Abstract
Purpose
The objective of this study was to discover a panel of microRNAs (miRNAs) as potential biomarkers for noninvasive prenatal testing (NIPT) of trisomy 21 (T21) and to predict the biological functions of identified biomarkers using bioinformatics tools.
Methods
Using microarray-based genome-wide expression profiling, we compared the expression levels of miRNAs in whole blood samples from non-pregnant women, whole blood samples from pregnant women with euploid or T21 fetuses, and placenta samples from euploid or T21 fetuses. We analyzed the differentially expressed miRNAs according to disease and tissue type (P value <0.05 and two-fold expression change). To predict functions of target genes of miRNAs, the functional annotation tools were used.
Results
We identified 299 miRNAs which reasonably separate the whole blood from the placenta. Among the identified miRNAs, 150 miRNAs were up-regulated in the placenta, and 149 miRNAs were down-regulated. Most of the up-regulated miRNAs in the placenta were members of the mir-498, mir-379, and mir-127 clusters. Among the up-regulated miRNAs in the placenta, mir-1973 and mir-3196 were expressed at higher levels in the T21 placenta than in the euploid placenta. The two miRNAs potentially regulate 203 target genes that are involved in development of brain, central nervous system, and nervous system. The genes are significantly associated with T21-related disorder such as congenital abnormalities, mental disorders, and nervous system diseases.
Conclusions
Our study indicates placenta-specific miRNAs that may be potential biomarkers for NIPT of fetal T21 and provides new insights into the molecular mechanisms of T21 via regulation of miRNAs.
Keywords: Noninvasive test, microRNA, Trisomy 21, Placenta
Introduction
MicroRNAs (miRNAs) are short (19 ~ 25 nucleotides), single-stranded, and non-coding RNAs [1]. The number of miRNAs known in humans is approaching 1000 and continues to rise (www.mirbase.org). They have emerged as key post-transcriptional regulators that inhibit gene expression by perfect complementary binding to target mRNA degradation or imperfect binding in the 3′ untranslated region (UTR) to inhibit translation [2, 3]. Moreover, abnormal miRNA expression has been reported to be involved in the occurrence and development of various diseases, such as cancer, cardiovascular disease, mental retardation, fetal growth restriction, and trisomy 21 (T21) [4–10].
Recently, the search for placenta-specific miRNAs in maternal blood has started. Approaches such as microarray, deep sequencing, and quantitative RT-PCR have been used to detect placenta-specific miRNAs in maternal blood [11–14]. The miRNAs originating in the trophoblast of the placenta are released into maternal circulation via exosomes [15, 16] and are more stable in maternal circulation compared to mRNA [17, 18]. Therefore, miRNAs have been proposed as a promising class of molecular biomarkers for noninvasive prenatal tests (NIPT) of placenta-related diseases.
T21 is the most common aneuploidy, referred to as Down syndrome. It has a high survival rate, affecting 1 in 800 to 1500 newborns [19] and is associated with a number of deleterious phenotypes, including cognitive impairment, leukemia, heart disease, and neurodegeneration [20]. Therefore, prenatal detection of T21 is considered the most common and important aspect of prenatal genetic testing in clinical practices.
Up to date, expression changes of miRNAs in T21 has primarily been investigated with miRNAs derived from human chromosome 21 (hsa21), because T21 is caused by an extra copy of all or part of hsa21. In previous studies, hsa21-derived miRNAs have been proven to be correlated with the complex and variable phenotypes of T21 [6–8, 21–24]. However, changes in miRNA expression that affect phenotypes can occur in the entire genome, i.e., an miRNA can potentially regulate a large number of protein-coding genes, and multiple miRNAs can regulate a single target gene [25]. Therefore, the genome-wide expression patterns of miRNAs must be investigated to identify miRNAs that are differentially expressed in T21. Moreover, miRNAs have disease specific characteristics [4–10, 21] and exhibit different expression pattern according to tissue type and the presence or absence of pregnancy [12, 26, 27]. Therefore, all of disease, tissue, and pregnancy-specific characteristics having miRNAs should be considered for identification of miRNAs as potential biomarkers for NIPT of fetal diseases using maternal blood. However, until now, placenta-specific miRNAs for NIPT have been identified based on the tissue-specific characteristics between placenta and maternal blood [11–14]. Moreover, the genome-wide expression profiling of placenta-specific miRNAs presenting in maternal blood for NIPT of fetal T21 has not yet been reported.
The goal of this study was to discover a panel of genome-wide placental miRNAs as potential biomarkers for the NIPT of fetal T21 using microarray-based expression profiling. We applied various bioinformatics tools to explore the biological function of the identified candidate miRNAs.
Materials and methods
Study subjects
This study was conducted according to the principles expressed in the Declaration of Helsinki. Pregnant women with euploid or T21 fetuses and non-pregnant women who attended the Department of Obstetrics and Gynecology, Cheil General Hospital, Korea were recruited between March 2011 and December 2012. All women included in this study were of Korean origin. Pregnant women were women who received prenatal care at Cheil General Hospital and non-pregnant women were women who had regular checkups. None of the participants had a history of preexisting hypertension, diabetes mellitus, liver disease, or chronic kidney disease. Institutional review board approval for this study was obtained from the Ethics Committee at Cheil General Hospital (#CGH-IRB-2011-85). All patients provided written informed consent for the collection of samples and subsequent analysis.
Sample processing and RNA isolation
First trimester maternal peripheral blood samples were collected into PAXgene™ Blood RNA Tube just before obstetric procedures such as chorionic villus sampling. All placenta samples were obtained at chorionic villus sampling. All samples were stored in liquid nitrogen until analysis. Total RNA was extracted using a mirVana miRNA Isolation Kit (Ambion, TX), and RNA concentrations were determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., DE). The integrity of the RNA was evaluated by microfluidic electrophoresis using an RNA 6000 Nano kit (Agilent Technologies, CA) and 2100 Bioanalyzer (Agilent Technologies). RNA integrity numbers (RIN) ≥ 7.0 were considered for microarray analysis.
Cytogenetic Analysis for detection of T21
Chromosomal analyses of fetal chorionic villus samples were carried out using standard protocols [28]. Cells from chorionic villus samples were cultured in the AmnioMAX-C100 culture medium (Invitrogen, CA). Metaphase chromosomes were stained using the GTG banding method, and 20 metaphases per sample were analyzed.
