Abstract
Reprogramming somatic cells from one cell fate to another can generate specific neurons suitable for disease modeling. To maximize the utility of patient-derived neurons, they must model not only disease-relevant cell classes but also the diversity of neuronal subtypes found in vivo and the pathophysiological changes that underlie specific clinical diseases. Here, we identify five transcription factors that reprogram mouse and human fibroblasts into noxious stimulus-detecting (nociceptor) neurons that recapitulate the expression of quintessential nociceptor-specific functional receptors and channels found in adult mouse nociceptor neurons as well as native subtype diversity. Moreover, the derived nociceptor neurons exhibit TrpV1 sensitization to the inflammatory mediator prostaglandin E2 and the chemotherapeutic drug oxaliplatin, modeling the inherent mechanisms underlying inflammatory pain hypersensitivity and painful chemotherapy-induced neuropathy. Using fibroblasts from patients with familial dysautonomia (hereditary sensory and autonomic neuropathy type III), we show that the technique can reveal novel aspects of human disease phenotypes in vitro.
Directed differentiation from pluripotent stem cells and lineage reprogramming of fibroblasts can both be used to derive a wide range of different neuronal subtypes1,2. While the known sequence of morphogen exposure and consequent molecular changes in the development of specific neurons can guide directed differentiation strategies, the selection of transcription factors for lineage reprogramming from fibroblasts remains essentially empirical. No single transcription factor has proved essential for driving cell fates in all neuronal reprogramming studies to date, despite the fact that specific factors such as Ascl1 or Ngn2 seem particularly potent in deriving a range of different neuronal subtypes3. Brn2, Ascl1 and Myt1l (abbreviated BAM) generate generic neurons on their own4 and specific neuronal subtypes when combined with additional factors5,6. Moreover, the developmental stage at which a particular transcription factor acts in vivo may determine whether that factor facilitates or inhibits the patterning of reprogrammed neurons4,5.
Nociceptors are the first-order neurons in the pain sensory transduction pathway and play the critical initial step in the detection of noxious stimuli (nociception) and the development of inflammatory and neuropathic pain7. Nociceptor neurons employ a host of highly specific ionotropic receptors and ion channels, including TrpV1, TrpA1, TrpM8 and P2X3 receptors to transduce stimuli, as well as slow, tetrodotoxin (TTX)-resistant sodium channels (NaV1.8 and NaV1.9) that generate their characteristic broad action potentials8. Efforts to derive nociceptors using a small molecule-based directed differentiation strategy from human neural crest precursors have produced neurons that recreate some but not all of these characteristic receptors and channels9.
Mutations in nociceptor-specific membrane proteins underlie a wide range of pain diseases, including rare but severe channelopathies due to NaV1.7 or TrpA1 mutations10, common small fiber neuropathies due to activating mutations in NaV1.7 or NaV1.811,12, as well as a variety of pain-predisposing polymorphisms13,14; however, the biological effects of these mutations on nociception have not been studied in human sensory neurons. Nociceptors normally activate only following intense, potentially damaging stimuli in order to provide a protective warning of imminent tissue injury. However, they also have the remarkable capacity to become sensitized after exposure to inflammatory mediators15,16 or by chemotherapeutic drugs17, resulting in a reduced activation threshold so that innocuous stimuli can generate a pain response. Pain hypersensitivity can play a physiologically useful role in minimizing further injury and in promoting healing once damage has occurred; however, such transient sensitization, when it persists, promotes the development of chronic pain.
Nociceptor neuron development occurs through dorsalization within the neural tube18, followed by neural crest induction and migration19 and then nociceptor specification within the still-multipotent neural crest lineage20. The generation of nociceptor progenitors expressing the TrkA neurotrophin receptor (Ntrk1) and postnatal nociceptors expressing TrpV1 requires the basic helix-loop-helix transcription factor Ngn1 (Neurogenin1), which is normally present from approximately days E9–E13 in the embryonic mouse21. Although developing nociceptors express multiple Trk-family receptors, maturing nociceptors express only TrkA. Brn3a (POU4F1) promotes Runx1 expression, which together with Isl1 (Islet 1) and Klf7 maintain TrkA expression in developing nociceptors22–25. A subset of nociceptors that become the peptidergic subclass of nociceptors maintain TrkA expression and express calcitonin gene-related peptide (CGRP, CalcA) and substance P. For non-peptidergic nociceptors, most of which bind isolectin B4, the glial cell derived neurotrophic factor (GDNF) receptor Ret replaces TrkA in a process dependent on Runx1, and loss of Runx1 markedly reduces TrpV1 expression23.
We set out to produce nociceptor neurons through transcription-mediated lineage conversion of fibroblasts. From an initial set of 12 factors, we find that expressing five factors is sufficient to generate functional mouse nociceptor neurons. In a direct comparison between the induced nociceptors and primary adult mouse nociceptors, we show that the induced neurons mimic bona fide nociceptors with regard to the function of the specific individual receptors and channels, such as TrpA1, TrpM8, P2X3 and NaV1.8, as well as with regard to the population diversity. We demonstrate that the induced neurons also model inflammatory peripheral sensitization, a critical process that underlies transient pain hypersensitivity and contributes to the pathological transition to chronic pain, as well as sensitization following exposure to the chemotherapeutic drug oxaliplatin. Finally, we derive human nociceptor neurons from patients with familial dysautonomia (FD) and show that these neurons reveal potentially disease-relevant phenotypes in vitro.
RESULTS
Selection and Optimization of Transcription Factors
We first developed nociceptor reporter mice by taking advantage of an existing TrpV1 Cre-driver26 and floxed tdTomato mice to generate TrpV1-Cre+/−::tdTomato+/− reporter mice, from which we obtained mouse embryonic fibroblasts (MEFs). Activation of the tdTomato reporter signaled the conversion of the MEFs to TrpV1-expressing cells (Supplementary Fig. 1). We began with nine transcription factors selected to promote lineage conversion to nociceptors in combination with the three BAM factors (12 total, Table 1). These factors were chosen using a combination of the prior literature, transcription factor expression profiles in FACS-sorted adult mouse nociceptors (NaV1.8-positive) as compared to proprioceptors (parvalbumin-positive) (Chiu et al, submitted), an expression profile similarity to NaV1.8 in the BioGPS database27, and postnatal dorsal root ganglion (DRG) expression in the Allen Brain Atlas28.
Table 1.
Candidate transcriptions factors for lineage conversion to nociceptor neurons. Lit, literature; Exp, transcriptome of sorted nociceptors compared to proprioceptors; BioGPS27.
| Gene | Source | Family | Role in Reprogramming/ Sensory System |
|---|---|---|---|
| Ascl1 | Lit | Basic helix-loop-helix / achaete-scute | Neuronal lineage reprogramming4 |
| Drgx (Drg11) | Lit | Helix-turn-helix / Paired box | Survival of peptidergic and non-peptidergic nociceptors48 |
| Ebf1 | Exp | Zinc-finger | Downstream of Ngn49 |
| Etv1 | Exp | Helix-turn-helix/tryptophan clusters | Proprioceptive marker24 |
| Isl2 | Exp, BioGPS | Homeo-domain / LIM region | Unknown |
| Klf7 | Lit, Exp, BioGPS | Zinc-finger / Krueppel like | TrkA maintenance25 |
| Myt1L | Lit | Zinc-finger | Neuronal lineage reprogramming4 |
| Ngn1 | Lit | Basic helix-loop-helix | TrkA and subsequent TrpV1 expression21 |
| Pknox2 | Exp | Homeo-domain / TALE | Unknown |
| Pou4f1 (Brn3a) | Lit | Homeo-domain / POU (Class IV) | Neuronal lineage reprogramming4 |
| Runx1 | Lit | β-scaffold / Runt | Non-peptidergic identify and TrpV1 expression23 |
| Tlx3 | Lit | Helix-turn-helix / homeo-domain | Glutamatergic identity50 |
As expected, there was no baseline activation of the tdTomato reporter in MEFs (not shown). Staining of MEFs for neuronal precursor markers using antibodies to Nestin, Sox1, and Ki67, as well as for neuron-specific class III β-tubulin (Tuj1), were all negative (Supplementary Fig. 2). After transducing the fibroblasts with a combination of all 12 individual retroviruses containing the selected transcription factors, we detected a small number of tdTomato-positive cells with a neuronal morphology after two weeks (Supplementary Fig. 3a). In order to identify those transcription factors that were either critical for or inhibitory to lineage reprogramming into TrpV1-expressing cells, we then sequentially eliminated each factor, one at a time. Surprisingly, the omission of some transcription factors strongly supported by the literature for a role in promotingTrpV1 expression, such as Runx1, did not eliminate TrpV1 reporter expression (Supplementary Fig 3b). In fact, omission of Brn3a led to a marked increase in the number of tdTomato-positive cells (Supplementary Fig. 3c). We identified from this iterative process three factors that were critical to the TrpV1 lineage reprogramming process in that their omission led to a near complete absence of tdTomato and neuronal Class III β-tubulin (Tuj1)-positive cells bearing a neuronal morphology: Ascl1, Myt1l and Klf7 (Supplementary Fig. 3d–f).
