1 | Cleavage and polyadenylation specificity factor,putative [Schistosoma mansoni] | CSR | * | Suppressor of IKBKE 1 | IPS |
| 38573 | AAA39573.1 | MHC H2-D-q alpha-chain, partial [Mus musculus] | PDR (HT) | * | MHC class I homology | NCBI |
| 40314 | GAA48275.1 | Lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog [Clonorchis sinensis] | CSR | * | TNF-α homology | IPS |
| 40900 | XP_002577301.1 | Hypothetical protein [Schistosoma mansoni] | PDR (HT)/PSR | * | TGF-β stimulation | NCBI/GeneDB |
| 42935/76935 | GAA51525.1 | CD2 antigen cytoplasmic tail-binding protein 2 [Clonorchis sinensis] | PDR (HT)/PDR (HT) | * | Proline binding | NCBI/NCBI |
| 44058 | XP_002578816.1 | Bone morphogenetic protein antagonist noggin [Schistosoma mansoni] | CSR | * | TGF-β antagonist | IPS |
| 44260 | 3Q5T_A | Chain A, V BetaV BETA HOMODIMERIZATION-Based Pre-Tcr Model Suggested By Tcr Beta Crystal Structures [Mus musculus] | PDR (HT) | T-cell receptor homology | NCBI | |
| 47252 | XP_002575081.1 | Suppressor of cytokine signaling [Schistosoma mansoni] | CSR | * | Suppressor of cytokine signalling | IPS |
| 49819 | GAA49058.1 | Activin receptor type-1, partial [Clonorchis sinensis] | CSR | * | TGF-β receptor homology | IPS |
| 53490/55117 | XP_003946490.1 | PREDICTED: h-2 class I histocompatibility antigen, D-D alpha chain-like isoform 7 [Mus musculus] | PDR (VT)/PDR (HT) | * | MHC class I homology | IPS/IPS |
| 54791 | AAA39576.1 | MHC H2-K-d transplantation antigen H2-Kd, partial [Mus musculus] | PDR (VT) | * | MHC class I homology | NCBI |
| 55207 | XP_002570016.1 | Protein kinase [Schistosoma mansoni] | CSR | * | TGF-β receptor homology | IPS |
| 56418 | XP_002570128.1 | Bone morphogenetic protein antagonist noggin [Schistosoma mansoni] | CSR | * | TGF-β antagonist | IPS |
| 56437 | NP_001077023.1 | Transforming growth factor-beta receptor-associated protein 1 homolog [Danio rerio] | CSR | TGF-β receptor homology | IPS | |
| 58384 | 1404428A | Cytotoxic T lymphocyte [Murid herpesvirus 1] | PDR (HT) | * | MHC class I homology | IPS |
| 58628 | GAA54758.1 | TNF receptor-associated factor 4 [Clonorchis sinensis] | CSR | TNF receptor homology | IPS | |
| 58983 | NM_001033288.3 | Somatomedin B and thrombospondin, type 1 domain containing (Sbspon), mRNA [Mus musculus] | PDR (VT) | * | TGF-β stimulation | UniProt |
| 59522 | XP_003946048.1 | PREDICTED: h-2 class I histocompatibility antigen, K-D alpha chain-like isoform 4 [Mus musculus] | PDR (VT) | * | MHC class I homology | IPS |
| 59632 | BAE26952.1 | Unnamed protein product [Mus musculus] | CSR | * | CD48 homology | NCBI/IPS |
| 59650 | CAE82020.1 | Unnamed protein product [Mus musculus] | PDR (VT) | * | MHC class I homology | NCBI |
| 60918 | GAA43145.2 | Protein DVR-1 [Clonorchis sinensis] | CSR | * | TGF-β homology | IPS |
| 61208 | AAA39567.1 | H-2D cell surface glycoprotein, partial [Mus musculus] | PDR (HT) | * | MHC class I homology | IPS |
| 64619 | BAE20821.1 | Unnamed protein product [Mus musculus] | PDR (HT) | * | T-cell receptor homology | IPS |
| 65009 | 1503111B | H2Dd gene [Mus musculus] | PDR (HT) | * | MHC class I homology | IPS |
| 70639 | CCD82741.1 | T-cell immunomodulatory protein [Schistosoma mansoni] | PDR (HT) | * | Modulator of T-cell function | UniProt |
| 72283 | CAX74995.1 | LIN1-like protein [Schistosoma japonicum] | PDR (HT) | * | Proline binding | NCBI |
| 73151 | CAA24128.1 | H2-Ld [Mus musculus] | PDR (VT) | * | MHC class I homology | IPS |
| 73578 | EDL36365.1 | mCG8696 [Mus musculus] | PDR (HT) | * | T-cell receptor homology | IPS |
| 73673 | EDL76957.1 | Transforming growth factor, beta receptor II, isoform CRA_b [Rattus norvegicus] | CSR | * | TGF-β receptor homology | IPS |
| 76453 | BAE34022.1 | Unnamed protein product [Mus musculus] | CSR | * | TGF-β receptor homology | IPS |
| 77913 | NP_034683.1 | Interleukin-18 receptor accessory protein precursor [Mus musculus] | CSR | * | IL-18 receptor accessory protein homology | IPS |
| 79120 | Smp_060190.1 | ctg4a [Schistosoma mansoni] | PDR (HT) | * | Toll like receptor 4 (TLR4) regulation | NCBI/GeneDB |
Virulence and immunomodulation-related transcripts of F. hepatica (VIRs) with accession and description of the corresponding sequences, identified by the transcript subgroups, are listed. The predicted immunomodulatory property function and the possibility for the host-parasite interaction (HPI) for VIRs are shown. Data resources for the categorisation and/or prediction of function are indicated. Most proteins inferred from VIRs (n = 63) were predicted to be possibly located in the extracellular space except a VIR protein which is possibly located at the cell plasma membrane. Accesion with description was obtained from the blast searches in the NCBI protein database or GeneDB (for only #79120). Asterisk (*) indicates potential association with extracellular localisation. Double asterisk (**) indicates potential association with plasma membrane location. No asterisk in the HPI column indicates the signs of cytoplasmic locations for the related transcripts except #3866, which showed the signs of being located in mitochondria and/or nucleus. IPS: InterProScan.
Figure 5.
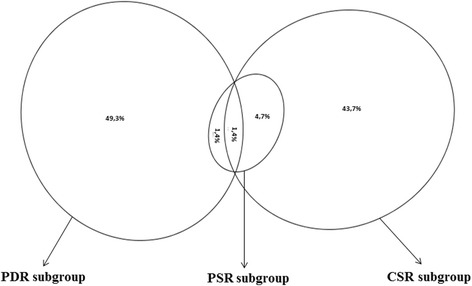
Relative abundances of the virulence and immunomodulation related transcripts determined by the transcript subgroups. The Venn diagram shows percentage distribution of the virulence and immunomodulation related transcripts (VIRs, n = 71), determined by the exclusive homology with the pathogen databases (PDR subgroup), observation of the signs of positive selection (PSR subgroup) and/or cytokine signaling relation (CSR subgroup) among the immunomodulation categorised transcripts. This diagram indicates that VIRs showing the signs of positive selection (Ka/Ks > 1; in PSR subgroup) are evolved towards cytokine signaling. The Venn diagram was drawn using eulerAPE v3 (http://www.eulerdiagrams.org/eulerAPE/) [118].
A number of VIRs (n = 15) showed sequential similarities with MHC I and an important part of VIRs indicated the relationship with TGF-β signaling based on sequence homologies with TGF-β, TGF-β receptor or other proteins associated with stimulation or inhibition of TGF-β (Figure 6). The other sequence homologies were associated with various immunomodulatory molecules including T-cell receptor, toll/interleukin-1 receptor, TNF receptor, cluster of differentiations (i.e. CD48, CD59, CD 147), IL-18 receptor accessory protein, interleukin-4/interleukin-13, TNF-α, modulators of T-cell function, suppressors of cytokine signaling and of IKBKE 1, or molecules involved in other immunomodulation-related mechanisms (including IL-10 stimulation, leukocyte mediated cytotoxicity, proline binding or macrophage migration inhibition, toll like receptor 4 regulation; Figure 6). The majority of VIRs (n = 64) individually were potentially located at extracellular space (n = 63) or localised within plasma membrane (n = 1) (Table 2), possibly indicating direct interactions of the parasite proteins with host immune system.
Figure 6.
