Abstract
Background
Tumour relapse is recognized to be the prime fatal burden in patients affected by head and neck squamous cell carcinoma (HNSCC), but no discrete molecular trait has yet been identified to make reliable early predictions of tumour recurrence. Expression of cell surface proteoglycans (PGs) is frequently altered in carcinomas and several of them are gradually emerging as key prognostic factors.
Methods
A PG expression analysis at both mRNA and protein level, was pursued on primary lesions derived from 173 HNSCC patients from whom full clinical history and 2 years post-surgical follow-up was accessible. Gene and protein expression data were correlated with clinical traits and previously proposed tumour relapse markers to stratify high-risk patient subgroups.
Results
HNSCC lesions were indeed found to exhibit a widely aberrant PG expression pattern characterized by a variable expression of all PGs and a characteristic de novo transcription/translation of GPC2, GPC5 and NG2/CSPG4 respectively in 36%, 72% and 71% on 119 cases. Importantly, expression of NG2/CSPG4, on neoplastic cells and in the intralesional stroma (Hazard Ratio [HR], 6.76, p = 0.017) was strongly associated with loco-regional relapse, whereas stromal enrichment of SDC2 (HR, 7.652, p = 0.007) was independently tied to lymphnodal infiltration and disease-related death. Conversely, down-regulated SDC1 transcript (HR, 0.232, p = 0.013) uniquely correlated with formation of distant metastases. Altered expression of PGs significantly correlated with the above disease outcomes when either considered alone or in association with well-established predictors of poor prognosis (i.e. T classification, previous occurrence of precancerous lesions and lymphnodal metastasis). Combined alteration of all three PGs was found to be a reliable predictor of shorter survival.
Conclusions
An unprecedented PG-based prognostic portrait is unveiled that incisively diversifies disease course in HNSCC patients beyond the currently known clinical and molecular biomarkers.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1336-4) contains supplementary material, which is available to authorized users.
Keywords: Proteoglycans, Squamous cell carcinoma, Biomarker, NG2/CSPG4, Tumour relapse
Background
Head and neck squamous cell carcinomas (HNSCC) have an estimated frequency of 38,160 new cases in the US (updated to August, 2014) [1] and an estimated occurrence of more than 442,000 new cases worldwide according to GLOBOCAN 2012 [2,3], thereby representing the primary lethal cancer entity in patients with head and neck tumours. Loco-regional relapsing is the most severe clinical problem encountered in these tumours, while the pre-operative presence of lymphnodal infiltration is a recognized prognostic factor [4,5]. Especially in patients presenting smaller primary lesions, occult secondary lesions in lymphnodes significantly complicate the clinical management of these individuals [6-13]. The currently adopted methods to predict disease recurrence, such as staging and grading, are too arbitrary and do not allow for a sufficiently accurate clinical stratification of the patients [14,15]. This deficit calls upon the need to identify distinct molecular markers that more reliably would predict disease progression, recurrence and metastasis formation, and many such have been proposed over the last decade (Table 1). Thus far, however, only three such markers have been considered as meaningful, i.e. HPV infection, TP53 mutation status and overexpression of EGFR [16-20], but their full independence from clinical parameters is still dubious.
Table 1.
Previously proposed prognostic biomarkers in HNSCC 1
| Biomarker2 | Clinical outcome | Method of detection | N. of cases/%/type of modulation | Annotation |
|---|---|---|---|---|
| ADAM17 | Lymph nodal metastasis/Loco-regional relapse | IHC/WB | 50/46/Up | None |
| CD44 | OS/DFS | IHC | 138/59/Down | None |
| E-cadherin | Recurrence/OS | IHC | 50/20/Up 112/59/Down | None |
| EGFR | OS | IHC | 109/73/Up 59/58/Up | None |
| Estrogen-R2 | OS | IHC/nPCR + sequencing | 67/51/Up | Laryngeal/hypopharingeal cancer |
| FHIT | OS/DFS | IHC | 53/61/Down | None |
| GLUT1 | OS | IHC | 40/26/up | Poor radiation response |
| HIF1A | OS/DFS | IHC | 85/63/Down | None |
| Keratin-18 | OS | IHC | 308/54/Up | None |
| Keratin-8 | OS | IHC | 308/54/Up | None |
| Laminin γ2 | DSS | DNA Microarray | 119/NS/Up | None |
| MCM5 | OS | IHC | 97/61/Up | None |
| MET | OS | IHC | 69/82/Up | None |
| Moesin | OS | IHC | 103/NS/Up | Cytoplasmic expression pattern |
| Mucin-1 | OS/DFS/Lymphnodal metastasis | IHC | 206/39/Up | Within 5-years follow-up |
| Mucin-4 | OS/DFS/Lymphnodal metastasis/Loco-regional relapse | IHC | 150/41/Up | Within 5-years follow-up |
| p21 | OS | IHC | 192/71/Down | None |
| p27 | DFS | IHC | 192/80/Down | Only in patients with lymphnodal infiltration |
| p57 | OS | IHC | 67/87/Down | None |
| p63 | OS | IHC | 62/NS/Up | None |
| P-cadherin | Disease recurrence/Loco-regional relapse/OS | IHC | 50/20/Down 67/45/Down 108/16/Down | None |
| Podoplanin | DSS | IHC | 35/56/Up | None |
| Rb | DFS | IHC | 220/49/Down | Only in p53+/pRb− patients |
| RUNX3 | OS | IHC/WB | 108/46/Down | None |
| S100A2 | DFS/Cervical metastasis | RT-PCR + seq/IHC | 135/26/Down 52/NS/Down | Nuclear expression pattern |
| SPARC | OS/DFI | DNA Microarray/IHC | 62/NS/Up | None |
| STAT1 | OS | IHC | 89/NS/Up | None |
| Survivin 3α | OS | RT-PCR | 97/NS/Up | Only in lymphnodes |
| TERT | OS | IHC | 62/NS/Up | None |
| Ezrin | OS | IHC | 47/85/Up | Cytoplasmic expression pattern |
1Specifically referred to oral and oropharyngeal squamous cell carcinoma;
2Alterations of TP53, CCND1 and FGFR4 genes are not included;
Abbreviations:OS, Overall Survival; DFS, Disease Free-Survival; DSS, Disease Specific-Survival; DFI, Disease Free-Interval; IHC, Immunohistochemistry; WB, Western Blotting; nPCR, nested Polymerase Chain Reaction; NS, Not Specified.
One class of molecules with the potential of acting as clinically relevant factors in HNSCC, especially for oral cavity and oropharynx cancer, is that comprising cell surface-associated proteoglycans (PGs). In fact, changes in their relative expression are progressively being associated with neoplastic transformation, propagation of local tumour masses, and formation of distant metastases. This not only in HNSCC, but also in numerous epithelial and non-epithelial tumour types. Both PGs produced by the HNSCC cells themselves and PGs associated with the intra-lesional tumour stroma may play critical roles in the control of HNSCC growth, dissemination and therapeutic refraction, and may therefore be contemplated as putative biomarkers as well as therapeutic targets. There are currently 15 cell surface PGs known in the human genome with the most representative ones belonging to either the transmembrane syndecan group, i.e. syndecan-1-4 (SDC1-SDC4) [21-25], or the GPI-anchored glypican group, i.e. glypican-1-6 (GPC1-GPC6) [23,26-28]. The unique structural traits of cell surface PGs enable them to modulate directly and/or indirectly several facets of the tumour cell phenotype and behavior, including growth kinetics, invasiveness and metastatic ability.
Previously documented, representative examples of the implication of diverse PGs expressions for disease outcome are afforded by the recently consolidated tumour-suppressing effect of GPC5 in lung carcinomas arising in “never smokers” [29-31], as well as by the well-established prognostic/predictive up-regulation of GPC1 in pancreatic cancer [32,33]. As a corollary, GPC3 is a recognized prognostic/predictive factor and therapeutic target in hepatocellular carcinoma [34-37]. SDC1, the only PG for which there is some documentation in oral squamous cell carcinoma, seems to be associated with the differentiation status of the tumour cells [38-40]. Clinical correlation of SDC1 expression with disease status specifically refers to its modulation in epithelial neoplastic cells [41-47] and tumour stroma [48], while the PG has been proposed to influence migration and invasion of oral squamous cell carcinoma cells in vitro by interacting with the β1 integrin subunit and the laminin β1 chain [48].
NG2/CSPG4 has been proposed to impact on tumourigenesis and evidence has been accrued suggesting that NG2/CSPG4 alone is able to confer metastatic potential to cancer cells by serving as a multivalent mediator of the cancer cell-host microenvironment interactions and by enhancing drug resistance and protecting cells from stress-induced programmed cell death [49,50]. In an increasing number of tumours, prognostic implications of NG2/CSPG4 are being unveiled and these discoveries accentuate the potential of the PG as a therapeutic target. Recently, a direct link between methylation and CSPG4 expression in HNSCC HPV-negative/stage IVa subgroup were proved, where high protein expression and low promoter methylation were significantly associated with an adverse progression-free and overall survival [51].
Based upon previously accrued information about the role of PGs in cancer and the currently available experimental evidences along this line, we have addressed the possibility that the pattern of expression of individual PGs, or groups of PGs, may act as either pro- or anti-tumourigenic and thereby be predictive, or indicative, of a discrete disease course in oral cavity HNSCC disease course.
Methods
Patients
Patients from whom surgical specimens were evaluated were treated surgically at the S. Orsola-Malpighi Hospital, at the Bellaria University Hospital in Bologna and at the Maxillo-Facial Surgery Division, Department of Head and Neck Surgery of the University of Parma. A total of 173 surgical specimens of primary oral cavity HNSCC were collected after informed consent obtained from each enrolled patients, all of them in adulthood (Additional file 1: Table S1; Additional file 2: Figure S1). Patients were referred to adjuvant radiation therapeutic treatment according to the guidelines defined by the National Comprehensive Cancer Network (NCCN) Clinical Practice (Version 2.2014; www.nccn.org). Clinical data were collected within the 2 years-post surgical follow-up every 6 months (Additional file 3; Additional file 2: Figure S1). The present study has been approved by the local ethics committees (Comitato Etico Provinciale di Parma –Parma University Hospital e Comitato Etico Provinciale di Bologna-Bologna University Hospital) and was conducted in compliance with the Helsinki Declaration’s Ethical Principles for Medical Research Involving Human Subjects.
RNA extraction and qPCR
Total RNA from healthy specimens and 119 neoplastic specimens were extracted using Trizol® according to the manufacturer’s instructions and in combination with Qiagen RNAeasy Mini Kit (Qiagen). Total RNA (1 μg) was reverse-transcribed with the QuantiTect® Reverse Transcription Kit (Qiagen). Each TaqMan Low Density Array was designed for quantification of the human PGs. The assays were chosen among the TaqMan Gene Expression Assay library (Additional file 3) and the cards were run on ABI PRISM 7900 HT Fast Real-Time PCR System (Applied Biosystems Inc., Foster City, CA, USA). Changes in gene expression levels were calculated using the “relative quantification method”. Relative gene expression fold-change were expressed as Log_2(2^-ΔΔCt) and to visualize the obtained expression profiles we used heatmap graphing by EPCLUST – Expression Profile data CLUSTering and analysis software (www.bioinf.ebc.ee/EP/EP/EPCLUST/) [52]. The data presented herein have been deposited in NCBI’s Gene Expression Omnibus [53] and are accessible through GEO Series accession number GSE33788 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33788) (Additional file 3).
