Abstract
The ortho-carboxylic acid substituted bisanilinopyrimidine 1 was identified as a potent hit (Aurora A IC50 = 6.1 ± 1.0 nM) from in-house screening. Detailed structure activity relationship (SAR) studies indicated that polar substituents at the para position of the B-ring are critical for potent activity. X-ray crystallography studies revealed that compound 1 is a type-I inhibitor that binds the Aurora kinase active site in a DFG-in conformation. Structure activity guided replacement of the A-ring carboxylic acid with halogens and incorporation of fluorine at the pyrimidine 5-position led to highly potent inhibitors of Aurora A that bind in a DFG-out conformation. B-ring modifications were undertaken to improve the solubility and cell permeability. Compounds such as 9m with water-solubilizing moieties at the para-position of the B-ring inhibited the autophosphorylation of Aurora A in MDA-MB-468 breast cancer cells.

Introduction
The Aurora kinases are a family of serine-threonine kinases that play an important role in the regulation of cell division. They are over expressed in a variety of cancer types and are implicated in many aspects of tumor development. The three members of the Aurora family Aurora A, B and C share a high degree of structural homology in their kinase domain but each kinase plays a different role in the control of mitosis. Aurora A and B have received the most attention to date as anticancer targets.1 Aurora A regulates the cell cycle and is associated with late S-phase and entry into the M phase. It is essential for many processes including centrosome maturation, and separation, chromosome alignment and mitotic spindle formation.2 Aurora A is frequently over-expressed in tumors and has characteristics of an oncogene.3, 4 Aurora B plays important roles in M phase to ensure correct chromosome-microtubule alignment and attachment and chromosomal cytokinesis. Aurora B is also over expressed5 in many cancer types but does not have oncogenic properties. Both Aurora A6-10 and Aurora B11-14 have been validated as targets for anticancer drugs in pre-clinical models. Intense activity has led to the development of numerous inhibitors15-17 that display selectivity to either Aurora A or B, or that are dual inhibitors. Many have been or are now in clinical development.18-22 Selected examples are shown in Figure 1. The pyrimidine derivative VX-680 (Tozasertib) developed by Vertex and Merck is a potent pan-Aurora inhibitor.23 Clinical development was halted after QTC prolongation was observed. As one of the first reported Aurora inhibitors VX-680 has been an important tool for studying Aurora biology. Other compounds remain in clinical trials including the orally bioavailable Aurora A selective inhibitor MLN805424-26 and its second generation analog MLN823727-30 (Figure 1). The AstraZeneca quinazoline AZD-1152 (Barasertib)31, 32 in phase II trials is a potent and selective Aurora B inhibitor administered via intravenous infusion. The 3-aminopyrazole PHA-73935833-36 (Figure 1) developed by Nerviano and Pfizer is in Phase II trials for CML and metastatic prostate cancer. The development of Aurora inhibitors continues to attract attention37-41 and ultimately will lead to therapeutic benefit in the clinic. The bisanilinopyrimidines are a class of compounds that has shown unusual high selectivity for Aurora A over Aurora B.42 The pyrimidine scaffold43,44,45 has been used by many groups to develop novel Aurora kinase inhibitors.
Figure 1.
Structures of selected Aurora inhibitors evaluated in the clinic. Inhibition data is shown for Aurora A and B.
In this report we describe our efforts in identifying novel and highly potent Aurora A inhibitors using the bisanilinopyrimidine scaffold. To this end, we screened our in-house 20,000 membered ChemDiv library using a Z-lyte assay and identified the bisanilinopyrimidine bis-carboxylic acid 1 (HLM008598, Figure 2) as a hit (IC50 = 0.075 ± 0.039 μM, in-vitro inhibitory activity). Optimization of compound 1 was undertaken initially via SAR guided focused library synthesis followed by rational design based on co-crystal structures of 1 and related analogs bound to Aurora A.