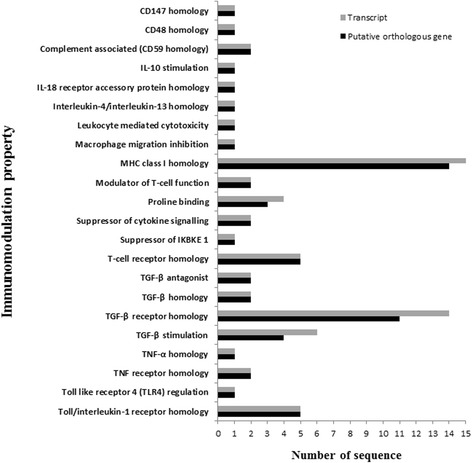
Distribution of putative virulence and immunomodulation-related orthologous genes by the immunomodulation property. The number of the virulence and immunomodulation-related transcripts and the corresponding putative orthologous genes based on the predicted immunomodulation property are demonstrated. The number of the putative orthologous genes showing MHC I homology (n = 14) was the highest, which was followed by the others showing sequence homologies with TGF-β receptor (n = 11), T-cell or Toll/interleukin-1 receptors (n = 5 for both receptors), and the relation with TGF-β stimulation (n = 4).
Gene ontology categories related to whole transcriptome, VR group/subgroups and VIR set
Gene ontology categories (i.e. biological process, molecular function and cellular component) for whole transcriptome of the liver fluke, in comparison with VR group, its subgroups (CSR, PDR, PSR) and VIR set are shown in Table 3. The relative transcript abundances of the cellular (GO:0009987; 31.27%) and metabolic processes (GO:0008152; 26.68 %) were found to be higher, in comparison to other biological processes in the total transcriptome, and this pattern was similar for VR group and its PDR and PSR subgroups. However, the proportions of the transcripts in response to stimulus (GO:0050896; 21.82%) and immune system process (GO:0002376; 9.09%) were the highest within VIR set than those within VR group and its subgroups. Furthermore, VIR set has higher sequence proportions in molecular transducer activity (GO:0060089; 20%) and receptor activity (GO:0004872; 6.15%), compared to VR group and its subgroups except CSR subgroup. The relative abundance in the GO cellular compartmant category within VIR set was skewed to membrane (GO:0016020; 68.29%).
Table 3.
Gene ontology for the liver fluke transcripts in different classifications using blast2GO
| Gene ontology category | Description | Transcript classification | |||||
|---|---|---|---|---|---|---|---|
| CSR subgroup | PDR subgroup | PSR subgroup | VR group | VIR set | WT | ||
| Biological process (GO:0008150) | Biological adhesion (GO:0022610) | - | 5 (1) | - | 5 (0.65) | - | 109 (0.55) |
| Biological regulation (GO:0065007) | 23 (18.25) | 37 (7.39) | - | 67 (8.71) | 27 (16.36) | 1272 (6.43) | |
| Cell killing (GO:0001906) | - | 4 (0.8) | 20 (6.43) | 4 (0.52) | 4 (2.42) | 4 (0.02) | |
| Cellular component organization or biogenesis (GO:0071840) | - | 20 (3.99) | 14 (4.5) | 28 (3.64) | - | 608 (3.07) | |
| Cellular process (GO:0009987) | 24 (19.05) | 141 (28.14) | 101 (32.48) | 215 (27.96) | 24 (14.55) | 6191 (31.27) | |
| Developmental process (GO:0032502) | 2 (1.59) | 3 (0.6) | 1 (0.32) | 5 (0.65) | 2 (1.21) | 73 (0.37) | |
| Immune system process (GO:0002376) | 2 (1.59) | 13 (2.59) | - | 15 (1.95) | 15 (9.09) | 30 (0.15) | |
| Localization (GO:0051179) | - | 22 (4.39) | 18 (5.79) | 32 (4.16) | - | 1289 (6.51) | |
| Locomotion (GO:0040011) | - | - | - | - | - | 31 (0.16) | |
| Metabolic process (GO:0008152) | 8 (6.35) | 131 (26.15) | 81 (26.05) | 176 (22.89) | 9 (5.45) | 5282 (26.68) | |
| Multicellular organismal process (GO:0032501) | - | 1 (0.2) | 3 (0.96) | 3 (0.39) | - | 43 (0.22) | |
| Multi-organism process (GO:0051704) | - | 5 (1) | - | 4 (0.52) | - | 7 (0.04) | |
| Reproduction (GO:0000003) | - | 3 (0.6) | 1 (0.32) | 2 (0.26) | - | 4 (0.02) | |
| Response to stimulus (GO:0050896) | 23 (18.25) | 31 (6.19) | 13 (4.18) | 57 (7.41) | 36 (21.82) | 1011 (5.11) | |
| Signaling (GO:0023052) | 21 (16.67) | 14 (2.79) | 11 (3.54) | 38 (4.94) | 21 (12.73) | 751 (3.79) | |
| Single-organism process (GO:0044699) | 23 (18.25) | 71 (14.17) | 48 (15.43) | 118 (15.34) | 27 (16.36) | 3091 (15.61) | |
| Molecular function (GO:0003674) | Antioxidant activity (GO:0016209) | - | 3 (0.68) | 3 (1.17) | 3 (0.5) | - | 30 (0.2) |
| Binding (GO:0005488) | 27 (48.21) | 264 (59.86) | 129 (50.39) | 341 (56.93) | 35 (53.85) | 7815 (51.94) | |
| Catalytic activity (GO:0003824) | 12 (21.43) | 127 (28.8) | 89 (34.77) | 176 (29.38) | 13 (20) | 5296 (35.2) | |
| Electron carrier activity (GO:0009055) | - | - | 1 (0.39) | 1 (0.17) | - | 39 (0.26) | |
| Enzyme regulator activity (GO:0030234) | 12 (2.72) | 3 (1.17) | 13 (2.17) | - | 300 (1.99) | ||
| Metallochaperone activity (GO:0016530) | - | - | - | - | - | 4 (0.03) | |
| Molecular transducer activity (GO:0060089) | 13 (23.21) | 2 (0.45) | 3 (1.17) | 15 (2.5) | 13 (20) | 143 (0.95) | |
| Nucleic acid binding transcription factor activity (GO:0001071) | 7 (1.59) | 4 (1.56) | 7 (1.17) | - | 120 (0.8) | ||
| Protein binding transcription factor activity (GO:0000988) | - | - | 1 (0.39) | 1 (0.17) | - | 44 (0.29) | |
| Receptor activity (GO:0004872) | 4 (7.14) | 1 (0.23) | 1 (0.39) | 5 (0.83) | 4 (6.15) | 92 (0.61) | |
| Structural molecule activity (GO:0005198) | - | 21 (4.76) | 12 (4.69) | 25 (4.17) | - | 521 (3.46) | |
| Transporter activity (GO:0005215) | - | 4 (0.91) | 10 (3.91) | 12 (2) | - | 643 (4.27) | |
| Cellular component (GO:0005575) | Cell (GO:0005623) | - | 92 (35.52) | 58 (36.02) | 119 (34.2) | 6 (14.63) | 3056 (34.02) |
| Cell junction (GO:0030054) | - | - | - | - | - | 30 (0.33) | |
| Extracellular matrix (GO:0031012) | 1 (0.39) | 1 (0.62) | 1 (0.29) | - | 28 (0.31) | ||
| Extracellular region (GO:0005576) | 1 (6.25) | 5 (1.93) | 2 (1.24) | 6 (1.72) | 1 (2.44) | 48 (0.53) | |
| Macromolecular complex (GO:0032991) | - | 50 (19.31) | 32 (19.88) | 64 (18.39) | 6 (14.63) | 1866 (20.77) | |
| Membrane (GO:0016020) | 15 (93.75) | 45 (17.37) | 21 (13.04) | 70 (20.11) | 28 (68.29) | 1693 (18.85) | |
| Membrane-enclosed lumen (GO:0031974) | - | 1 (0.39) | 3 (1.86) | 3 (0.86) | - | 142 (1.58) | |
| Organelle (GO:0043226) | - | 60 (23.17) | 44 (27.33) | 80 (22.99) | - | 2099 (23.37) | |
| Synapse (GO:0045202) | - | 1 (0.39) | - | 1 (0.29) | - | 14 (0.16) | |
| Virion (GO:0019012) | - | 4 (1.54) | - | 4 (1.15) | - | 6 (0.07) | |
Gene ontology categories (parental 2) for the whole transcriptome (WT) and VR group with its subgroups (CSR, PDR, PSR) and VIR set are shown. The relative transcript abundances of response to stimulus (GO:0050896; 21.82%) and immune system process (GO:0002376; 9.09%) were predominant in VIR set while the proportional transcript abundances for the cellular process (GO:0009987) and metabolic process (GO:0008152) were remarkably higher within WT, VR group, PDR and PSR subgroups (around 19-32%). The relative transcript abundances of molecular transducer (GO:0060089) and receptor activities (GO:0004872) appeared to be much higher within VIR set (20%, 6.15%, respectively) and CSR subgroup (23.21%, 7.14%, respectively), compared to the other subgroups, VR group and WT. The proportional transcript abundance for the membrane (GO:0016020) was predominant within VIR set (68.29%) and CSR subgroup (93.75%), but the cellular part (GO:0005623) (around 34.02-36.02%) was with the highest transcript ratio within VR group and its PDR and PSR subgroups and WT. Numerical values and brackets indicate the total number of transcript sequences and the relative transcript abundance (%) within each transcript classification, respectively.
