Abstract
Purpose
Recent studies have shown that genetic abnormalities may be responsible for most unknown cases of male infertility. Human Nsun7 gene, which is located on chromosome4, has a role in sperm motility by encoding the putative methyltransferase Nsun7 protein. The aim of the present study was to investigate the mutations of exon4 in the Nsun7 gene, which is associated with sperm motility defect.
Methods
Semen samples including those of fertile normospermic (normal), infertile oligospermic (with normal sperm motility), and infertile asthenospermic (with reduced sperm motility) men were collected from the Omid and Fatemezahra IVF centres (Babol, Iran). These samples were then analysed on the basis of World Health Organization guidelines using the general phenol–chloroform DNA extraction method. Exon4 was amplified using Sun-F/Sun-R primers. Samples from asthenospermic men, which showed different patterns of movement on single-strand conformation polymorphism compared with normal and oligospermic samples, were identified and subjected to sequencing for further identification of possible mutations.
Results
Analysis of extracted sperm proteins showed that the rate of Nsun7 decreased. Likewise, direct sequencing of PCR products, along with their analysis, confirmed the deletion mutation of adenine in location 11337 of the Nsun7 gene in asthenospermic men. Comparison of normal and mutant protein structures of Nsun7 indicated that the A11337-deletion of the exon4 resulted in the valine residues-157 with GTA-codon in normospermic replaced with TAG-early stop codon in asthenospermic samples, causing an abortive protein product with amino acid sequence shorter than normal. The secondary structure of the protein, the protein folding, and ligand binding sites were changed, indicating the impairment of the protein function.
Conclusions
Because the Nsun7 gene products have a role in sperm motility, it will lead to impairment in the activity of the protein and motility of sperm flagella as well as male infertility if a mutation occurs in this gene.
Keywords: Nsun7 gene, Deletion mutation, Protein folding, Sperm motility, Male infertility
Introduction
Infertility is a major medical and social problem that affects approximately 10–15 % of couples worldwide [1]. Infertility is a disease caused by a complex interaction between genetic and environmental factors in men [2–5]. About half of the infertility factors consist of male ones [6, 7]. Despite the fact that almost 50 % of male infertility cases are not yet diagnosed [8], genetic abnormalities have a large proportion in infertile men [9, 10]. It was recently discovered that genetic disorders cause 15–30 % of male infertility [11, 12]. The first role of fertility is transmission of genes from parents to children. Thus, any disorder in parental genes can affect natural fertility [13]. Sperm dysfunction is one of the most important causes of male infertility which may result in gene mutations [14]. Therefore, for a motile sperm, the number of genes and resulting proteins is essential [15]. Low sperm motility is a primary case of male infertility [16]. Consequently, the normal sperm flagella are important for normal sperm motility and male fertility [13], with any deficit in the Nsun7 gene resulting in impairments in flagella movement and sperm motility in infertile male mice [16]. Transversion mutation in the exon 7 of the Nsun7 gene is associated with low sperm motility in human and male infertility [17]. In humans, Nsun7 encodes a protein with 718-amino acid and 3697-bp belonging to the methyltransferase superfamily. Since Nsun7 can have a methyl transferase function and a role in sperm motility, Nsun7 protein is involved in mitochondrial rRNA processing in post-meiotic sperm. Therefore, any dysfunction in this gene results in a general lack of protein needed for optimum sperm performance (ExPASy/Swiss-Prot). The Nsun7 gene (NOP2/Sun domain family, member 7), which encodes putative methyltransferase Nsun7 (EC 2.1.1), has 12 exons and 2157 nucleotides of the Nsun7 gene in a conserved domains database (CDD). This gene is located on chromosome 4, chr4 40446671-40506755 (+): 4p14 forward strand. As motility is one of the most important characteristics of a successful sperm, mutations in the Nsun7 gene have been linked to infertility in mice and humans due to impaired sperm motility [16, 17]. Therefore, Nsun7 gene products (exon 4) can play a role in sperm motility and, similarly, any deficiency in these products can cause infertility.
As no study has been carried out on Nsun7 exon4, in the present study, we investigated exon 4 mutations located on the domain protected by Sun and associated with infertility and human sperm motility in idiopathic asthenospermic (infertile men with defect sperm motility), oligospermic (infertile men with normal sperm motility) and normospermic (fertile men as control group) men.
Materials and methods
Semen sample collection
Semen samples were collected from infertile men (asthenospermic, 60 samples; oligospermic, 30 samples) and fertile men (normospermic as control group, 30 samples) ranging in age from 24 to 38 from the infertility clinics Fatemeh Zahra and Omid Hospital (Babol, Iran) from 2012 to 2013. Semen samples were collected with the approval of the Medical Research Ethics Committee of the Faculty of Babol University of Medical Sciences. Semen samples were obtained by masturbation into a sterile container after sexual abstinence for 3 to 5 days. Before semen analysis, informed consents were obtained from the participants. The participants were also asked to fill out a questionnaire on smoking habits, alcohol use, use of other substances and drugs, and history of orchitis, testicular trauma, sexually-transmitted diseases, varicocele, surgery for inguinal hernia, and cryptorchism. All patients and controls were asked to attach a copy of all their previous medical records to their questionnaires.