MicroRNA quantification
Total RNA (100 ng) from each sample was labeled and hybridized on Human miRNA array 8x60K v16.0 (Agilent Technologies) according to the manufacturer’s recommendations. The raw intensity of the array was scanned and extracted using BeadScan, with the data corrected by background subtraction in the Genome Studio module. Expression data were extracted from the scanned images using Feature Extraction software, version 10.7 (Agilent Technologies) and analyzed using the R statistical environment [29]. The background was adjusted by subtracting the median background values from the median expression values obtained via the Feature Extraction software, followed by log base 2 transformations. Quartile normalization was applied to the data using the aroma-light R package (v. 2.2.0) (http://www.bioconductor.org/packages/release/bioc/html/aroma.light.html). To detect outlier samples, inter-array correlation (IAC) was performed using the Cluster R package (v. 1.14.4). Subsequently, rank products statistical analysis was applied to the data, using the RankProd R package (v. 2.38.0). To obtain a single expression value for each probe set, the median expression value was calculated for multiple probe sets corresponding to unique miRNA.
Data analysis
The expression levels of miRNAs were considered statistically significant between the blood and placenta groups for P values less than 0.05; miRNAs were checked for candidates with at least two-fold expression changes. Among the candidates, increased miRNAs in T21 placentas compared with euploid placentas (P value <0.05 with two-fold expression change) were selected as potential biomarkers for NIPT of fetal T21. To examine the functions of the miRNAs identified as potential biomarkers for NIPT of fetal T21, miRNA target prediction was performed using miRBase (version 20, release June 2013) [30], which integrates six established target prediction tools: DIANA-MICROT [31], MICRORNA.ORG [32], MIRDB [33], RNA22 [34], TARGET MINER [35], and TARGETSCAN [36]. We analyzed miRNA-target gene interactions simultaneously identified by at least three prediction programs on the 3′ UTR of all known human genes for further analysis. To investigate whether the predicted target genes of candidate miRNAs were located on has21 and related to T21, a total of 584 genes of hsa21 (NCBI Map Viewer, Release 106) were analyzed using the VENNY tool [37]. To investigate whether miRNAs selected as potential biomarkers for NIPT of fetal T21 were possibly regulating genes for T21, the functional annotation tools provided by the WebGestalt database (http://bioinfo.vanderbilt.edu/webgestalt/) were used. Gene ontology (GO) analysis and disease-associated analysis of target genes were performed. The interactive network of selected target genes was predicted using the Search Tool for the Retrieval of Interacting Genes (STRING v. 9.05) database. The target genes were considered as the seed molecules to obtain direct and indirect protein-protein interactions. This database provides information on both experimental and predicted interactions from varied sources based on their neighborhood, gene fusion, co-occurrence, co-expression, experiments, and literature mining. We constructed an interactive network of target genes based on a high confidence score of 0.7 and extracted interactions with high level of confidence.
Statistical analysis
The clinical characteristics of the study population were analyzed using the Mann-Whitney U-test and Kruskal-Wallis test for continuous variables and the χ2-test for categorical variables. The accuracy for detecting fetal T21 placentas was analyzed based on the miRNAs identified as potential biomarkers for NIPT of fetal T21. Receiver operating characteristic (ROC) curve analysis was performed to assess the optimal cutoff value. The optimal cutoff was set at a specificity of 100 %. Overall accuracy was estimated according to the area under the ROC curve (AUC). In addition, the sensitivity, positive predictive value (PPV), negative predictive value (NPV), and odds ratio (OR) were calculated with 95 % confidence intervals (CI) using the EpiMax Table Calculator. In all tests, a threshold of P < 0.05 was set for statistical significance. Statistical analyses were performed using the Statistical Package for Social Sciences 12.0 (SPSS Inc., IL).
Results
Clinical characterization of the study population
The clinical characteristics of the study groups are shown in Table 1. In total, whole blood samples from non-pregnant women (n = 2), whole blood samples from pregnant women (n = 5), euploid fetal placenta samples from pregnant women (n = 5), and T21 fetal placenta samples from pregnant women (n = 4) were used for the genome-wide miRNA expression profiling. Five whole blood samples from pregnant women and five euploid fetal placenta samples were paired. At blood sampling, age and body mass index were no different among all study groups (P > 0.05). Gestational age, gravidity, nulliparity, and gender ratio of the fetuses were also not different between pregnant women carrying T21 fetuses and pregnant women carrying euploid fetuses (P > 0.05).
Table 1.
Clinical characteristics of the study population
| Characteristics | Pregnant women carrying trisomy 21fetuses | Pregnant women carrying euploid fetuses | Non pregnant women | P value |
|---|---|---|---|---|
| (n = 4) | (n = 5) | (n = 2) | ||
| At blood sampling | ||||
| Age (years) | 32.0 | 35.0 | 29.0 | 0.185a |
| (30.0–35.5) | (32.0–37.0) | (28.0–30.0) | ||
| Body mass index (kg/m2) | 21.3 | 21.0 | 21.5 | 0.492a |
| (19.3–24.0) | (20.6–21.4) | (21.0–22.0) | ||
| Gestational age (weeks) | 12.5 | 12.5 | – | 0.902b |
| (11.9–12.9) | (11.3–13.0) | |||
| Gravidity (n) | 2.5 | 3.0 | 0.905b | |
| (1.5–4.5) | (2.0–4.0) | |||
| Nullipara (%) | 25 | 20 | 0.858 | |
| Gender-ratio of fetus (male:female) | 2:2 | 3:2 | – | 0.764 |
Values are medians with interquartile range in parentheses
aKruskal-Wallis test
bMann-Whitney U test
Identification of miRNA clusters specific to placenta
In this study, a total of 1349 miRNAs including 1205 human miRNAs and 144 human viral miRNAs were profiled. To identify miRNA clusters specific to the placenta, we analyzed miRNAs isolated from whole blood samples of non-pregnant and pregnant women and placenta samples of euploid and T21 fetus. We identified 299 miRNAs that reasonably separated the whole blood samples from the placenta samples. Among the identified total miRNAs, 150 miRNAs were up-regulated in the placenta samples (Table 2) and 149 miRNAs were down-regulated (Table 3). Among the 150 up-regulated miRNAs in the placenta samples, 48 miRNAs (32.0 %) and 27 miRNAs (18.0 %) were located on hsa19 and hsa14, respectively, and accounted for half of the total miRNA up-regulated in the placentas (Fig. 1). Among 48 miRNAs located on hsa19, 39 miRNAs were members of the mir-498 cluster. Among 27 miRNAs located on hsa14, 19 and 7 miRNAs were members of the mir-379 cluster and mir-127 cluster, respectively (Table 2). However, miRNAs identified as decreased in the placenta were distributed on a variety of chromosomes (Fig. 1). Among the miRNAs of hsa21 such as cause chromosome of T21, only one miRNA (hsa-let-7c) was more down-regulated in the placenta than in the maternal blood (Table 3 and Fig. 1).