Tissue microarray (TMA) construction
Tissue specimens form a total of 163 patients, which were independently assured to contain representative areas of the neoplastic lesions, were selected for TMA construction according to a previously described procedure [54,55]. Cases were considered representative when at least 50% of the section was composed of neoplastic cells. For each case, the core portion of the section with the highest percentage of tumour cells was used for analysis (Additional file 3).
Immunohistochemistry
Details on the antibodies used, characteristics of control tissues and experimental procedures are reported in Additional file 3. Relative antigen expression was assessed semi-quantitatively according to the arbitrary scoring: “-” = no positive cells were detected, “+” <10% of cells were positive, ≥10% “++” <50% of cells were positive, ≥50% “+++” <90% of cells were positive, and “++++” ≥90% of cells were positive.
Statistical and bioinformatic analyses
Demographic data, presence of recognized risk factors for development of HNSCC, clinical diagnostic parameters, gene expression and protein distribution patterns for the PGs GPC1-6, SDC1-4 and NG2/CSPG4 were comparatively evaluated for their potential correlation with the following disease outcomes: loco-regional recurrence, lymphnodal metastasis, distant metastasis, disease-related deaths and probability of incurring into one or more of these clinical outcomes. Estimation of influence of each variable considered for the above disease outcomes was analyzed independently with both the Log-rank and Wilcoxon’s rank test. Survival rate was estimated using the Kaplan-Meier method from the time of surgery to the end of the follow-up. Cox’s multivariate proportional hazards regression method was used to extract a parsimonious set of independent variables. All analyses were performed using the Statgraphics Centurion XVI software (StatPoint Technologies, Inc, Virginia, USA). P values <0.05 were considered to be significant (Additional file 3).
Results
Transcriptional profiles of PGs in primary oral cavity HNSCC lesions
Analyses of the relative mRNA expression levels of the eleven prevalent cell surface-associated PGs conducted on a total of 119 primary oral cavity HNSCC lesions revealed that 3 of the PGs, including NG2/CSPG4, GPC2 and GPC5, were de novo expressed in neoplastic cells, i.e. were not detectable in the healthy control tissues, but were detectable in cancer cells. These were transcribed in 71% (NG2/CSPG4), 36% (GPC2) and 72% (GPC5) of the lesions, respectively. The remaining 8 PGs, for which transcripts were expressed at a frequency of 84% (GPC1), 86% (GPC3), 88% (GPC4), 70% (GPC6), 100% (SDC1), 94% (SDC2), 93% (SDC3) and 95% (SDC4) of the tumour cases, respectively, were found to be differently modulated. Thus, SDC2, SDC3 and SDC4 were up-regulated in 79-84% of the patients, whereas GPC4 was enhanced in 11% and GPC3 in 57% of the specimens. However, GPCs were more frequently down-regulated (GPC3, 22%; GPC4, 21%; GPC1, 24%; and GPC6 30%) than SDCs (SDC1, 8%; SDC2, 8%; SDC3, 11%; and SDC4, 7%; Figure 1a; Additional file 4: Table S2).
Figure 1.
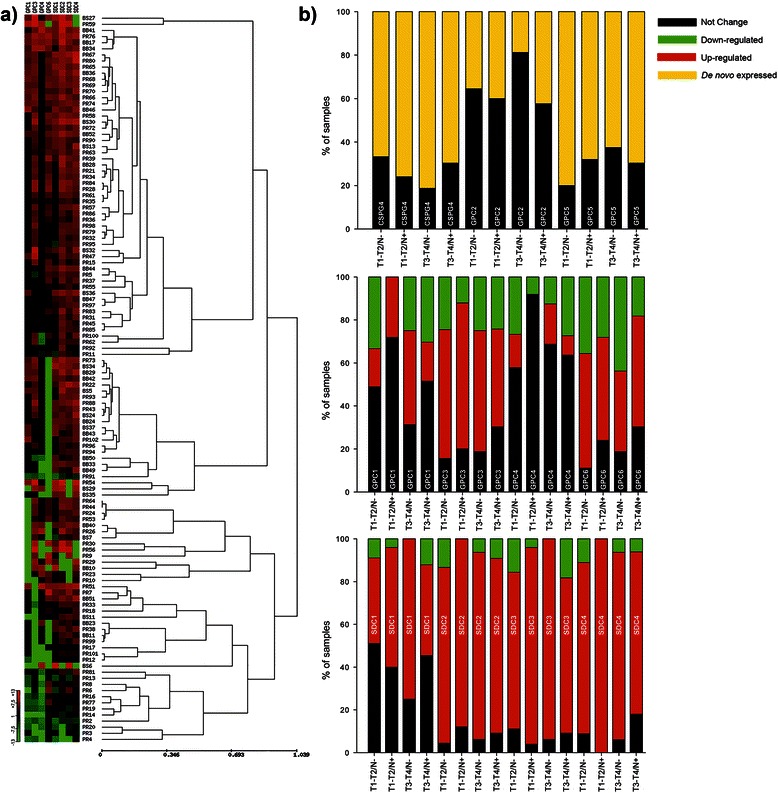
Cell surface-associated PGs are differentially expressed in primary lesions of oral cavity HNSCC patients. (a) Heat map and hierarchical clustering of the relative expression levels (columns) of the 8 most modulated cell surface-associated PGs in primary lesions of 119 oral cavity HNSCC patients (rows): red, up-regulation; green, down-regulation, black: no change in comparison to the healthy epithelial tissue. Distance between clusters was calculated as reported in Additional file 3. (b) PG expression profiles in oral cavity HNSCC specimens derived from T1-T2-N-, T1-T2-N+, T3-T4-N- and T3-T4-N+ graded tumours. SDC1-4, syndecans 1–4; GPC1-6, glypicans 1–6.
We next compared the PG expression patterns exhibited by discrete groups of patients differentiated by tumour staging, i.e. T1-T2/N- versus T1-T2/N+ and T3-T4/N- versus T3-T4/N+. GPC2 was expressed in a mere 19% of the T3-T4/N- classified lesions, whereas it was a two-fold more frequently transcribed in patients belonging to the other three classes (T1-T2/N-, 36%, T1-T2/N+, 40% and T3-T4/N+, 42%). GPC5 and NG2/CSPG4 were detectable in samples of a large proportion of patients, ranging from 62% in T3-T4/N- to 80% in T1-T2/N-, but their relative expression levels did not discriminate between the above patient subsets (Figure 1b). GPC1 was similarly differently expressed in the distinct groups of patients with a 1.5-fold higher frequency in the T3-T4/N- patients compared to the other patient subsets. GPC4 was down-regulated in about 27% of the T1-T2/N- and T3-T4/N+ subgroups, and <13% in the T1-T2/N+ and T3-T4/N- patient subgroups (Figure 1b). Syndecans were generally up-regulated in most of the lesions (Figure 1b), with SDC4 showing enhanced expression in 100% of T1-T2/N+ patients.
Immunolocalization of PGs in oral cavity HNSCC lesions
Intralesional distribution of PGs was further examined in oral cavity HNSCC lesions and control healthy tissue using empirically validated, pre-selected antibodies against each of the PGs (Figure 2; Figure 3; Additional file 5: Figure S2). The percentage of cases in which GPCs could be disclosed on the epithelial neoplastic cells varied from 18% (30 out of 163 cases) for GPC3 to 72% for GPC1 (118 out of 163 cases; Table 2). Relative frequency of expression was in the order: GPC1 > GPC4 (41%; 67 out of 163 cases) > GPC6 (37%; 61 out of 163 cases) > GPC3. GPCs were often detected within the cytoplasm as well as on the cell membrane, consistent with their thoroughly described internalization and recycling patterns. The hybrid cell membrane/cytoplasmic distribution of these PGs was characteristically observed for GPC1, GPC3 and GPC4, with GPC1 being most strongly associated with these two cellular compartments in keratinizing neoplastic cells (Additional file 5: Figure S2). GPC3 was entirely absent in healthy tissue, while GPC4 showed a widespread distribution both on normal epithelial cells and in the intralesional stromal compartment (Additional file 5: Figure S2). GPC6 appeared to be preferentially retained within intracellular vesicles (Figure 2), as deduced by the appearance of GPC6-positive granules throughout the cytoplasm of neoplastic cells. This seemed rather specific for tumour cells since it was not observed in healthy epithelial cells (Additional file 5: Figure S2). GPC1 and GPC6 were rarely seen in the intralesional stroma of oral cavity HNSCC lesions (13 out of 163 and 16 out of 163 of the cases, respectively), whereas GPC3 was consistently absent from this compartment and GPC4 showed a somewhat more frequent expression in stromal cells (19%; 31 out of 163 of the cases; Table 2; Figure 2).
Figure 2.
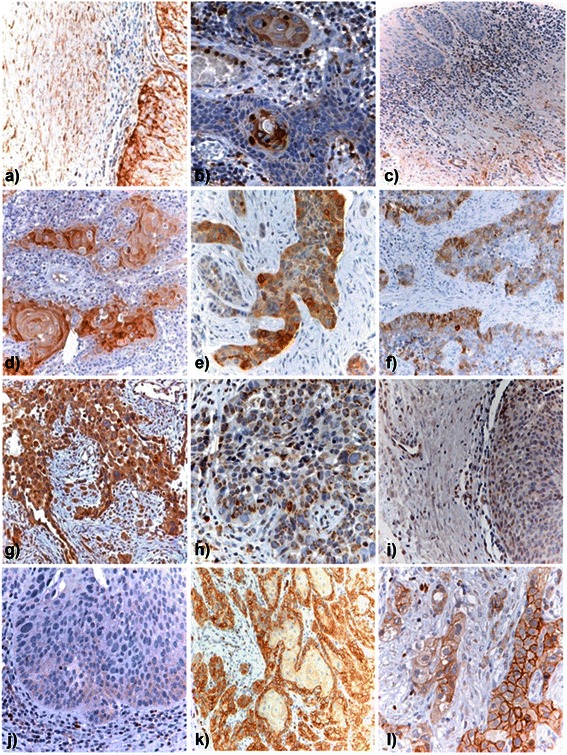
In situ immunolocalization of GPCs and NG2/CSPG4 in oral cavity HNSCC primary lesions. Representative patterns of GPC and NG2/CSPG4 distribution in oral cavity HNSCC lesions. (a, b) representative views of GPC1 expression in lesions with different degrees of keratinizing neoplastic cells. (c, d) representative images of GPC1 expression in stromal cells of pre-malignant lesions (c) and lack of expression in the stromal cells of HNSCC tissue (d). GPC3 was detected in neoplastic cells (e), but not stromal fibroblasts (f), whereas GPC4 (g) and GPC6 (h) were primarily found to be associated with the neoplastic cells. (i) Shows the lack of expression of GPC6 in the intralesional stroma. NG2/CSPG4 was found to be abundantly expressed in both well- (k) and moderately-differentiated (l) oral cavity HNSCC lesions, whereas it was similarly absent from potentially pre-malignant lesions (j).