Semen sample analysis
Routine analysis of semen was carried out within 1 h according to World Health Organization guidelines [18]. Samples classified to fertile or normospermic men and infertile with sperm motility <50 % as an asthenospermic men and infertile with sperm concentration less than 20×106 as an oligospermic men. Standard semen parameters analysis showed in (Table 1).
Table 1.
Standard semen parameters analyses in normospermic, oligospermic and asthenospermic men
| Semen parameters | Normospermic (n = 30) | Asthenospermic (n = 60) | Oligospermic (n = 30) |
|---|---|---|---|
| Volume (ml) | 3.067 ± 0.88 | 2.29 ± 1.01** | 2.48 ± 1.01** |
| Sperm count (× 106 ml−1) | 96.67 ± 8.16 | 47.75 ± 22.97* | 14.55 ± 20.63* |
| Sperm motility (%) | 62.66 ± 7.98 | 33.50 ± 4.66* | 58.70 ± 4.88* |
| Sperm morphology (%) | 27.86 ± 9.34 | 24.48 ± 4.42* | 22.58 ± 2.61* |
Values expressed as mean ± SD. C: Controls (n = 30), A: asthenospermic (n = 60), O: oligospermic (n = 30). * p-value <0.001, ** p-value <0.05. Semen parameter in patient groups compared with control group. p-value <0.05 was considered statistically significant
Human sperm DNA extraction
Total human sperm DNA was extracted with few changes according to the general phenol–chloroform DNA extraction by Green and Sambrook (2012) methods. An aliquot of 10–20 × 106 spermatozoa was incubated at 56 °C and for 2 h in a lysis buffer. The lysate was extracted by phenol/chloroform (1:1, v/v), DNA was precipitated with isopropanol (1:1, v/v) Incubated at −20 °C overnight, then Washed with 75 % ethanol and the pellet DNA was dissolved in 50 μl ddH2O. Extracted total DNA, were separated by Agarose gel at Voltage 90 V for 1.5 h, with general 1X TBE (Tris-Borat-EDTA) buffer. DNA bands stained with 1 μg/ml ethidium bromide and visualized by UV-transilluminators. All of these performed by Green and Sambrook (2012) methods [19].
Synthesis of oligonucleotide primers
The forward (Sun4-F) and reverse (Sun4-R) primers (5′- CCAGATCATTTGAGCAGTCTTATT and 5′- GGTTCTCTACTTCTTGAACTTCTGA, respectively) were designed using Oligo7 software for exon 4 of the Nsun7 gene (Accession no. NM-024677 for Homo sapiens NOP2 mRNA; and Accession no. 224589816 for 40751914–40812002H. sapiens chromosome 4) in NCBI gene bank. Oligonucleotide primers were synthesised by MWG Biotech AG Company, Germany.
Polymerase chain reaction
The amplification of exon 4 performed in 50 μl final volume reaction mixture contain; 20 ng of template DNA, 5 μl of 10X PCR buffer [100 mM KCl, 20 mM Tris–Cl; pH 8.8 at 25 °C, 0.1 mM DTT, 50 % Glycerol, 0.5 % Tween 20, 0.5 % NP-40], 1.5 μM MgCl2, 0.3 μM of dNTP, 0.5 μM of each primers (Sun4-F/Sun4-R) and 0.25 U Taq DNA polymerase. PCR amplification was carried out for 35 cycles in a DNA thermal cycler (Eppendorf Master Cycler Gradient, Germany) using the thermal profile of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and primer extension at 72 °C for 1 min. After confirmation of Taq-PCR products, PCR for DNA sequencing performed with Pfu DNA polymerase PCR products which hold by used in 1X PCR buffer [10 mM KCl, 20 mM Tris–Cl pH 8.8 at 25 °C, 0.1 % Triton X100, 10 mM (NH4)2SO4, 0.1 mg/ml BSA], 2 mM MgSO4, 0.2 mM each of dNTP, 0.5 mM of each primer, 20 ng of template DNA and 1.25U Pfu DNA polymerase.
Electrophoresis of the PCR products
Exon4 PCR Products, were separated on an agarose gel at 70 V for 3 h, with 1X TBE buffer and DNA bands were visualized by transillumination under UV light after staining with 1 μg/ml ethidium bromide at 25 °C for 5 min. Agarose gel electrophoresis and gel staning performed by Green and Sambrook (2012) methods.
Single-strand conformation polymorphism
A 1-μl aliquot of exon 4 PCR products were mixed with 9 μl single-strand conformation polymorphism (SSCP) dye (containing 9.5 ml formamide, 400 μl of 20 mM EDTA · Na2 (pH 8.0) and 100 μl of 1 % bromophenol blue dissolved in absolute ethanol) and placed in a 95 °C water bath for 5 min. The samples were then placed on ice, for 2 min, before being loaded on a 6 % polyacrylamide gel (= SSCP gel), containing 20 ml polyacrylamide 30 %, 5 ml of 10× TBE, 350 μl of 10 % APS and 10 μl of 10 % TEMED, made up to 100 ml total volume with double-distilled water. Gel electrophoresis was performed at 200 V for 10 h at 4 °C with 0.5× TBE buffer before staining with AgNO3, as described previously [19].