Figure 2.
a. Compound 1 (hit) identified from in-house ChemDiv library as an Aurora A inhibitor. b. Synthetic modifications for library synthesis.
The bisanilinopyrimidine carboxylic acid 1 was re-synthesized in-house to confirm the structure and activity against Aurora A and B. It has not previously been reported as an Aurora kinase A inhibitor. The general synthetic route used for preparation of compound 1 (Figure 2a) and 2,4-bisanilinopyrimidine focused libraries 3 and 4 from readily available building blocks is shown in Scheme 1. The displacement of the 4-chloro group of 2,4-dichloropyrimidine by various anilines is already widely reported in the literature.42,46, 47,48 The hit 1 was resynthesized by hydrolysis (method h, Scheme 1) of the methyl ester 3w, which was prepared from 2a and methyl para-aminobenzoate (method m, Scheme 2). Attempts to directly synthesize 1 from 2a and para-aminobenzoic acid via method a resulted in formation of decarboxylated by-product 3b (1:3b ca. 5:1, observed by NMR and HPLC-MS analysis). Attempts to purify the mixture were not successful. For the library synthesis of analogs of 1 the 2,4-dichloropyrimidine was initially reacted with the requisite commercially available anilines using literature reported protocols46, 49, 50 to obtain the intermediate library 2 (Scheme 1) with A-ring substituents such as carboxylic acid, carboxylic acid ester, amide groups and chlorine functional groups at the ortho-position. In this step, aqueous hydrochloric acid was used predominantly as the solvent with microwave assisted heating (method a) to obtain the required analogs 2. The ortho-chloro analogs 2k and 2l were synthesized in good yields using aqueous acid at room temperature (method b) as shown in the Scheme 1.46 The 5- and 6-aminopyrimidine building blocks 2h and 2i were obtained from commercially available 5- and 6-amino-2,4-dichloropyrimidine starting materials respectively in moderate yields. The library 2 members were further functionalized under acid catalysis (methods d-g) to provide the library 3 in good yields.46 Although the pyrimidine scaffold features in many kinase inhibitors, only one member of library 3, i.e. compound 3c has been reported as a kinase inhibitor.50 Intermediates 2h and 2i were converted to 3u and 3v using methods f and e respectively to obtain ethyl esters as intermediates. Similarly,ethyl esters 3s and 3t were obtained from 2m and 2n respectively using method f. Intermediates 3s-3v were hydrolyzed (method h) to obtain 4a-4d as final compounds.
Scheme 1.
Synthetic route to bisanilinopyrimidine carboxylic acid library. Reagents and conditions; method a: HCl (0.1 M, aq., 1-3 mL/mmol), microwave, 100 °C, 30 min. method b: HCl (0.1 M, aq., 3 mL/mmol), r.t., 3-5 days. method c: HCl (0.1 M, aq., 1.5 mL/mmol), sealed tube, 100 °C, 24 h. method d: HCl (0.1 M, aq., 3-6 mL/mmol), microwave, 160 °C, 15 min. method e: EtOH: HCl (1 M, aq., 1:1, 4 mL/mmol), microwave, 160 °C, 15 min.-1 h. method f: (i) HCl (4 M in dioxane, 0.5 mL/mmol), 2-butanol (3 mL/mmol), sealed tube, 120 °C, overnight (24 h), 72% or (ii) EtOH, sealed tube, 120 °C, overnight – 4 days. method g: THF: HCl (1 M, aq.) (1:2, 6 mL/mmol), microwave, 160 °C, 15 min. method h: THF:NaOH (2 M, aq.) (1:2, 4-7 mL/mmol), sealed tube, 85-100 °C, 0.5-16 h.
Scheme 2.