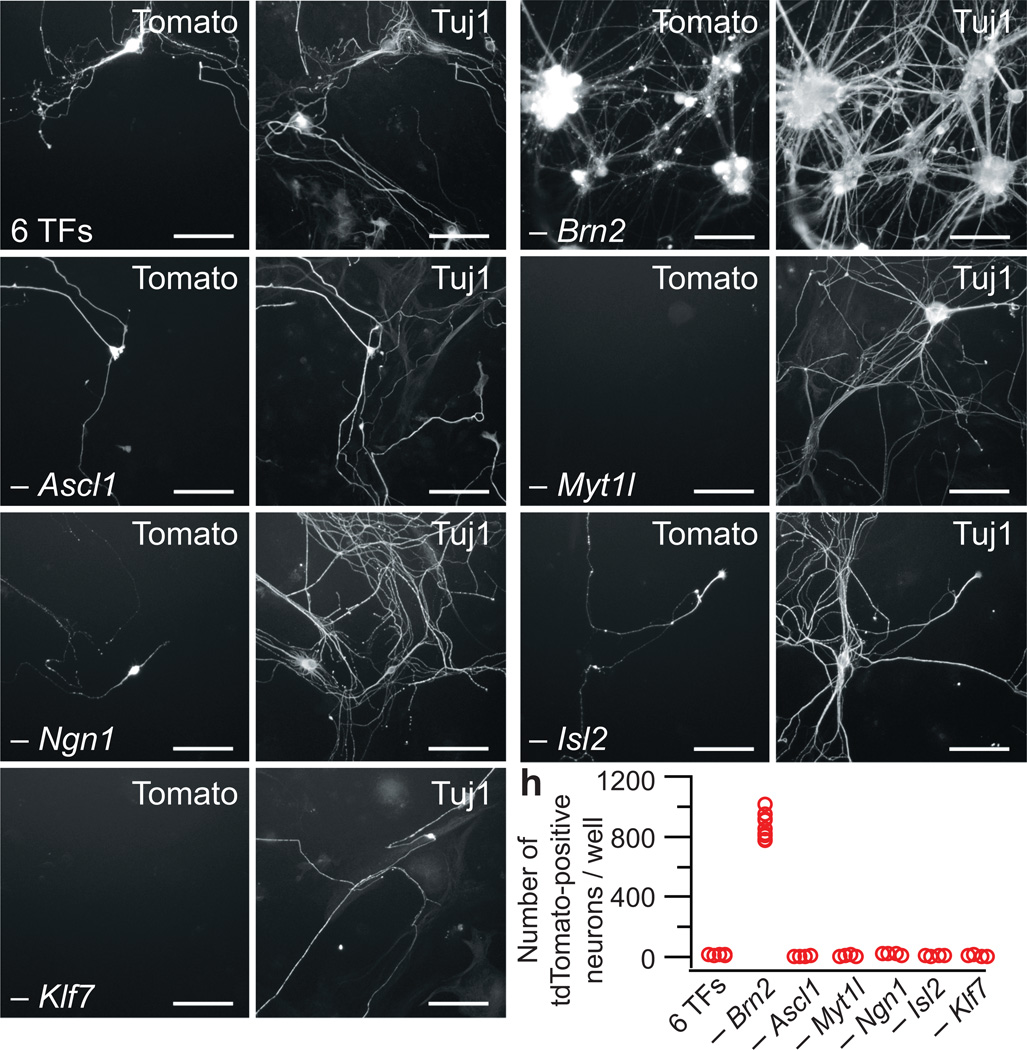
When we combined the three BAM factors with Isl2, Ngn1, and Klf7, we again observed only a small number of tdTomato, Tuj1-positive cells (Fig. 1a). Because prior studies5 and our initial drop out experiments detected specific factors that could inhibit the lineage reprogramming process, we performed single factor dropouts from these six factors and found that omission of Brn2 led to a striking increase in the number of tomato-positive neurons (Fig. 1b), giving a yield of approximately 14% of plated fibroblasts that were both tdTomato- and Tuj1-positive (less than 0.1% were tdTomato-positive but Tuj1-negative). Removal of any other factor from the six sharply reduced the number of tdTomato-positive neurons (Fig. 1c–h). Next, we evaluated Ngn1 alone and in combination with the BAM factors; however, the yield was much lower than with the optimized five factor combination (Supplementary Fig. 4). Indeed, further removal of any of the five factors resulted in a marked decrease in tdTomato, Tuj1-positive cells (Supplementary Fig. 5).
Figure 1.
Combinations of transcription factors result in nociceptor production. (a) Few tdTomato, Tuj1-positive neurons are produced by the combination of six factors (6 TFs): Brn2, Ascl1, Myt1l, Ngn1, Isl2 and Klf7. (b) Removal of Brn2 markedly increases the number of tdTomato, Tuj1-positive neurons. (c–g) Omission of Ascl1 (c), Myt1l (d), Ngn1 (e), Isl2 (f) or Klf7 (g) from the six factors disrupts the generation of nociceptor neurons. Representative images for each transcription factor drop out were taken from n=4 wells from two separate transductions. Scale bars: 100 µm. (h) Quantification of the single factor dropout studies from n=4 wells from each of two separate independent transductions for each category.
Molecular Characterization of Induced Mouse Nociceptors
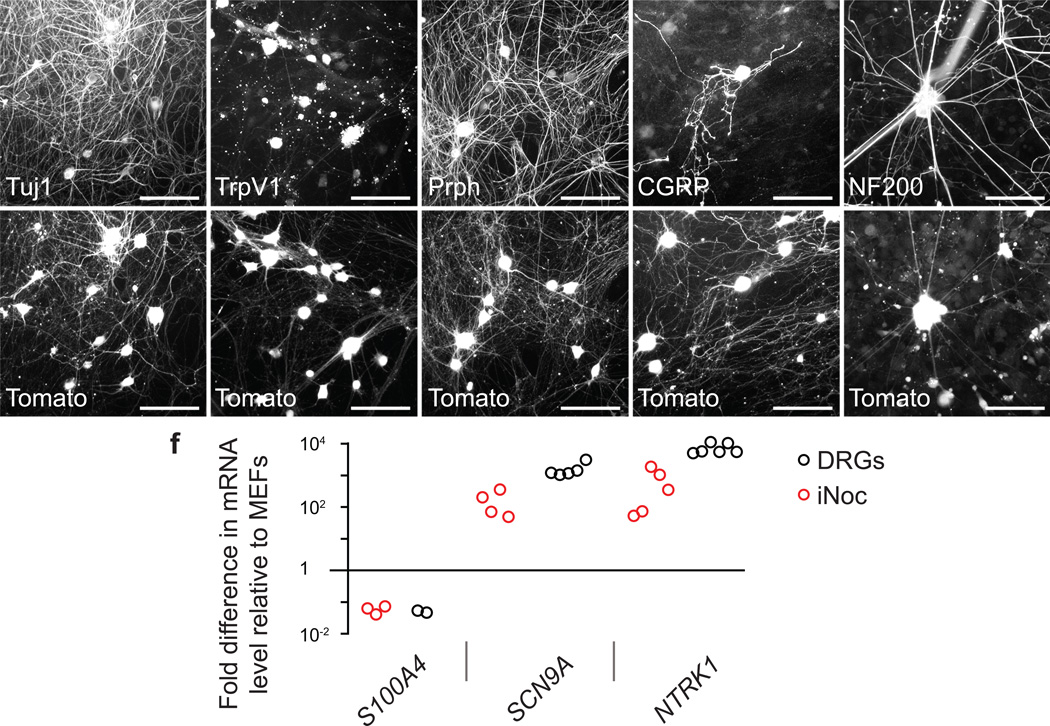
To determine if tdTomato-positive reprogrammed neurons phenocopied bona fide nociceptors, we evaluated the expression of protein markers specific for nociceptor neurons. Nearly all tdTomato-positive neurons stained for the pan-neuronal marker Tuj1 and had a neuronal-like morphology with many long branching axons, and most Tuj1-positive neurons were tdTomato-positive (Fig. 2a). Staining with an anti-TrpV1 antibody confirmed the translation of the TrpV1 protein in the vast majority of tdTomato-positive neurons (Fig. 2b). In mouse dorsal root ganglia, most TrpV1-expressing neurons are C-fibers that express the marker peripherin (Prph)29, while only a small percentage of A-δ fibers are also TrpV1-positive26. In the induced neurons, a majority of tdTomato-positive neurons expressed peripherin (66.9 ± 4.1%, n=16 wells from 4 separate transductions) (Fig. 2c) and many CGRP (22.3 ± 6.6%, n=4 wells from 2 separate transductions) (Fig 2d); however, a smaller number of cells stained for the intermediate filament NF200, a marker, in this context, of myelinated A-δ nociceptors (Fig. 2e). In contrast, the derived nociceptors did not stain for smooth muscle actin (SMA), a marker of muscle, despite reports of TrpV1 expression in muscle30 (Supplementary Fig. 6a,b). Furthermore, neurons derived from the three BAM factors did not express nociceptor markers, consistent with their high specificity (Supplementary Fig. 6c–g).
Figure 2.
Induced nociceptors (iNoc) express characteristic nociceptor genes. (a,b) Tuj1 (a) and TrpV1 (b) expression in fibroblast-derived nociceptor neurons. (c) Most induced nociceptors stain for the C-fiber marker peripherin (Prph). (d) A number of induced nociceptors express the peptidergic-marker CGRP. (e) A small number of induced nociceptors express the intermediate filament marker NF200 found in myelinated fibers. Representative images were selected from immunostaining that was repeated in n=4 wells from two independent transductions. Scale bars: 100 µm. (f) RT-qPCR data showing expression levels of nociceptor-specific genes in 50-picked tdTomato-positive primary adult mouse nociceptors (DRGs, black circles) and 50-picked tdTomato-positive induced nociceptors (red circles), relative to their levels in MEFs, from a minimum of two independent biological replicates (biological replicates represented as independent circles).