Human sperm protein extraction
Total human sperm protein was extracted by Green and Sambrook (2012) methods. Approximately (10 -15×106) sperm cells were centrifuged in 12,750 g for 2 min, and the supernatant was discarded. The precipitate was solved in 200 ml 1x PBS (pH = 7.3). The resulting solution was exposed to sonication for 10 min in the W400. The solution was centrifuged in 12,750 g for 5 min and then upper phase was transferred to a clean Eppendorf.
SDS-poly acrylamide gel
After the extraction of protein, samples were dispersed in 20 μl loading buffer (containing SDS, DTT, bromophenol blue and Tris–Cl). The stacking and resolving gels contained 8 and 10 % acrylamide/bis (30: 8), respectively.
DNA sequencing and analysis
Direct sequencing of the purified PCR products has done with the Seq Lab Co. Gottingen, Germany. DNA and amino acid sequences analyzed using different bioinformatics programs and tools. For example: Oligo7 for primer designing; Chromas as a tools for sequence analysis from Chromas sequence chromatogram viewer (http://www.technelysium.com.au/chromas_lite.html); Blast, nucleotide blast, protein blast tools from free version software of the National Center for Biotechnology Information (NCBI: http://www.ncbi.nlm.nih.gov/); ClustalW2 tool used from European Bioinformatics Institute (EBI server services: http://www.ebi.ac.uk/); Prot param, Prot Scale, Swiss-Prot and Scan prosite tools used from the Expert Protein Analysis System (ExPASy server services: http://web.expasy.org/protparam/); The secondary structure of the protein was predicted using Bioinf server (http://bioinf.cs.ucl.ac.uk/psipred/result/27a50f56-09fc-11e4-b01f-00163e110593); Information regarding the experimentally determined structures of Nsun7 protein was obtained from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/home/home.d); the three-dimensional structure of the protein was predicted using I-Tas.SER (http://zhanglab.ccmb.med.umich.edu/I-Tas.SER); Accelrys DS Visualiser 1.7 was used to investigate the three-dimensional structure and folding of proteins; and molecular docking was evaluated using Molegro Virtual Docker MVD 5.0 (Molegro 2011) to predict protein–ligand interactions.