Synthetic route to bisanilinopyrimidine library 6 with halogens (F, Cl, Br, and I), CN, non-polar groups (H, Ph) and polar hydrophobic groups (OCF3, CF3 and OMe) on the A-ring. Reagents and conditions: method i: n-BuOH, DIPEA, 125 °C. method j: n-BuOH, Na2CO3, 100 °C. method k: DMF, NaH, r.t., overnight (14-16 h). method l: EtOH (1 drop of 1M HCl), microwave, 160 °C, 15 min., 56-75%,. method m: EtOH or MeOH, 150 °C, 20 min., microwave. method n: Cat. HCl, THF, reflux 14 h. method o: THF, NaOH (1.8 M aq., 5 equivalents), THF, reflux, 14 h., method b: HCl (0.1 M, aq., 3 mL/mmol), r.t., 3-5 days.
The high in-vitro potency observed with compounds 3n (with 5-fluoropyrimidine moiety), 3l and 3o (ortho-chloro analogs, Scheme 1) prompted us to further investigate this series. The synthesis of library 6 with hydrophobic groups (Cl, Br, I, CF3, phenyl) and hydrogen-bond acceptor groups (OMe, CN, OCF3) on the A-ring is described in Scheme 2. Synthesis of the key intermediates 5a-i with F, Cl, Br, I, OCF3, CF3, OMe, CN and phenyl (Scheme 2) was achieved from commercially available 2,4-dichloropyrimidine building blocks using literature reported protocols.50-52 Intermediates 5j and 5k were synthesized via alkylation of 2l using C22CO3 in acetonitrile (Scheme 2).51, 52 These intermediates were directly reacted via nucleophilic aromatic substitution with anilines to obtain the final library 6 possessing halogens (F, Cl, Br and I), polar groups (CN), non-polar groups (Ph, H) and polar hydrophobic groups (OCF3, CF3, OMe) in the ortho-position of the A-ring in moderate to high yields (methods l, m or n in Scheme 2). The majority of the library members 6 also readily precipitated under the reaction conditions and the purity of final compounds tested against Aurora A inhibitory activity was determined as > 95% by HPLC (high performance liquid chromatography). The analog 6k with para-carboxylic acid and meta-OH (i.e. salicylic acid moiety) was obtained from its precursor methyl ester 6u via basic hydrolysis (method o in Scheme 2).
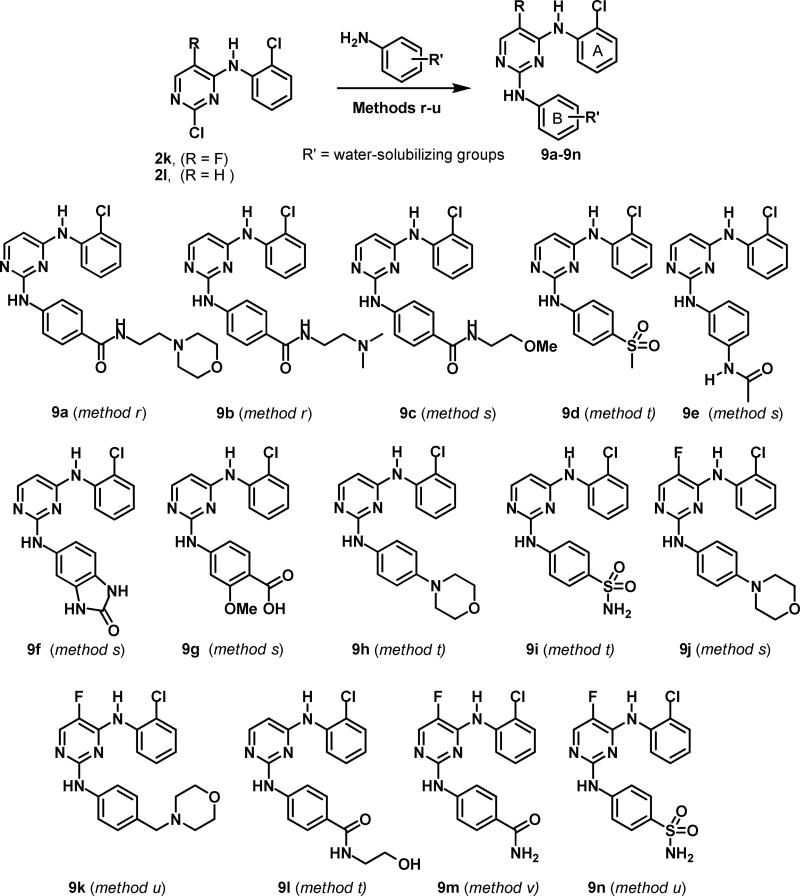
Using the synthetic routes and protocols shown in Schemes 1 and 2, we were able to explore detailed invitro SAR toward Aurora A inhibition. Furthermore, we designed and synthesized new molecules exploiting the structures of compounds 1, 3l, 3n and 3o complexed with Aurora A to develop potent Aurora A inhibitors with desirable drug-like properties for in vitro and in vivo studies.53 Compounds 3l, 3o (Scheme 1), with ortho-chloro and para-carboxylic acid groups were further modified via introduction of water-solubilizing groups to improve solubility and cell permeability (Scheme 4). The solubilizing group was attached via an amide of the