Biological pathways related to whole transcriptome, VR group/subgroups and VIR set
A total of 96 different KEGG biological pathways (195 enzyme types) were determined in whole transcriptome of the parasite, in which the purine (map00230; 16.62%) and pyrimidine metabolisms (map00240; 7.93%) were the biological pathways with higher abundant transcript numbers, where the relative abundances of the other pathways, in transcript number, were less than 5% (Additional file 7). The pathway with the highest transcript number was the purine (map00230; around 12-16%) metabolism within PSR and PDR subgroups; however, the relative transcript abundance for the pyrimidine metabolism (map00240) within PDR subgroup was approximately twice than that within PSR subgroup (Table 4). Aminobenzoate degradation (map00627), beta-Alanine metabolism (map00410), glycine, serine and threonine metabolism (map00260) were uniquely identified in PDR subgroup while butanoate metabolism (map00650) and pentose phosphate pathway (map00030) were found specific to PSR subgroup among the transcript subgroups (Table 4).
Table 4.
Comparisons of the biological pathways of the liver fluke transcripts in different classifications
| Biological pathway | Transcript classification | |||||
|---|---|---|---|---|---|---|
| CSR subgroup | PDR subgroup | PSR subgroup | VR group | VIR set | WT | |
| Amino sugar and nucleotide sugar metabolism (map00520) | - | 2 (3.7) | 1 (2.63) | 2 (2.78) | - | 49 (2.22) |
| Aminoacyl-tRNA biosynthesis (map00970) | - | 3 (5.56) | 1 (2.63) | 3 (4.17) | - | 86 (3.89) |
| Aminobenzoate degradation (map00627) | - | 3 (5.56) | - | 3 (4.17) | - | 18 (0.82) |
| Arachidonic acid metabolism (map00590) | - | 2 (3.7) | 2 (5.26) | 2 (2.78) | - | 8 (0.36) |
| beta-Alanine metabolism (map00410) | - | 1 (1.85) | - | 1 (1.39) | - | 3 (0.14) |
| Butanoate metabolism (map00650) | - | - | 1 (2.63) | 1 (1.39) | - | 13 (0.59) |
| Glutathione metabolism (map00480) | - | 2 (3.7) | 3 (7.89) | 3 (4.17) | - | 50 (2.26) |
| Glycine, serine and threonine metabolism (map00260) | - | 2 (3.7) | - | 2 (2.78) | - | 24 (1.09) |
| Glycosaminoglycan degradation (map00531) | - | 1 (1.85) | 1 (2.63) | 1 (1.39) | - | 4 (0.18) |
| Inositol phosphate metabolism (map00562) | - | 1 (1.85) | 2 (5.26) | 2 (2.78) | - | 23 (1.04) |
| Nicotinate and nicotinamide metabolism (map00760) | - | 2 (3.7) | 1 (2.63) | 3 (4.17) | - | 48 (2.17) |
| Oxidative phosphorylation (map00190) | - | 1 (1.85) | 2 (5.26) | 3 (4.17) | - | 78 (3.53) |
| Pentose phosphate pathway (map00030) | - | - | 2 (5.26) | 2 (2.78) | - | 72 (3.26) |
| Purine metabolism (map00230) | - | 7 (12.96) | 6 (15.79) | 9 (12.5) | - | 367 (16.62) |
| Pyrimidine metabolism (map00240) | - | 5 (9.26) | 2 (5.26) | 7 (9.72) | - | 175 (7.93) |
Biological pathways, identified at least for one of the transcript classifications, are shown. Most transcripts within VR group and its PSR and PDR subgroups were related to purine metabolism (map00230; around 12-17%), similar to the profile of WT. The relative transcript abundance for the pyrimidine metabolism (map00240) within PSR subgroup was found approximately half in PDR subgroup. Aminobenzoate degradation (map00627), beta-Alanine metabolism (map00410), glycine, serine and threonine metabolism (map00260) were uniquely identified in PDR subgroup among the other subgroups. Butanoate metabolism (map00650) and pentose phosphate pathway (map00030) were only identified in PSR subgroup in comparison with the other subgroups. No any biological pathway was determined in CSR subgroup and VIR set.
Discussion
In this study, we applied detailed in silico analyses to determine the virulence and immunomodulation-related genes of the liver fluke through a comparative assessment of the transcriptome profile with current NGS technology (HiSeq 2000, Illumina). The observed GO categories of F. hepatica transcripts in terms of biological processes were mostly concordant with the previous classification [6]. However, some GO categories, including signaling (GO:0023052; biological process), receptor activity (GO:0004872; molecular function) and membrane (GO:0016020; cellular component) were firstly described in the present study. This indicates overall more comprehensive coverage of total transcriptome profile of this parasite. Beside GO analysis approach with blast2GO, the manual inspection approach, including analysis of all the detected protein motifs, significantly increased the number of the categorised transcript into various functional terms that were previously used in the related studies [24,25,27].
Comparative transcriptome profile analysis of the parasite transcriptome with the sequences from non-parasitic organisms (i.e. Dugesiidae species and C. elegans) revealed that 51.87% of the obtained parasite transcripts shared significant homology, and the remaining (48.13%) were possibly be lineage- or genus-specific. A small part of the transcriptome (3.46%) showed the sequence homology with the pathogen related databases but not with the non-parasite related databases. The important strategy for picking out candidate transcripts with virulence was based on the identification of the transcripts with Ka/Ks > 1 with the assumption of the possible fast evolutionary pattern of parasitism associated genes [18]. This strategy has been successfully employed to identify virulence and immunomodulation-related genes of other parasitic organisms such as Plasmodium and Theileria species [30-33]. In our study, as a result of Ka/Ks analysis of 12,394 orthologous transcripts, a total of 418 F. hepatica transcripts were found to be with Ka/Ks > 1, hinting at a faster evolutionary rate because of likely involvement of these genes in the process of parasitism. More detailed analysis of the transcriptome with the motifs of proteins known to be associated with cytokine signaling was useful for the elucidation of other important genes from F. hepatica. The similar evolutionary characteristics of previously described virulence genes including cathepsin protease L1 and L2 [34,35], cathepsin protease B [36], thioredoxin/peroxiredoxin [13], glutathione S-transferase (sigma, mu, omega classes) [37-39], protein disulfide-isomerase [40] and 14-3-3 protein [41] boosts the authenticity of parasitism associated genes identified in this study.
The presence of more transcripts likely encoding immunomodulatory proteins within the pathogen database related subgroup (PDR subgroup), in comparison to the positive selection related subgroup (PSR subgroup), suggests a possible role for nucleotide diversity at the lineage/genus for the establishment of parasitism. Because of more frequent extracellular localisation of the transcripts within PDR subgroup and the cytokine signaling related subgroup (CSR subgroup) relative to PSR subgroup, it could be postulated that these gene products could directly interact with host proteins. Pyrimidine metabolism and other aminoacid related metabolisms such as aminobenzoate degradation (map00627), beta-Alanine metabolism (map00410), glycine, serine and threonine metabolism (map00260), specific to PDR subgroup, could hint a possible lineage/genus specific nucleotide diversification.
Through the comparative transcriptome analysis of the liver fluke sequences, a set of virulence-related transcripts (n = 71, corresponding to 64 putative genes) were found likely to possess immunomodulatory functional characteristics. To our knowledge, all the virulence and immunomodulatory genes of F. hepatica identified hitherto have not been reported, with the exception of two putative genes (i.e. #7694 and #32989) which are related to CD59 [42].
The proportion of transcripts with receptor activity (GO:0004872) was higher in VIR set, in comparison with VR group and its PDR and PSR subgroups, possibly denoting that VIRs are coevolved with host proteins due to direct interactions. The skewed proportionality in abundance of membrane (GO:0016020) in VIR set supports this further.
Majority of VIRs and their corresponding genes showed sequence similarity with host immune receptors (TGF-β receptor; n = 10, toll/interleukin-1 receptor; n = 5, T-cell receptor; n = 5 and MHC class I; n = 14) or cytokines (TGF-β; n = 3, interleukin-4/interleukin-13; n = 1 and TNF-α; n = 1) that these host molecules are known to be involved in CD4+ T-helper cell differentiation and regulation of the subsequent immune responses [43].
The identification of the transcripts sequentially similar to TGF-β cytokine (i.e. #10264 and #60918), TGF-β receptor (i.e. #11610/#45296/#64440, #19626/#20908, #20661, #26180, #26955, #37746, #49819, #55207, #56437, #73673 and #76453) or TGF-β antagonists (i.e. #44058 and #56418) could be important for controlling TGF-β cytokine levels. Additionally, the other genes that share sequence similarity with somatomedin B and thrombospondin (type 1) (i.e. #58983), hypothetical proteins containing thrombospondin (type 1) domain (i.e. #34645, #40900; Ka/Ks > 1), proposed to stimulate TGF-β expression [44,45], or bone morphogenetic protein antagonist noggin proteins, known to inhibit of the effects of TGF-β (i.e. #44058 and #56418) [46], could play roles in regulating the activities of TGF-β.