Table 2.
miRNAs increased in the placenta compared with the blood
| cχ2-testSystematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-205-5p | 1.14E-17 | 107 | miR-517-5p* | 1.92E-11 | 19 | miR-141-3p | 2.08E-08 | 53 | miR-3692-5p | 9.2E-06 | 2 | miR-193a-3p | 6.52E-04 | 4 |
| miR-526b-5p* | 1.45E-17 | 341 | miR-214-3p | 1.99E-11 | 20 | miR-542-5p | 2.18E-08 | 8 | miR-27b-3p | 9.47E-06 | 12 | miR-135b-5p | 7.10E-04 | 2 |
| miR-518e-5p* | 2.78E-17 | 189 | miR-224-5p | 2.74E-11 | 19 | miR-520c-3p* | 2.51E-08 | 10 | miR-30d-5p | 9.69E-06 | 3 | miR-1181 | 7.33E-04 | 5 |
| miR-517a-3p* | 5.57E-17 | 2536 | miR-503 | 3.38E-11 | 42 | miR-154-3p† | 2.64E-08 | 5 | miR-4284 | 1.01E-05 | 6 | miR-1183 | 8.75E-04 | 8 |
| miR-512-3p* | 6.96E-17 | 451 | miR-199a-3p | 5.07E-11 | 53 | miR-519e-3p* | 2.83E-08 | 11 | miR-125b-5p | 1.34E-05 | 10 | miR-1469 | 1.10E-03 | 3 |
| miR-515-5p* | 7.09E-17 | 442 | miR-381† | 7.45E-11 | 11 | miR-218-5p | 3.16E-08 | 8 | miR-518a-5p* | 1.44E-05 | 4 | miR-4253 | 1.15E-03 | 4 |
| miR-525-5p* | 1.03E-16 | 706 | miR-521* | 9.7E-11 | 127 | hsa-let-7e-5p | 3.42E-08 | 5 | miR-205-3p | 1.59E-05 | 3 | miR-198 | 1.16E-03 | 3 |
| miR-518e-3p* | 1.46E-16 | 258 | miR-200c-3p | 1.47E-10 | 37 | miR-199b-5p | 3.71E-08 | 9 | miR-224-3p | 2E-05 | 4 | miR-3605-5p | 2.08E-03 | 3 |
| miR-520h* | 9.79E-16 | 131 | miR-455-3p | 1.89E-10 | 11 | miR-520d-3p* | 4.95E-08 | 10 | miR-34a-5p | 2.18E-05 | 5 | miR-3926 | 2.42E-03 | 2 |
| miR-376c† | 1.06E-15 | 78 | miR-30a-5p | 2.35E-10 | 63 | miR-143-3p | 5.36E-08 | 5 | miR-299-3p† | 3.26E-05 | 4 | miR-3663-3p | 2.90E-03 | 4 |
| miR-517b* | 1.28E-15 | 1352 | miR-519b-3p* | 2.69E-10 | 64 | miR-323a-3p† | 5.53E-08 | 4 | miR-654-3p† | 3.36E-05 | 4 | miR-1275 | 3.39E-03 | 3 |
| miR-516b-5p* | 4.19E-15 | 905 | miR-193b-3p | 5.04E-10 | 7 | miR-487a† | 6.13E-08 | 4 | miR-450a-5p | 4.46E-05 | 4 | miR-3622b-5p | 3.42E-03 | 3 |
| miR-518c-5p* | 4.31E-15 | 172 | miR-377-3p† | 5.29E-10 | 23 | miR-493-5p# | 9.99E-08 | 7 | miR-149-3p | 5.02E-05 | 3 | miR-662 | 3.52E-03 | 3 |
| miR-520a-5p* | 5.82E-15 | 121 | miR-335-5p | 5.42E-10 | 12 | miR-518c-3p* | 1.67E-07 | 20 | miR-372 | 5.21E-05 | 4 | miR-514b-5p | 3.71E-03 | 2 |
| miR-522-3p* | 6.4E-15 | 64 | miR-518f-5p* | 5.46E-10 | 16 | miR-100-5p | 1.98E-07 | 24 | miR-520a-3p* | 5.7E-05 | 4 | miR-1226-5p | 3.72E-03 | 3 |
| miR-518a-3p* | 7.44E-15 | 90 | miR-934 | 1.06E-09 | 2 | miR-551b-3p | 2.49E-07 | 11 | miR-371a-5p | 7.71E-05 | 11 | miR-520f* | 3.73E-03 | 3 |
| miR-519d* | 9.01E-15 | 291 | miR-99b-5p | 1.51E-09 | 21 | miR-758† | 6.26E-07 | 3 | miR-99b-3p | 9.04E-05 | 3 | miR-422a | 4.86E-03 | 3 |
| miR-518b* | 1.22E-14 | 194 | miR-519c-3p* | 1.88E-09 | 39 | miR-379-5p† | 8.23E-07 | 4 | miR-3194-5p | 9.24E-05 | 7 | miR-10b-3p | 5.19E-03 | 5 |
| miR-1323* | 1.63E-14 | 435 | miR-518f-3p* | 2.14E-09 | 10 | miR-433# | 8.44E-07 | 2 | miR-543† | 1.03E-04 | 3 | miR-3132 | 5.29E-03 | 2 |
| miR-516a-5p* | 2.37E-14 | 109 | miR-299-5p† | 3.03E-09 | 7 | miR-542-3p | 9.22E-07 | 7 | miR-3196 | 1.19E-04 | 6 | miR-340-5p | 6.73E-03 | 2 |
| miR-520g* | 4.46E-14 | 53 | miR-524-3p* | 3.09E-09 | 10 | miR-136-3p# | 1.28E-06 | 6 | miR-3654 | 1.97E-04 | 2 | miR-3934 | 6.84E-03 | 2 |
| miR-410† | 5.11E-14 | 19 | miR-30a-3p | 3.31E-09 | 5 | miR-455-5p | 1.54E-06 | 4 | miR-376b† | 2.74E-04 | 3 | miR-4322 | 8.11E-03 | 3 |
| miR-517c-3p* | 1.72E-13 | 111 | miR-370 | 4.11E-09 | 5 | miR-3659 | 1.97E-06 | 6 | miR-452-5p | 2.88E-04 | 2 | miR-3610 | 8.19E-03 | 3 |
| miR-127-3p# | 2.01E-13 | 23 | miR-4287 | 4.27E-09 | 12 | miR-411-5p† | 2.63E-06 | 4 | miR-3911 | 3.61E-04 | 4 | miR-718 | 8.64E-03 | 3 |
| miR-431-5p# | 2.09E-13 | 16 | miR-483-3p | 4.7E-09 | 10 | miR-337-5p# | 3.13E-06 | 7 | miR-3682-3p | 3.76E-04 | 3 | miR-548q | 1.16E-02 | 3 |
| miR-376a-3p† | 6.64E-13 | 42 | miR-523-3p* | 5.51E-09 | 12 | miR-21-5p | 3.34E-06 | 3 | miR-135a-3p | 4.02E-04 | 2 | miR-1225-5p | 1.55E-02 | 5 |
| miR-515-3p* | 1.7E-12 | 141 | miR-424-3p | 8.78E-09 | 3 | miR-1973 | 4.24E-06 | 6 | miR-31-5p | 4.08E-04 | 3 | miR-134† | 3.19E-02 | 3 |
| miR-495† | 1.76E-12 | 21 | miR-382-5p† | 1.01E-08 | 4 | miR-27a-3p | 4.31E-06 | 12 | miR-1471 | 4.32E-04 | 3 | miR-125a-3p* | 3.65E-02 | 2 |
| miR-498* | 2.88E-12 | 43 | miR-152 | 2.03E-08 | 3 | miR-92b-5p | 4.53E-06 | 6 | miR-221-3p | 4.54E-04 | 4 | miR-638 | 4.28E-02 | 4 |
| miR-512-5p* | 1.46E-11 | 64 | miR-432-5p# | 2.07E-08 | 5 | miR-24-3p | 6.76E-06 | 7 | miR-4314 | 5.26E-04 | 4 | miR-483-5p | 4.89E-02 | 2 |
#, members of the mir-127 cluster; †, members of the mir-379 cluster; *, members of the mir-498 cluster
Table 3.