Results
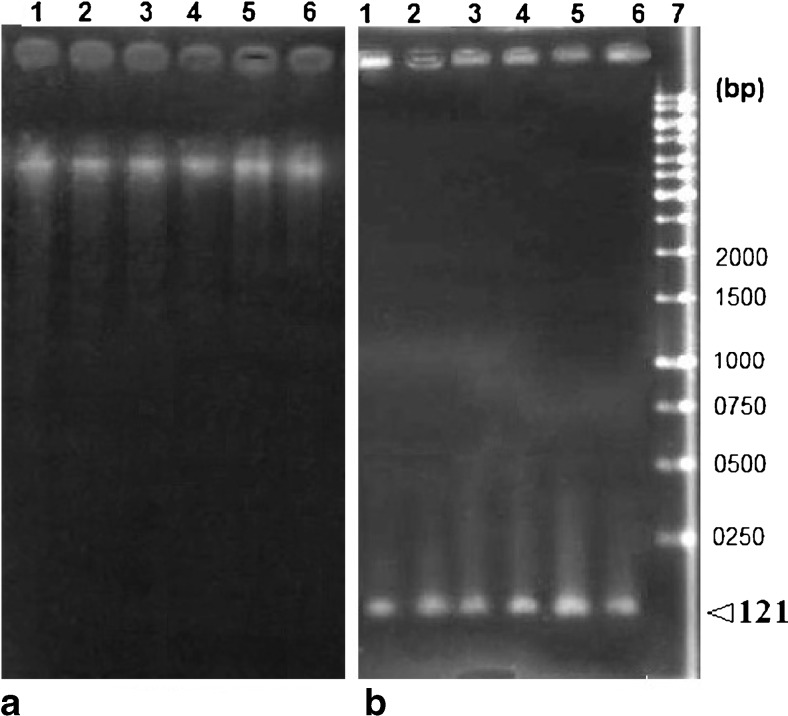
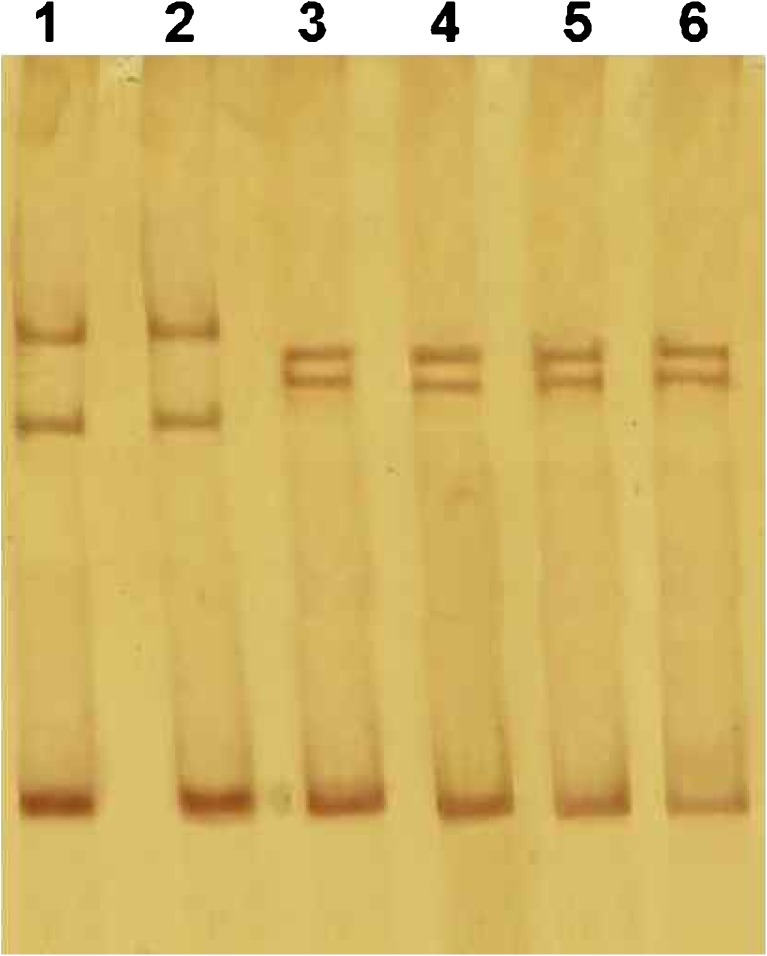
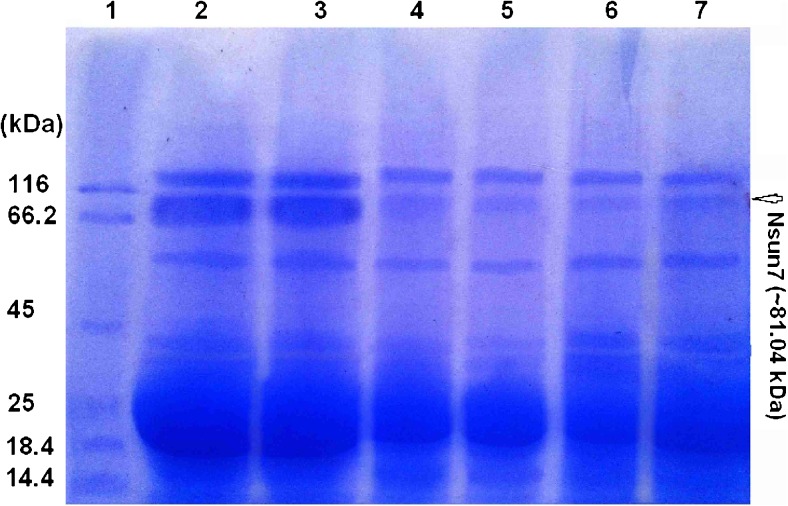
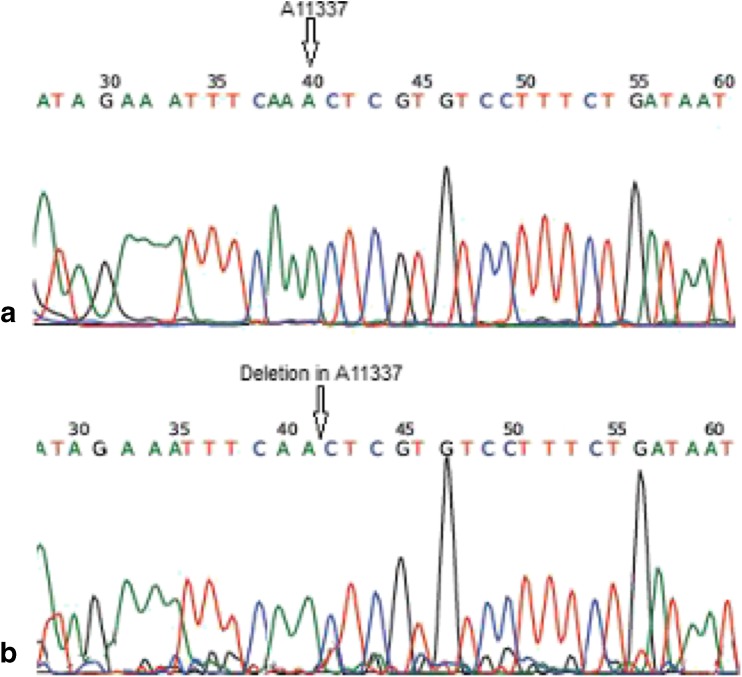
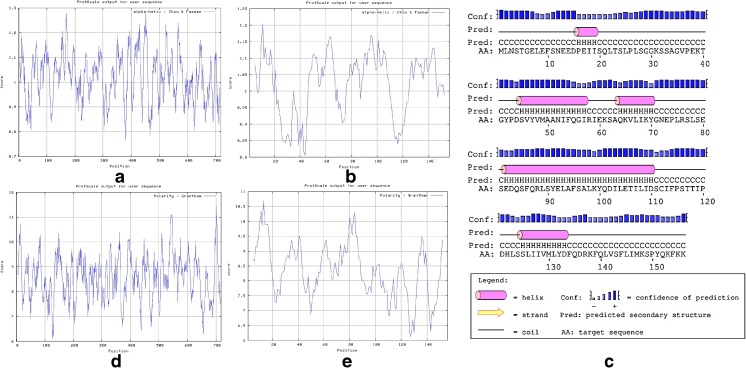
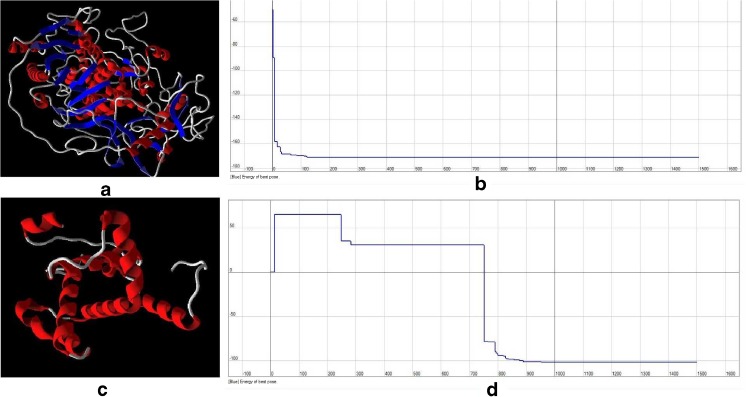
All parameters (count, Total count, motility and morphology of spermatozoa) were strongly significant in normospermic compared with asthenospermic men, with sperm motility being significantly lower in asthenospermic compared with normospermic and oligospermic men. Sperm parameters of semen samples were analysed (Table 1). Total human sperm DNA extracted was analysed both quantitatively and qualitatively using the method proposed by Green and Sambrook (2012) and gel electrophoresis (Fig. 1a). The exons 4 PCR fragments separated on an agarose gel, results were verified and qualified PCR products (Fig. 1b). Then the amplified fragments were loaded onto a 6 % polyacrylamide gel (as an SSCP gel). After gel staining, it was apparent that there was a significant difference in the migration of bands between asthenospermic and normospermic, and oligospermic men samples (Fig. 2). To further identify mutations, asthenospermic samples deviated from the normospermic samples (in the migration of bands) were evaluated and sequenced on an SSCP gel. After staining the human sperm proteins, the results indicated that the intensity of bands in astenospermic samples was different from that of the normospermic samples (Fig. 3). To identify mutations, different samples of normal-with-astenospermic were determined on SDS-PAGE. Direct PCR products sequencing was analysed using Chromas sequence chromatogram viewer and aligned by ClustalW2 (Fig. 4). The comparison of the direct sequencing of exon 4 in asthenospermic (Fig. 4b) with