Our results showed that sequence homology with receptor like genes was not limited to TGF-β receptor, some other parasite genes were found to have sequential similarities with toll/interleukin-1 receptor (i.e. #1584, #6556, #6733, #16002 and #38341), TNF receptor (i.e. #37409 and #58628) or IL-18 receptor accessory protein (i.e. #77913) that all of which are known to be involved in controlling proinflammatory responses [47-50].
Putative parasite genes (n = 14) with sequence homologies to MHC class I receptor had the highest proportion of all identified parasitism genes. The predicted protein sequences of the related transcripts (i.e. #38312, #53490, #55117, #65009, #22528, #61208, #13771, #73151, #59522, #38573 and #58384) cover the region of alpha 1 and 2 domains, but not of alpha 3 domain of MHC class 1 molecule. We speculate that the parasite peptides may be presented to host immune cells (i.e. CD8 T-cells and NK cells) by alpha 1 and 2 domains, but the cytotoxic effects of immune cells could be potentially blocked because of the absence of alpha 3 domain. This proposed mechanism may result in suppression of subsequent pro-inflammatory responses including related CD4+ T-cell differentiation (possibly Th1) and cytotoxic cell killing mechanisms, known to be harmful to the liver fluke [51-53].
Another interesting finding was the presence of a number of putative genes with T-receptor homology for a number of putative genes (i.e. #27939, #34729, #44260, #65009 and #73578). The imitation of T-cell receptor by the parasite may interefere with the presentation of the parasite’s antigen to T-cells and subsequent cellular and humoral responses. Taken together with the observed homologies related to T-cell receptor and the proteins of TGF-β signaling accentuates the importance of the stimulation of Th2 type responses for the success of parasitism as suggested by another study conducted for Trichuris suis [54].
Some of the parasite transcripts encoding interleukin-4/interleukin-13 conserved site (IPR018096) (i.e. #12300) or showing sequential similarity with TNF-α (i.e. #40314) cytokine may possibly act in driving immune responses to Th2 and Th1, respectively. The other transcript (i.e. #31945) encoding a protein motif (IPR015535), known to induce the expression of IL-10 cytokine (an element of T-reg cells), could be important in balancing the Th1 and Th2 responses [55].
The identification of a transcript sharing sequence similarity with CD147 (basigin) (i.e. #23314) was another noteworthy finding. CD147 is a membrane protein of suppressed T-reg cells and associated with negative regulation of T-reg associated cytokine signaling and T-cell activation (via impaired expression of IL-2 receptor α-chain CD25) [56,57]. We suggest that F. hepatica transcripts which coevolved with CD147 could be important for the regulation of T-reg associated responses. The other cluster of differentiation (CD) similarity was found to be associated with CD48 (i.e. #59632), stimulating various regulatory factors in B and T lymphocytes through binding molecules such as CD2 and CD244 (2B4) [58].
Two of F. hepatica transcripts, #10702 and #79120, were found to share high level of sequential similarities with macrophage inhibition factor and ctg4a (protein canopy homolog-related 3), respectively. Both proteins are known to be involved in the activation of macrophages during inflammation [59,60]. In relation to this, three genes, #42935/#76935, #72283, #3866, were associated with binding proline (GYF super family, # cl00072) [61,62] which is secreted by Th2 type immune response-related alternatively activated macrophages promoting the development of fibrosis [63]. Taken together, the parasite proteins appear to be involved in the regulation of the macrophage activation and control of the development of excessive fibrotic tissue, in concordance with observations in clinical studies [64,65].
There are other identified transcripts which showed sequential similarities with a neutrophil protein (i.e. #5195) [66], molecules known to suppressors of cytokine signaling (i.e. #11419, #47252) [67] and of IKBKE1 [1IKK-epsilon and TBK1, influencing type I IFN production (#38525)] [68] or modulators of T-cell (i.e. #7920 and #70639) [69]. These genes could be other components of the immunomodulatory mechanism induced by the parasite, but which specific immune responses they stimulate can not be predicted with the available data.
Conclusions
By comparative analysis of total transcriptome of F. hepatica with publicly open databases, a number of putative genes (n = 62), which are potentially critical for virulence through immunomodulation or associated mechanisms and firstly described in this study. The majority of these genes appeared to be lineage- or genus-specific, suggesting a modus operandi through the enhancement of sequential diversity for genes encoding proteins which are likely to be at the frontline for the establishment of parasitism. In addition, the nucleotide diversity stemming from positive selectional pressure was found to be associated with cytokine signaling mechanisms by relying on the observed homologies with known genes such as toll/interleukin-1 receptor, TGF-β receptor, CD147 and a S. mansoni orthologous protein containing thrombospondin (type 1) domain (associated with TGF-β stimulation). A significant percentage of the transcripts have a remarkable level of sequential similarity with host immune receptors and cytokines, which are known to be part of array of immunological responses through CD4+ T-helper differentiation, indicating modulation of host immune system via controlling cellular responses associated with the T-helper heterogeneity (T-reg, Th1, Th2 and Th17 in particular). In conclusion, the blockage of the effects of aforementioned parasite proteins with RNAi or other strategies (e.g. vaccine or drug) would be a good approach in the fight against fasciolosis. This may even be important in promoting the efficacy of other immunoprophylactic molecules which are experimentally tested in the prevention of fasciolosis. Apart from the dealing with fasciolosis, synthetic versions of the identified virulence and immunomodulatory genes reported in this study could be important to control undesired pro-inflammatory responses, by considering immunoregulatory effects of the parasite and current therapeutic approaches in the treatment of autoimmune defects (e.g. helminth therapy) [70-73]. Studies including the synthesis of the indicated genes in prokaryotic and eukaryotic systems and evaluating their immunological effects in vitro and in vivo are underway. The present approach can be used for other studies with similar purposes.
Methods
Parasites
Adult liver flukes were collected from the bile ducts of naturally infected cattle in an abottoir (Tuzla, İstanbul) and immediately placed in a 50 ml tube containing warm PBS (Biochrom) as previously described [74]. Intact liver flukes were washed with warm PBS and placed in a flask containing culture medium (DMEM, Invitrogen) and gentamycin (50 μg as a final concentration) (Invitrogen). The flask containing the parasites was kept in a suitable enviroment (37°C, 5% CO2) for 2 hours for the regurgitation of all contents from the parasites’ digestive tracts as previously described [6]. After the flukes were washed with PBS (37°C), each fluke was placed in a seperate cryogenic tube (#1620-2700, Seal-Rite), snap-frozen using liquid nitrogen and kept −80°C until use.
RNA extraction
Total RNA from whole body of each fluke was extracted by using RNeasy Protect Mini Kit (Qiagen) with an on-column DNAse step (Macherey–Nagel) as previously described [75]. Purity and quantity of the extracted total RNA were analysed using a spectrophotometer (NANODROP 1000, Thermo Scientific) and a fluorescence based system (Qubit 2.0 Fluorometer using Qubit RNA BR assay kit, Invitrogen), respectively [75]. Quality of RNA for each extraction was analysed by a microfluidics capillary based electrophoretic system (Agilent 2100 Bioanalyzer using Agilent RNA 6000 Nano Kit (Agilent Technologies) according to the manufacturer’s recommendations except heat-denaturation which breaks 28S rRNA and prevent determination of RNA integrity number (RIN) [75]. Among several RNA extractions, the sample with the highest RIN number (RIN = 10) was used for sequencing.
Next generation RNA-sequencing (RNA-seq) of whole transcriptome of F. hepatica
RNASeq library was prepared from 1.25 μg of total RNA with TruSeq RNA Sample Preparation kit (Illumina) according to the kit’s user guide (Part#15026495 Ref. D). Briefly, mRNA was denaturated (65°C for 5 min, 4°C hold), eluted, fragmented and primed (elution 1; 80°C for 2 min, 25°C hold, elution 2-frag-prime; 94°C for 8 min, 4°C hold). Double strand (ds) complementary DNA (cDNA) was synthesised using a reverse transcriptase enzyme (SuperScript II Reverse Transcriptase, Invitrogen) and the other required reagents supplied by the kit. After the end repair, 3′ end adenylation and adapter ligation steps, cDNA fragments were enriched with PCR amplification as described by the user guide. All cDNA clean up steps were performed using Agencourt AMPure XP beads (Beckman Coulter) and a magnetic stand (Agencourt Bioscience Corporation). Quality and quantitative parameters of the library were determined by the Agilent High Sensitivity DNA kit (#5067-4626) and KAPA Library Quantification Kit (#KK4844, KAPA Biosystems) using the Agilent 2100 Bioanalyzer and a quantitative PCR system (iQ5, Biorad) according to the manufacturers’ instructions, respectively. After ds DNA fragments were denatured with NaOH (0.05 N as final concentration), a bridge PCR amplification for the cluster generation from single-molecule DNA templates was performed on the inside surface of a flow cell (Illumina) by an automated instrument (cBot, Illumina, user guide; Part#15006165 Rev. F) using TruSeq PE Cluster Kit V3 (Illumina, user guide; Part#15023336 Rev. B). Paired-end sequencing was performed with a current next generation sequencing instrument, HiSeq 2000 (Illumina, user guide; Part# 15011190 Rev. H) using TruSeq SBS Kit v3 (200 cycles, Illumina, user guide; Part#15023333 Rev. B).