Figure 3.
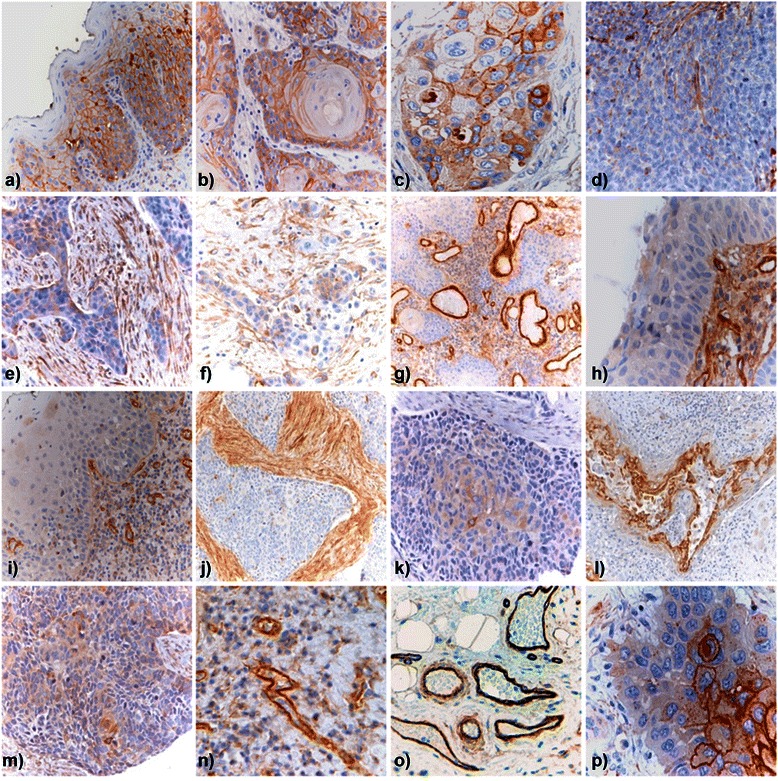
Immunodetection of SDCs in oral cavity HNSCC primary lesions. Representative view of the SDC1 expression pattern, inversely correlating with the overall differentiation status of the tumour (a, displatyc tissue; b, c, well-differentiated; d, poorly differentiated), while being particularly abundant in the center of neoplastic nests (p) and in the stromal compartment (e-f). SDC2 was seen strongly associated with tumour vessels (g, n, o) and was the only PG to be widely expressed in the different degree of dysplastic tissue (h-j). SDC3 (k) and SDC4 (m) immunolocalized in the epithelial tumour cells, but not in the stromal compartment (l, SDC3; m, SDC4).
Table 2.
Patterns of the in situ distribution of PGs in HNSCC lesions (% of cases)
| PG | Tumor cell positivity1 | Overall staining intensity2 | Subcellular localization3 | Stromal expression | ||||
|---|---|---|---|---|---|---|---|---|
| GPC1 | - | (27.5) | - | (27.6) | Membrane | - | - | (92.0) |
| + | (29.2) | + | (33.1) | Cytoplasmic | - | + | (6.7) | |
| ++ | (33.1) | ++ | (30.1) | Both | (72.4) | ++ | (0.6) | |
| +++ | (7.4) | +++ | (6.7) | +++ | (0.6) | |||
| ++++ | (2.5) | ++++ | (2.5) | ++++ | - | |||
| GPC3 | - | (81.6) | - | (81.6) | Membrane | - | - | (100.0) |
| + | (14.1) | + | (14.1) | Cytoplasmic | - | + | - | |
| ++ | (3.1) | ++ | (4.3) | Both | (18.4) | ++ | - | |
| +++ | (1.2) | +++ | - | +++ | - | |||
| ++++ | - | ++++ | - | ++++ | - | |||
| GPC4 | - | (58.9) | - | (59.5) | Membrane | - | - | (81.0) |
| + | (20.9) | + | (14.1) | Cytoplasmic | (40.5) | + | (5.5) | |
| ++ | (19.0) | ++ | (25.8) | Both | - | ++ | (12.9) | |
| +++ | (1.2) | +++ | (0.6) | +++ | (0.6) | |||
| ++++ | - | ++++ | - | ++++ | - | |||
| GPC6 | - | (62.2) | - | (62.6) | Membrane | - | - | (90.2) |
| + | (30.1) | + | (31.9) | Cytoplasmic | (37.4) | + | (9.8) | |
| ++ | (7.4) | ++ | (5.5) | Both | - | ++ | - | |
| +++ | - | +++ | - | +++ | - | |||
| ++++ | - | ++++ | - | ++++ | - | |||
| SDC1 | - | (8.6) | - | (8.6) | Membrane | (42.9) | - | (79.1) |
| + | (31.3) | + | (25.8) | Cytoplasmic | (37.4) | + | (9.2) | |
| ++ | (33.7) | ++ | (50.3) | Both | (10.4) | ++ | (10.4) | |
| +++ | (22.1) | +++ | (10.4) | +++ | (1.2) | |||
| ++++ | (4.3) | ++++ | (4.9) | ++++ | - | |||
| SDC2 | - | (88.3) | - | (88.3) | Membrane | (1.8) | - | (26.4) |
| + | (6.7) | + | (8.0) | Cytoplasmic | (8.6) | + | (20.2) | |
| ++ | (4.3) | ++ | (3.1) | Both | - | ++ | (25.8) | |
| +++ | (0.6) | +++ | (0.6) | +++ | (21.5) | |||
| ++++ | - | ++++ | - | ++++ | (6.1) | |||
| SDC3 | - | (65.0) | - | (65.0) | Membrane | - | - | (100.0) |
| + | (27.0) | + | (28.2) | Cytoplasmic | - | + | - | |
| ++ | (6.1) | ++ | (6.7) | Both | (34.4) | ++ | - | |
| +++ | (1.8) | +++ | - | +++ | - | |||
| ++++ | - | ++++ | - | ++++ | - | |||
| SDC4 | - | (80.4) | - | (80.4) | Membrane | - | - | (100.0) |
| + | (16.0) | + | (19.6) | Cytoplasmic | - | + | - | |
| ++ | (3.7) | ++ | - | Both | (17.8) | ++ | - | |
| +++ | - | +++ | - | +++ | - | |||
| ++++ | - | ++++ | - | ++++ | - | |||
| NG2/CSPG4 | - | (34.2) | - | (34.2) | Membrane | (62.6) | - | (96.7) |
| + | (38.8) | + | (36.8) | Cytoplasmic | - | + | (3.3) | |
| ++ | (21.7) | ++ | (19.7) | Both | - | ++ | - | |
| +++ | (2.6) | +++ | (7.2) | +++ | - | |||
| ++++ | (2.6) | ++++ | (2.0) | ++++ | - | |||
1PG expression was assessed semi-quantitatively according to the arbitrary scoring: “-”, no positively staining cells were detected; “+”, <10% of cells were positive; “++”, ≥10% and <50% of positive cells; “+++”, ≥50% and <90% of positive cells; “++++”, ≥90% of positive cells;
2Refers to the average staining intensity within the examined lesion, according to the arbitrary scoring: “-” = absent; “+”, faint; “++”, weak; “+++”, moderate; “++++”, strong;
3Immunostaining was prevalently cell membrane-associated (“Membrane”) or diffuse cytoplasmatic (“Cytoplasmic”).
HNSCC lesions showed variable expression of SDCs with a relative frequency of positive cases decreasing in the order: SDC1 > SDC3 > SDC4 > SDC2 (Table 2). In fact, a total of 149 cases out of 163 lesions that were evaluated for the in situ expression of the SDC1/CD138 protein had epithelial neoplastic cells presenting the PG on the cell surface, or in intracellular locations (Figure 3). In contrast, a mere 57 (35%), 32 (19.6%), and 19 (12%) out of 163 examined lesions had epithelial neoplastic cells staining positively for respectively SCD2, SDC3 and SDC4 (Table 2). The relative number of cancer cells that expressed these PGs in each lesion markedly differed and a similar divergence was seen in terms of subcellular localization of the molecules. Thus, neoplastic cells with plasma membrane-associated SDC1 were mainly keratinizing cells located at the center of the neoplastic nests (Figure 3). Fibroblasts of the tumour stroma that surrounded the neoplastic nests were positive for SDC1 in 34 out of 163 (21%) of the tumours. In 14 out of 163 of the lesions (8.6%), SDC2 was immunolocalized within the cytoplasm of neoplastic cells, while it appeared widespread in the stromal cells of the majority of the lesions (73.6%, 120 out of 163 of the cases; Figure 3) and was particularly enriched in lesions containing desmoplastic stroma. Intriguing was the fact that in 100% of the lesions, SDC2 could be observed in the wall of both normal and intra-lesional blood vessels, suggesting that it was associated with both endothelial and neovascular pericytes (Figure 3). In contrast to SDC1 and SDC2, both SDC3 and SDC4 were undetectable in the healthy epithelium, or the tumour stroma, but could be immunolocalized both subcellularly and on defined portions of the cell surface of neoplastic cells, with a particular concentration in focal plaque-like structures (Figure 3; Additional file 5: Figure S2). Finally, the diversity of SDCs expressions in oral cavity HNSCC lesions was even more remarkable when considering the relative distribution of these PGs in the stromal compartment. In this case, the frequency of occurrence of the PGs was largely reversed with respect to that seen in the cancer cells and decreased in the order: SDC2 > SDC1 > SDC3 = SDC4 (Table 2). Deviating from the pronounced intracellular distribution of SDC3 and SDC4 was that of NG2/CSPG4 which showed an exclusive cell membrane localization in all samples in which the PG could be disclosed (63%). NG2/CSPG4 was rarely detected in the stromal compartment (5 out of 163; Table 2; Figure 2), where, if occurring, was concentrated on the membrane of basal cells (Additional file 5: Figure S2).
Altered expression of discrete PGs correlates with disease outcome
All demographic and clinical-pathological traits of the patients were initially compared by univariate analysis of the cumulative PG expression data, except for patient categories comprised of less than 13 patients (independently defined as a cut-off level of “statistical” exclusion). In these correlation analyses we considered five primary disease outcomes, including loco-regional tumour recurrence, lymphnodal metastasis, distant metastases, disease-related death and a situation in which at least one of the former disease outcomes was manifested (Table 3).
Table 3.