normospermic (Fig. 4a) samples with Chromas showed the deletion mutation in exon 4 of the Nsun7 gene in human asthenospermic men with poor motility. This mutation was not found in the fertile or normal group and oligospermic men. Moreover, both were blast with ClustalW2, showing the deletion mutation of Adenine in the 11337 position of the Nsun7 gene (Fig. 5a). Primary structure of amino acid sequences of the Nsun7 (Putative methyltransferase Nsun7) proteins in normospermic and asthenospermic was blast with ClustalW2 (Fig. 5b). Due to the A11337-Deletion mutation in exon 4, three nucleotide patterns of the codons were disturbed (Fig. 5b). In the 157th amino acid of the Nsun7 protein, the valin with codon GTA in normospermic and oligospermic samples was converted to stop-codon with codon TAG in asthenospermic samples (Fig. 5b). Even if the protein (with 156 amino acids) was produced, the analysis with ProtParam and Protscale servers revealed that the secondary structure of the Nsun7 protein (in asthenospermic samples) differs from normospermic and oligospermic samples (Fig. 6). The physicochemical characteristics of the Nsun7 protein such as polarity, aliphatic, molecular formula, molecular mass, isoelectric point (pI) and instability index were changed (Table. 2). Likewise, using the Bioinf server showed that the number and structure of the alpha-helix are changed (Fig. 6a–b) and there is no beta-strand in asthenospermic samples (Figs. 6c and 7c). The Protein folding prediction, ligand binding site, energy and protein–ligand interactions for the normal and mutant Nsun7 protein were analysed using Molegro Virtual Docker MVD 5. 0 (Fig. 7). Results from Molegro Virtual Docker indicated that there is no beta-strand and that the correct ligand-binding site in the mutant protein has changed. Protein folding had changed in the produced protein of the asthenospermic with deletion mutation in exon 4 (Fig. 7). Thus, this mutation indicates the impairment of protein function.