Because specific antibodies do not exist for many quintessential nociceptor proteins, we utilized quantitative RT-PCR to compare nociceptor marker mRNA levels in tdTomato-positive induced nociceptors and tdTomato-positive adult mouse nociceptors relative to levels in MEFs (Fig. 2f). For this analysis, we used patch pipettes to pick tdTomato-positive induced and primary mouse neurons, as well as MEFs, and plotted the levels of specific transcripts in induced and primary nociceptors relative to MEFs. The fibroblast marker S100A4 was expressed at a similar very low level in both the induced and primary nociceptors, consistent with a non-fibroblast identity of the induced nociceptors. NaV1.7 (Scn9a), which is found in nociceptor and autonomic peripheral neurons, was present in both the induced and primary nociceptors, as was TrkA (NTRK1), which is turned on in developing nociceptors and persists in the peptidergic subset of mature nociceptors, although the expression of NaV1.7 and TrkA in the induced neurons was several fold less than in the primary DRGs. Together, these immunohistochemistry and PCR data suggest that the induced neurons express a complement of bona fide nociceptor-specific markers.
Functional Properties of Induced Mouse Nociceptors
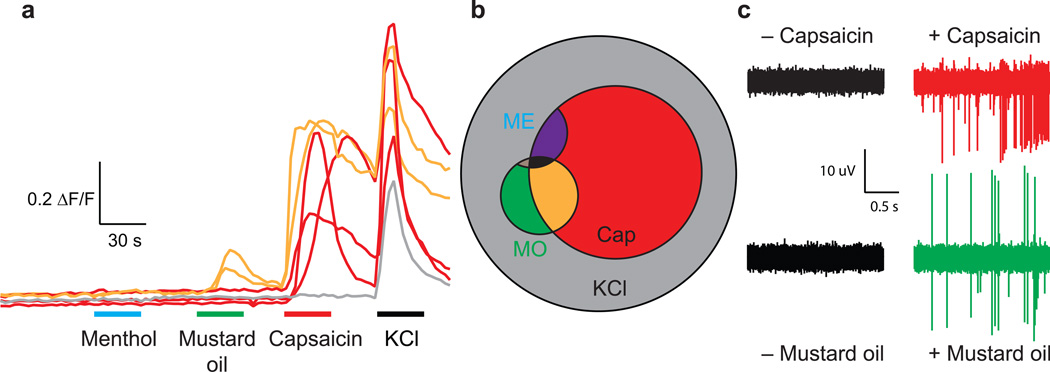
In order to investigate the functional properties of the induced nociceptors, we performed calcium imaging with a battery of agonists and evaluated the number of responders within the tdTomato-positive population with a stable baseline and response to potassium chloride (KCl, which activates voltage-gated calcium channels through depolarization and serves as a measure of neuronal functional integrity) (Fig. 3a). We chose concentrations of agonists for TrpM8 (250 µM menthol), TrpA1 (100 µM mustard oil) and TrpV1 (1 µM capsaicin) that only activated their respective cognate receptors31. 39% of the cells responded to capsaicin, 9% to mustard oil and 3% to menthol (Fig. 3a,b; n=227 tdTomato-positive cells that responded to KCl). We observed occasional cells that responded to both mustard oil and capsaicin, a single cell that responded to menthol and mustard oil but not capsaicin, and one cell that responded to all three agonists. We did not observe any tdTomato-positive cells that responded to menthol alone, but we did identify a small number of tdTomato-negative cells that responded to menthol but not the other Trp agonists (Supplementary Fig. 7a). In contrast, 0/50 KCl-responding neurons derived from the BAM factors alone responded to capsaicin (not shown). Using the same experimental procedure, we then asked how the frequencies of the different combinations of receptors within individual neurons compared between induced nociceptors and adult mouse nociceptors. In tdTomato-positive primary DRG neurons dissected and cultured from adult TrpV1::tdTomato mice, we found that 36% of the neurons responded to capsaicin, 2.5% to mustard oil and 2.5% to menthol (Supplementary Fig. 7b,c; n=249 tdTomato-positive cells that responded to KCl). Thus, the nociceptor lineage reprogramming not only yielded physiologically functional TrpV1, TrpA1 and TrpM8 proteins in the induced neurons, but the frequencies and combinations of the different receptors in the induced neurons closely mimicked those of adult mouse nociceptors.
Figure 3.
Induced neurons respond to different Trp channel agonists. (a) Sample calcium imaging responses to sequential application of menthol (250 µM), mustard oil (100 µM), capsaicin (1 µM), and potassium chloride (40 mM) in a single dish of induced tdTomato-positive derived nociceptors. Traces are representative recordings from n=227 tdTomato-positive / KCl-responding cells cultured in 19 dishes from 3 independent transductions (b) Venn diagram showing subgroups of tdTomato-positive cells that responded to KCl (40 mM, grey), capsaicin (Cap, 1 µM, red, 39%), mustard oil (MO, 100 µM, lower small circle, green, 9%) and menthol (ME, 250 µM, upper small partial circle, blue, 3%; note that no tdTomato-positive cells responded to menthol alone) (c) Sample electrodes from extracellular multi-electrode array recordings of induced neurons before (left) and after (right) the application of capsaicin (1 µM, upper) and mustard oil (100 µM, lower). Sample recordings for each agonist are indicative of results from three experiments across two independent transductions, in which all replicates showed an increase in firing after agonist application.
While calcium imaging provides detailed information about calcium entry through Trp channels, it does not evaluate whether activation of these channel evokes action potential firing in the neurons. By culturing the induced neurons on extracellular multi-electrode arrays, we found that both capsaicin and mustard oil application evoked robust action potential firing from the induced neurons (Fig. 3c; 3/3 arrays for capsaicin and 3/3 arrays for mustard oil).
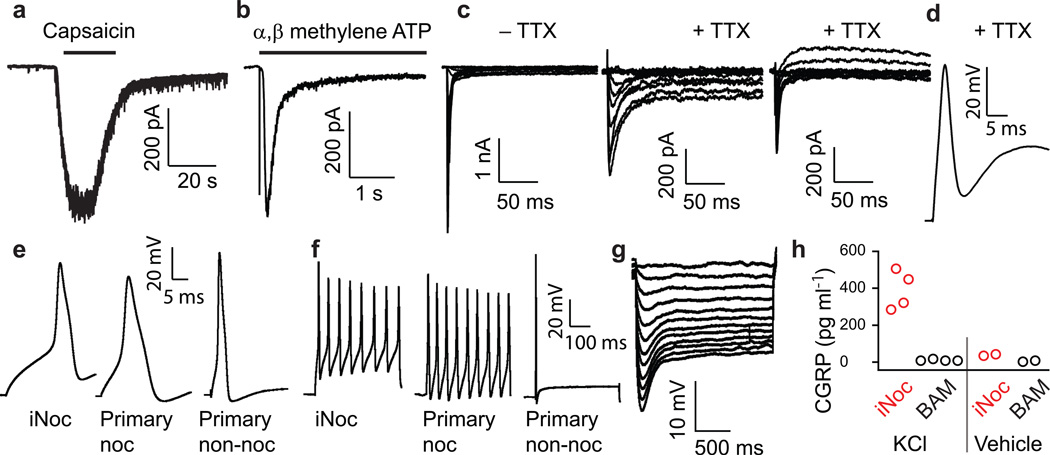
We next used whole-cell patch clamp recordings to define the electrophysiological properties of the induced nociceptors and found that capsaicin (1 µM) elicited inward currents in 6/11 tdTomato-positive induced neurons, consistent with but somewhat higher than the calcium imaging results (Fig. 4a). The P2X3 subtype of ionotropic purinergic receptors is expressed specifically in nociceptor neurons32. Application of the P2X3-specific agonist α, β-methylene-ATP (30 µM) elicited rapidly-adapting inward currents in 8/16 neurons (Fig. 4b) that were blocked completely by A-397491, a specific P2X3 antagonist, in 4/4 neurons (not shown)33.
Figure 4.
Whole-cell patch clamp recordings of tdTomato-positive induced nociceptors. (a) Current recording in response to treatment with 1 µM capsaicin (6/11 induced neurons responded). (b) Current recording following the application of 30 µM α, β-methylene-ATP (8/16 induced neurons responded). (c) Inward currents following step depolarization before (left) and after (middle) the application of 300 nM tetrodotoxin (TTX) (14/15 induced neurons had TTX-resistant sodium currents greater than 50 pA). Right panel shows a different neuron without a persistent TTX-resistant sodium current (d) Action potential firing elicited by depolarizing current in the presence of 300 nM TTX (7/12 cells fired single TTX-resistant action potentials with peak greater than 0 mV). (e, f) Examples of individual action potentials (e) and trains (f) elicited from induced nociceptors (iNoc), tdTomato-positive primary adult nociceptors (Primary noc) and tdTomato-negative primary adult non-nociceptors (Primary non-noc) (12/13 induced neurons fired tonically). (g) Examples of sag depolarizations in response to hyperpolarizing current injections in induced nociceptors (11/17 induced neurons produced a sag depolarization). (h) CGRP was released from induced nociceptors (5F), but not BAM-derived neurons, in response to KCl (80 mM), but not vehicle. Mean (SEM) for 5F and BAM following KCl stimulation are 390.4 (52.5) and 10.3 (2.6) pg/ml (n=4; Mann-Whitney U-test p=0.03).