Univariate analysis of PG expression in relation to known prognostic indicators
| Prognostic indicator/PG | N.of cases | Clinical outcomes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loco-regional recurrence | Lymphnodal metastasis | Distant metastasis | Disease-related death | Any of the clinical outcomes1 | |||||||||
| % cases | p value | % cases | p value | % cases | p value | % cases | p value | % cases | p value | ||||
| Prognostic indicator | |||||||||||||
| Sex | Male | 99 | 14.1 | 0.342 | 15.2 | 0.043 | 10.1 | 0.847 | 22.2 | 0.155 | 36.4 | 0.094 | |
| Female | 74 | 7.0 | 5.4 | 9.5 | 13.5 | 24.3 | |||||||
| Age | ≤45 yrs | 17 | 11.8 | 0.689 | 11.8 | 0.964 | 0.0 | - | 5.9 | 0.146 | 17.6 | 0.184 | |
| >45 yrs | 156 | 12.2 | 10.9 | 10.9 | 19.9 | 32.7 | |||||||
| Smoking | No | 66 | 13.6 | 0.689 | 9.1 | 0.466 | 9.1 | 0.718 | 12.1 | 0.109 | 25.8 | 0.253 | |
| Yes | 105 | 11.4 | 12.4 | 10.5 | 21.9 | 34.3 | |||||||
| Alcohol2 | No | 83 | 10.8 | 0.521 | 8.4 | 0.231 | 4.8 | 0.026 | 6.0 | <0.001 | 21.7 | 0.012 | |
| Yes | 88 | 13.6 | 13.6 | 14.8 | 29.5 | 39.8 | |||||||
| Familial cancer history | No | 144 | 12.5 | 0.732 | 10.4 | 0.615 | 9.7 | 0.880 | 18.8 | 0.876 | 31.3 | 0.994 | |
| Yes | 27 | 11.1 | 14.8 | 11.1 | 18.5 | 33.3 | |||||||
| Precancerous lesions | No | 121 | 12.4 | 0.869 | 5.8 | 0.003 | 9.9 | 0.970 | 18.2 | 0.869 | 28.1 | 0.305 | |
| Yes | 51 | 11.8 | 21.6 | 9.8 | 19.6 | 37.3 | |||||||
| Tumor site | OC | 156 | 12.2 | 0.530 | 11.5 | 0.204 | 10.3 | 0.776 | 17.9 | 0.858 | 31.4 | 0.399 | |
| OP | 14 | 7.1 | 0.0 | 7.1 | 21.4 | 21.4 | |||||||
| OC+OP | 3 | 33.3 | 33.3 | 0.0 | 33.3 | 66.7 | |||||||
| T classification | T1 | 51 | 3.9 | 0.007 | 7.8 | 0.326 | 2.0 | 0.073 | 5.9 | 0.038 | 17.6 | 0.018 | |
| T2 | 60 | 8.3 | 8.3 | 16.7 | 21.7 | 30.0 | |||||||
| T3 | 17 | 23.5 | 17.6 | 5.9 | 23.5 | 47.1 | |||||||
| T4 | 45 | 22.2 | 15.6 | 11.1 | 26.7 | 42.2 | |||||||
| T1-T2 | 111 | 6.3 | 0.001 | 8.1 | 0.065 | 9.9 | 0.957 | 14.4 | 0.043 | 24.3 | 0.005 | ||
| T3-T4 | 62 | 22.6 | 16.1 | 9.7 | 25.8 | 43.5 | |||||||
| N classification | Negative | 97 | 9.3 | 0.079 | 5.2 | 0.001 | 5.2 | 0.016 | 9.3 | <0.001 | 19.6 | <0.001 | |
| Positive | 76 | 15.8 | 18.4 | 15.8 | 30.3 | 46.1 | |||||||
| Differentiation Degree | Well | 20 | 15.0 | 0.441 | 0.0 | 0.223 | 5.0 | 0.392 | 15.0 | 0.952 | 20.0 | 0.44 | |
| Moderate | 50 | 16.0 | 12.0 | 14.0 | 18.0 | 36.0 | |||||||
| Poor | 78 | 7.7 | 12.8 | 7.7 | 16.7 | 30.8 | |||||||
| Radiotherapy | No | 83 | 8.4 | 0.082 | 4.8 | 0.005 | 3.6 | 0.007 | 8.4 | <0.001 | 20.5 | 0.002 | |
| Yes | 90 | 15.6 | 16.7 | 15.6 | 27.8 | 41.1 | |||||||
| PG (mRNA) 3 | |||||||||||||
| SDC1 | ↓ / = | 61 | 11.5 | 0.553 | 6.6 | 0.396 | 4.9 | 0.016 | 11.5 | 0.036 | 21.3 | 0.013 | |
| ↑ | 58 | 13.8 | 10.3 | 19.0 | 25.9 | 41.4 | |||||||
| SDC2 | ↓ / = | 19 | 5.3 | 0.253 | 5.3 | 0.519 | 5.3 | 0.329 | 10.5 | 0.314 | 15.8 | 0.104 | |
| ↑ | 100 | 14.0 | 9.0 | 13.0 | 20.0 | 34.0 | |||||||
| SDC3 | ↓ / = | 24 | 8.3 | 0.405 | 8.3 | 0.896 | 4.2 | 0.193 | 20.8 | 0.855 | 25.0 | 0.352 | |
| ↑ | 95 | 13.7 | 8.4 | 13.7 | 17.9 | 32.6 | |||||||
| SDC4 | ↓ / = | 19 | 5.3 | 0.253 | 5.3 | 0.519 | 5.3 | 0.329 | 15.8 | 0.780 | 26.3 | 0.601 | |
| ↑ | 100 | 14.0 | 9.0 | 13.0 | 19.0 | 32.0 | |||||||
| GPC1 | = | 62 | 12.9 | 0.803 | 6.5 | 0.502 | 12.9 | 0.668 | 17.7 | 0.940 | 32.3 | 0.625 | |
| ↓ / ↑ | 57 | 12.3 | 10.5 | 10.5 | 19.3 | 29.8 | |||||||
| GPC2 | = | 76 | 10.5 | 0.393 | 6.6 | 0.363 | 10.5 | 0.596 | 21.1 | 0.335 | 27.6 | 0.307 | |
| De novo | 43 | 16.3 | 11.6 | 14.0 | 14.0 | 37.2 | |||||||
| GPC3 | ↓ | 26 | 7.7 | 0.437 | 0.0 | 0.219 | 11.5 | 0.378 | 11.5 | 0.303 | 23.1 | 0.624 | |
| = | 25 | 20.0 | 12.0 | 4.0 | 12.0 | 32.0 | |||||||
| ↑ | 68 | 11.8 | 10.3 | 14.7 | 23.5 | 33.8 | |||||||
| GPC4 | = | 81 | 11.1 | 0.598 | 7.4 | 0.647 | 13.6 | 0.365 | 21.0 | 0.308 | 33.3 | 0.348 | |
| ↓ / ↑ | 38 | 15.8 | 10.5 | 7.9 | 13.2 | 26.3 | |||||||
| GPC5 | = | 33 | 12.1 | 0.995 | 9.1 | 0.851 | 6.1 | 0.241 | 15.2 | 0.609 | 30.3 | 0.986 | |
| De novo | 86 | 12.8 | 8.1 | 14.0 | 19.8 | 31.4 | |||||||
| GPC6 | ↓ | 36 | 19.4 | 0.351 | 8.3 | 0.994 | 8.3 | 0.495 | 16.7 | 0.838 | 36.1 | 0.824 | |
| = | 24 | 8.3 | 8.3 | 8.3 | 16.7 | 29.2 | |||||||
| ↑ | 59 | 10.2 | 8.5 | 15.3 | 20.3 | 28.8 | |||||||
| NG2/CSPG4 | = | 34 | 2.9 | 0.029 | 8.8 | 0.914 | 8.8 | 0.487 | 14.7 | 0.401 | 20.6 | 0.077 | |
| De novo | 85 | 16.5 | 8.2 | 12.9 | 20.0 | 35.3 | |||||||
| PG (Protein) 4 | |||||||||||||
| SDC1 Tumor cells | Negative | 14 | 7.1 | 0.447 | 21.4 | 0.312 | 7.1 | 0.769 | 7.1 | 0.269 | 35.7 | 0.947 | |
| Positive | 149 | 13.4 | 10.7 | 9.4 | 19.5 | 31.5 | |||||||
| Stroma | Negative | 129 | 13.2 | 0.962 | 11.6 | 0.812 | 9.3 | 0.998 | 17.1 | 0.294 | 31.0 | 0.429 | |
| Positive | 34 | 11.8 | 11.8 | 8.8 | 23.5 | 35.3 | |||||||
| SDC2 Tumor cells | Negative | 143 | 13.3 | 0.783 | 11.9 | 0.875 | 8.4 | 0.303 | 18.9 | 0.792 | 30.8 | 0.385 | |
| Positive | 19 | 10.5 | 10.5 | 15.8 | 15.8 | 42.1 | |||||||
| Stroma | Negative | 43 | 9.3 | 0.277 | 2.3 | 0.015 | 2.3 | 0.062 | 2.3 | 0.001 | 14.0 | 0.002 | |
| Positive | 120 | 14.2 | 15.0 | 11.7 | 24.2 | 38.3 | |||||||
| SDC3 Tumor cells | Negative | 106 | 13.2 | 0.923 | 12.3 | 0.793 | 8.5 | 0.655 | 17.9 | 0.758 | 30.2 | 0.575 | |
| Positive | 56 | 12.5 | 10.7 | 10.7 | 19.6 | 35.7 | |||||||
| SDC4 Tumor cells | Negative | 131 | 15.3 | 0.071 | 13.0 | 0.329 | 9.2 | 0.957 | 17.6 | 0.588 | 32.8 | 0.649 | |
| Positive | 32 | 3.1 | 6.3 | 9.4 | 21.9 | 28.1 | |||||||
| GPC1 Tumor cells | Negative | 44 | 11.4 | 0.653 | 15.9 | 0.340 | 4.4 | 0.214 | 15.6 | 0.602 | 31.8 | 0.825 | |
| Positive | 108 | 13.6 | 10.2 | 10.7 | 18.9 | 32.2 | |||||||
| Stroma | Negative | 149 | 11.4 | 0.007 | 12.1 | 0.884 | 8.4 | 0.319 | 16.8 | 0.012 | 30.9 | 0.104 | |
| Positive | 13 | 30.8 | 7.7 | 15.4 | 38.5 | 46.2 | |||||||
| GPC3 Tumor cells | Negative | 133 | 12.8 | 0.805 | 12.0 | 0.841 | 9.6 | 0.584 | 18.4 | 0.834 | 33.1 | 0.605 | |
| Positive | 30 | 13.3 | 10.0 | 6.3 | 15.6 | 26.7 | |||||||
| GPC4 Tumor cells | Negative | 96 | 13.5 | 0.969 | 10.2 | 0.493 | 9.2 | 0.928 | 17.3 | 0.716 | 33.3 | 0.794 | |
| Positive | 66 | 12.1 | 13.0 | 8.7 | 18.8 | 30.3 | |||||||
| Stroma | Negative | 131 | 13.0 | 0.860 | 11.5 | 0.692 | 9.2 | 0.900 | 18.3 | 0.771 | 32.8 | 0.810 | |
| Positive | 31 | 12.9 | 12.9 | 9.7 | 19.4 | 29.0 | |||||||
| GPC6 Tumor cells | Negative | 101 | 12.9 | 0.860 | 6.9 | 0.010 | 5.9 | 0.052 | 15.8 | 0.222 | 27.7 | 0.102 | |
| Positive | 61 | 13.1 | 19.7 | 14.8 | 23.0 | 39.3 | |||||||
| Stroma | Negative | 146 | 11.6 | 0.058 | 12.3 | 0.599 | 10.3 | - | 18.5 | 0.844 | 32.2 | 0.828 | |
| Positive | 16 | 25.0 | 6.3 | 0.0 | 18.8 | 31.3 | |||||||
| NG2/CSPG4 Tumor cells | Negative | 52 | 15.4 | 0.665 | 9.6 | 0.406 | 9.6 | 0.923 | 17.3 | 0.563 | 34.6 | 0.967 | |
| Positive | 100 | 12.0 | 14.0 | 10.0 | 21.0 | 33.0 | |||||||
| T group/NG2/CSPG4 mRNA5 | T1-T2/= | 21 | 0 | <0.001 | |||||||||
| T1-T2 / De novo | 49 | 6.1 | |||||||||||
| T3-T4/= | 13 | 7.7 | |||||||||||
| T3-T4/De novo | 36 | 30.6 | |||||||||||
| Precancerous lesions/SDC2 stroma | -/- | 29 | 0 | 0.001 | |||||||||
| -/+ | 84 | 8.3 | |||||||||||
| +/- | 14 | 7.1 | |||||||||||
| +/+ | 35 | 28.6 | |||||||||||
| N status6/SDC1 mRNA | Negative/↓/= | 31 | 0 | 0.004 | |||||||||
| Negative/↑ | 30 | 10.0 | |||||||||||
| Positive/↓/= | 30 | 10.0 | |||||||||||
| Positive/↑ | 28 | 28.6 | |||||||||||
| N status/SDC2 stroma | -/- | 33 | 0 | <0.001 | |||||||||
| -/+ | 57 | 14.0 | |||||||||||
| +/- | 10 | 10.0 | |||||||||||
| +/+ | 63 | 33.3 | |||||||||||
| N status/SDC1 mRNA | Negative/↓/= | 31 | 3.2 | <0.001 | |||||||||
| Negative/↑ | 57 | 33.3 | |||||||||||
| Positive/↓/= | 10 | 40.0 | |||||||||||
| Positive/↑ | 63 | 50.0 | |||||||||||
| PGs pattern | |||||||||||||
| SDC1 mRNA ↑ NG2/CSPG4 mRNA de novo + SDC2 stroma | Yes | 36 | 16.7 | 0.271 | 11.1 | 0.405 | 25 | <0.001 | 33.3 | 0.002 | 50 | <0.001 | |
| No | 73 | 12.3 | 8.2 | 4.1 | 11 | 23.3 | |||||||
1This refers to the situation in which patients manifested at least one of the four adopted clinical outcomes within the follow-up period;
2Excessive alcohol consumption was based upon self-provided information;
3PG transcript expression was defined as “↓”, down-regulated; “↑”, up-regulated; “=”, not changed; and “De novo”, de novo expressed, when compared to a healthy mucosal tissues pool that was used as sample calibrator;
4Protein expression data are reported as detectable or non-detectable by indirect immunohistochemistry;
5Univariate analyses combining the prognostic indicators that were deemed to be independent poor predictors of each of the five clinical outcomes as accomplished through the Cox proportional hazard model;
6 N status positive or negative is according to N classification AJCC staging system;
p values <0,05 were considered to be significant (in bold); p values within ≥ 0,05 and <0,06 were considered borderline and were included in the following multivariate regression model; p value was not calculated where a monotone likelihood was established.