Fig. 1.
Total genomic DNA of spermatozoa and PCR products of the Nsun7 (exon 4): (a) Total genomic DNA of spermatozoa: Lane 1, total genomic DNA of normal spermatozoa; lane 2, total genomic DNA of spermtozoa from an oligospermic sample; lanes 3–6, total genomic DNA of spermatozoa from an astenospermic sample. (b) PCR products of the Nsun7 (exon 4) subjected to agarose gel electrophoresis: lane 1, exon 4 PCR product (121 bp) from total spermatozoa DNA of a normospermic sample; lane 2, exon 4 PCR product (121 bp) from total spermatozoa DNA of a oligospermic sample; lanes 3–6, exon 4 PCR product (121 bp) from total spermatozoa NA of an asthenospermic sample; lane 7, 1-kb DNA ladder (SM0311; Fermentas Company. (http://www.fermentas.com)
Fig. 2.
Single-strand conformation polymorphism (SSCP) of exon 4 PCR products of the NOP2/Sun domain family, member 7 (Nsun7) gene; Lane 1, normospermic; lanes 2, oligospermic; Lanes 3–6, asthenospermic
Fig. 3.
SDS-Poly acrylamide gel of extracted proteins of the total spermatozoa: Lane 1, Protein ladder; Lane 2, normospermic; lanes 3, oligospermic; Lanes 4–7, asthenospermic
Fig. 4.
Direct sequencing of exon 4 of the NOP2/Sun domain family, member 7 (Nsun7) gene in asthenospermic men: (a) in the wild-type Nsun7 gene, ‘A’ is at nucleotide position 11337. (b) Sequence of a sample from an asthenospermic infertile man with poor sperm motility showing a Deletion ‘A’ which was observed in samples from asthenospermic infertile men
Fig. 5.
Comparison of the primary structure of the amino acid sequences of Nsun7 proteins (b) and Blast analysis of exon 4 sequences in all asthenospermic (A) and normospermic (N) samples (a). The asterisks indicate similarity; different symbols indicate conversion of the valine (V) residue to stop-codon
Fig. 6.
Comparison of the secondary structure of the Nsun7 protein using Protscale server. Alpha-helix in normospermic (a), Alpha-helix in asthenospermic (b), the secondary structure of the Nsun7 protein in asthenospermic (c), Polarity in normospermic (d), Polarity in asthenospermic (e). Revealed that there were changes in the secondary structure of the Nsun7 protein in asthenospermic sample and there is no beta strand in sample with deletion mutation in exon4
Table 2.
Comparisons of the physicochemical properties of Nsun7 protein with Protparam server in normospermic and asthenospermic men
| Sample | Amino acid chain | Molecular formula | Molecular mass | Isoelectric point (pI) | Aliphatic index | Instability index |
|---|---|---|---|---|---|---|
| Normospermic | 718 | C3614H5809N987O1070S26 | 81,040.41 kDa | 8.85 | 90.68 | 50.39 |
| Asthenospermic | 156 | C797H1254N194O244S5 | 17,618.1 kDa | 4.91 | 95.00 | 49.73 |
Fig. 7.
Comparisons of the binding energy and three-dimensional structure of the Nsun7 protein and ligand (S-adenosyl-l-methionine) in (a, b) normospermic and (c, d) asthenospermic men. (a, c) Schematic diagram of protein folding of the Nsun7. (b, d) Binding energy and binding mode of the Nsun7 ligand (S-adenosyl-l-methionine) to the active site region of the Nsun7 protein
Discussions
Although about 50 % of male infertility causes are unknown [20], impaired male reproductive activity may lead to infertility. In recent years, male infertility has increased due to DNA damages and deficits in the sperm function [21]. An important criterion in dysfunction and low sperm motility is mutation in the genetic material of sperm [22]. Recent reports have shown that genetic disorders affecting spermatogenesis may be responsible for most unknown male infertility cases [23]. Harris et al. (2007) asserted that there are mutations in the sperm of infertile male mice with low sperm motility. Furthermore, poor progressive motility is characterized by links to the rigidity of the middle part and apparent defects of the mitochondrial sheath. Likewise, Khosronezhad et al. (2014) showed that transversion mutation in exon 7 of the Nsun7 gene caused protein to be more hydrophobic. In their study, while the protein folding was normal, the ligand-binding site and protein-ligand interactions were changed, resulting in the impairment of the Nsun7 protein function. Thus, according to the previous findings and the role of the Nsun7 gene in sperm motility and male fertility, we investigated exon 4 mutations located in the CDD region. Accordingly, the occurrence of the A-11337-deletion mutation in exon 4 of the Nsun7 gene of asthenospermic men was significantly higher than that in normospermic and oligospermic men (Fig. 4a and b). Comparison of normal and mutant protein structures of Nsun7 indicated that the deletion mutation in adenine caused a disorder in the 3nt pattern of codons and a reading frame shift with early termination. Accordingly, the A11337-deletion mutation in exon 4 resulted in the valin with codon GTA in normospermic and oligospermic samples (amino acid NO: 157) being converted to stop-codon with codon TAG in asthenospermic samples. The A11337-deletion mutation is a nonsense mutation that leads to the amino acid sequence being shorter than normal (Fig. 5a and b). Therefore, the protein was produced with a shorter than normal amino acid chain, and hence it was in a dysfunctional state. Analysis of the extracted sperm proteins showed that the Nsun7 protein with molecular weight 81.04 kD had a