B-ring para-carboxylic acid as this part of the molecule is solvent exposed in the co-crystal complexes53 of 1 (see Figure 3) and 3o with Aurora A. The building blocks 8a-c were synthesized as shown in the Scheme 3 via acylation of commercially available para-nitrobenzoyl chloride with amines possessing water-solubilizing groups (morpholine, N,N-dimethylamine, and methyl ether) followed by the hydrogenation of the nitro group in high yields.54 The final bisanilinopyrimidines with water-solubilizing groups 9 (Scheme 4) were prepared by reaction of intermediates 2k and 2l (Scheme 1) with appropriate anilines in moderate yields and > 95% HPLC purity. The library 9 with water-solubilizing moieties in the B-ring and an orthochloro A-ring comprises novel compounds that have not previously been reported as kinase inhibitors.
Scheme 4.
Synthesis of bisanilinopyrimidines with water-solubilizing groups in the B-ring. Reagents and conditions; method r: Conc. HCl, iso-PrOH, 170 °C, microwave, 20 min., method s: EtOH, 150 °C, microwave, 20 min., method t: EtOH, cat. HCl, 180 °C or 160 °C or 140 °C microwave, 15 min., method u: X-Phos (10 mmol%), bis(dibenzylideneacetone) palladium(0) (10 mmol%), K2CO3, tert-BuOH, reflux,18 h., method v: MeOH, 100 °C, seal tube, 6 h.
Figure 3.
a. Cartoon representing key binding interactions of hit (compound 1) with Aurora A. b. X-ray structure of compound 1 bound to Aurora A active site (compound 1, cyan: carbon, red: oxygen, and blue: nitrogen, PDB ID 3UP7). c. Compound 1 co-crystallized with Aurora A shows hinge region (green), DFG-in (magenta) activation loop (blue) open conformation (compound 1, yellow: carbon; red: oxygen and blue: nitrogen). d. Overlay of ADP (carbons shown green) and compound 1 (carbons shown cyan). e. ADP bound to Aurora A active site (PDB ID 4DEE).
Scheme 3.
Synthesis of building blocks with water-solubilizing groups. Reagents and conditions: method p: 10% Pd/C, EtOH, H2, r.t., overnight. method q: H-Cube, 10% Pd/C, MeOH, 30 bar, 4 loops, rt.
Synthesis of the para-tetrazole aniline building block 11 was achieved as shown in the Scheme 5 and used to generate compounds 12 (derivatives of 3l, 3n and 3o shown in the Scheme 1). The starting material 4-nitrobenzonitrile was reacted with sodium azide (condition b, Scheme 5) to obtain intermediate 10 in high yield.55 Hydrogenation of intermediate 10 using Pd-C as the catalyst provided the required building block 11 in good yield (condition c). The compound 11 was next coupled with intermediates 2k, 2l (Scheme 1) or 2o to provide the final compounds 12a-d (Scheme 5) with > 95% purity (determined by HPLC). The final compounds 12 with a tetrazole moiety in the B-ring are reported for the first time.