De novo assembly of sequence reads and annotation of the contiguous sequences
Sequencing images were generated by HiSeq 2000 and the image analysis step was performed by RTA (Real Time Analysis) software (Illumina). The base calling step was performed with RTA or OLB (Off-Line Basecaller) softwares (Illumina). Cluster intensities and noise estimates were used in the analysis. The base sequence from each cluster, a confidence level for each base, and the filtering parameter (whether the read passes filtering) were given as the output for the base calling. The output files (with bcl extension) were converted to compressed fastq files by analysis software, CASAVA 1.8.2 (Illumina). The reads with quality score less than 33 were eliminated and the first 13 bases of each read were trimmed because of the insufficient quality. Reads with length less than 15 bases were removed before the assembly step. A bioinformatic programme, VELVET 1.2.08 [76], was used for the first step of the assembly and shortPaired run was applied where k-mer length was set to 31, insertion length was set to 400, expected coverage was set to 25, coverage cutoff was set to 2, and minimum contig length was set to 100. The output of this step was used as the input for the second step of the assembly process where OASES 0.2.28 [77] programme was applied. As similar to the previous process, insertion length with the value of 400 and coverage cutoff with the value of 2 were used for OASES 0.2.28 run. Contiguous sequences (contigs) were annotated with blast analyses (E value < 10−5). Nucleotide sequences of the contigs were searched aganist publicly available non-redundant protein and nucleotide databases from NCBI using standalone blastx and blastn programs, respectively [24]. The contigs with E values (E value < 10−5) in the blastn analysis were searched against the same database with standalone tblastx programme to predict the correct frame of the contigs. Nucleotide sequences of the remaining unannotated contigs were searched against Schistosoma mansoni database [n = 11,810 for protein (obtained from Martin Aslett, The Wellcome Trust Sanger Institute, United Kingdom), n = 11,912 for mRNA, downloaded from GeneDB (www.genedb.org [78]) on 20 January 2014], S. japonicum database [n = 12,657; v3, both protein and nucleotide, n = 13,469; v4, both protein and nucleotide, n = 17,401; cDNAs, downloaded from Chinese National Human Genome Center (CHGC) at Shanghai, The Schistosoma japonicum Genome Project (http://www.chgc.sh.cn/japonicum/Resources.html) on 21 January 2014], S. haematobium and S.mansoni databases [ShaeEgypt; n = 13,073 for protein and nucleotide, SmanPuertoRico; n = 3,897for nucleotide/n = 3,896 for protein, downloaded from SchistoDB (http://SchistoDB.net) [79] on 23 January 2014]. All the blast results were extracted without cutoff parameters using Blast Parser (v1.2.6.14; http://geneproject.altervista.org/) and annotated contigs were termed transcripts. Nucleotide sequences of the identified transcripts were conceptually translated into amino acid sequences using Transeq (http://www.ebi.ac.uk/Tools/st/emboss_transeq/) [80] based on the blast matching frames. The transcript N50 value was determined as previously described [81,82].
Investigation of sequence homology of F. hepatica transcripts with the specialised databases
To detect non-parasitism homology of the liver fluke transcripts, sequence data of Dugesiidae family and of Caenorhabditis elegans were used, because 1) Species of Dugesiidae family and C. elegans are known free-living (non-parasitic) organisms, 2) both Dugesiidae and Fasciolidae families taxonomically belong to same phylum, platyhelminths (flatworms; http://www.ncbi.nlm.nih.gov/), 3) a large number of nucleotide sequences of Dugesia sp. and of Schmidtea sp. in the Dugesiidae family is publicly available [DNA Data Bank of Japan (DDBJ); www.ddbj.nig.ac.jp], 4) C. elegans is a well studied free-living organism and a comprehensive sequence information for this organism is publicly available (WormBase; http://www.wormbase.org). Nucleotide sequences of Dugesia sp. (n = 72,225) and of Schmidtea sp. (n = 82,784) were downloaded from the DDBJ resource on 10 and 11 February 2014, respectively. In addition, a small number of nucleotide sequences (n = 125) for other organisms belonging to the Dugesiidae family, including Cura sp., Girardia sp., Neppia sp., Romankenkius sp. were downloaded from the same resource (11 February 2014). Protein coding nucleotide sequences (cds) and protein sequences of C. elegans (c_elegans.PRJNA13758.current.* and c_elegans.PRJNA13758.current_development.*, n = 26,769 and n = 26,983, respectively) were obtained from the ftp site of WormBase (http://www.wormbase.org). Nucleotide sequences of all the F. hepatica transcripts were searched against nucleotide sequences of the Dugesiidae species and protein sequences of C. elegans using the standalone tblastx and blastx, respectively (E value < 10−5).
To determine pathogen database related liver fluke transcripts, all F. hepatica transcript sequences were searched against protein sequences of the helminth secretome database (HSD; including secretory databases for nematodes; n = 16,460, trematodes; n = 1,409, cestodes; n = 1,123, and a collection for experimentally determined excretory/secretory proteins; n = 1,485), obtained from Gagan Garg [11,12], and a vaccine related pathogen sequence resource, Vaccine Investigation and Online Information Network (Violin; http://www.violinet.org) [15], including Protegen (Protective Antigens; n = 350 for nt, n = 591 for protein) [83,84], VirmugenDB (A Database of Virulent Genes used for Development of Live Attenuated Vaccines; n = 174 for nt, n = 216 for protein) [85] and DNAVaxDB (DNA vaccine; n = 642 for nt, n = 326 for protein) [86], downloaded on 19 February 2014 (E value < 10−5 for blastx, E value < 10−7 for tblastx). Biological functions of some of the virulence-related transcripts (HTs and VTs) which could not be categorised with the InterProScan search were predicted based on the manual inspection of the information from the blast2GO searches (blastx, tblastx and blastp; GO DB version: 2013–09; E value < 10−5), public resources (UniProt, NCBI and GeneDB) and the referred publications. For the InterProScan and NCBI database searches, the information was obtained from the European Bioinformatics Institute - InterPro (http://www.ebi.ac.uk/interpro/) [87] and the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd/) [88], respectively.
Detection of nonsynonymous/synonymous substitution rate of F. hepatica transcripts
Protein coding sequences of F. hepatica transcripts were obtained using GETORF (http://emboss.bioinformatics.nl/cgi-bin/emboss/getorf), with the consideration of the frame sense and longest sequence length (n > 30), and translated into amino acid sequences with Transeq (Jemboss; v1.5) [89]. The F. hepatica orthologous Dugesiidae sequences were determined with the blastx search based on the parasite’s translated amino acid sequences (E value < 10−3). Protein coding sequences of the Dugesiidae sequences were determined by GETORF with the same parameters and the sequences were trimmed from the ends to excise error letters (until the end letter, leaving the minimum sequence length of 18). The sequence alignment for each orthologous transcript was carried out with ClustalW (v2.1) [90] using ParaAT (Parallel Alignment and back-Translation; version 1.0) [91]. The ratio for the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) for the aligned transcripts were estimated using KaKs_Calculator (version 1.2) [92], with the MYN method (a modified version of the Yang-Nielsen algorithm [93,94]. For the remaining F. hepatica transcripts that are not orthologous to Dugesiidae or Ka/Ks ratio was not calculable, the Ka/Ks analysis was performed using the F. hepatica orthologous C. elegans sequences with the same approach. The P value for Ka/Ks ratio was calculated by the Fisher’s exact test by KaKs_Calculator and Ka/Ks ratio with the P value less than 0.05 was accepted statistically significant.
Functional categorisation
Translated amino acid sequences of all the F. hepatica transcripts were analysed with InterProScan 5.0 [95] using blast2GO [96] (version 2.7.0/2.7.1). All available applications in the InterProScan configuration were run, which were; blastProDom (scanning the families in the ProDom database [97]), FPrintScan (scanning the fingerprints in the PRINTS database [98]), HMMPIR (scanning the hidden markow models (HMMs) in the PIR Protein Sequence Database [99]), HMMPFAM (scanning the HMMs in the PFAM protein families database [100,101]), SMART (scanning the HMMs in the SMART domain/domain families database [102]), HMMTigr (scanning the HMMs in the TIGRFAMs protein families database [103]), ProfileScan (scanning PROSITE profiles [104]), PaternScan (scanning PROSITE profiles with a new version of the PROSITE pattern search software [104,105]), HAMAP (scanning HAMAP profiles [106]), SuperFamily (scanning a library of profile HMMs that represent all proteins of known function [107]), SignalPHMM (predicting signal peptides and/or anchors [108]), TMHMM (predicting transmembrane helices in proteins [109]), HMMPanther (scanning the HMMs in the Panther database [110-112]), Gene3D (scanning a large collection of CATH protein domain assignments for ENSEMBL genomes and UniProt sequences [113]), Phobius (predicting combined transmembrane protein topology and signal peptide [114,115]), and Coils (predicting coiled coil regions in proteins [116]). The parasite transcripts were categorised in biological function based on the InterProScan information, considering the order of protein family, domain and functional site (conserved site, active site, binding site or repeat).