miRNAs decreased in the placenta compared with the blood
| Systematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold | Systematic name | P value | Fold |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-486-5p | 7.22E-17 | 2600 | miR-664-3p | 6.56E-09 | 11 | miR-29b-1-5p | 1.35E-06 | 3 | miR-1238 | 5.87E-05 | 5 | miR-933 | 6.42E-04 | 3 |
| hsa-let-7i-5p | 1.02E-12 | 84 | miR-320c | 1.21E-08 | 4 | miR-19b-3p | 1.36E-06 | 6 | miR-425-3p | 6.03E-05 | 4 | miR-195-5p | 6.62E-04 | 4 |
| hsa-let-7d-5p | 1.12E-12 | 93 | miR-320a | 1.4E-08 | 4 | miR-18b-5p | 1.42E-06 | 4 | miR-18a-5p | 7.24E-05 | 6 | miR-30c-5p | 7.53E-04 | 3 |
| miR-150-5p | 1.16E-12 | 684 | miR-296-5p | 1.5E-08 | 5 | miR-331-3p | 1.72E-06 | 12 | miR-32-3p | 7.6E-05 | 3 | miR-671-5p | 7.72E-04 | 4 |
| miR-144-5p | 1.16E-12 | 57 | miR-15a-5p | 2.1E-08 | 22 | miR-532-3p | 2.37E-06 | 35 | miR-584-5p | 8.67E-05 | 3 | miR-132-5p | 8.01E-04 | 2 |
| miR-16-5p | 3.31E-12 | 38 | miR-107 | 2.25E-08 | 11 | miR-19a-3p | 2.84E-06 | 12 | miR-625-3p | 9.49E-05 | 3 | miR-326 | 8.51E-04 | 3 |
| miR-92a-3p | 3.58E-12 | 58 | miR-4323 | 3.68E-08 | 4 | miR-3171 | 3.11E-06 | 3 | miR-3923 | 9.63E-05 | 2 | miR-196b-5p | 9.04E-04 | 2 |
| miR-16-2-3p | 8.59E-12 | 6 | miR-183-5p | 5.29E-08 | 15 | miR-3613-5p | 3.22E-06 | 4 | miR-19b-2-5p | 1.01E-04 | 3 | miR-1285-3p | 9.45E-04 | 3 |
| miR-4306 | 1.4E-11 | 77 | miR-20b-5p | 5.86E-08 | 6 | miR-550a-3p | 3.51E-06 | 25 | miR-1825 | 1.29E-04 | 5 | miR-181d | 1.08E-03 | 3 |
| hsa-let-7g-5p | 1.48E-11 | 71 | miR-501-5p | 7.37E-08 | 7 | miR-3647-3p | 3.92E-06 | 4 | miR-502-5p | 1.40E-04 | 2 | miR-18b-3p | 1.26E-03 | 2 |
| miR-197-3p | 1.55E-11 | 18 | miR-17-5p | 1.24E-07 | 5 | miR-190b | 4.12E-06 | 3 | miR-3115 | 1.45E-04 | 2 | miR-140-5p | 1.36E-03 | 2 |
| miR-423-5p | 1.93E-11 | 20 | miR-3924 | 1.61E-07 | 3 | miR-302a-3p | 4.38E-06 | 3 | miR-500a-5p | 1.45E-04 | 5 | miR-3145-3p | 1.60E-03 | 2 |
| miR-25-3p | 2.89E-11 | 76 | miR-142-3p | 1.86E-07 | 17 | miR-1294 | 4.44E-06 | 2 | miR-1277-3p | 1.56E-04 | 2 | miR-193a-5p | 2.18E-03 | 3 |
| miR-186-5p | 4.6E-11 | 15 | miR-320b | 1.91E-07 | 4 | miR-4307 | 4.8E-06 | 4 | miR-1282 | 1.57E-04 | 2 | miR-3679-3p | 2.47E-03 | 2 |
| miR-484 | 6.25E-11 | 33 | miR-574-3p | 2.01E-07 | 5 | miR-548b-5p | 6.06E-06 | 4 | miR-4318 | 1.69E-04 | 5 | miR-26a-2-3p | 2.77E-03 | 2 |
| miR-342-5p | 9.7E-11 | 12 | miR-192-5p | 2.08E-07 | 22 | miR-3676-3p | 7.24E-06 | 5 | miR-625-5p | 1.93E-04 | 8 | miR-1249 | 3.18E-03 | 2 |
| hsa-let-7c* | 1.75E-10 | 32 | miR-106b-5p | 2.24E-07 | 7 | miR-3129-5p | 7.94E-06 | 3 | miR-128 | 1.97E-04 | 12 | miR-1234 | 3.30E-03 | 3 |