Scheme 5.
Synthesis of bisanilinopyrimidine tetrazole derivatives. Reagents and conditions: a: 12a: Ethanol, microwave, 150 °C, 40 min., 37%; 12b: Ethanol, microwave, 170 °C, 40 min., 33%; 12c: Ethanol, microwave, 160 °C, 40 min., 36%; 12d: Ethanol, microwave, 150 °C, 40 min., 25%; b: 10a and 10b: NaN3, Et3N·HCl, toluene, 100 °C, 15 h, 92% and 95%; c: 11a and 11b: H2, Pd/C, methanol, r.t., 20 h, 93% and 98%.
Results and discussion
The high throughput screening (HTS) hit 1 was identified from our in-house 20,000 compound ChemDiv library as a potent and highly selective inhibitor for Aurora A (in vitro IC50 = 0.075 ± 0.039 μM) over Aurora B (in vitro IC50 = 5.4 ± 1.8 μM) using the Z-LYTE™ assay using LRRASLG as an Aurora substrate.56, 57 We verified the dose response curve of the hit (compound 1) using a coupled enzyme assay58 (DiscoveRx) which measures ADP formation from the Aurora A phosphorylation of the same synthetic peptide LRRASLG, as described under methods. The determination of dose response curve and IC50 value of the compound 1 using this coupled assay revealed in vitro Aurora A potency in the range of 6.1 ± 1.0 nM and we used this assay to establish the SAR described in this study.
The bis-anilinopyrimidine scaffold, but not specifically compound 1, has previously been reported for inhibitors of Aurora kinase42, 44 as well as other kinases such as JNK150, FAK59, ephrin type-B receptor 4 kinase,60 CDK2 and CDK4.61 For an HTS hit, compound 1 displayed an unusually high potency in the range of the most active Aurora A inhibitors reported to date. The suitability of this scaffold to focused library synthesis and availability of crystallization-grade protein prompted us to pursue the improvement of 1 by SAR studies and structure-based design.
In the beginning of this work, SAR studies were initiated while attempts were being made to co-crystallize compound 1 with Aurora A. Focused library synthesis based on 1 (Figure 2) was first undertaken varying 4 points of molecular diversity (R1, R2, R3 and R4, see Figure 2b) by systematically replacing or introducing the functional groups in the A and B-rings. Replacement of the B-ring para-carboxylic acid in 1 by hydrogen or a morpholino as in 3b or 3c resulted in 13- and 10-fold loss of inhibitory activity respectively, suggesting that the carboxylic acid is critical (Entries 3 and 4, Table 1). Replacing both carboxylic acid moieties in 1 by hydrogens as in 3f (Entry 7, Table 1) resulted in 70-fold loss of potency further emphasizing the importance of the carboxylic acid moieties. In addition, replacement of both carboxylic acid moieties by corresponding methyl esters as shown in 3e (Entry 6, Table 1) was detrimental and resulted in over 4000-fold loss of potency demonstrating the importance of the negative charge of the acid moieties in the binding region. However, compound 3g (Entry 8, Table 1) with carboxamide in ortho- and para-positions of the A and B-rings respectively was only 6-fold less potent than the compound 1. Our SAR data showed the positions of both carboxylic acid moieties in A and B-rings are critical for activity. Moving the B-ring para-carboxylic acid moiety to ortho-position as in 3d demonstrated 5000-fold loss of potency (Entry 5, Table 1, IC50 = 31,300 nM). The compound 3q (Entry 18, Table 1, IC50 = 18.3 nM) with meta-carboxylic acid in the B-ring was less detrimental with 3-fold loss of in vitro potency. Furthermore, moving the A-ring carboxylic acid moiety in compound 1 from the ortho to para position as in 3i (Entry 10, Table 1) resulted in 42-fold loss of potency. Replacement of the B-ring para-carboxylic acid by ortho-amide (compound 3a) was also detrimental for Aurora A inhibitory activity resulting in greater than 1000-fold loss of inhibitory activity (Entries 2, Table 1). These observations suggested that the ortho-position of the A-ring and para-position of the B-ring are critical for inhibition of enzymatic activity and focused library synthesis.