Abbreviations: OC, oral cavity; OP, oropharynx.
In order to test whether there is a relationship between PGs transcript and protein expressions and clinicopathological parameters, a Chi-Square test was applied and just three such correlation resulted statistically significant: N classification and SDC2 stromal positivity, p = 0.002; alcohol consumption and SDC1 mRNA up-regulation, p = 0.021, and presence of precancerous lesion and SDC1 mRNA up-regulation, p = 0.016.
Although radiation therapy and excessive alcohol consumption independently correlated with one or more of the above clinical outcomes (Table 3), these parameters were not considered in the multivariate logistic regression analyses because of being potentially confounding indicators. The first because almost all patients presenting lymphnodal infiltrations had been routinely subjected to radiation therapy, the second because, despite of its well-recognized importance as a risk factor in HNSCC, the admission of this habit was measured by a self-provided questionnaire and no details were available on the accuracy of the information provided by the patients. A further consideration is that self-reported excessive alcohol intake is often denied, causing underestimation of the cohort of patient that may fall under this “risk category”.
Advanced T classification (p = 0.007), T3-T4 grouping (p = 0.001; Figure 4), positive NG2/CSPG4 transcript expression (p = 0.029; Figure 4), or GPC1 positivity in stromal cells (p = 0.007) were all conditions strongly associated with a high loco-regional tumour relapse rate (Table 3). Stromal GPC6 expression could, however, not be included as a parameter in the multivariate logistic regression model due to the low number of cases contained within this category and the borderline statistical significance in univariate analyses (p = 0.058). Application of the Cox proportional hazard model revealed that T3-T4 classification of the tumour (HR, 6.36, p = 0.001) and de novo expression of NG2/CSPG4 mRNA (HR, 6.76, p = 0.017) were independent, robust prognostic factors for local tumour recurrence (Table 4; Figure 5). If combining T-grouping and mRNA expression of NG2/CSPG4, the probability to develop a secondary loco-regional lesion was further increased (p < 0.001; Table 3; Figure 4).
Figure 4.
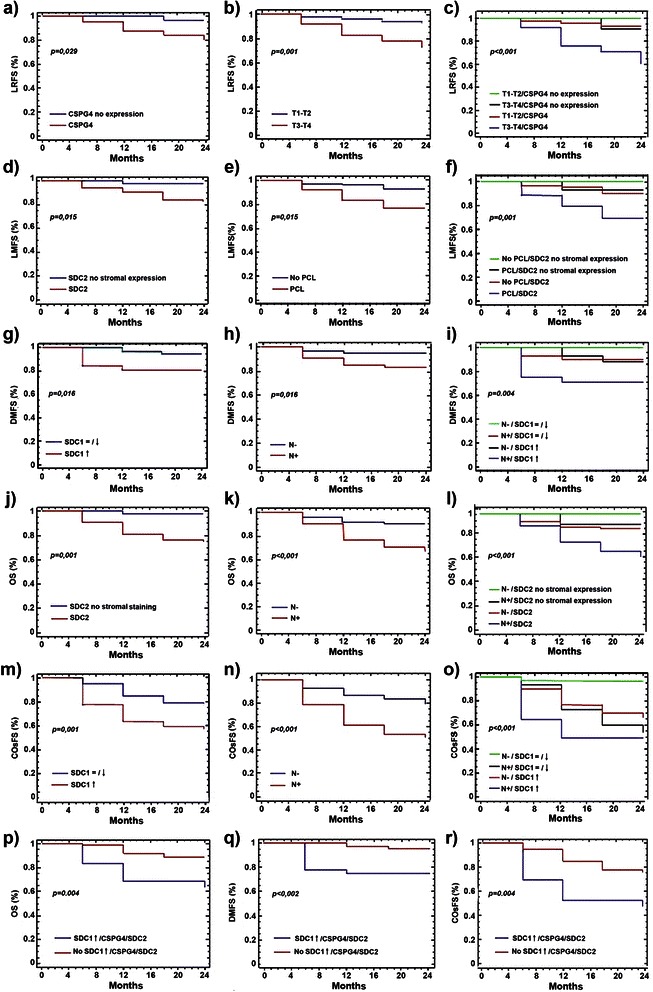
Differential PG expression correlates with clinical outcome. Survival and probability curves for the following correlations: (a) loco-regional relapse vs de novo expression of NG2/CSPG4, (b) loco-regional relapse vs T group classification; (c) loco-regional relapse vs coincident NG2/CSPG4 expression and advanced T classification; (d) lymphnodal metastases vs enhanced SDC2 expression in stromal cells; (e) lymphnodal metastases vs manifestation of precancerous lesions; (f) lymphnodal metastases vs the combination of both previous prognostic indicators; (g) distant metastases vs up-regulated SDC1 expression; (h) distant metastases vs infiltration of cervical lymph nodes; (i) distant metastases vs the combination of both previous prognostic indicators; (j) overall survival vs enhanced SDC2 expression in the stromal compartment; (k) overall survival vs N classification; (l) overall survival vs the coincidence of both previously indicated events; (m) the occurrence of any of the clinical outcomes vs up-regulated SDC1 expression; (n) the occurrence of any of the clinical outcomes vs N classification; (o) the occurrence of any of the clinical outcomes vs the combination of both above listed events; (p) overall survival, (q) distant metastases and (r) the occurrence of any of the clinical outcomes vs the combination of SDC1 up-regulation, de novo expression of NG2/CSPG4 and stromal enhancement of SDC2 positivity. Abbreviations: LRFS, loco-regional relapse-free survival; LMFS, lymphnodal metastasis-free survival; DMFS, distant metastasis-free survival; OS, overall survival, COsFS, clinical outcomes-free survival.
Table 4.
Multivariate analyses of the prognostic implication of altered PG expression for the different clinical outcomes 1 Estimated regression coefficient and confidence interval;
| Prognostic indicator/PG | Estimate1(95% CI) | SE2 | p value 3 | HR4 | Clinical outcome |
|---|---|---|---|---|---|
| T classification | |||||
| T25 | 0.359 (0.153/0.565) | 0.105 | 0,012 | 1.432 | Loco-regional relapse |
| T3 | 1.857 (1.556/2.158) | 0.153 | 6.404 | ||
| T4 | 2.160 (1.956/2.364) | 0.104 | 8.671 | ||
| T3-T4 vs T1-T2 | 1.850 (1.691/2.009) | 0.081 | 0.001 | 6.360 | Loco-regional relapse |
| NG2/CSPG4 mRNA6 | |||||
| De novo expression vs no expression | 1.911 (1.735/2.087) | 0.090 | 0.017 | 6.760 | Loco-regional relapse |
| Precancerous lesions | |||||
| Presence vs Absence | 1.328 (1.184/1.471) | 0.073 | 0.005 | 3.773 | Lymphnodal metastases |
| SDC2 stroma | |||||
| Positive vs Negative | 2.035 (1.885/2.184) | 0.076 | 0.007 | 7.652 | Lymphnodal metastases |
| SDC2 stroma | |||||
| Positive vs Negative | 2.160 (2.022/2.298) | 0.070 | 0.003 | 8.671 | Disease-related deaths |
| N classification | |||||
| Positive vs Negative | 1.477 (1.326/1.628) | 0.077 | 0.012 | 4.380 | Distant metastasis |
| N classification | |||||
| Positive vs Negative | 1.089 (0.967/1.211) | 0.062 | 0.005 | 2.971 | Disease-related deaths |
| N classification | |||||
| Positive vs Negative | 1.164 (1.003/1.325) | 0.082 | <0.001 | 3.203 | Any of the clinical outcomes7 |
| SDC1 mRNA | |||||
| ↓ /= vs ↑ | −1.460 (−1.612/-1.309) | 0.077 | 0.013 | 0.232 | Distant metastasis |
| SDC1 mRNA | |||||
| ↓ /= vs ↑ | −0.845 (−1.007/-0.684) | 0.082 | 0.012 | 0.429 | Any of the clinical outcomes |
| SDC1 mRNA up-regulation + NG2/CSPG4 mRNA de novo expression + SDC2 stroma | |||||
| Single condition vs combination of all 3 conditions8 | −1.924 (−2.088/-1.760) | 0.084 | 0.002 | 0.146 | Distant metastasis |
| −1.302 (−1.459/-1.146) | 0.080 | 0.004 | 0.272 | Disease-related deaths | |
| −1.014 (−1.194/-0.833) | 0.092 | 0.003 | 0.363 | Any of the clinical outcomes |
1Estimated regression coefficient and confidence interval;
2Standard error of estimated regression coefficient;
3p value <0,05 were considered to be significant;
4Hazard Ratio estimated from Cox proportional hazard regression model;
5Compared to T1 stage;
6PG transcript expression (↓, down-regulated; ↑, up-regulated; =, not changed; De novo expression, de novo expressed in comparison to a healthy mucosal tissues pool that was used as sample calibrator) could be grouped according to the trend of each PG gene in relation to the clinical outcomes;
7Is referred to a patient that had at least one of the other outcomes within the follow-up;
8Refers to the comparison between a situation in which all three indicated conditions were manifested (“combination of all 3 conditions”) versus either condition alone or the combination of any two conditions;
Abbreviations: CI, Confidence Interval; HR, Hazard Ratio; SE, standard error.