Perhaps the most nociceptor-specific functional marker is the TTX-resistant NaV1.8 sodium channel, which produces a portion of the current in the nociceptor action potential upstroke34. In voltage-clamp, we found that depolarizing voltage steps elicited inward sodium currents both before and after the application of 300 nM TTX (Fig. 4c; 14/15 recorded induced nociceptors had TTX-resistant sodium currents greater than 50 pA). In agreement with our expression studies, the slow channel kinetics of the TTX-resistant currents are typical for NaV1.8 as opposed to the fast NaV1.5 cardiac sodium channel, which is present in developing embryonic nociceptors35. Furthermore, five of the 14 neurons with TTX-resistant sodium currents also exhibited a persistent sodium component, which previous studies have found to be due to NaV1.934,36 (Fig. 4c). The ability to generate action potentials in the presence of TTX is a feature of nociceptors but not of other DRG or central neurons. The induced neurons fired single TTX-resistant action potentials that overshot 0 mV in 7/12 neurons (Fig. 4d). NaV1.8 is responsible for the characteristic broad action potential shape of the nociceptor action potential8, which we found to be a property of the induced neurons (mean action potential width 3.32 ± 0.33 ms; n=13); as expected, adult primary tdTomato-positive nociceptors fired broad action potentials, but not large tdTomato-negative primary non-nociceptor DRG neurons (Fig. 4e). In addition to differences in action potential morphology, the firing pattern of nociceptor neurons to prolonged depolarizing currents is tonic, compared to the phasic firing of most large A-β DRG neurons37. Induced nociceptors fired tonic action potential trains in response to depolarizing current steps in 12/13 cells, consistent with the tonic firing found in tdTomato-positive adult primary mouse nociceptors, and in contrast to the single action potentials elicited in non-nociceptor, large tdTomato-negative adult DRG neurons (Fig. 4f).
While hyperpolarization-activated cyclic nucleotide-sensitive (HCN) currents are not specific for nociceptor neurons, they play an important role within these cells in neuropathic and inflammatory pain38, and thus their presence may be important for disease-modeling. We found that the induced nociceptors produced typical sag depolarizations in response to hyperpolarization (Fig. 4g) in 11/17 tdTomato-positive induced neurons, consistent with ZD7288-sensitive HCN currents recorded in voltage clamp (2/2, not shown).
A critical function of peptidergic neurons, most of which express TrpV17, is to release neuropeptides such as CGRP and Substance P. To assess the fidelity of the induced nociceptors in this capacity, we measured CGRP levels in supernatant following a depolarizing stimulus and found that induced nociceptors, but not BAM-derived neurons, released CGRP after KCl stimulation (Fig. 4h; n=4; Mann-Whitney U-test p=0.03). The concentrations of CGRP released by the induced neurons were comparable to those released by primary DRG neurons (Supplementary Fig. 8), thus indicating that the induced neurons have synaptic vesicle release mechanisms in place.
Induced Nociceptors Model Inflammatory Sensitization
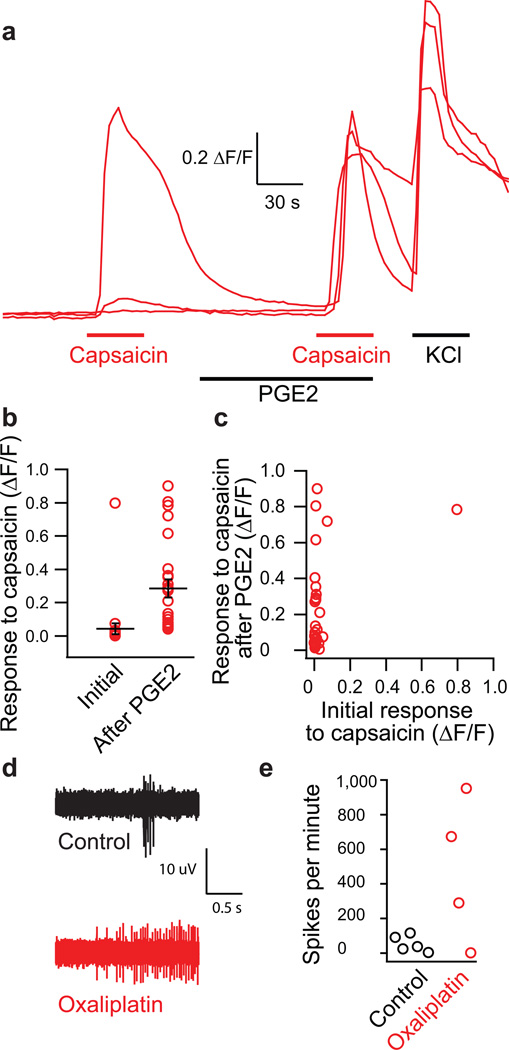
The transition from high-threshold baseline nociception to low-threshold clinical pain hypersensitivity commonly involves peripheral sensitization of nociceptors. For the induced nociceptors to be valuable in vitro models of in vivo pathophysiology, they must replicate not only the specific functional channels and receptors of the cells but also the process of sensitization that leads to pathological pain. Prostaglandin E2 (PGE2) activates the PKA pathway and sensitizes the TrpV1 receptor, reducing its threshold and decreasing desensitization15,16. In the tdTomato-positive induced neurons, a low concentration (300 nM) of capsaicin rarely yielded a detectable response (mean change in fluorescence absorption ratio of 0.028 ± 3.0*10−3) (Fig. 5a,b). However, after treatment with PGE2 (1 µM) for two minutes, a second identical capsaicin (300 nM) application yielded a mean response of 0.18 ± 6.0*10−3 (n=41 cells; paired t-test p=1.5×10−4). Plotting the magnitudes of the initial capsaicin and PGE2-sensitized capsaicin responses revealed that although the majority of neurons exhibited small or undetectable initial responses to capsaicin, they produced robust signals after PGE2 sensitization (Fig. 5c).
Figure 5.
Sensitization of induced nociceptors treated with the inflammatory mediator PGE2 and the chemotherapeutic drug oxaliplatin. (a) Sample calcium imaging recordings of induced nociceptors treated with 300 nM capsaicin before and after treatment with 1 µM PGE2 from recordings of n=41 tdTomato-positive / KCl-responding cells. (b) Plot of individual and mean response amplitudes for initial and PGE2-sensitized capsaicin treatments (paired t-test p=1.5×10−4). (c) Plot of initial versus PGE2-sensitized capsaicin response amplitudes for individual induced neurons. (d) Sample traces from extracellular multi-electrode array recordings of induced neurons in response to 300 nM capsaicin following 10 minute exposure to vehicle control (n=5 MEAs) or oxaliplatin (50 µM; n=4 MEAs) on induced neurons from two separate transductions. (e) Quantification of spikes per minute from induced nociceptors in response to capsaicin alone (control) and capsaicin following oxaliplatin treatment. Error bars are ± SEM.
TrpV1 sensitization also may contribute to painful chemotherapy-induced neuropathy due to oxaliplatin17. Using MEA recording, we compared capsaicin responses in induced nociceptors treated with either 50 µM oxaliplatin or vehicle control, and found marked sensitization in the oxaliplatin-treated nociceptors (Fig. 5d,e).
Induction of Human Nociceptors
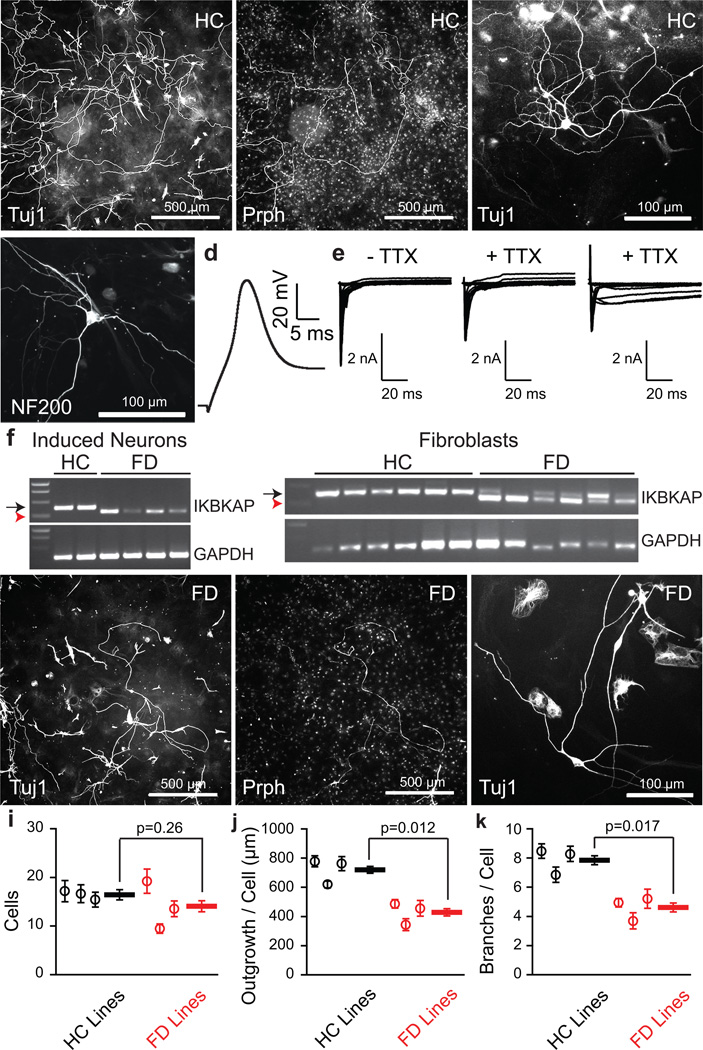
To derive nociceptors from human fibroblasts, we initially included NeuroD1 in the nociceptor induction protocol, as this transcription factor was important in prior human lineage reprogramming studies39. However, we found that the reprogramming efficiency, was greater without NeuroD1 (five factors) than with NeuroD1 (six factors) (20.7 ± 1.4 cells per field for five factors; 9.7 ± 1.1 cells per field for six factors, n=6 wells/group; t-test p=1.0×10−4) (Supplementary Fig. 9). Furthermore, more neurons exhibited larger sodium currents (67% of patched five factor neurons had peak transient sodium currents greater than 500 pA, versus 29% of six factor neurons) and five factor neurons were healthier (resting Vm −49.3 ± 2.2 mV, n=33 five factor neurons; Vm −37.3 ± 3.2, n=20 six factor neurons; Mann-Whitney U-test p-value = 0.001). Using healthy control (HC) subject fibroblasts, the 5 factors yielded Tuj1-positive neurons at an efficiency of 5% of plated fibroblasts, and 16% of the Tuj1-positive neurons were also peripherin-positive (Fig. 6a,b), efficiencies that were somewhat lower than the mouse induced nociceptors. A small number of the Tuj1-positive neurons were NF200-positive (Fig. 6c). We recorded from the neurons using whole-cell patch clamp. Although we did not have a reporter for a particular neuronal subtype, the induced human neurons fired broad action potentials (mean action potential width 3.88 ± 0.41 ms; n=17; Fig. 6d), consistent with functional nociceptors. In 38 voltage clamp recordings, we applied TTX to neurons with a large total sodium current (greater than 1 nA) and detected TTX-resistant sodium currents in 10/10 neurons (Fig. 6e). As in both our mouse induced nociceptors and primary mouse and human nociceptors34,36, the induced human neurons had different combinations of slow- and persistent TTX-resistant sodium currents, consistent with NaV1.8 and NaV1.9 contributions, respectively (Fig. 6e).