Table 1.
Synthetic Modifications, structure activity relationship studies and in vitro activities of bisanilinopyrimidine libraries 3 and 4 against Aurora A.

| ||||||
|---|---|---|---|---|---|---|
| Entry | Compound | R1 | R2 | R3 | R4 | In vitro IC50 |
| 1 | 1 | H | H | ortho-CO2H | para-CO2H | 6.1 ± 1.0 nM |
| 2 | 3a | H | H | ortho-CO2H | ortho-CONH2 | 9.0 ± 6.8 μM |
| 3 | 3b | H | H | ortho-CO2H | H | 79.4 ± 18 nM |
| 4 | 3c | H | H | ortho-CO2H | para-morpholine | 57.6 ± 5.4 nM |
| 5 | 3d | H | H | ortho-CO2H | ortho-CO2H | 31.3 ± 5.9 μM |
| 6 | 3e | H | H | ortho-CO2Me | para-CO2Me | 24.6 ± 6.0 μM |
| 7 | 3f | H | H | H | H | 423 ± 66 nM |
| 8 | 3g | H | H | ortho-CONH2 | para-CONH2 | 38.2 ± 8.8 nM |
| 9 | 3h | H | H | H | para-CO2H | 10 ± 1.6 nM |
| 10 | 3i | H | H | para-CO2H | para-CO2H | 256 ± 38 nM |
| 11 | 3j | CH3 | H | ortho-CO2H | para-CO2H | 281 ± 59 nM |
| 12 | 3k | H | CH3 | ortho-CO2H | para-CO2H | >50 μM |
| 13 | 3l | H | H | ortho-Cl | para-CO2H | 2.5 ± 0.3 nM |
| 14 | 3m | F | H | ortho-CO2H | All H | 11.3 ± 1.7 nM |
| 15 | 3n | F | H | ortho-CO2H | para-CO2H | 3.9 ± 0.5 nM |
| 16 | 3o | F | H | ortho-Cl | para-CO2H | 0.8 ± 0.16 nM |
| 17 | 3p | F | H | ortho-Cl | H | 19.9 ± 2.2 nM |
| 18 | 3q | H | H | ortho-CO2H | meta-CO2H | 18.3 ± 3.4 nM |
| 19 | 3r | F | H | ortho-CO2H | meta-CO2H | 5.1 ± 1.1 nM |
| 20 | 4c | Cl | Cl | ortho-CO2H | para-CO2H | 1.49 ± 0.196 μM |
| 21 | 4d | Cl | H | ortho-CO2H | para-CO2H | 3.17 ± 0.51 nM |
| 22 | 4a | NH2 | H | ortho-CO2H | para-CO2H | 376 ± 64 nM |
| 23 | 4b | H | NH2 | ortho-CO2H | para-CO2H | >50 μM |
The X-ray co-crystal structure of 1 bound to Aurora A supports the above findings (Figure 3). Analysis of this structure shows the para-carboxylic acid group of the B-ring forming key H-bond interactions with the solvent exposed residues Arg137 and Arg220 (Figures 3a and 3b). As expected of a bisanilinopyrimidine, compound 1 is a type I62 kinase inhibitor that targets the ATP binding site (Figure 3c). Compound 1 binds to Aurora A in an active DFG-in conformation, as shown in Figure 3c (DFG-motif shown in magenta). The activation loop (shown blue in Figure 3c) is oriented away from the ATP binding site. The pyrimidine scaffold and the amine moiety of the B-ring establish H-bonding with the hinge region (residues 211-213, Figures 3a and 3b). Analysis of ADP/ATP bound to Aurora A indicated highly conserved residues Lys162, Asp274 and Glu181 undergoing electrostatic interactions with ADP phosphate moieties in the active site (Figure 3e). Compound 1 bound to Aurora A shows these key residues in the active site are now in contact with the ortho-COOH moiety of the A-ring (Figure 3b). The ortho-carboxylic acid of the A-ring in compound 1 is in the vicinity (3.5 Å) to form an electrostatic interaction with Lys162 (Figure 3b). The carboxylic acid moiety of the Asp274 of the DFG motif (Figures 3a and 3b) is also in close distance (3.2 Å) with Lys162. These key interactions contribute to the high in-vitro potency of the compound 1. Clearly the apparent