| miR-142-5p | 2.48E-10 | 9 | miR-29a-5p | 2.43E-07 | 4 | miR-135a-5p | 9.11E-06 | 3 | miR-548am-5p | 2.25E-04 | 4 | miR-342-3p | 4.08E-03 | 4 |
| miR-98 | 2.9E-10 | 29 | miR-942 | 2.85E-07 | 6 | miR-1244 | 9.28E-06 | 3 | miR-146a-5p | 2.54E-04 | 4 | miR-2110 | 4.51E-03 | 2 |
| miR-223-3p | 3.3E-10 | 54 | miR-20a-5p | 3.24E-07 | 4 | miR-548d-5p | 1E-05 | 4 | miR-30e-5p | 2.67E-04 | 3 | miR-374b-5p | 6.01E-03 | 4 |
| miR-185-5p | 3.64E-10 | 74 | miR-302b-3p | 3.33E-07 | 2 | miR-30c-1-3p | 1.03E-05 | 2 | hsa-let-7f-1-3p | 2.70E-04 | 2 | miR-151a-3p | 6.27E-03 | 4 |
| hsa-let-7b-5p | 5.46E-10 | 55 | miR-194-5p | 3.75E-07 | 20 | miR-223-5p | 1.48E-05 | 2 | miR-1228-3p | 2.77E-04 | 4 | miR-634 | 8.51E-03 | 2 |
| miR-140-3p | 5.57E-10 | 8 | miR-190a | 5E-07 | 4 | miR-4313 | 1.79E-05 | 4 | miR-501-3p | 3.99E-04 | 3 | miR-3138 | 8.74E-03 | 2 |
| miR-766-3p | 9.08E-10 | 8 | miR-548t-5p | 7.27E-07 | 4 | miR-3149 | 2.04E-05 | 2 | miR-23c | 4.28E-04 | 3 | miR-210 | 1.77E-02 | 4 |
| miR-454-3p | 1.19E-09 | 5 | miR-324-3p | 7.48E-07 | 8 | miR-924 | 2.53E-05 | 2 | miR-122-5p | 4.52E-04 | 2 | miR-1281 | 2.16E-02 | 3 |
| miR-320e | 1.52E-09 | 25 | miR-550a-5p | 7.8E-07 | 4 | miR-191-3p | 3.08E-05 | 5 | miR-3180-5p | 4.79E-04 | 3 | miR-130b-3p | 2.39E-02 | 3 |
| miR-320d | 1.89E-09 | 4 | miR-548m | 1.31E-06 | 4 | miR-215 | 3.82E-05 | 14 | miR-4310 | 5.14E-04 | 2 | miR-595 | 2.70E-02 | 2 |
| hsa-let-7f-5p | 3.07E-09 | 18 | miR-96-5p | 1.32E-06 | 10 | miR-1265 | 4.85E-05 | 2 | miR-1237 | 5.27E-04 | 4 | miR-3653 | 3.69E-02 | 3 |
| miR-7-5p | 4.41E-09 | 9 | miR-182-5p | 1.33E-06 | 16 | miR-423-3p | 5.42E-05 | 7 | miR-1225-3p | 5.77E-04 | 4 | miR-3651 | 4.50E-02 | 3 |
| hsa-let-7a-5p | 5.79E-09 | 7 | miR-553 | 1.34E-06 | 3 | miR-93-3p | 5.8E-05 | 8 | miR-302c-3p | 6.37E-04 | 3 |
*, miRNA located on has 21
Fig. 1.
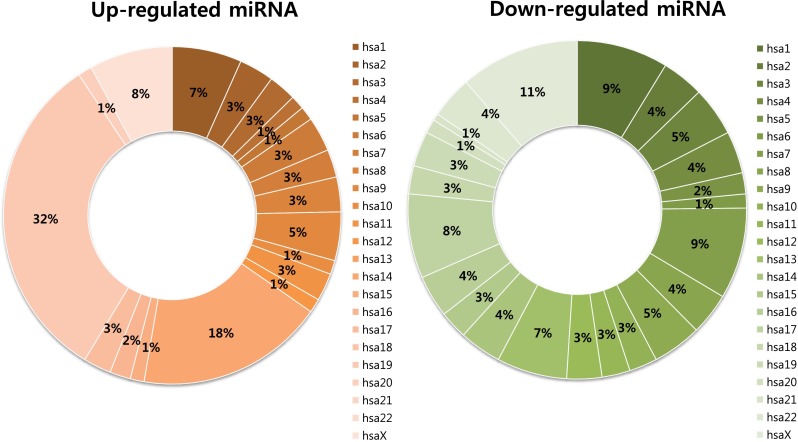
Chromosomal distribution of miRNAs differentially expressed in placenta
Identification of placental miRNAs for NIPT of fetal T21
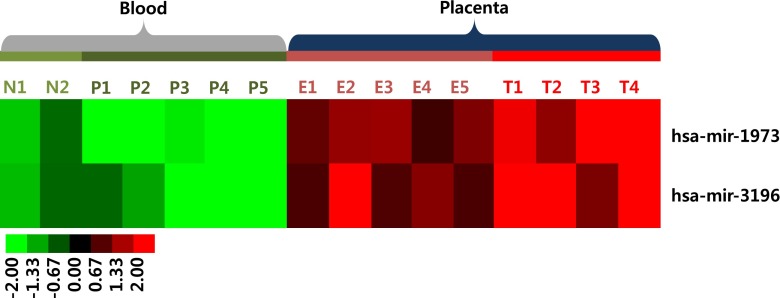
The cutoff value for miRNAs for differentiating the T21 placenta from the euploid placenta was set at 100 % specificity. Among 150 placental up-regulated miRNAs, mir-1973 and mir-3196 were significantly increased in the T21 placenta as compared to the euploid placenta (P < 0.05, Fig. 2). mir-1973 was located on hsa4 (117220881-117220924 [+]) and mir-3196 was located on hsa20 (61870131-61870194 [+]). The two miRNAs did not belong to any miRNA clusters. Both mir-1973 and mir-3196 had sensitivity of 75 %, PPV of 100 %, NPV of 83.3 %, and AUC of 0.900 (95 % CI: 0.732–0.998) with a SE of 0.068 (P < 0.05). Therefore, these two miRNAs were selected as potential biomarkers for NIPT of fetal T21.
Fig. 2.