Figure 6.
Human fibroblast-derived neurons for human disease modeling. (a) Low magnification of Tuj1 (left) and peripherin (Prph, right) staining of healthy control (HC)-derived neurons. Scale bars: 500 µm. (b) High magnification of Tuj1 staining of HC-derived neurons. Scale bar: 100 µm. (c) NF200-positive cell derived from HC fibroblasts. (d) Current recording of an action potential train from a HC-derived neuron (17/33 induced neurons with peak Na current > 500 pA fired at least one action potential with peak greater than 0 mV). (e) Total (left) and TTX-resistant (middle) sodium currents from a single HC-derived neuron. Right panel shows persistent TTX-resistant sodium current recordings from a separate HC-derived neuron characteristic of Nav1.9. (f) RT-PCR for IKBKAP and GAPDH from single human induced neurons (left) and single human fibroblasts (right) show normal (black arrow) and abnormally spliced (red arrowhead) transcripts. Full-length gels are presented in Supplementary Fig. 10. (g) Low magnification of Tuj1 (left) and peripherin (right) staining of neurons derived from a patient with Familial Dysautonomia (FD). Scale bars: 500 µm. (h) High magnification of Tuj1 staining of FD-derived neurons. Scale bar: 100 µm. For all images, representative images were selected from human neurons generated in n=6 wells from three separate transductions. (i) Quantification of Tuj1-positive neurons in HC and FD-derived nociceptors (random intercept mixed-effects model p=0.26). (j) Neurite outgrowth per cell for HC and FD-derived Tuj1-positive nociceptors (random intercept mixed effects model p=0.012). (k) Number of branches per cell for HC and FD-derived Tuj1-positive nociceptors (random intercept mixed effects model p=0.017). For i–k, images were analyzed from three pairs of age-matched HC and FD patient lines from each of three separate transductions (n=20 wells/line). Error bars are ± SEM.
In order to evaluate the potential of the human neurons for disease modeling, we reprogrammed fibroblasts from three HC and three unrelated, age-matched subjects with familial dysautonomia (FD, hereditary sensory and autonomic neuropathy type III, Riley-Day syndrome), due to a homozygous donor splice site mutation that results in deletion of intron 20 from the I-κ-β kinase complex-associated protein (IKBKAP) RNA40. We found that single FD-derived neurons picked using patch pipettes exclusively expressed the abnormally spliced transcript, something not previously identified, while the HC-derived neurons expressed only the normal transcript (Fig. 6f). FD fibroblasts expressed a mixture of abnormally spliced and normal transcripts, consistent with prior studies41,42, while HC fibroblasts expressed only the normal transcript (Fig. 6f; Supplementary Fig. 10).
Although we detected peripherin-positive, Tuj1-positive neurons from all HC and FD subjects (Fig. 6g,h), the neurons from FD subjects showed a trend toward decrease in number (Fig. 6i; 16.5 ± 1.1 HC neurons/well, n=60 wells; 14.1 ± 1.1 FD neurons/well, n=60 wells; difference between HC neurons/well and FD neurons/well 2.3 ± 1.5, n=60 wells; random intercept mixed-effects model p=0.26) and a robust reduction in neurite outgrowth per cell (Fig. 6j; 725 ± 24 µm per HC neuron, n=60 wells; 433 ± 25 µm per FD neuron, n=60 wells; difference between HC neuron outgrowth per cell/well and FD neuron outgrowth per cell/well 291.3 ± 32.6 µm, n=60 wells; random intercept mixed-effects model p=0.012), as well as number of branches per neuron (Fig. 6k; 7.9 ± 0.3 branches per HC neuron, n=60; 4.7 ± 0.3 branches per FD neuron, n=60 wells; difference between HC braches per neuron/well and FD branches per neuron/well 3.3 ± 0.4, n=60 wells; random intercept mixed-effects model p=0.017) compared to HC-derived neurons.
DISCUSSION
The subjective nature of pain as a human experience confounds its clinical study, raises questions about the relevance of animal models and complicates the development of effective treatments43. Furthermore, limited physiological studies of primary human nociceptors highlight differences between human and rodent nociceptors, including the function of individual channels and receptors as well as their distribution within different nociceptor subtypes44, and thus emphasize the importance of investigating human nociception using human nociceptors. Modeling key mechanistic aspects of human pain processing with derived human cells may enable phenotypic screens for analgesics based on basal and sensitized neurons from chronic pain subjects. Such approaches would improve upon current drug screens that employ heterologously-expressed targets in non-neuronal cells and consequently do not reflect the native scaffolding and molecular signaling present in human neurons, the pathophysiological changes that drive clinical pain and genetic backgrounds that may increase pain susceptibility.
We find that a small number of transcription factors can quite efficiently convert fibroblasts into neurons that express the key specific functional receptors found in bona fide adult nociceptors. While TrpV1 is expressed in a tiny fraction of central neurons30, NaV1.8 and TrpA1 are not expressed within the central nervous system. The collective expression of subsets of these markers defines specific subpopulations7, and indeed to a first approximation, our neurons recreate the combinatorial patterns that define the diversity of TrpV1-expressing nociceptive neuronal cohorts found in primary mouse nociceptors. Interestingly, we seem not to have derived a single nociceptor type but instead have engineered multiple subtypes of cells with properties and frequencies similar to those found in vivo. Three possible explanations will need to be investigated in future studies: first, there may be an autonomous program driving the native lineage diversity that is replicated by our transcription factors; second, non-cell autonomous communication among neurons could influence their identify; third, different identities could reflect different relative levels of transcription factor expression.
While the transcription factor combinations that facilitate induction of specific fates have typically incorporated factors known to play defined roles in the development of those neurons1, some of the factors we used do not have any well-defined developmental role. We chose Isl2 due to its strong differential expression in FACS-sorted nociceptors compared to proprioceptors and an expression pattern similarity to NaV1.827, although nothing is known about its role in nociceptor development. Interestingly, the in situ expression of Isl2 appears much more nociceptor-specific than that of Isl127,28, for which a role in nociceptor development and TrpV1 expression has been documented24, although another report found broader RNA expression of Isl245. Our results also raise questions as to why specific transcription factors facilitate or inhibit the reprogramming both to neuronal and nociceptor lineages. In contrast with prior studies4,5,39, we found that Brn2 inhibited lineage reprogramming of mouse fibroblasts to neurons, and NeuroD1 decreased the efficiency and quality of human neurons. With regard to the nociceptor lineage, the roles of Brn3a and Klf7 in maintaining TrkA expression during embryonic development appear similar in the literature22,25. However, we found that Brn3a markedly inhibits lineage reprogramming to nociceptors while Klf7 promotes it. Runx1, which is active and critical in the later embryonic stages of nociceptor development, does not facilitate the reprogramming process. Ngn1 has a well-characterized role in nociceptor development and eventual TrpV1 expression21, but is involved much earlier in development (E9–E13) compared to the other studied factors20. Thus, while developmental studies may inform the choice of transcription factors for lineage reprogramming, reprogramming studies may themselves provide insight into important developmental pathways and their regulators, recognizing of course that reprogramming may not recapitulate transcription factors expressed in normal development.
The intricacy and specificity of primary nociceptor neuron physiology and the fortunate ability to culture adult primary sensory neurons provide an unusual and well-controlled opportunity to evaluate how closely lineage-reprogrammed neurons resemble the molecular expression, function and maturity of primary adult neurons. We found that the reprogrammed neurons produced functional TrpV1, TrpA1 and TrpM8-expressing neurons in similar relative percentages to those found in primary tdTomato-positive adult mouse nociceptors. In addition, the induced neurons yielded not only functional TTX-resistant action potentials, but also the broad action potential morphology and phasic firing pattern characteristic of nociceptors. Indeed, their function globally was remarkably close to that of adult primary nociceptors by every measure we made, although we cannot exclude other contributions, such as neuronal maturity, to firing pattern and action potential morphology. Future studies will explore the synaptic capacity, neurotrophin dependence, and requirement for sustained viral transgene expression in the derived nociceptors.