Cytokine signaling association
The InterProScan information for the observed protein motifs were manually inspected in terms of the relationship with cytokine signaling at the publicly available database of the European Bioinformatics Institute-InterPro (http://www.ebi.ac.uk/interpro/). The parasite transcripts that are potentially associated with cytokine signaling on the basis of protein family, domain or functional site were reported and termed CSRs (cytokine signaling transcripts).
Subcellular localisation analysis
Subcellular localisations of all the predicted F. hepatica protein sequences were analysed by WoLF PSORT (the value for the ‘k used for kNN’ was set to 32) [117].
Identification of virulence- and virulence and immunomodulation-related F. hepatica transcripts
The liver fluke transcripts that showed sequential homology with the non-parasite related databases but not showed signs of positive selection and/or cytokine signaling association were termed non-virulence-related transcripts (NVTs). The virulence-related transcripts (VRs) were predicted based on the following criteria; 1) observation of the exclusive homology with the data of the pathogen databases (PDRs), including transcripts sequentially homologous to HSD (termed HTs) and Violin (termed VTs) but not NVTs, 2) demonstration of the signs of positive selection (Ka/Ks > 1; PSRs), 3) detection of the predicted functional protein site which is related to cytokine signaling (proven by protein family, domain or funcitonal site information; CSRs). Some of the virulence-related transcripts which could not be categorised with the InterProScan data were categorised using the information from the Gene Ontology, UniProt, NCBI and referred publications. The transcripts specific to immunomodulation category was determined on the basis of sequential identity level to known immunomodulatory proteins. The immunodulation categorised PDRs and PSRs and all CSRs were termed virulence and immunomodulation-related transcript(s) [VIR(s)].
Gene ontology and biological pathway analyses
Gene ontology categories at parental level 2 (i.e. biological process, molecular function and cellular component) (http://geneontology.org/), and KEGG biological pathways (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) were analysed for the detected protein motifs (family, domain or funcitonal site) using the blast2GO software.
Acknowledgments
The authors would like to thank Dr. Gagan Garg and Prof. Shoba Ranganathan (Department of Chemistry and Biomolecular Sciences and ARC Centre of Excellence in Bioinformatics, Macquarie University, Sydney NSW 2109, Australia) for kindly providing the helminth secretome database and Martin Aslett (The Wellcome Trust Sanger Institute, United Kingdom) for the supply of S. mansoni protein database (GeneDB). The authors are thankful to Dr. Gearóid Patrick Sayers for reviewing the manuscript in language particularly. This study was supported by the TUBITAK-BILGEM-UEKAE grant (K030-T439 to M.Ş.S. and B.Y.) under the Republic of Turkey Ministry of Development Infrastructure Grant to establish Advanced Genomics and Bioinformatics Research Center (2011 K120020).
Abbreviations
- NGS
Next generation sequencing
- RNA-seq
RNA sequencing
- RNAi
RNA interference
- HSD
Helminth secretome database
- ES
Excretory/secretory
- Violin
Vaccine Investigation and Online Information Network
- DDBJ
DNA Data Bank of Japan
- NCBI
National Center for Biotechnology
- UniProt
Universal Protein Resource
- CHGC
Chinese National Human Genome Center
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NVT(s)
Non-virulence-related transcript(s)
- HT(s)
HSD related transcript(s)
- VT(s)
Violin related transcript(s)
- VIR(s)
Virulence and immunomodulation-related transcript(s)
- TNFs
Tumor necrosis factors
- Th1
T-helper 1
- Th2
T-helper 2
- Ka
The number of nonsynonymous substitutions per non-synonymous site
- Ks
The number of synonymous substitutions per synonymous site
- TGF-β
Transforming growth factor beta
- TLR4
Toll-like receptor 4
Additional files
Annotation process of F. hepatica contigs. Blast searches for the annotation of the contigs are indicated subsequently. The blastn annotated sequences were subjected to tblastx search to predict the sequence frame. pt; protein sequence, nt; nucleotide sequence, NCBI; http://www.ncbi.nlm.nih.gov, GeneDB; http://www.genedb.org, SchistoDB; http://SchistoDB.net, CHGC; http://www.chgc.sh.cn/japonicum/Resources.html.
Nonsynonymous/synonymous substitution rate statistics for the F. hepatica transcripts. Ka/Ks ratios and the related statistics for a total of 12,394 transcript pairs (Dugesiidae species or C. elegans orthologous F. hepatica transcripts; E value < 10−3) are demonstrated.
Distribution of the virulence-related F. hepatica transcripts detected by the different methods. The virulence-related liver fluke transcripts, determined by the exclusive pathogen database homology (PDR; including HT, VT, or HT/VT), positive selection (PSR) and/or cytokine signaling relation (CSR), are shown.
Functional categorisation of the liver fluke transcripts. A total of 20,160 transcripts which were categorised in various functions are listed. Of these, majority (93.23%) was categorisable by the InterProScan determined functional protein motifs and the rest was categorised by other means [gene ontology (GO), data search in other resources such as NCBI, UniProt, GeneDB and referred publications]. The InterProScan accession number for each related transcript is provided.
Subcellular localisation signals at individual transcript level. Predicted subcellular localisations for a total of 40,255 transcripts are listed. Numerical values after the subcellular localisation description indicate the calculated number of nearest neighbours to the query sequence. cysk; cytoskeleton, cysk_plas; cytoskeleton & plasma membrane, cyto; cytosol, cyto_mito; cytosol & mitochondria, cyto_nucl; cytosol & nuclear, cyto_pero; cytosol & peroxisome, cyto_plas; cytosol & plasma membrane, E.R.; endoplasmic reticulum, E.R._golg; endoplasmic reticulum & golgi apparatus, E.R._mito; endoplasmic reticulum & mitochondria, extr; extracellular, extr_plas; extracellular & plasma membrane, golg; golgi apparatus, lyso; lysosome, mito; mitochondria, mito_nucl; mitochondria & nuclear, mito_pero; mitochondria & peroxisome, nucl; nuclear, pero; peroxisome, plas; plasma membrane.
Functional and descriptional details of VIRs. The blast comparisons and E value are provided for the assigned accession number and description from the reference databases (NCBI or GeneDB) for each VIR. Accession and description of the homologous molecules in the specialised secondary databases are provided. The detected InterProScan numbers and Ka/Ks ratios are listed for the related transcripts. Subcellular location signs for VIRs are demonstrated individually.
Biological pathways of the liver fluke at individual transcript level. Biological pathways and enzyme types with enzyme ID, detected by the KEGG search, are demonstrated individually. EC: Enzyme Commission.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OH designed the study, collected the liver flukes and extracted total RNA. OH and BY carried out the sample preparation and RNA-seq. MA, PK, and MŞS performed the assembly of sequence reads and carried out the blastx annotation for the NCBI database. OH performed the other annotation processes and data analysis procedures including categorisation of the transcripts, identification of VRs (CSRs, PDRs and PSRs) and VIRs. OH wrote the manuscript and BY reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Orçun Haçarız, Email: orcun.hacariz@tubitak.gov.tr.
Mete Akgün, Email: mete.akgun@tubitak.gov.tr.
Pınar Kavak, Email: pinar.kavak@tubitak.gov.tr.
Bayram Yüksel, Email: bayram.yuksel@tubitak.gov.tr.
Mahmut Şamil Sağıroğlu, Email: mahmut.sagiroglu@tubitak.gov.tr.