dual-band range between 116 and 66.2 kD of the protein marker (Fig. 3). As shown in Fig. 3, the intensity of protein bands in infertile men (asthenospermic) samples with A-11337-Deletion mutation leading to stop-codon in exon 4 is less compared with that of normal fertile (normospermic) and infertile without A-11337-deletion mutation (oligospermic) samples. Consequently, it showed that the rate of the Nsun7 protein was reduced in infertile ones with deletion mutation (asthenospermic) samples. Since there was a mutation in exon 4, the stop-codon was created, with the protein chain synthesis being ended and defective protein being produced or decomposed. Investigation of the secondary structure of the defective protein with protscale and bioinf servers revealed that there were changes in the structure and number of the alpha helix (Fig. 6a and b), with no beta strands in asthenospermic samples (Fig. 6c). The physicochemical properties of the Nsun7 protein and aliphatic index, as an important factor estimating its resistance against heat, was determined using the protparam server (Table 2), revealing that the physicochemical properties of the Nsun7 protein differed between normospermic and asthenospermic samples. Since mutant protein does not contain any Trp residues, experience shows that this could result in more than 10 % error in the computed extinction coefficient. Investigation of the polarity of the Nsun7 protein using the scale Polarity/Grantham revealed that there were changes in the polarity of the normal and mutant proteins (Fig. 6d and e). Thus, polarity and the secondary structure of the Nsun7 protein differed between normospermic and asthenospermic samples, having an impact on the function of the Nsun7 protein. Since the Nsun7 protein results in the motility of sperm flagellum, the failure to produce protein would lead to the impairment in activity and motility of sperm flagella, leading to sperm motility deficit and infertility. Indeed, analysis of the three-dimensional structure of the Nsun7 protein (Fig. 7) indicated that due to the deletion mutation, the amino acid sequence became shorter, with the protein being partially reproduced, changing the protein folding. Furthermore, there is no beta-strands (the second form of regular secondary structure in proteins) in protein with deletion mutations (Figs. 6c and 7c) which could have an effect on the protein folding and proper functioning (Fig. 7a–c). Since the mutant protein is incomplete, part of the protein attached to the ligand is not produced and the ligand cannot connect to the protein properly. The binding mode of the Nsun7 ligand (S-adenosyl-l-methionine) to the active site region of the Nsun7 protein was changed and if ligand was binding to protein, the energy of the ligand binding and protein-ligand interactions was changed (Fig. 7b–d). Considering the fact that the methyltransferase domains of the incomplete Nsun7 protein are not produced correctly, methyltransferase has not performed correctly and, hence, the protein function has been impaired. The deficiently-produced protein would lead to impairment in the protein activity and sperm flagella motility, causing sperm motility deficit and infertility. In conclusion, sperm motility is the most remarkable index of fertility potential and is correlated with the Nsun7 gene because it has a role in mitochondrial rRNA processing for sperm motility. The present study has demonstrated that the frequency of occurrence and proportion of deletion mutation of adenine in exon 4 in location 11337 of Nsun7 gene is significantly higher in men with poor sperm motility than that in men with normal sperm motility. The A-11337 is a deletion mutation, which is apparently correlated with changes in the protein folding and sperm motility in asthenospermic infertile men. As these mutations cause the amino acid chain to be shorter and hence an incomplete Nsun7 protein, there are changes in the secondary structure of the Nsun7 protein, protein folding, rate of the Nsun7 protein and protein function. Furthermore, changes in deficiently-produced protein were compared with normal result in changes to protein folding and impair methyltransferase activity. Because Nsun7 has a role in mitochondrial rRNA processing in post-meiotic sperm, protein dysfunction leads to a general lack of translation of mRNA and proteins needed for sperm motility, causing sperm motility deficits and male infertility. Since this mutation is located in an area where it is highly conserved evolutionarily, it affects the activity of the protein in asthenospermic men, which can cause deficiency in sperm motility. Because motility is one of the most important characteristics of a successful spermatozoon and Nsun7 gene products (exon 4) play a role in sperm motility, any deficiency causes infertility. Hence, the A11337-Deletion mutation in exon 4 of the Nsun7 gene can be used as an indicator of infertility. It is essential to clarify the molecular basis of these mutations in infertile men with poor sperm motility for better diagnosis and management of infertility treatment (ART).
Acknowledgments
Thanks to Dr. Seyed Golam Ali Jorsaraei, from Fatemeh Zahra Hospital (Babol Iran), for sample preparation and persons who helped prepare the manuscript and helped us to revise the paper.