Patient-derived neurons would have optimal utility as a drug screening tool if the derived neurons replicate the sequence of pathophysiological events that result in specific clinical diseases. Reprogrammed nociceptors may be particularly useful as an in vitro model for pain, because the pain sensitization process mimicked by the induced nociceptors is one of the major factors that drive the transition to pathological pain. The development of a fluorescent nociceptor-specific marker will enable further optimization of human induced nociceptor maturation as well as more extensive molecular and physiological characterization of the human neurons. Whether nociceptors reprogrammed from individuals with chronic pain or peripheral neuropathy will reveal phenotypic differences compared to control subject-derived nociceptors can be evaluated in future research.
Analysis of FD patient tissue samples40,41 and FD iPSC-derived neural precursors42 have consistently shown the presence of both abnormally and normally spiced transcripts in the same sample. The detection of both splice forms could reflect either the presence of both normal and aberrant splicing in the affected neuronal types or simply the heterogeneity of affected and unaffected tissue types in the samples. Our finding that the normal IKBKAP splice variant is apparently absent in the FD-derived neurons indicates the latter is likely the case and may have implications regarding the extent of splice correction necessary for disease treatment, and may explain the observation in a mouse FD model that small amounts of IKAP are sufficient to revert the phenotype46. The reduction in outgrowth and branching in familial dysautonomia compared to control-derived neurons may reflect the presence of processes similar to those responsible for the progressive decrease in unmyelinated sensory neurons that is observed clinically40 and the loss of TrkA-positive neurons in a mouse model47. The ability to obtain FD nociceptors from patient fibroblasts will facilitate future studies to examine the mechanisms of disease and to screen and evaluate potential treatments. Our experiments illustrate how derived neurons with major features of primary nociceptors can be generated and employed as a model for “pain a dish”.
Online Methods
A methods checklist is available with the supplementary materials.
Fibroblasts
TrpV1-Cre+/−::tdTomato+/− transgenic mice were generated by crossing TrpV1-Cre+/+ mice with tdTomato+/+ reporter mice on a C57Bl6 background (both from Jackson Laboratories). Mouse embryonic fibroblasts (MEFs) were harvested from TrpV1::tdTomato embryos, of either gender, at E12.5, passaged once and frozen at −120°C. Human fibroblasts (all from Coriell Institute) were obtained from three healthy control subjects (GM00969, 2 year old Caucasian female; GM03348, 10 year old Caucasian male; GM00316, 12 year old Caucasian male) and from three age-matched subjects with familial dysautonomia (GM04663, 2 year old Caucasian female; GM04959, 10 year old Caucasian female; GM04899, 12 year old Caucasian female). The use of human lines was approved under the Boston Children’s Hospital Institutional Review Board.
Viruses and transductions
Complimentary DNAs for the 9 candidate factors (obtained from the Dana Farber/ Harvard Cancer Center DNA Resource Core except Ngn1, Tlx3 and Runx1, which were obtained from Q. Ma) were each cloned into the pMXs retroviral expression vector modified to contain a WRE using Gateway technology (Invitrogen). 293T cells were co-transfected with individual viruses and pHDMG and pIKLMV packaging plasmids using Lipofectamine 2000 (Life Technologies). Media was changed to new DMEM (GIBCO), 20% FBS (Invitrogen), 50 U/mL Penicillin/Streptomycin (CellGro) after 16 hours. At that time, fibroblasts were thawed and plated on 24-well plates (25K cells/well), 6-well plates (150K cells/well), 35mm dishes (150K cells/well), or p515A multi-electrode array (MEA) probes (Alpha Med Scientific) (12K cells/MEA) that were previously coated with poly-D-lysine (Sigma), gelatin (Cell Signaling) and laminin (Sigma). Viruses were harvested 24 hours later, concentrated approximately 5 fold using Amicon ultra centrifugal filter units (Millipore) and applied to fibroblasts with 5 µg/ml polybrene (Sigma) (Day 0, transduction). Cortical mouse glia obtained from P0–P2 C57Bl6 mice were added on Day 2 for all but the calcium imaging experiments. Media was switched on Day 4 to N3 media: DMEM/F-12 (GIBCO), N2 and B27 supplements (Life Technologies), glutaMAX (Invitrogen), pen/strep, FGF (20 ng/mL, Millipore) with 5% FBS, along with the neurotrophic factors BDNF, CNTF, GDNF (R&D Systems) at 10 ng/ml each. The TGFβ-Inhibitor RepSox (7.5 µM; Millipore), which has been shown to improve survival of different neuronal types over long-term culture (Ichida and Eggan, unpublished), was added for calcium imaging and human transductions. Media was changed every two days, and on Day 10, NGF was also added to the media (50 ng/mL; Invitrogen).
Immunohistochemistry
Cells were fixed with 4% paraformaldehyde (PFA), washed three times with 1× PBS, incubated in blocking buffer (1% Blocking Reagent (Roche), 0.5% BSA, 0.1% TritonX-100) for one hour at room temperature and stained with primary antibodies overnight at 4°C in blocking buffer. The next day the cells were washed three times with 1× PBS, stained with secondary antibodies for one hour at room temperature and washed three times with 1× PBS before imaging, which was performed using the microscope setup described below.
Primary antibodies included: mouse anti-β tubulin III (Sigma T8660, 1:1000, validated51), rabbit anti-peripherin (Millipore AB1530, 1:800, validated52), rabbit anti-TrpV1 (Alomone Labs ACC-030, 1:200, validated53), rabbit anti-CGRP (Calbiochem/Millipore PC205L, 1:300, validated54), chicken anti-neurofilament, heavy chain (Millipore AB5539, 1:1000, validated55), mouse anti-Nestin (Abcam ab6142, 1:500, validated56), mouse anti-smooth muscle actin (Sigma A5228, 1:300, validated57), goat anti-Sox1 (Santa Cruz #SC17317, 1:50, validated58), mouse anti-Ki67 (Sigma P6834, 1:500, validated59). Secondary antibodies included: goat anti-chicken AlexaFluor 568 (Life Technologies A11041), goat anti-chicken AlexaFluor 488 (Life Technologies A11039), goat anti-mouse AlexaFluor 488 (Life Technologies A11029), goat anti-mouse AlexaFluor 568 (Life Technologies A11031), goat anti-rabbit AlexaFluor 488 (Life Technologies A11008), goat anti-rabbit AlexaFluor 568 (Life Technologies A11011), donkey anti-goat AlexaFluor 488 (Life Technologies A11055).
Primary DRG culture
DRGs were dissected from adult TrpV1-Cre::tdTomato mice (12–13 weeks) into Hank’s balanced salt solution (HBSS) (Life Technologies). DRG were dissociated in 1 mg ml−1 collagenase A plus 2.4 U ml−1 dispase II (enzymes, Roche Applied Sciences) in HEPES-buffered saline (Sigma) for 90 min at 37 °C and then triturated down to single cell level using glass Pasteur pipettes of decreasing size. DRGs were the centrifuged over a 10% BSA gradient and plated on laminin-coated cell culture dishes (Sigma). DRGs were cultured 24 hours in B27-supplemented neurobasal-A medium plus 50 ng/ml NGF (Invitrogen), 2 ng/ml GDNF (Sigma), 10uM arabinocytidine (Sigma) and penicillin/streptomycin (Life Technologies).
qPCR
To compare expression levels of select genes in TrpV1-tdTomato-positive induced nociceptors, TrpV1-tdTomato-positive primary DRGs and TrpV1-tdTomato MEFs, individual tdTomato-positive neurons and fibroblasts were picked using a micropipette. RNA was harvested from sets of 50 cells with the RNeasy Micro Kit (Qiagen) and reverse transcribed with SuperScript VILO cDNA synthesis kit (Life Technologies). Quantitative PCR was completed using mouse-specific TaqMan Gene Expression Assays (Life Technologies) and the TaqMan Gene Expression Master Mix (Life Technologies). A minimum of two technical replicates for each of three biological replicates (independent cell collections) were completed for each gene.
Single cell RT-PCR
Single human induced nociceptors were picked using individual patch pipettes and placed into Single Transcript Amplification (RT-STA) mixture from the CellsDirect One-Step qRT-PCR Kit (Life Technologies) using primers for normally and aberrantly spliced IKBKAP39 and GAPDH. RT-STA reaction products were used for PCR using the same IKBKAP and GAPDH primers and resulting products were run on 1% agarose gels.
Calcium imaging
Cells were loaded with Fura2-AM (10ug/mL, Molecular Probes) by incubating at room temperature for one hour and then de-stained for 15 minutes in saline. For primary DRGs from adult TrpV1-Cre+/−::tdTomato+/− mice, cells were imaged after 24 hours in culture using an identical protocol. Cells were imaged using a Nikon Eclipse Ti microscope with a Xenon lamp, Andor DL-604M camera and standard 340 nM and 380 nM filters controlled by a Ludl Mac6000 shutter using Nikon Elements software. Exposure times were 300–600 ms and images were taken every three seconds. One minute of baseline imaging was recorded prior to the addition of the agonists, during which control vehicle was applied after 30 seconds (not shown). Menthol (250 µM) was applied at one minute, followed by Mustard oil (100 µM) at two minutes and Capsaicin (1 µM) at three minutes and finally KCl (40 mM) at four minutes. For Trp channel experiments, each agonist was applied for 20 seconds and then washed out with external solution. In the PGE2 sensitization experiments, capsaicin (300nM) was applied for 20 seconds after two minutes of recording, followed immediately by PGE2 (1µM) for two minutes, a conditioned capsaicin (300nM) application for 20 seconds and KCl (40 mM) after 4.5 minutes. Analysis of tdTomato positive cells was performed using custom Matlab (Mathworks) software to include cells that responded to KCl (1.5 × baseline), had a stable baseline during control vehicle application and a response to agonist with an amplitude of at least 10% of baseline, with subsequent agonist responses required to be both at least 10% of initial baseline and 10% above a second baseline value obtained during the immediately preceding wash period.