Figure 5.
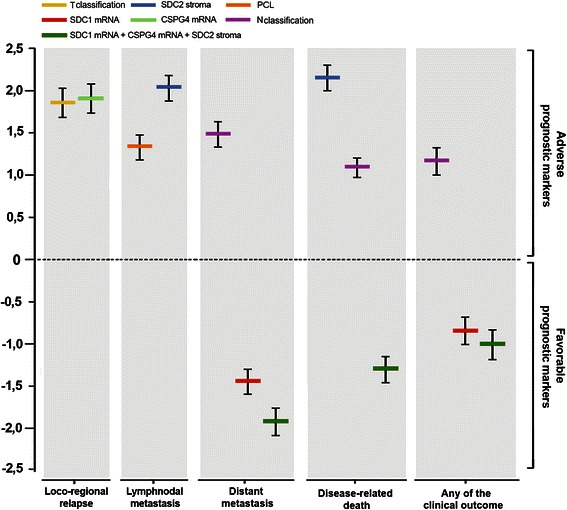
Cox proportional hazard analysis. Plot overview of Cox proportional hazard estimated regression coefficients of the resulting independent prognostic factors for loco-regional recurrence, lymphnodal metastases, distant metastases, disease-related death and the occurrence of any of the clinical outcomes.
Significant correlations with post-surgical lymphnodal metastases were further disclosed between gender (p = 0.043), the presence of precancerous lesions (p = 0.003; Figure 4), the occurrence of lymphnodal infiltration at time of surgery (p = 0.001), stromal expression of SDC2 (p = 0.015; Figure 4) and altered GPC6 protein expression in tumour cells (p = 0.01; Table 3). As predictable, “sentinel lymphnodes” at diagnosis significantly influenced the later appearance of more prominent lymphnodal lesions, but this factor was not considered in the multivariate logistic regression model because of its unuttered prognostic implication. Multivariate analysis corroborated that the occurrence of precancerous lesions (HR, 3.773, p = 0.005), and more incisively the presence of SDC2 in the stromal compartment (HR, 7.652, p = 0.007), but not GPC6 expression or gender of the patient, were independent prognostic markers for post-surgery secondary infiltrations of lymphnodes (Table 4; Figure 5). If we then accounted for both a history of precancerous lesions and SDC2 stromal expression, the probability to develop post-surgical lymphnodal infiltration was significantly increased (p = 0.001; Table 3; Figure 4).
Contrary to the above associations, univariate logistic analyses revealed that the N classification , at time of surgical removal of the primary tumour mass, and up-regulated transcription of SDC1 (Figure 4) were significantly associated with the formation of distant metastases ( p = 0.016 for both correlations; Table 3). The frequency of GPC6 expression in neoplastic cells did not satisfy the limit of significance (p = 0.052), but could be a potentially interesting indicator to take into account in future investigations on larger cohorts of patients. Conversely, multivariate analyses reinforced the impact of N classification (HR, 4.38, p = 0.012) and down-regulated or unaltered SDC1 expression (HR, 0.232, p = 0.013) as independent factors predicting the formation of distant metastases, albeit with opposite trends, (Table 4; Figure 5). When we next considered the combination of the unfavourable conditions represented by lymphnodal infiltration and up-regulation of SDC1 transcription, we unfolded a significantly increased probability to develop distant metastases (p = 0.004; Table 3; Figure 4). Noteworthy was also the fact that 91% of patients with up-regulated SDC1 transcription that developed distant metastases within the follow-up period invariably succumbed the disease.
Advanced T classification (p = 0.038), T3-T4 grouping (p = 0.043), positive N classification (p < 0.001; Figure 4), up-regulation of SDC1 transcription (p = 0.036) and stromal expression of SDC2 or GPC1 (p = 0.001 and p = 0.012 respectively; Table 3, Figure 4) were also found to be strongly associated with disease-related death. Cervical lymphnodal involvement (HR, 2.971, p = 0.005) and, more markedly, synthesis of SDC2 in the stromal cells (HR, 8.671, p = 0.003), established two independent predictors of survival (Table 4; Figure 5). The combination of these two conditions further decreased the survival probability of the patients (p < 0.001, Table 3; Figure 4).
In the evaluation of situations in which patients presented at any of the mentioned clinical outcomes, we similarly found a tight correlation between advanced T classification (p = 0.018), T3-T4 grouping (p = 0.005), positive N classification (p < 0.001; Figure 4), up-regulation of SDC1 mRNA (p = 0.013; Figure 4) and SDC2 stromal reactivity (p = 0.002). Finally, in multivariate analyses, N involvement (HR, 3.203, p < 0.001) and down-regulation or unaltered SDC1 expression (HR, 0.429, p = 0.012), but not T classification or SDC2 detection in stromal cells, were independent factors with opposite trends for the prediction of poor prognosis (Table 4; Figure 5). Even in this case the combination of positive N classification and up-regulation of SDC1 mRNA expression significantly increased the probability of the patients to incur into a dismal disease course (p < 0.001;Table 3; Figure 4).
We finally evaluated the disease course in patients scoring positively for the 3 dismal prognostic indicators, i.e. de novo expression of NG2/CSPG4, stromal abundance of SDC2 and up-regulation of SDC1 mRNA, which, in an independent manner, associated with one or more of the adverse clinical outcomes. This conditions was found in 36 of the 173 patients (21%) and within this patient subgroup 17% and 25%, respectively, developed loco-regional secondary lesions or distant metastases. Lymphnodal metastasis was observed in 11% of the patients, whereas 33% of them succumbed to the disease. Survival analyses revealed a strong association between the PG pattern analyzed and the presence of distant metastases (p < 0.002), disease-related deaths (p = 0.004) and a cumulative bad prognosis (p < 0.004). The Cox regression model ascertained that patients not expressing simultaneously the three bad independent disease course markers resulted to have a best prognosis in terms of putative development of distant metastases (HR, 0.146, p = 0.002), for survival (HR, 0.272, p = 0.004) and for incurring into any of the unfavourable clinical events under consideration (HR, 0.363, p = 0.003) (Table 4; Figure 5).
Discussion
Despite the relatively high incidence of oral cavity HNSCC, very few reliable prognostic and/or predictive molecular markers are currently available for the routine clinical management of the patients. In light of this deficiency, we have explored the possibility that variation in the expression of cell surface PGs, widely recognized to be key factors in the control of tumour progression [33,56-59], could afford more effective means of prognosticating patients affected by these tumours. Indeed, we find that, upon neoplastic transformation, epithelial cells of the oral cavity and oropharynx modify their transcriptional/translational rates of virtually all currently known cell surface PGs. This led us to conclude that transformation-dependent modulation of PG synthesis may be part of the globally altered pattern of gene expression in these cells, as well as contribute to the cancer cell’s acquisition of a defined repertoire a cell surface molecules capable of dictating their malignant behavior.
SDCs are widely recognized to undergo malignancy-associated changes in their expressions in several types of carcinomas, including those of thyroid, breast, colon, skin, stomach and urogenital tract, and SDC1 is recognized to be the best documented prognostic biomarker [39,41,43,40,46,60-64]. Its expression pattern frequently correlates with the differentiation status of the cells and thereby with their malignancy degree [47,54,65-68]. This characterizing trait of the SDC1 tumour-associated expression was corroborated here, along with its widespread distribution in neoplastic HNSCC lesions.
Although much less studied, SDC2 has also been reported to be associated with malignant carcinoma lesions in various anatomical sites/organs including head and neck [63,69,70]. In this study, transcription/translation of SDC2 was found to be more prominent in the intralesional stroma than in the neoplastic cells and, hence, the PG showed an expression pattern that was complementary to SDC1. Notably, SDC2 was also observed to be strongly enriched in neovascular structures, where it appeared to be associated with both endothelial cells and pericytes. Our present mapping study is the first to reveal a de novo expression of SDC3 and SDC4 in oral cavity HNSCC and the accumulation of SDC4 in areas of cell-cell contact [22,24,25] within such lesions. Intracellular abundance of SDCs in HNSCC cells may reflect the incapacity of the cells to complete the post-translational processing of these PGs and/or their transport and intercalation into the cell membrane, or an accentuated internalization and intracellular recycling process.
Several GPCs were also found to be misexpressed in oral cavity HNSCC lesions, albeit with frequencies that were generally lower than those seen for SDCs. GPC1, known to be highly expressed in pancreatic and breast carcinomas [32,33], was found to be the prevalent GPC of these lesions, alongside with GPC3, which has independently been reported to be up-regulated in several other tumour types and has a recognized value as prognostic factor and putative therapeutic target in hepatocellular carcinomas [34,35,37]. In this context, it is, however, worth noting that GPC3 has also been proposed to act as a potential tumour suppressor in certain neoplasia, showing a putative transformation-dependent silencing of the glypican [30,71-73].
When we applied univariate and multivariate meta-analytical methods to correlate the observed PG expression patterns with clinically relevant disease outcomes we unveiled striking associations. Appearance of NG2/CSPG4, a PG with a precedent prognostic impact in numerous solid tumours [74-82], was found to tightly correlate with loco-regional tumour recurrence and, hence, was disclosed to be the first ever to be described molecular relapse predictor in oral cavity HNSCC. Enhanced expression of GPC1 in the stromal compartment of these lesions also closely correlated with tumour recurrence and paralleled the more predictable prognostic implication of tumour staging. Beside its prognostic role in pancreatic cancer, there is currently no other indication that altered expression of GPC1 may influence the course of any tumour type. Another crucial finding of this study was the close association betweenSDC2 up-regulation in the intralesional stromal compartment and the overall survival of the patients carrying such SDC2-rich primary lesions. Even in this case, the present study provides the first evidence for such a prognostic relationship in any cancer type and, similarly to the potential of NG2/CSPG4, emphasizes that SDC2 may serve as a putative target for prevention and/or treatment of relapsing oral cavity HNSCC.