Footnotes
Capsule
Our results indicate that there is an association between the exon 4 mutation in Nsun7 gene and sperm motility defect with male infertility in an Iranian population, hence the A11337-Deletion mutation in exon 4 of the Nsun7 gene can be used as an indicator of infertility.
References
- 1.Gnoth C, Godehardt E, Frank-Herrmann P, Friol K, Tigges J, Freundl G. Definition and prevalence of subfertility and infertility. Hum Reprod. 2005;20(5):1144–7. doi: 10.1093/humrep/deh870. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Laboratory manual for the examination of human semen and semencervical mucus interaction. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- 3.Zhou DX, Wang HX, Zhang J, Gao XL, Zhao WB. Di-n-butylphthalate (DBP) exposure induces oxidative damage in testes of adult rats. Syst Biol Reprod Med. 2010;6(6):413–9. doi: 10.3109/19396368.2010.509902. [DOI] [PubMed] [Google Scholar]
- 4.Zhou DX, Wang HX, Zhang J. Di-n-butyl phthalate (DBP)exposure induces oxidative stress in epididymis of adult rats. Toxicol Ind Health. 2011;27:65–71. doi: 10.1177/0748233710381895. [DOI] [PubMed] [Google Scholar]
- 5.Zhou DX, Zhang J, Wang HX. Assessment of the potential reproductive toxicity of long-term exposure of adult male rat’s tolow-dose formaldehyde. Toxicol Ind Health. 2011;27(7):591–8. doi: 10.1177/0748233710393401. [DOI] [PubMed] [Google Scholar]
- 6.Kretser DM, Baker HW. Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metab. 1999;84(10):3443–50. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 7.Irvine DS. Epidemiology and aetiology of male infertility. Hum Reprod. 1998;3:33–44. doi: 10.1093/humrep/13.suppl_1.33. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Malekpour M, Al-Madani N, Kahrizi K, Zanganeh M, Lohr NJ. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet. 2007;44:233–40. doi: 10.1136/jmg.2006.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamsi MB, Kumar K, Dada R. Genetic and epigenetic factors: role in male infertility. Indian J Urol. 2011;27:110–20. doi: 10.4103/0970-1591.78436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14:734–45. doi: 10.1016/S1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 11.Vogt PH. Molecular genetic of human male infertility from genes to new therapeutic perspectives. Curr Pharm Des. 2004;10:1–30. doi: 10.2174/1381612043453261. [DOI] [PubMed] [Google Scholar]
- 12.Liska F. Selected genetic aspects of male infertility-what animal models tell us. Folia Biol (Praha) 2003;49:129–41. doi: 10.14712/fb2003049040129. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert SF. Developmental biology. 6th ed, Chaptet 7 & 19, Sunderland (MA), Sinauer Associates. 2000.
- 14.Spiropoulos J, Turnbull DM, Chinnery PF. Can mitochonderial DNA mutations cause sperm dysfuunction? Mol Hum Reprod. 2002;8:719–21. doi: 10.1093/molehr/8.8.719. [DOI] [PubMed] [Google Scholar]
- 15.Turner RM. Moving to the beat: a review of mammalian sperm motility regulation. Reprod Fertil Dev. 2006;18:25–38. doi: 10.1071/RD05120. [DOI] [PubMed] [Google Scholar]
- 16.Harris T, Marquez B, Suare S, Schimenti J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the sun domain-containing family of putative RNA methyltransferases. Biol Reprod. 2007;77:376–82. doi: 10.1095/biolreprod.106.058669. [DOI] [PubMed] [Google Scholar]
- 17.Khosronezhad N, Hosseinzadeh-Colagar A, Jorsaraei G. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermic men. Reprod Fertil Develop. 2014 doi: 10.1071/RD13371. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization . Laboratory manual for the examination of human semen and semen–cervical mucus interaction. 4. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 19.Green MR, Sambrook J. Molecular cloning: a laboratory manual. 4. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 20.Jing Z, Dang-xia Z, Hai-xu W, Zhao T. An association study of SPO11 gene single nucleotide polymorphisms with idiopathic male infertility in Chinese Han population. J Assist Reprod Genet. 2011;28:731–6. doi: 10.1007/s10815-011-9571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hull MG, Glazener CM, Kelly NJ, Conway DI, Foster PA, Hinton RA, et al. Population study of causes, treatment and outcome of infertility. Br Med J (Clin Res Ed) 1985;291(6510):1693–7. doi: 10.1136/bmj.291.6510.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hargreave TB. Genetic basis of male fertility. Br Med Bull. 2000;56:650–7. doi: 10.1258/0007142001903454. [DOI] [PubMed] [Google Scholar]
- 23.Nishimune Y, Tanaka H. Infertility caused by polymorphisms or mutations in spermatogenesis-specific genes. J Androl. 2006;27:326–34. doi: 10.2164/jandrol.05162. [DOI] [PubMed] [Google Scholar]