References
- 1.Gonzales Santana B, Dalton JP, Vasquez Camargo F, Parkinson M, Ndao M. The diagnosis of human fascioliasis by enzyme-linked immunosorbent assay (ELISA) using recombinant cathepsin L protease. PLoS Negl Trop Dis. 2013;7 doi: 10.1371/journal.pntd.0002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carnevale S, Cabrera MG, Cucher MA, di Risio CA, Malandrini JB, Kamenetzky L, et al. Direct, immunological and molecular techniques for a fasciolosis survey in a rural area of San Luis, Argentina. J Parasit Dis. 2013;37:251–9. doi: 10.1007/s12639-012-0175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yılmaz B, Köklü S, Gedikoğlu G. Hepatic mass caused by Fasciola hepatica: a tricky differential diagnosis. Am J Trop Med Hyg. 2013;89:1212–3. doi: 10.4269/ajtmh.13-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mas-Coma S, Agramunt VH, Valero MA. Neurological and ocular fascioliasis in humans. Adv Parasitol. 2014;84:27–149. doi: 10.1016/B978-0-12-800099-1.00002-8. [DOI] [PubMed] [Google Scholar]
- 5.Brockwell YM, Elliott TP, Anderson GR, Stanton R, Spithill TW, Sangster NC. Confirmation of Fasciola hepatica resistant to triclabendazole in naturally infected Australian beef and dairy cattle. Int J Parasitol Drugs Drug Resist. 2013;4:48–54. doi: 10.1016/j.ijpddr.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young ND, Hall RS, Jex AR, Cantacessi C, Gasser RB. Elucidating the transcriptome of Fasciola hepatica - a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol Adv. 2010;28:222–31. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Lyons RE, Johnson AM. Gene sequence and transcription differences in 70 kDa heat shock protein correlate with murine virulence of Toxoplasma gondii. Int J Parasitol. 1998;28:1041–51. doi: 10.1016/s0020-7519(98)00074-5. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, et al. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Załuga J, Stragier P, Baeyen S, Haegeman A, Van Vaerenbergh J, Maes M, et al. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics. 2014;15:392. doi: 10.1186/1471-2164-15-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bello-Orti B, Aragon V, Pina-Pedrero S, Bensaid A. Genome comparison of three serovar 5 pathogenic strains of Haemophilus parasuis: insights into an evolving swine pathogen. Microbiology. 2014;160(Pt 9):1974–84. doi: 10.1099/mic.0.079483-0. [DOI] [PubMed] [Google Scholar]
- 11.Garg G, Ranganathan S. In silico secretome analysis approach for next generation sequencing transcriptomic data. BMC Genomics. 2011;12(Suppl 3):S14. doi: 10.1186/1471-2164-12-S3-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg G, Ranganathan S. Helminth secretome database (HSD): a collection of helminth excretory/secretory proteins predicted from expressed sequence tags (ESTs) BMC Genomics. 2012;13(Suppl 7):S8. doi: 10.1186/1471-2164-13-S7-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly S, O’Neill SM, Sekiya M, Mulcahy G, Dalton JP. Thioredoxin peroxidase secreted by Fasciola hepatica induces the alternative activation of macrophages. Infect Immun. 2005;73:166–73. doi: 10.1128/IAI.73.1.166-173.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn RJ, Mannion C, Golden O, Hacariz O, Mulcahy G. Experimental Fasciola hepatica infection alters responses to tests used for diagnosis of bovine tuberculosis. Infect Immun. 2007;75:1373–81. doi: 10.1128/IAI.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Racz R, Sayers S, Lin Y, Todd T, Hur J, et al. Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res. 2014;42(Database issue):D1124–32. doi: 10.1093/nar/gkt1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaxter M, Koutsovoulos G. The evolution of parasitism in Nematoda. Parasitology. 2014;25:1–14. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson AP. Genome evolution in trypanosomatid parasites. Parasitology. 2014;28:1–17. doi: 10.1017/S0031182014000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland TE, Logan N, Rückerl D, Humbles AA, Allan SM, Papayannopoulos V, et al. Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat Immunol. 2014;15:1116–25. doi: 10.1038/ni.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maizels RM, Nussey DH. Into the wild: digging at immunology’s evolutionary roots. Nat Immunol. 2013;14:879–83. doi: 10.1038/ni.2643. [DOI] [PubMed] [Google Scholar]
- 21.Zarowiecki M, Berriman M. What helminth genomes have taught us about parasite evolution. Parasitology. 2014;8:1–13. doi: 10.1017/S0031182014001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frech C, Chen N. Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashida K, Hara Y, Abe T, Yamasaki C, Toyoda A, Kosuge T, et al. Comparative genome analysis of three eukaryotic parasites with differing abilities to transform leukocytes reveals key mediators of Theileria-induced leukocyte transformation. MBio. 2012;3:e00204–12. doi: 10.1128/mBio.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haçarız O, Sayers G, Baykal AT. A proteomic approach to investigate the distribution and abundance of surface and internal Fasciola hepatica proteins during the chronic stage of natural liver fluke infection in cattle. J Proteome Res. 2012;11:3592–604. doi: 10.1021/pr300015p. [DOI] [PubMed] [Google Scholar]
- 25.Haçarız O, Baykal AT, Akgün M, Kavak P, Sağıroğlu MŞ, Sayers GP. Generating a detailed protein profile of Fasciola hepatica during the chronic stage of infection in cattle. Proteomics. 2014;14:1519–30. doi: 10.1002/pmic.201400012. [DOI] [PubMed] [Google Scholar]
- 26.Haçarız O, Sayers G, Mulcahy G. A preliminary study to understand the effect of Fasciola hepatica tegument on naïve macrophages and humoral responses in an ovine model. Vet Immunol Immunopathol. 2011;139:245–9. doi: 10.1016/j.vetimm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 2009;8:1891–907. doi: 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith RE, Spithill TW, Pike RN, Meeusen EN, Piedrafita D. Fasciola hepatica and Fasciola gigantica: cloning and characterisation of 70 kDa heat-shock proteins reveals variation in HSP70 gene expression between parasite species recovered from sheep. Exp Parasitol. 2008;118:536–42. doi: 10.1016/j.exppara.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Chambers E, Ryan LA, Hoey EM, Trudgett A, McFerran NV, Fairweather I, et al. Liver fluke β-tubulin isotype 2 binds albendazole and is thus a probable target of this drug. Parasitol Res. 2010;107:1257–64. doi: 10.1007/s00436-010-1997-5. [DOI] [PubMed] [Google Scholar]
- 30.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall N, Carlton J. Comparative genomics of malaria parasites. Curr Opin Genet Dev. 2005;15:609–13. doi: 10.1016/j.gde.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Carlton J, Silva J, Hall N. The genome of model malaria parasites, and comparative genomics. Curr Issues Mol Biol. 2005;7:23–37. [PubMed] [Google Scholar]
- 33.Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–3. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- 34.Dalton JP, Neill SO, Stack C, Collins P, Walshe A, Sekiya M, et al. Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int J Parasitol. 2003;33:1173–81. doi: 10.1016/s0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 35.Stack CM, Caffrey CR, Donnelly SM, Seshaadri A, Lowther J, Tort JF, et al. Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica. J Biol Chem. 2008;283:9896–908. doi: 10.1074/jbc.M708521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckham SA, Law RH, Smooker PM, Quinsey NS, Caffrey CR, McKerrow JH, et al. Production and processing of a recombinant Fasciola hepatica cathepsin B-like enzyme (FhcatB1) reveals potential processing mechanisms in the parasite. Biol Chem. 2006;387:1053–61. doi: 10.1515/BC.2006.130. [DOI] [PubMed] [Google Scholar]
- 37.Chemale G, Morphew R, Moxon JV, Morassuti AL, Lacourse EJ, Barrett J, et al. Proteomic analysis of glutathione transferases from the liver fluke parasite, Fasciola hepatica. Proteomics. 2006;6:6263–73. doi: 10.1002/pmic.200600499. [DOI] [PubMed] [Google Scholar]
- 38.LaCourse EJ, Perally S, Morphew RM, Moxon JV, Prescott M, Dowling DJ, et al. The Sigma class glutathione transferase from the liver fluke Fasciola hepatica. PLoS Negl Trop Dis. 2012;6 doi: 10.1371/journal.pntd.0001666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morphew RM, Eccleston N, Wilkinson TJ, McGarry J, Perally S, Prescott M, et al. Proteomics and in silico approaches to extend understanding of the glutathione transferasesuperfamily of the tropical liver fluke Fasciola gigantica. J Proteome Res. 2012;11:5876–89. doi: 10.1021/pr300654w. [DOI] [PubMed] [Google Scholar]
- 40.Salazar-Calderón M, Martín-Alonso JM, Castro AM, Parra F. Cloning, heterologous expression in Escherichia coli and characterization of a protein disulfide isomerase from Fasciola hepatica. Mol Biochem Parasitol. 2003;126:15–23. doi: 10.1016/s0166-6851(02)00269-4. [DOI] [PubMed] [Google Scholar]
- 41.Hernández-González A, Valero ML, del Pino MS, Oleaga A, Siles-Lucas M. Proteomic analysis of in vitro newly excysted juveniles from Fasciola hepatica. Mol Biochem Parasitol. 2010;172:121–8. doi: 10.1016/j.molbiopara.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Toet H, Rathinasamy V, Young ND, Gasser RB, Beddoe T, et al. First insight into CD59-like molecules of adult Fasciola hepatica. Exp Parasitol. 2014;144:57–64. doi: 10.1016/j.exppara.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunobiology. 6th Edition. Garland Science Publishing; 2005.