Expression pattern of miRNA candidates for noninvasive prenatal testing of fetal trisomy 21. miRNAs were clustered using the Pearson uncentered distance metric with average linkage. Each column represents an individual sample and each row represents an individual miRNA. Expression levels of miRNAs are shown in red (up-regulated) and green (down-regulated), with brighter shades indicating higher fold differences. N: non-pregnant, P: pregnant, E: euploid fetus, T: trisomy 21 fetus
Exploratory in silico pathway analysis of placental miRNA for NIPT of fetal T21
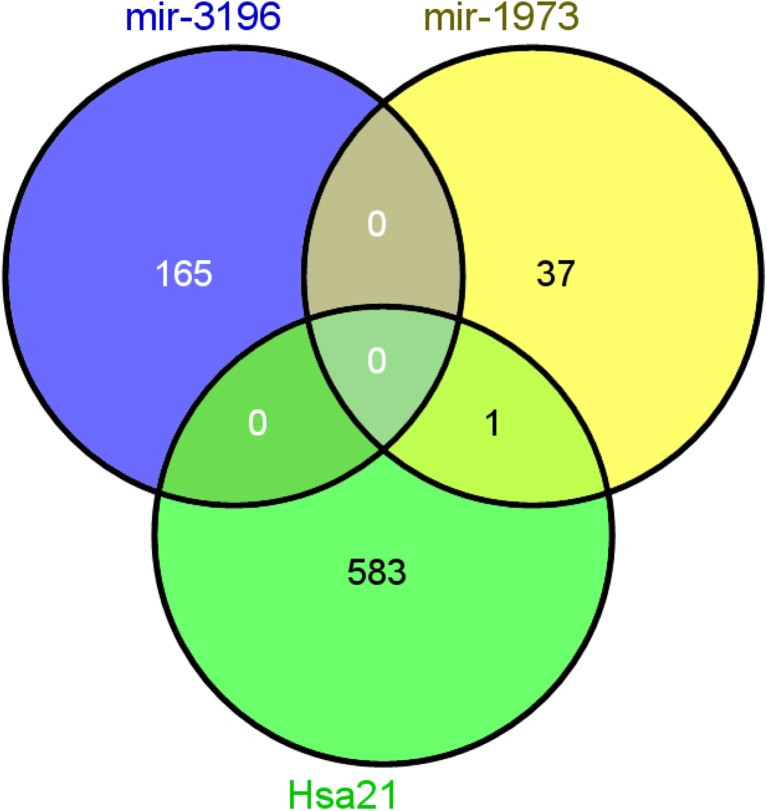
We used six established target prediction tools to obtain the list of Entrez genes predicted to be targeted by mir-1973 and mir-3196 identified as potential biomarkers for NIPT of fetal T21. Target genes of the two miRNAs were simultaneously identified by at least three prediction programs. Genes predicted to be targeted by mir-1973 and mir-3196 were 38 and 165, respectively. There was no gene targeted by both miRNAs (Fig. 3). Among the target genes, the KCNJ15 gene encoding the potassium inwardly-rectifying channel, subfamily J, member 15 was located on hsa21; the gene is predicted to be regulated by mir-1973 (Fig. 3).
Fig. 3.
Hsa21-derived target genes of mir-1973 and mir-3196
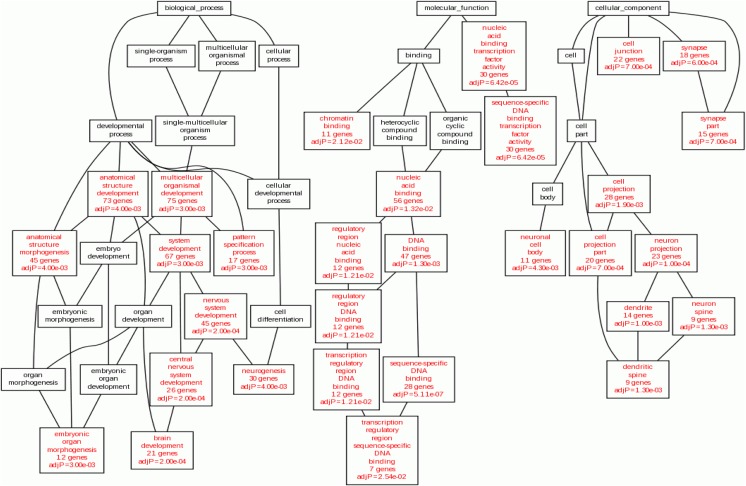
We performed GO analysis and disease association analysis of total target genes of the two miRNAs. Target genes of miRNAs were analyzed in categories of biological process (BP), cellular component (CC), and molecular function (MF) by GO analysis (Fig. 4). In the BP category, the most statistically significant associations with target genes were found in brain development, central nervous system development, and nervous system development (adjP = 0.0002 in all, Fig. 4). Sequence-specific DNA binding transcription factor activity (adjP = 5.11e-07) in the MF category and neuron projection (adjP = 0.0001) in the CC category were the most significantly associated with the target genes (Fig. 4). Disease association of target genes is shown in Table 4, and the most statistically significant association with target genes was found in congenital abnormalities (adjP = 6.38e-08). More than 10 target genes were significantly associated with various disorders such as mental disorders, schizophrenia, Nelson syndrome, nervous system diseases, and human immunodeficiency virus.
Fig. 4.
Gene ontology (GO) analysis of target genes of mir-1973 and mir-3196. Significant processes in GO term are shown in red
Table 4.
Diseases association with target genes of mir-1973 and mir-3196
| Disease | Gene symbol | rawP | adjP |
|---|---|---|---|
| Congenital Abnormalities | GDAP1, GLI3, ALX4, ADAMTS10, TBX1, BBS5, CYP4F22, COL5A1, SOX3, PAX2, NR5A1, KCNJ15, KAL1, MAFB, HOXD13, DDB1, ZIC1 | 9.12E-09 | 6.38E-08 |
| Mental Disorders | SYNGAP1, GABRP, PER1, SNCB, VGF, GRIN1, ABCA2, GRIN2D, CPLX1, NALCN, DLGAP3, NEUROD2, NRGN, NEUROG1, SLITRK1 | 6.5E-08 | 2.27E-07 |
| Schizophrenia | GABRP, TBX1, GRIN1, NRG2, GRIN2D, CPLX1, NALCN, NRGN, MBNL2, SPTBN4, NEUROG1 | 1.02E-06 | 2.38E-06 |
| Syndrome | GLI3, ALX4, WIPF3, ADAMTS10, TBX1, BBS5, SLC29A3, COL5A1, PAX2, KAL1, MEN1, SLITRK1 | 5.60E-05 | 9.80E-05 |
| Nelson syndrome | CDK5R2, C2orf16, WIPF3, BBS5, ABCF2, TMEM163, OTX1, BZW2, ZMIZ2, CTNNA2, REPIN1 | 3.00E-04 | 4.00E-04 |
| Nervous System Diseases | GDAP1, DTNA, SNCB, SOX3, LRRK2, ABCA2, ARC, SLITRK1, ATP1A3, ZIC1 | 1.50E-03 | 1.80E-03 |
| Human immunodeficiency virus | EIF4G1, IL8, PIK3R2, GRIN1, B3GALT2, MAN1C1, GRIN2D, DDB1, KPNB1, TUBA4A | 2.70E-03 | 2.70E-03 |
rawP: p value from hypergeometric test
adjP: p value adjusted by the multiple test adjustment (Benjamini and Hochberg test)
An interactive network of genes targeted by mir-1973 and mir-3196
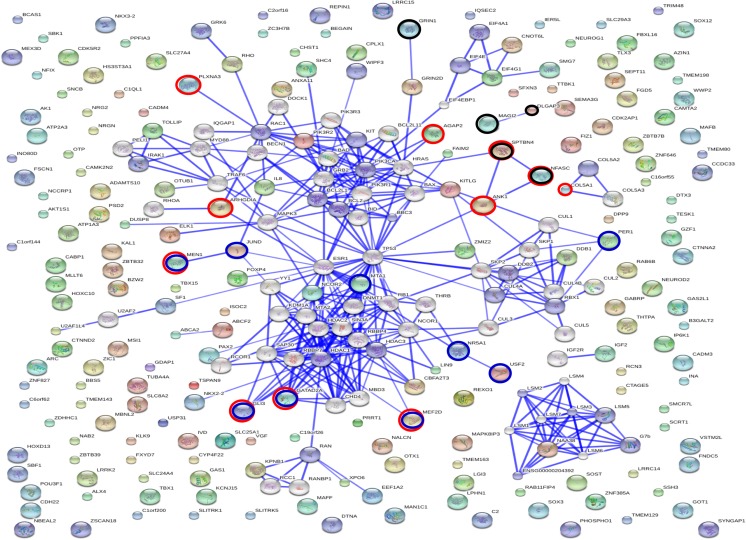
Based on total 203 genes targeted by mir-1973 and mir-3196, we constructed an interactive signaling network of target genes (Fig. 5). The biological interaction network has biological significance beyond statistical data, suggesting that 72 of the target genes identified in this study were an integral part of the dynamic complex of signaling under a high confidence score of 0.7. These genes are involved in “neuron projection” (GOTERM_CC P = 0.0001, black circle), “nervous system development” (GOTERM_BP P = 0.0002; red circle), and “sequence-specific DNA binding” (GOTERM_MF P = 5.11e-07, blue circle) (Fig. 5).