CGRP ELISA
Induced nociceptors, BAM-derived neurons and primary DRGs were exposed to KCl (20 mM, 40 mM, 60 mM, or 80 mM), capsaicin (0.1 µM), or vehicle for 10 minutes at 37°C. The supernatants were collected and analyzed using the Rat CGRP Enzyme Immunoassay Kit (Bertin Pharma/Cayman Chemical, #589001). Plates were read at 405nm for 0.1s on a Wallac Victor2 1420 Multilabel Counter (Perkin Elmer), and data were analyzed using the Wallac 1420 Workstation.
MEA recording
TrpV1-Cre+/−::tdTomato+/− MEFs were plated on poly-D-lysine/laminin coated p515A probes (Alpha Med Scientific) at typical densities of 12,000 cells per probe, transduced with retroviruses and cultured for four weeks. Recordings from 64 extracellular electrodes were made using a Med64 (Alpha Med Scientific) MEA recording amplifier with a head stage that maintained a temperature of 37°C. Data were sampled at 20 kHz, digitized, and analyzed using Mobius software (Alpha Med Scientific) with a 2 kHz 9-pole Bessel low pass filter using a sodium-based extracellular solution: 135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES 10, pH 7.4. The probes were recorded for one minute before the application of the agonists to obtain a baseline and two minutes after the application of capsaicin (1 µM concentration) or mustard oil (100 µM), which were applied at a 10× concentration at the edge of the well (far from the electrodes). Three replicates for each agonist, capsaicin and mustard oil, were completed from two separate transductions. For oxaliplatin sensitization experiments, cells were treated with either 50 µM oxaliplatin or vehicle control for 10 minutes and then recorded for one minute during treatment with 300 nM capsaicin.
Patch electrophysiology
Whole-cell patch recordings were performed on induced tdTomato-positive nociceptors, derived from TrpV1-Cre+/−::tdTomato+/− MEFs, four–five weeks post-transduction and assessed for responses to capsaicin and α,β-methylene ATP (30 µM, Sigma), total and TTX (300 nM, Sigma) -resistant sodium currents, HCN depolarizating current sags and action potentials. Whole-cell current-clamp and voltage-clamp recordings were performed using a Multiclamp 700B (Molecular Devices) at room temperature (21–23°C). Data were sampled at 20 kHz and digitized with a Digidata 1440A A/D interface and recorded using pCLAMP 10 software (Molecular Devices). Data were low-pass filtered at 2 kHz. Patch pipettes were pulled from borosilicate glass capillaries on a Sutter Instruments P-97 puller and had resistances of 2–4 MΩ. The pipette capacitance was reduced by wrapping the shank with Parafilm and compensated for using the amplifier circuitry. Series resistance was 5–10 MΩ and compensated by at least 80%.
For voltage-clamp recordings, voltages were elicited by 200-ms depolarizing steps from a holding potential of −80 mV to test potentials ranging from −100 mV to 30 mV in 10 mV increments. Responses to capsaicin (1 µM) and α,β-methylene ATP (30 µM) were measured in voltage clamp at a holding potential of −80 mV. Electrode drift was measured at the end of each recording and was typically 1–2 mV. The potassium-based intracellular solution contained 150 mM KCl, 2 mM MgCl2, 10 mM HEPES, 4 mM MgATP, 0.3 mM NaGTP, 10 mM Na2PhosCr, 1mM EGTA, pH 7.4. For isolation of voltage-gated sodium currents internal KCl was replaced by CsCl to block potassium currents and 100 µM CdCl2 was applied to block calcium currents. 300 nM TTX was used to block TTX-sensitive voltage-gated sodium channels. HCN currents were measured by sequential hyperpolarizing steps in current clamp with an increment of −10 pA steps.
Quantification of cell number, axon length and axonal branching
All quantifications of cell count and morphological properties were performed by a researcher blind to genotype/condition. Reprogramming efficiency and percentage of TrpV1::tdTomato-positive, Tuj1-positive, peripherin-positive and CGRP-positive neurons were determined using MetaXpress software. The number of TrpV1::tdTomato-positive (mouse, minimum of 3 wells from each of three separate transdifferentiations) and Tuj1-positive (human, minimum of 12 wells from each of three separate transdifferentiations) neurons was divided by the number of fibroblasts plated to calculate the reprogramming efficiency. Axon length and branching were quantified from the six age-matched human fibroblast lines (three HC and three FD) with images from eight wells (two transductions) and four wells (one transduction) of each line (20 wells total/line).
To quantify the number of TrpV1-tomato-positive neurons resulting from infection of different combinations of transcription factors, tomato-positive neurons were counted by hand in at least four separate wells for each condition from two separate transductions and averaged (Fig. 1, Supplementary Fig. 4). Due to the variability of retroviral transductions with large numbers of viruses, we did not quantify experiments with transductions of 11–12 viruses at once (Supplementary Fig. 3). For comparison of 5 Factors ± NeuroD1, images were taken of 6 wells of each condition from one transduction.
Statistical Analyses
Figures show mean ± SEM for all analyses; all tests are two-tailed using a significance threshold of 0.05. We used a Mann-Whitney U test to evaluate CGRP release after log transformation to equalize variance between groups (Fig. 4h). This gave a W statistic of 16 (4 assays/group) and p-value of 0.03. We used a paired t-test to evaluate baseline and PGE2-sensitized capsaicin responses in the induced mouse nociceptors (Fig. 5a). This gave a T statistic of 4.61 with 20 degrees of freedom (21 cells) and a p-value of 9.7*10−4. For comparison of transdifferentiation efficiency using 5 vs 6 factors, we used an unpaired t-test to compare average number of cells/field in 6 wells/group and got a T statistic of 6.1 with 10 degrees of freedom and p-value 1.0*10−4 (Supplementary Fig. 9).
For morphological analyses of human HC and FD-derived neurons, we performed analyses of cell number, axonal outgrowth, and number of branches (Fig. 6). Distribution of cell number and number of branches were not normal (Shapiro-Wilk test). We looked at the distribution of all the measures graphically and found that they are approximately normal. Variance of cell number, axon outgrowth, and number of branches were not different between HC and FD-derived neurons (Levene test). Using random intercept model and taking matching of cell lines into account provided FD disease effect estimates (standard errors) compared to HC: −2.3 (1.5) cell numbers, p=0.2632; −291.3 (32.6) µM axon outgrowth/cell, p=0.0123; −3.3 (0.4) branches/cell, p=0.0165. Post-hoc analysis to compare individual lines was performed using Tukey-Kramer for the three pairs of lines. Comparisons of FD line 1 to matched HC line 1 gave estimate of differences as 2.1 (cells), −292.3 (outgrowth), −3.5 (branches) respectively. Comparisons of FD line 2 to matched HC line 2 gave estimate of differences as −7.2 (cells), −274.7 (outgrowth), −3.2 (branches) respectively. Comparisons of FD line 3 to matched HC line 3 gave estimate of differences as −1.9 (cells), −307.1 (outgrowth), −3.1 (branches) respectively. However, the model with interaction parameter for cell line type and disease group only provided generalized inverse estimates of the standard errors. Hence, the confidence intervals and p-values for these individual cell lines are not reported. All statistical analyses were performed in software R ver 3.1.0 and SAS ver 9.1 (Cary, NC). No power analyses were used to pre-determine sample sizes, but our sample sizes are similar to those used by others in the field.
Supplementary Material
Acknowledgments
We thank M. Costigan for assistance with RT-PCR and A. Yekkirala and J. Sprague for help with calcium imaging, Q. Ma and E. Turner for constructs, J. Gardner and J. McNeish for helpful advice and support, and K. Wainger for assistance with figure preparation. We also thank the Boston Children’s Hospital IDDRC Molecular Genetics Core Facility for RNA Bioanalyzer analyses and the Harvard Medical School ICCB Screening Facility for assistance with ImageXpress and MetaXpress analyses. This research was supported by National Institute of General Medical Sciences T32 GM07592 and National Institute of Neurological Disorders and Stroke 1K08-NS082364 (B.J.W.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (J.T.O.), GlaxoSmithKline Regenerative Medicine DPU (C.J.W.), NINDS NS038253 (C.J.W.) and the Dr. Miriam and Sheldon G. Adelson Medical Foundation (C.J.W.).
Footnotes
Author Contributions
B.J.W. conceived, designed, and performed lineage reprogramming experiments and physiological experiments, analyzed data and wrote the manuscript. E.D.B. designed, performed and analyzed reprogramming, qPCR, single cell RT-PCR, immunohistochemistry, and CGRP ELISA experiments, and wrote the manuscript. J.T.O. performed and optimized induced nociceptor technique. C.M. performed and analyzed physiological studies and edited the manuscript. S.L. performed CGRP ELISA and single cell RT-PCR assays. W.A.S. performed reprogramming and immunohistochemistry experiments. A.J.W performed initial cloning and transduction experiments. J.K.I provided essential advice for nociceptor reprogramming strategy and edited the manuscript. I.M.C gave critical advice regarding the genetic reporter, choice of transcription factors, performed cell sorting experiments and edited the manuscript. L.B advised and performed molecular biology experiments. E.H. performed image quantification and analysis. C.B. assisted with reprogramming and immunohistochemistry. N.T. assisted with human motor neuron culture and together with C.B. performed culture and characterization using initial approaches. K.K. performed statistical modeling of human nociceptor data. L.L.R advised regarding reprogramming experiments and edited the manuscript. K.E. provided advice and reagents for reprogramming and edited the manuscript. C.J.W designed experiments, interpreted findings and wrote the manuscript.