Conclusion
The present study provides the first evidence that altered expression of cell surface-bound PGs is strongly links to the formation and progression of oral cavity HNSCC. Elective modulation of PG expression in primary oral cavity HNSCC lesions correlates, in a predictive manner, with several clinical outcomes and may therefore serve as an adjunct in the molecular diagnosis of these tumours. More specifically, enhanced expression of SDC2 in the tumour stroma significantly correlates with overall survival and is indicative of lymphonodal metastasis, whereas aberrant increased of SDC1 transcription is indicative of the presence of distant metastases. Strikingly, up-regulation of NG2/CSPG4 is tightly linked to loco-regional recurrence of the tumour, underscoring the potential of this biomarker to forcefully predict the clinical course of oral cavity HNSCC patients.
Acknowledgements
We are indebted to Dr. Alice Dallatomasina and Dr. Mirca Lazzaretti for technical assistance and to Prof. S.A. Pileri, University of Bologna, for assistance in the preparation of TMAs. The work was supported by grants from RER (Programma di ricerca Regione-Università 2007–2009; Area La Ricerca Innovativa to MPF, ES and RP); AIRC, Associazione Italiana per la Ricerca sul Cancro (IG 2009 to RP), the Italian Ministry of University, Education and Research, MIUR (PRIN 2008 to RP) and by Asse POR-FESR “Creazione Tecnopoli” (Regione Emilia-Romagna).
Additional files
Patient demographic and clinic-pathological features.
Overview of the distribution of clinical outcomes manifested by oral cavity HNSCC patients during the entire follow-up period subdivided in 6-months time intervals.
Supplemental and detailed Materials and methods.
Listing of the individual relative PG expression data.
Representative immunostaining on healthy control tissue sections: GPC1 (a), GPC3 (b), GPC4 (c), GPC6 (d), NG2/CSPG4 (e), SDC1 (f), SDC2 (g), SDC3 (h) and SDC4 (i).
Footnotes
Anna Farnedi and Silvia Rossi contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conception and design: AF, SR, NB and RP. Development of methodology: EMS, TP, ES, CM, RC, MPF and RP. Acquisition of data: AF, RS, NB, MG, DL, MTM, EL and AAS. Analysis and interpretation of data: AF, RS, NB, MG, LM, EL, MPF and RP. Writing, review and/or revision of the manuscript: AF, SR, NB and RP. Administrative, technical, or material support: all co-author. Study supervision: TP, ES, MPF and RP. All authors read and approved the final version of the manuscript.
Contributor Information
Anna Farnedi, Email: farnedi@hotmail.com.
Silvia Rossi, Email: silvia.rossi1@unipr.it.
Nicoletta Bertani, Email: nicoletta.bertani@unipr.it.
Mariolina Gulli, Email: mariolina.gulli@unipr.it.
Enrico Maria Silini, Email: enricomaria.silini@unipr.it.
Maria Teresa Mucignat, Email: mtmucignat@cro.it.
Tito Poli, Email: tito.poli@unipr.it.
Enrico Sesenna, Email: enrico.sesenna@unipr.it.
Davide Lanfranco, Email: lanfranco82@yahoo.it.
Lucio Montebugnoli, Email: lucio.montebugnoli@unibo.it.
Elisa Leonardi, Email: elisa.leonardi4@unibo.it.
Claudio Marchetti, Email: claudio.marchetti@unibo.it.
Renato Cocchi, Email: roberto.cocchi@ausl.bologna.it.
Andrea Ambrosini-Spaltro, Email: andreaambrosini@yahoo.it.
Maria Pia Foschini, Email: mariapia.foschini@unibo.it.
Roberto Perris, Email: roberto.perris@unipr.it.
References
- 1.DeSantis C, Lin C, Mariotto A, Siegel R, Stein K, Kramer J, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–71. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataran I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 4.Dragovic AF, Caudell JJ, Spencer SA, Carroll WR, Nabell LA, Bonner JA. Locoregional failure and the risk of distant metastasis after modern radiotherapy for head and neck cancer. Head Neck. 2013;35:381–7. doi: 10.1002/hed.22977. [DOI] [PubMed] [Google Scholar]
- 5.Kang CJ, Liao CT, Hsueh C, Lee LY, Lin CY, Fan KH, et al. Outcome analysis of patients with well-differentiated oral cavity squamous cell carcinoma. Oral Oncol. 2011;47:1085–91. doi: 10.1016/j.oraloncology.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Reibel J. Prognosis of oral pre-malignant lesions: significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med. 2003;14:47–62. doi: 10.1177/154411130301400105. [DOI] [PubMed] [Google Scholar]
- 7.Brinkman BM, Wong DT. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr Opin Oncol. 2006;18:228–33. doi: 10.1097/01.cco.0000219250.15041.f8. [DOI] [PubMed] [Google Scholar]
- 8.Brennan M, Migliorati CA, Lockhart PB, Wray D, Al-Hashimi I, Axéll T, et al. Management of oral epithelial dysplasia: a review. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103:S19.e1–S19.e12. doi: 10.1016/j.tripleo.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Hu S, Wong DT. Oral cancer proteomics. Curr Opin Mol Ther. 2007;9:467–76. [PubMed] [Google Scholar]
- 10.Scully C, Bagan JV. Recent advances in oral oncology 2008; squamous cell carcinoma imaging, treatment, prognostication and treatment outcomes. Oral Oncol. 2009;45:e25–30. doi: 10.1016/j.oraloncology.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Coughlin A, Resto VA. Oral cavity squamous cell carcinoma and the clinically n0 neck: the past, present, and future of sentinel lymph node biopsy. Curr Oncol Rep. 2010;12:129–35. doi: 10.1007/s11912-010-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Naaj IA, Leiser Y, Shveis M, Sabo E, Peled M. Incidence of oral cancer occult metastasis and survival of T1-T2N0 oral cancer patients. J Oral Maxillofac Surg. 2011;69:2674–9. doi: 10.1016/j.joms.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Woolgar JA. Pathology of the N0 neck. Br J Oral Maxillofac Surg. 1999;37:205–9. doi: 10.1054/bjom.1999.0035. [DOI] [PubMed] [Google Scholar]
- 14.Søland TM, Brusevold IJ. Prognostic molecular markers in cancer - quo vadis? Histopathology. 2013;63:297–308. doi: 10.1111/his.12184. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira LR, Ribeiro-Silva A. Prognostic significance of immunohistochemical biomarkers in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2011;40:298–307. doi: 10.1016/j.ijom.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licitra L, Perrone F, Tamborini E, Bertola L, Ghirelli C, Negri T, et al. Role of EGFR family receptors in proliferation of squamous carcinoma cells induced by wound healing fluids of head and neck cancer patients. Ann Oncol. 2011;22:1886–93. doi: 10.1093/annonc/mdq756. [DOI] [PubMed] [Google Scholar]
- 18.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 19.Granata R, Miceli R, Orlandi E, Perrone F, Cortelazzi B, Franceschini M, et al. Tumour stage, human papillomavirus and smoking status affect the survival of patients with oropharyngeal cancer: an Italian validation study. Ann Oncol. 2012;23:1832–7. doi: 10.1093/annonc/mdr544. [DOI] [PubMed] [Google Scholar]
- 20.Wittekindt C, Wagner S, Mayer CS, Klussmann JP. Basics of tumour development and importance of human papilloma virus (HPV) for head and neck cancer. GMS Curr Top Otorhinolaryngol Head Neck Surg. 2012;11:Doc09. doi: 10.3205/cto000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirn-Safran C, Farach-Carson MC, Carson DD. Multifunctionality of extracellular and cell surface heparan sulfate proteoglycans. Cell Mol Life Sci. 2009;66:3421–34. doi: 10.1007/s00018-009-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambaerts K, Wilcox-Adelman SA, Zimmermann P. The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr Opin Cell Biol. 2009;21:662–9. doi: 10.1016/j.ceb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 24.Xian X, Gopal S, Couchman JR. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 2010;339:31–46. doi: 10.1007/s00441-009-0829-3. [DOI] [PubMed] [Google Scholar]
- 25.Choi Y, Chung H, Jung H, Couchman JR, Oh ES. Syndecans as cell surface receptors: Unique structure equates with functional diversity. Matrix Biol. 2011;30:93–9. doi: 10.1016/j.matbio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108:497–501. doi: 10.1172/JCI200113712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veugelers M, De Cat B, Ceulemans H, Bruystens AM, Coomans C, Dürr J, et al. Glypican-6, a New Member of the Glypican Family of Cell Surface Heparan Sulfate Proteoglycans. J Biol Chem. 1999;274:26968–77. doi: 10.1074/jbc.274.38.26968. [DOI] [PubMed] [Google Scholar]
- 28.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell CA, Xu G, Filmus J, Busch S, Brody JS, Rothman PB. Oligonucleotide microarray analysis of lung adenocarcinoma in smokers and nonsmokers identifies GPC3 as a potential lung tumour suppressor. Chest. 2002;121:6S–7. doi: 10.1378/chest.121.3_suppl.6S. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Xu GL, Borczuk AC, Busch S, Filmus J, Capurro M, et al. The heparan sulfate proteoglycan GPC3 is a potential lung tumour suppressor. Am J Respir Cell Mol Biol. 2003;29:694–701. doi: 10.1165/rcmb.2003-0061OC. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Sheu CC, Ye Y, De Andrade M, Wang L, Chang SC, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–30. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleeff J, Ishiwata T, Kumbasar A, Friess H, Büchler MW, Lander AD, et al. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102:1662–73. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda K, Maruyama H, Guo F, Kleeff J, Itakura J, Matsumoto Y, et al. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61:5562–9. [PubMed] [Google Scholar]
- 34.Zhu ZW, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander AD, et al. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48:558–64. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, et al. Glypican-3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumour marker. Biochem Biophys Res Commun. 2003;306:16–25. doi: 10.1016/S0006-291X(03)00908-2. [DOI] [PubMed] [Google Scholar]
- 36.Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, et al. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res. 2004;64:2418–23. doi: 10.1158/0008-5472.CAN-03-2191. [DOI] [PubMed] [Google Scholar]
- 37.Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, et al. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100:1403–7. doi: 10.1111/j.1349-7006.2009.01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soukka T, Pohjola J, Inki P, Happonen RP. Reduction of syndecan-1 expression is associated with dysplastic oral epithelium. J Oral Pathol Med. 2000;29:308–13. doi: 10.1034/j.1600-0714.2000.290704.x. [DOI] [PubMed] [Google Scholar]