Fig. 5.
Interaction networks of target genes of mir-1973 and mir-3196. The list of the identified target genes was subjected to STRING (v. 9.05) analysis to reveal functional interactions. Each node represents a protein, and each edge represents an interaction. Black, red, and blue circles show genes that are involved in neuron projection, nervous system development, and sequence-specific DNA binding, respectively
Discussion
In this study, we profiled miRNAs from the whole blood of non-pregnant and pregnant women and the placentas of normal and T21 fetuses. We found that 299 miRNAs were significantly differentially expressed between blood and placenta. Among the identified miRNAs, most miRNAs increased specific to the placenta were members of mir-498 clusters, mir-379 clusters, and mir-127 clusters that are known to be placenta-related miRNA clusters. In particular, we found the expression pattern of the two miRNAs (mir-1973 and mir-3196) in a list of candidate miRNAs for NIPT was significantly different in the placenta according to the presence or absence of T21, and both miRNAs were up-regulated in the placentas of T21 fetuses. Therefore, we suggest that the two miRNAs might serve as potential miRNA-based biomarkers for NIPT of fetal T21. Target genes of the two miRNAs were significantly associated with biological processes such as brain development, central nervous system development, and nervous system development. Moreover, they were associated with T21-related disorders such as congenital abnormalities, mental disorders, and nervous system diseases. These results suggest that T21-specific placental miRNAs might affect important mechanisms of neurological dysfunction and various complications associated with T21.
One of the most interesting features of circulating miRNAs is their high stability, which enables them to resist enzymatic degradation, freeze-thaw cycles and extreme pH conditions [17, 18]. Due to these characteristics, miRNAs have been proposed to be potential blood-based biomarkers for the noninvasive detection of tumors [17]. Similarly, circulating placental miRNAs have been investigated as biomarkers for NIPT. Recent studies have demonstrated a placenta-specific miRNA profile including numerous miRNAs within the miR-379 cluster on hsa14 and the miR-498 and miR-127 clusters on hsa19 [27]. These miRNAs originating in the trophoblast of the placenta are released into the maternal circulation via exosomes [15, 16]. Therefore, they enable noninvasive detection in maternal plasma [11–14]. Up to date, various placenta-specific miRNAs such as circulating biomarkers for NIPT have been selected based on tissue-specific characteristics of miRNAs that are differentially expressed in blood and placenta. However, miRNAs are differentially expressed according to tissue type and presence and absence of disease and pregnancy [12, 27, 38 ~]. Therefore, pregnancy and T21-specific characteristics of miRNAs should be considered in the development of miRNAs for NIPT of fetal T21. Therefore, non-pregnant women, pregnant women carrying euploid fetus, and pregnant women carrying T21 fetus were selected for this study and whole blood samples from non-pregnant women, whole blood samples from pregnant women, euploid fetal placenta samples, and T21 fetal placenta samples were obtained. In this study, we identified placenta-specific miRNAs considering all the tissue-specific characteristics of blood and placenta as well as the pregnancy and T21-specific characteristics. Therefore, we suggest that the placenta-specific miRNAs identified in this study might be useful as noninvasive biomarkers in the detection of fetal T21.
T21, referred to as Down syndrome, is the most frequent survivable congenital chromosomal abnormality. Many studies of miRNAs in T21 have been reported [6–8, 22–24], with most studies focused the correlation of hsa21-derived miRNAs with T21 phenotypes. Recently, Kotlabova et al. reported hsa21-derived miRNAs in plasma samples from pregnancies bearing euploid and T21 fetuses [24]. They investigated expression changes of hsa21-derived miRNAs in maternal plasma and demonstrated that the concentrations and relative gene expression levels of hsa21-derived miRNAs did not differ between the two groups. Therefore, they suggested that analysis of hsa21-derived miRNAs had no benefit for screening programs and NIPT of T21. In our study, hsa21-derived miRNAs did not increase in the placenta as compared with the blood. Therefore, our results support the results previously reported by Kotlabova et al. [24]. These findings indicate that hsa21-derived miRNAs in maternal blood could originate from both the maternal blood and the placenta. Therefore, we also suggested that hsa21-derived miRNAs might not be appropriate for NIPT of fetal T21 using miRNAs in maternal blood, as previously suggested by Kotlabova et al. [24].