REFERENCES
- 1.Qiang L, Fujita R, Abeliovich A. Remodeling neurodegeneration: somatic cell reprogramming-based models of adult neurological disorders. Neuron. 2013;78:957–969. doi: 10.1016/j.neuron.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 3.Liu ML, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiang L, et al. Directed conversion of Alzheimer's disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair NT, Bean BP. Roles of tetrodotoxin (TTX)-sensitive Na+ current, TTX-resistant Na+ current, and Ca2+ current in the action potentials of nociceptive sensory neurons. J Neurosci. 2002;22:10277–10290. doi: 10.1523/JNEUROSCI.22-23-10277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers SM, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waxman SG. Channelopathic pain: a growing but still small list of model disorders. Neuron. 2010;66:622–624. doi: 10.1016/j.neuron.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, et al. Small-fiber neuropathy Nav1.8 mutation shifts activation to hyperpolarized potentials and increases excitability of dorsal root ganglion neurons. J Neurosci. 2013;33:14087–14097. doi: 10.1523/JNEUROSCI.2710-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han C, et al. Nav1.7-related small fiber neuropathy: impaired slow-inactivation and DRG neuron hyperexcitability. Neurology. 2012;78:1635–1643. doi: 10.1212/WNL.0b013e3182574f12. [DOI] [PubMed] [Google Scholar]
- 13.Sorge RE, et al. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med. 2012;18:595–599. doi: 10.1038/nm.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reimann F, et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A. 2010;107:5148–5153. doi: 10.1073/pnas.0913181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitchford S, Levine JD. Prostaglandins sensitize nociceptors in cell culture. Neurosci Lett. 1991;132:105–108. doi: 10.1016/0304-3940(91)90444-x. [DOI] [PubMed] [Google Scholar]
- 16.Lopshire JC, Nicol GD. Activation and recovery of the PGE2-mediated sensitization of the capsaicin response in rat sensory neurons. J Neurophysiol. 1997;78:3154–3164. doi: 10.1152/jn.1997.78.6.3154. [DOI] [PubMed] [Google Scholar]
- 17.Anand U, Otto WR, Anand P. Sensitization of capsaicin and icilin responses in oxaliplatin treated adult rat DRG neurons. Mol Pain. 2010;6:82. doi: 10.1186/1744-8069-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KJ, Jessell TM. The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- 19.Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- 20.Woolf CJ, Ma Q. Nociceptors--noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dykes IM, Lanier J, Eng SR, Turner EE. Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev. 2010;5:3. doi: 10.1186/1749-8104-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CL, et al. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, et al. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei L, et al. The zinc finger transcription factor Klf7 is required for TrkA gene expression and development of nociceptive sensory neurons. Genes Dev. 2005;19:1354–1364. doi: 10.1101/gad.1227705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavanaugh DJ, et al. Restriction of transient receptor potential vanilloid-1 to the peptidergic subset of primary afferent neurons follows its developmental downregulation in nonpeptidergic neurons. J Neurosci. 2011;31:10119–10127. doi: 10.1523/JNEUROSCI.1299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Website: ©2012 Allen Institute for Brain Science. Allen Spinal Cord Atlas [Internet] Available from: http://mousespinal.brain-map.org/.
- 29.Fornaro M, et al. Neuronal intermediate filament expression in rat dorsal root ganglia sensory neurons: an in vivo and in vitro study. Neuroscience. 2008;153:1153–1163. doi: 10.1016/j.neuroscience.2008.02.080. [DOI] [PubMed] [Google Scholar]
- 30.Cavanaugh DJ, et al. Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J Neurosci. 2011;31:5067–5077. doi: 10.1523/JNEUROSCI.6451-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bautista DM, et al. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006;124:1269–1282. doi: 10.1016/j.cell.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Chen CC, et al. A P2X purinoceptor expressed by a subset of sensory neurons. Nature. 1995;377:428–431. doi: 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis MF, et al. A-317491, a novel potent and selective non-nucleotide antagonist of P2X3 and P2X2/3 receptors, reduces chronic inflammatory and neuropathic pain in the rat. Proc Natl Acad Sci U S A. 2002;99:17179–17184. doi: 10.1073/pnas.252537299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. Sodium channels in normal and pathological pain. Annu Rev Neurosci. 2010;33:325–347. doi: 10.1146/annurev-neuro-060909-153234. [DOI] [PubMed] [Google Scholar]
- 35.Renganathan M, Dib-Hajj S, Waxman SG. Na(v)1.5 underlies the 'third TTX-R sodium current' in rat small DRG neurons. Brain Res Mol Brain Res. 2002;106:70–82. doi: 10.1016/s0169-328x(02)00411-4. [DOI] [PubMed] [Google Scholar]
- 36.Dib-Hajj SD, et al. Two tetrodotoxin-resistant sodium channels in human dorsal root ganglion neurons. FEBS Lett. 1999;462:117–120. doi: 10.1016/s0014-5793(99)01519-7. [DOI] [PubMed] [Google Scholar]
- 37.Nagy I, Urban L, Woolf CJ. Morphological and membrane properties of young rat lumbar and thoracic dorsal root ganglion cells with unmyelinated axons. Brain Res. 1993;609:193–200. doi: 10.1016/0006-8993(93)90873-l. [DOI] [PubMed] [Google Scholar]
- 38.Emery EC, Young GT, Berrocoso EM, Chen L, McNaughton PA. HCN2 ion channels play a central role in inflammatory and neuropathic pain. Science. 2011;333:1462–1466. doi: 10.1126/science.1206243. [DOI] [PubMed] [Google Scholar]
- 39.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slaugenhaupt SA, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuajungco MP, et al. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am J Hum Genet. 2003;72:749–758. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolf CJ. Overcoming obstacles to developing new analgesics. Nat Med. 2010;16:1241–1247. doi: 10.1038/nm.2230. [DOI] [PubMed] [Google Scholar]
- 44.Davidson S, et al. Human sensory neurons: Membrane properties and sensitization by inflammatory mediators. Pain. 2014 doi: 10.1016/j.pain.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, et al. Cloning and expression of a novel human gene, Isl-2, encoded a LIM-homeodomain protein. Mol Biol Rep. 2007;34:19–26. doi: 10.1007/s11033-006-9003-0. [DOI] [PubMed] [Google Scholar]
- 46.Dietrich P, Alli S, Shanmugasundaram R, Dragatsis I. IKAP expression levels modulate disease severity in a mouse model of familial dysautonomia. Hum Mol Genet. 2012;21:5078–5090. doi: 10.1093/hmg/dds354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George L, et al. Familial dysautonomia model reveals Ikbkap deletion causes apoptosis of Pax3+ progenitors and peripheral neurons. Proc Natl Acad Sci U S A. 2013;110:18698–18703. doi: 10.1073/pnas.1308596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebelo S, Chen ZF, Anderson DJ, Lima D. Involvement of DRG11 in the development of the primary afferent nociceptive system. Mol Cell Neurosci. 2006;33:236–246. doi: 10.1016/j.mcn.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Anderson DJ. Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr Opin Neurobiol. 1999;9:517–524. doi: 10.1016/S0959-4388(99)00015-X. [DOI] [PubMed] [Google Scholar]
- 50.Cheng L, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee A, Roach MC, Trcka P, Luduena RF. Increased microtubule assembly in bovine brain tubulin lacking the type III isotype of beta-tubulin. J Biol Chem. 1990;265:1794–1799. [PubMed] [Google Scholar]
- 52.Lysakowski A, Alonto A, Jacobson L. Peripherin immunoreactivity labels small diameter vestibular 'bouton' afferents in rodents. Hear Res. 1999;133:149–154. doi: 10.1016/s0378-5955(99)00065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belghiti M, et al. Potentiation of the transient receptor potential vanilloid 1 channel contributes to pruritogenesis in a rat model of liver disease. J Biol Chem. 2013;288:9675–9685. doi: 10.1074/jbc.M113.455162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maatkamp A, et al. Decrease of Hsp25 protein expression precedes degeneration of motoneurons in ALS-SOD1 mice. Eur J Neurosci. 2004;20:14–28. doi: 10.1111/j.1460-9568.2004.03430.x. [DOI] [PubMed] [Google Scholar]
- 55.Ghilardi JR, et al. Sustained blockade of neurotrophin receptors TrkA, TrkB and TrkC reduces non-malignant skeletal pain but not the maintenance of sensory and sympathetic nerve fibers. Bone. 2011;48:389–398. doi: 10.1016/j.bone.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat Cell Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- 57.Skalli O, et al. A monoclonal antibody against alpha-smooth muscle actin: a new probe for smooth muscle differentiation. J Cell Biol. 1986;103:2787–2796. doi: 10.1083/jcb.103.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panayi H, et al. Sox1 is required for the specification of a novel p2-derived interneuron subtype in the mouse ventral spinal cord. J Neurosci. 2010;30:12274–12280. doi: 10.1523/JNEUROSCI.2402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffmann S, et al. Retinoic acid inhibits angiogenesis and tumor growth of thyroid cancer cells. Mol Cell Endocrinol. 2007;264:74–81. doi: 10.1016/j.mce.2006.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.