- 39.Kurokawa H, Matsumoto S, Murata T, Yamashita Y, Tomoyose T, Zhang M, et al. Immunohistochemical study of syndecan-1 downregulation and the expression of p53 protein or Ki-67 antigen in oral leukoplakia with or without epithelial dysplasia. J Oral Pathol Med. 2003;32:513–21. doi: 10.1034/j.1600-0714.2003.00117.x. [DOI] [PubMed] [Google Scholar]
- 40.Vered M, Dayan D, Yahalom R, Dobriyan A, Barshack I, Bello IO, et al. Cancer-associated fibroblasts and epithelial-mesenchymal transition in metastatic oral tongue squamous cell carcinoma. Int J Cancer. 2010;127:1356–62. doi: 10.1002/ijc.25358. [DOI] [PubMed] [Google Scholar]
- 41.Inki P, Joensuu H, Grenman R, Klemi P, Jalkanen M. Association between syndecan-1 expression and clinical outcome in squamous cell carcinoma of the head and neck. Br J Cancer. 1994;70:319–23. doi: 10.1038/bjc.1994.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inki P, Larjava H, Haapasalmi K, Miettinen HM, Grenman R, Jalkanen M. Expression of syndecan-1 is induced by differentiation and suppressed by malignant transformation of human keratinocytes. Eur J Cell Biol. 1994;63:43–51. [PubMed] [Google Scholar]
- 43.Anttonen A, Kajanti M, Heikkilä P, Jalkanen M, Joensuu H. Syndecan-1 expression has prognostic significance in head and neck carcinoma. Br J Cancer. 1999;79:558–64. doi: 10.1038/sj.bjc.6690088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson LL, Wade Z, Hessler RB, Abdelsayed R, Rogers JB, Gourin CG. Quantitative analysis of syndecan-1 expression in dysplasia and squamous cell carcinoma of the oral cavity. Laryngoscope. 2007;117:868–71. doi: 10.1097/MLG.0b013e318033c810. [DOI] [PubMed] [Google Scholar]
- 45.Saussez S. In reference to quantitative analysis of syndecan-1 expression in dysplasia and squamous cell carcinoma of the oral cavity. Laryngoscope. 2008;118:944–5. doi: 10.1097/MLG.0b013e318165a9f1. [DOI] [PubMed] [Google Scholar]
- 46.Martínez A, Spencer ML, Brethauer U, Grez P, Marchesani FJ, Rojas IG. Reduction of syndecan-1 expression during lip carcinogenesis. J Oral Pathol Med. 2009;38:580–3. doi: 10.1111/j.1600-0714.2009.00761.x. [DOI] [PubMed] [Google Scholar]
- 47.Mukunyadzi P, Liu K, Hanna EY, Suen JY, Fan CY. Induced expression of syndecan-1 in the stroma of head and neck squamous cell carcinoma. Mod Pathol. 2003;16:796–801. doi: 10.1097/01.MP.0000081731.17549.53. [DOI] [PubMed] [Google Scholar]
- 48.Siqueira AS, Gama-de-Souza LN, Arnaud MV, Pinheiro JJ, Jaeger RG. Laminin-derived peptide AG73 regulates migration, invasion, and protease activity of human oral squamous cell carcinoma cells through syndecan-1 and beta1 integrin. Tumour Biol. 2010;31:46–58. doi: 10.1007/s13277-009-0008-x. [DOI] [PubMed] [Google Scholar]
- 49.Nicolosi PA, Dallatomasina A, Perris R: Theranostic impact of NG2/CSPG4 proteoglycan in cancer. Theranostic Journal, in press. [DOI] [PMC free article] [PubMed]
- 50.Benassi MS, Pazzaglia L, Chiechi A, Alberghini M, Conti A, Cattaruzza S, et al. NG2 expression predicts the metastasis formation in soft-tissue sarcoma patients. J Orthop Res. 2009;27:135–40. doi: 10.1002/jor.20694. [DOI] [PubMed] [Google Scholar]
- 51.Warta R, Herold-Mende C, Chaisaingmongkol J, Popanda O, Mock A, Mogler C, et al. Reduced promoter methylation and increased expression of CSPG4 negatively influences survival of HNSCC patients. Int J Cancer. 2014;135:2727–34. doi: 10.1002/ijc.28906. [DOI] [PubMed] [Google Scholar]
- 52.Brazma A, Vilo J. Gene expression data analysis. FEBS Lett. 2000;480:17–24. doi: 10.1016/S0014-5793(00)01772-5. [DOI] [PubMed] [Google Scholar]
- 53.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37(Database issue):D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimpfer A, Schonberg S, Agostinelli C, Pileri SA, Went P, Dirnhofer S. Construction and validation of a bone marrow tissue microarray. J Clin Pathol. 2007;60:57–61. doi: 10.1136/jcp.2005.035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tzankov A, Pehrs AC, Zimpfer A, Ascani S, Lugli A, Pileri S, et al. Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell-like types: a tissue microarray analysis of 90 cases. J Clin Pathol. 2003;56:747–52. doi: 10.1136/jcp.56.10.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Numa F, Hirabayashi K, Kawasaki K, Sakaguchi Y, Sugino N, Suehiro Y, et al. Syndecan-1 expression in cancer of the uterine cervix: association with lymph node metastasis. Int J Oncol. 2002;20:39–43. [PubMed] [Google Scholar]
- 57.Garusi E, Rossi S, Perris R. Antithetic roles of proteoglycans in cancer. Cell Mol Life Sci. 2012;69:553–79. doi: 10.1007/s00018-011-0816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MA, Colvin Wanshura LE, Yang J, Carlson J, Xiang B, Li G, et al. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res. 2011;24:1148–57. doi: 10.1111/j.1755-148X.2011.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoneda A, Lendorf ME, Couchman JR, Multhaupt HA. Breast and ovarian cancers: a survey and possible roles for the cell surface heparan sulfate proteoglycans. J Histochem Cytochem. 2012;60:9–21. doi: 10.1369/0022155411428469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pulkkinen JO, Penttinen M, Jalkanen M, Klemi P, Grénman R. Syndecan-1: a new prognostic marker in laryngeal cancer. Acta Otolaryngol. 1997;117:312–5. doi: 10.3109/00016489709117794. [DOI] [PubMed] [Google Scholar]
- 61.Kurokawa H, Zhang M, Matsumoto S, Yamashita Y, Tanaka T, Takamori K, et al. Reduced syndecan-1 expression is correlated with the histological grade of malignancy at the deep invasive front in oral squamous cell carcinoma. J Oral Pathol Med. 2006;35:301–6. doi: 10.1111/j.1600-0714.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 62.Chen CL, Ou DL. Expression of syndecan-1 (CD138) in nasopharyngeal carcinoma is correlated with advanced stage and poor prognosis. Hum Pathol. 2006;37:1279–85. doi: 10.1016/j.humpath.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 63.Máthé M, Suba Z, Németh Z, Tátrai P, Füle T, Borgulya G, et al. Stromal syndecan-1 expression is an adverse prognostic factor in oral carcinomas. Oral Oncol. 2006;42:493–500. doi: 10.1016/j.oraloncology.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Kim YJ, Go H, Wu HG, Jeon YK, Park SW, Lee SH. Immunohistochemical study identifying prognostic biomolecular markers in nasopharyngeal carcinoma treated by radiotherapy. Head Neck. 2011;33:1458–66. doi: 10.1002/hed.21611. [DOI] [PubMed] [Google Scholar]
- 65.Mikami S, Ohashi K, Usui Y, Nemoto T, Katsube K, Yanagishita M, et al. Loss of syndecan-1 and increased expression of heparanase in invasive esophageal carcinomas. Jpn J Cancer Res. 2001;92:1062–73. doi: 10.1111/j.1349-7006.2001.tb01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Contreras HR, Ledezma RA, Vergara J, Cifuentes F, Barra C, Cabello P, et al. The expression of syndecan-1 and −2 is associated with Gleason score and epithelial-mesenchymal transition markers, E-cadherin and beta-catenin, in prostate cancer. Urol Oncol. 2010;28:534–40. doi: 10.1016/j.urolonc.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 67.Wang H, Si JL, Zhang XZ, Qi YQ, Niu ZY, Zhou CH. Expression and clinical significance of syndecan-1 mRNA and HPA-1 mRNA in colorectal cancer detected with real-time fluorescent quantitative polymerase chain reaction. Chin J Cancer. 2010;29:288–93. doi: 10.5732/cjc.009.10502. [DOI] [PubMed] [Google Scholar]
- 68.Lee SH, Choi EJ, Kim MS, Park JW, Lee YS, Kim SY, et al. Prognostic significance of syndecan-1 expression in squamous cell carcinoma of the tonsil. Int J Clin Oncol. 2014;19:247–53. doi: 10.1007/s10147-013-0552-7. [DOI] [PubMed] [Google Scholar]
- 69.Huang X, Xiao DW, Xu LY, Zhong HJ, Liao LD, Xie ZF, et al. Prognostic significance of altered expression of SDC2 and CYR61 in esophageal squamous cell carcinoma. Oncol Rep. 2009;21:1123–9. doi: 10.3892/or_00000332. [DOI] [PubMed] [Google Scholar]
- 70.Popović A, Demirović A, Spajić B, Stimac G, Kruslin B, Tomas D. Expression and prognostic role of syndecan-2 in prostate cancer. Prostate Cancer Prostatic Dis. 2010;13:78–82. doi: 10.1038/pcan.2009.43. [DOI] [PubMed] [Google Scholar]
- 71.Lin H, Huber R, Schlessinger D, Morin PJ. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59:807–10. [PubMed] [Google Scholar]
- 72.Murthy SS, Shen T, De Rienzo A, Lee WC, Ferriola PC, Jhanwar SC, et al. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–6. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 73.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20:7408–12. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 74.Chang CC, Campoli M, Luo W, Zhao W, Zaenker KS, Ferrone S. Immunotherapy of melanoma targeting human high molecular weight melanoma-associated antigen: potential role of nonimmunological mechanisms. Ann N Y Acad Sci. 2004;1028:340–50. doi: 10.1196/annals.1322.040. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J Natl Cancer Inst. 2010;102:1496–512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, Planagumà J, et al. Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, et al. CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clin Cancer Res. 2012;18:5352–63. doi: 10.1158/1078-0432.CCR-12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cattaruzza S, Nicolosi PA, Braghetta P, Pazzaglia L, Benassi MS, Picci P, et al. NG2/CSPG4-collagen type VI interplays putatively involved in the microenvironmental control of tumour engraftment and local expansion. J Mol Cell Biol. 2013;5:176–93. doi: 10.1093/jmcb/mjt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, Wang Y, Yu L, Sakakura K, Visus C, Schwab JH, et al. CSPG4 in cancer: multiple roles. Curr Mol Med. 2010;10:419–29. doi: 10.2174/156652410791316977. [DOI] [PubMed] [Google Scholar]
- 80.Campoli M, Ferrone S, Wang X. Functional and clinical relevance of chondroitin sulfate proteoglycan 4. Adv Cancer Res. 2010;109:73–121. doi: 10.1016/B978-0-12-380890-5.00003-X. [DOI] [PubMed] [Google Scholar]
- 81.Kelly-Goss MR, Sweat RS, Stapor PC, Peirce SM, Murfee WL. Targeting pericytes for angiogenic therapies. Microcirculation. 2014;21:345–57. doi: 10.1111/micc.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Bie P, Leida Z, Xia Z, Bai L. NG2/CSPG4 proteoglycan as a novel prognostic indicator and therapeutic target in malignant cancer. J Stem Cell Res Ther. 2014;4:171. doi: 10.4172/2157-7633.1000238. [DOI] [Google Scholar]