miRNAs, as key post-transcriptional regulators of gene expression, are processed from precursor molecules, which are either transcribed from independent miRNA genes or are portions of introns of protein coding RNA polymerase II transcripts. Subsequently, mature miRNAs recognize their target mRNAs by base pairing interactions between nucleotides 2 and 8 of the miRNA and complementary nucleotides in the 3′-UTR of mRNAs and inhibit gene expression by targeting mRNAs for translational repression or destabilization [2, 3]. These miRNAs have the distinctive gene expression regulation activity as follows: A miRNA can potentially regulate a large number of protein-coding genes, while multiple miRNAs can regulate a single gene. Therefore, analysis of the genome-wide expression of miRNAs is indispensable to understanding their molecular regulating mechanisms. Recently, genome-wide miRNAs expression profiles of the T21 fetal cord blood mononuclear cells were reported, and found six up-regulated and 143 down-regulated miRNAs to be differentially expressed in T21 [8]. Among them, four hsa21-derived miRNAs including mir-99a, let-7c, mir-125b-2, and mir-155 were down-regulated in T21, while mir-802, mir-3648, and mir-3687 were up-regulated in T21. However, these miRNAs may be difficult to use as biomarkers for NIPT of fetal T21 because expression levels of hsa21-derived miRNAs do not differ in plasma samples from pregnancies bearing euploid and T21 fetuses [24]. Moreover, changes in hsa21-derived miRNAs between maternal blood and cord blood have not yet been fully investigated. In this study, we investigated genome-wide placental miRNAs as potential biomarkers for NIPT of fetal T21 and found mir-1973 and mir-3196 with disease-specific characteristics. These miRNAs were identified based on characteristics of miRNAs according to tissue type and presence and absence of disease. Therefore, our findings suggest that these miRNAs may be useful as potential biomarkers for the non-invasive detection of fetal T21. In particular, the target genes of the identified miRNAs were significantly associated with biological processes and pathogenesis of neurological abnormalities of T21, and these results were confirmed by interactive network construction and in silico pathway analysis. Hence, our findings suggest that aberrant expression of these miRNAs may be involved in various complications such as congenital abnormalities, mental disorders, and nervous system diseases observed in T21 patients. However, a lot of our results are based on databases of bioinformatics tools. Therefore, there are limitations as follows. First, some bias exists in bioinformatics tools used to identify the function of any given miRNA. Second, each miRNA has multiple functions, so some database may categorize a miRNA as having function A, while another database may assign it function B. Moreover, this study was limited by its small sample size and the inclusion of only Korean patients and was not supported by studying expression variation on more placentas. Therefore, further study of expression differences for the target miRNAs in a larger scale study within different ethnic populations will need to clarify the findings. Additionally, further studies for NIPT using fetal miRNA will need to consider genotypes of fetal miRNAs, maternal miRNAs, and paternal miRNAs, because familial genotype analysis could prove that increased miRNAs in maternal blood are from the fetus.
In conclusion, in this study, we identified a set of 299 differentially expressed miRNAs between placenta and blood. Among them, increased placenta-specific miRNAs may be potential markers for NIPT. Notably, we found two miRNAs to be biomarkers for NIPT of fetal T21, which clearly distinguished T21 placentas from euploid placenta. Furthermore, predicted target genes of the two miRNAs were associated with biological processes and complications of neuronal degeneration associated with T21. Our findings suggest that miRNA profiling, considering all the characteristics of tissue type and disease, may be a useful strategy in the development of NIPT of fetal T21. Together, our data provided novel information that might contribute to a better understanding of the molecular mechanisms and biological pathways implicated in T21.
Acknowledgments
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A111550). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank the following physicians and staff who took the time and effort to participate in this study: Joung Yeol Han, Jin Hoon Chung, Dong Wook Kwak, Jin Woo Kim, Bom Yi Lee, Ju Yeon Park, Eun Young Choi, Yeon Woo Lee, Ah Rum Oh, Shin Yeong Lee, and So Min Seo.
Conflict of interest
Authors’ conflict of interest disclosure: The authors stated that there are no conflicts of interest regarding the publication of this article. Research support played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Footnotes
Capsule Placenta-specific miRNAs are identified that may be biomarkers for the noninvasive prenatal diagnosis for trisomy 21.
Contributor Information
Hyun Mee Ryu, Phone: 82-2-2000-7175, Email: hmryu@yahoo.com.
So Yeon Park, Phone: 82-2-2000-7175, Email: paranip@yahoo.co.kr.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Szulwach KE, Jin P, Alisch RS. Noncoding RNAs in mental retardation. Clin Genet. 2009;75:209–19. doi: 10.1111/j.1399-0004.2008.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Shen Z, Xu Q, Huang X, Chen Q, Li D. Increased levels of microRNA-424 are associated with the pathogenesis of fetal growth restriction. Placenta. 2013;34:624–7. doi: 10.1016/j.placenta.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Siew WH, Tan KL, Babaei MA, Cheah PS, Ling KH. MicroRNAs and intellectual disability (ID) in Down syndrome, X-linked ID, and Fragile X syndrome. Front Cell Neurosci. 2013;7:41. doi: 10.3389/fncel.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Li W, Liu X, Chen H, Tan K, Chen Y, et al. Identification of dysregulated microRNAs in lymphocytes from children with Down syndrome. Gene. 2013;530:278–86. doi: 10.1016/j.gene.2013.07.055. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Li W, Liu X, Ma H, Tu Z, Dai Y. Analysis of microRNA expression profile by small RNA sequencing in Down syndrome fetuses. Int J Mol Med. 2013;32:1115–25. doi: 10.3892/ijmm.2013.1499. [DOI] [PubMed] [Google Scholar]
- 9.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–87. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Kasinski AL, Adams BD, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 12.Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56:1767–71. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 13.Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation – identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J Reprod Immunol. 2011;89:185–91. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci U S A. 2013;110:4255–60. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–29. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 16.Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–24. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham FG, Leveno KJ, Bloom SL, Hauth JC, Rouse DJ, Spong CY. Williams Obstetrics. 23rd ed. McGraw-Hill’s Medical; 2010. pp. 268.
- 20.Mégarbané A, Ravel A, Mircher C, Sturtz F, Grattau Y, Rethoré MO, et al. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med. 2009;11:611–6. doi: 10.1097/GIM.0b013e3181b2e34c. [DOI] [PubMed] [Google Scholar]
- 21.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 22.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elton TS, Sansom SE, Martin MM. Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins. RNA Biol. 2010;7:540–7. doi: 10.4161/rna.7.5.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotlabova K, Doucha J, Chudoba D, Calda P, Dlouha K, Hromadnikova I. Extracellular chromosome 21-derived microRNAs in euploid & aneuploid pregnancies. Indian J Med Res. 2013;138:935–43. [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 26.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–9. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97:51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Barch MJ, Knutsen T, Spurbeck JL. The AGT cytogenetics laboratory manual. 3. New York: Lippincott-Raven; 1997. [Google Scholar]
- 29.R Development Core Team, 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing (http://www.r-project.org. Access April 10, 2013).
- 30.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–6. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 35.Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25:2625–31. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 36.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel D. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveros JC. 2007. VENNY. An interactive tool for comparing lists with Venn Diagrams. Http://bioinfogp.cnb.csic.es/tools/venny/index.html (Access November 20, 2013).
- 38.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]