- 44.Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, et al. Activation of canonical Wnt signaling is required for TGF-β-mediated fibrosis. Nat Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–70. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 46.Hinck AP, Huang T. TGF-β antagonists: same knot, but different hold. Structure. 2013;21:1269–70. doi: 10.1016/j.str.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Neill L. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem Soc Trans. 2000;28:557–63. doi: 10.1042/bst0280557. [DOI] [PubMed] [Google Scholar]
- 48.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 49.Cheung H, Chen NJ, Cao Z, Ono N, Ohashi PS, Yeh WC. Accessory protein-like is essential for IL-18-mediated signaling. J Immunol. 2005;174:5351–7. doi: 10.4049/jimmunol.174.9.5351. [DOI] [PubMed] [Google Scholar]
- 50.Bouchery T, Kyle R, Ronchese F, Le Gros G. The differentiation of CD4(+) T-helper cell subsets in the context of helminth parasite infection. Front Immunol. 2014;5:487. doi: 10.3389/fimmu.2014.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tliba O, Moire N, Le Vern Y, Boulard C, Chauvin A, Sibille P. Early hepatic immune response in rats infected with Fasciola hepatica. Vet Res. 2002;33:261–70. doi: 10.1051/vetres:2002014. [DOI] [PubMed] [Google Scholar]
- 52.Haçarız O, Sayers G, McCullough M, Garrett M, O’Donovan J, Mulcahy G. The effect of Quil A adjuvant on the course of experimental Fasciola hepatica infection in sheep. Vaccine. 2009;27:45–50. doi: 10.1016/j.vaccine.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 53.Pleasance J, Wiedosari E, Raadsma HW, Meeusen E, Piedrafita D. Resistance to liver fluke infection in the natural sheep host is correlated with a type-1 cytokine response. Parasite Immunol. 2011;33:495–505. doi: 10.1111/j.1365-3024.2011.01305.x. [DOI] [PubMed] [Google Scholar]
- 54.Cantacessi C, Young ND, Nejsum P, Jex AR, Campbell BE, Hall RS, et al. The transcriptome of Trichuris suis–first molecular insights into a parasite with curative properties for key immune diseases of humans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol Rev. 2008;223:114–31. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]
- 56.Staffler G, Szekeres A, Schütz GJ, Säemann MD, Prager E, Zeyda M, et al. Selective inhibition of T cell activation via CD147 through novel modulation of lipid rafts. J Immunol. 2003;171:1707–14. doi: 10.4049/jimmunol.171.4.1707. [DOI] [PubMed] [Google Scholar]
- 57.Landskron J, Taskén K. CD147 in regulatory T cells. Cell Immunol. 2013;282:17–20. doi: 10.1016/j.cellimm.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Elishmereni M, Levi-Schaffer F. CD48: A co-stimulatory receptor of immunity. Int J Biochem Cell Biol. 2011;43:25–8. doi: 10.1016/j.biocel.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Liu A, Fang H, Dirsch O, Jin H, Dahmen U. Early release of macrophage migration inhibitory factor after liver ischemia and reperfusion injury in rats. Cytokine. 2012;57:150–7. doi: 10.1016/j.cyto.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 60.Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, et al. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinze M, Kofler M, Freund C. Investigating the functional role of CD2BP2 in T cells. Int Immunol. 2007;19:1313–8. doi: 10.1093/intimm/dxm100. [DOI] [PubMed] [Google Scholar]
- 62.Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273:245–56. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 63.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 64.Haçarız O, Sayers G, Flynn RJ, Lejeune A, Mulcahy G. IL-10 and TGF-beta1 are associated with variations in fluke burdens following experimental fasciolosis in sheep. Parasite Immunol. 2009;31:613–22. doi: 10.1111/j.1365-3024.2009.01135.x. [DOI] [PubMed] [Google Scholar]
- 65.Golbar HM, Izawa T, Juniantito V, Ichikawa C, Tanaka M, Kuwamura M, et al. Immunohistochemical characterization of macrophages and myofibroblasts in fibrotic liver lesions due to Fasciola infection in cattle. J Vet Med Sci. 2013;75:857–65. doi: 10.1292/jvms.12-0536. [DOI] [PubMed] [Google Scholar]
- 66.Lomax KJ, Leto TL, Nunoi H, Gallin JI, Malech HL. Recombinant 47-kilodalton cytosol factor restores NADPH oxidase in chronic granulomatous disease. Science. 1989;245:409–12. doi: 10.1126/science.2547247. [DOI] [PubMed] [Google Scholar]
- 67.Larsen L, Röpke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002;110:833–44. doi: 10.1034/j.1600-0463.2002.1101201.x. [DOI] [PubMed] [Google Scholar]
- 68.Nakatsu Y, Matsuoka M, Chang TH, Otsuki N, Noda M, Kimura H, et al. Functionally distinct effects of the C-terminal regions of IKKε and TBK1 on type I IFN production. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiscella M, Perry JW, Teng B, Bloom M, Zhang C, Leung K, et al. TIP, a T-cell factor identified using high-throughput screening increases survival in a graft-versus-host disease model. Nat Biotechnol. 2003;21:302–7. doi: 10.1038/nbt797. [DOI] [PubMed] [Google Scholar]
- 70.Wolff MJ, Broadhurst MJ, Loke P. Helminthic therapy: improving mucosal barrier function. Trends Parasitol. 2012;28:187–94. doi: 10.1016/j.pt.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson MW, Donnelly S, Dalton JP. Helminth defence molecules-immunomodulators designed by parasites! Front Microbiol. 2013;4:296. doi: 10.3389/fmicb.2013.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wammes LJ, Mpairwe H, Elliott AM, Yazdanbakhsh M. Helminth therapy or elimination: epidemiological, immunological, and clinical considerations. Lancet Infect Dis. 2014;14:1150–62. doi: 10.1016/S1473-3099(14)70771-6. [DOI] [PubMed] [Google Scholar]
- 73.Tanasescu R, Constantinescu CS. Helminth therapy for MS. Curr Top Behav Neurosci. 2014. in press. [DOI] [PubMed]
- 74.Haçarız O, Baykal AT. Investigation of the abundance of proteins secreted by Fasciola hepatica, which is exposed to environmental change in experimental studies, with an advanced proteomic approach. Turkiye Parazitol Derg. 2014;38:106–10. doi: 10.5152/tpd.2014.3443. [DOI] [PubMed] [Google Scholar]
- 75.Haçarız O, Sayers G. Fasciola hepatica - where is 28S ribosomal RNA? Exp Parasitol. 2013;135:426–9. doi: 10.1016/j.exppara.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 76.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–92. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA, et al. GeneDB–an annotation database for pathogens. Nucleic Acids Res. 2012;40(Database issue):D98–108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerlotini A, Aguiar ER, Yu F, Xu H, Li Y, Young ND, et al. SchistoDB: an updated genome resource for the three key schistosomes of humans. Nucleic Acids Res. 2013;41(Database issue):D728–31. doi: 10.1093/nar/gks1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(Web Server issue):W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yandell M, Ence D. A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13:329–42. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 82.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang B, Sayers S, Xiang Z, He Y. Protegen: a web-based protective antigen database and analysis system. Nucleic Acids Res. 2011;39(Database issue):D1073–8. doi: 10.1093/nar/gkq944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He Y, Xiang Z. Bioinformatics analysis of bacterial protective antigens in manually curated Protegen database. Procedia Vaccinol. 2012;6:3–9. [Google Scholar]
- 85.Racz R, Chung M, Xiang Z, He Y. Systematic annotation and analysis of “virmugens”-virulence factors whose mutants can be used as live attenuated vaccines. Vaccine. 2013;31:797–805. doi: 10.1016/j.vaccine.2012.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Racz R, Li X, Patel M, Xiang Z, He Y. DNAVaxDB: the first web-based DNA vaccine database and its data analysis. BMC Bioinformatics. 2014;15:S2. doi: 10.1186/1471-2105-15-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(Database issue):D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(Database issue):D348–52. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 90.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, et al. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 2012;419:779–81. doi: 10.1016/j.bbrc.2012.02.101. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–63. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Z, Li J, Yu J. Computing Ka and Ks with a consideration of unequal transitional substitutions. BMC Evol Biol. 2006;6:44. doi: 10.1186/1471-2148-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 97.Bru C, Courcelle E, Carrère S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33(Database issue):D212–5. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scordis P, Flower DR, Attwood TK. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15:799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- 99.Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, et al. The protein information resource. Nucleic Acids Res. 2003;31:345–7. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue):D302–5. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41(Database issue):D387–95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(Database issue):D344–7. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fuchs R. Predicting protein function: a versatile tool for the Apple Macintosh. Comput Appl Biosci. 1994;10:171–8. doi: 10.1093/bioinformatics/10.2.171. [DOI] [PubMed] [Google Scholar]
- 106.Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, et al. HAMAP in 2013, new developments in the protein family classification and annotation system. Nucleic Acids Res. 2013;41(Database issue):D584–9. doi: 10.1093/nar/gks1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–19. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 108.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 109.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 110.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Database issue):D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(Database issue):D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lees JG, Lee D, Studer RA, Dawson NL, Sillitoe I, Das S, et al. Gene3D: Multi-domain annotations for protein sequence and comparative genome analysis. Nucleic Acids Res. 2014;42(Database issue):D240–5. doi: 10.1093/nar/gkt1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 115.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 117.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Micallef L, Rodgers P. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]