disfavored interaction between the ortho-COOH moiety of 1 and the side chain carboxylic acid moiety of the Asp274 must be compensated by the presence of the Lys162. This is in consistent with the fact that compound 3f (Entry 7, Table 1) where both A and B-rings contain unsubstituted phenyl was 70-fold less active than parent compound 1, and the dimethyl ester 3e lost the in-vitro inhibitory activity (IC50 24.6 μM, Entry 6, Table 1). These observations further confirm that the key interactions observed from the X-ray structure are important for inhibitory activity (Figure 3b). The loss of inhibitory activity observed with 3d (Entry 5, Table 1) in our SAR studies is consistent with the X-ray structure of compound 1 bound to Aurora A where ortho-substituted B-ring causes steric clash with the main chain residues Ala213 and Pro214 (Figures 3a and 3b). However, the activity of compound 3q is retained with carboxylic acid in the meta-position, and improved when fluorine is added as in 3r (Entries 18 and 19 table 1). It is also likely that the meta-COOH is able interact with Arg220 and Arg137.
During further investigation of this series, several X-ray structures were obtained in order to improve our understanding of the binding modes of this class of compound with Aurora A (Figure 4). Substitution of both carboxylic acid groups in compound 1 by primary amides (Figure 4B) reduced the inhibitory activity 6-fold (Entry 8, 3g, Table 1). The biscarboxylic acid 3i (Figure 4A) where the A-ring ortho-carboxylic acid is moved to the para-position was 42-fold less potent than parent molecule (Table 1, Entry 10, IC50 = 256 nM). This decrease in inhibitory activity is probably due to lack of key binding interactions of the para-carboxyl or ortho-carboxamide groups in the A-ring with the Lys162 active site residue (Figure 4A and 4B, compounds 3i and 3g bound to Aurora A). In contrast, removal of the ortho-carboxylic group as in 3h (Entry 9, Table 1) retains activity reflecting both the loss of positive interaction with Lys162 and negative interaction with Asp274. The co-crystal structure of 3i (Figure 4) does not explain the differences in activities of 3h and 3i. Compound 13 (Figure 4C) where A-ring has a CF3 in the meta-position is a weaker binder (IC50 = 371 nM, see supporting information Table S3 for IC50 data of additional compounds) and has the trifluoromethyl group positioned in the region that binds the diphosphate group of ADP (see Figure 3e). Thus the presence of a hydrophobic group close to Asn261 and Glu260 does not contribute to binding affinity of Aurora A. Overall meta-substituents (not reported here) in the A-ring did not improve the activity.
Figure 4.
X-ray structures of compounds 3i (A), 3g (B) and compound 13 (C, see supplementary information) bound to Aurora A active site. (PDB IDs 4DEA, 4DED and 4DEB respectively). Compound 1 bound to Aurora A (D); R1 hydrogen is shown in yellow, Leu194, Leu210 and Ala160 shown as spheres to show the narrow space where R1 is occupying in the binding region.
The surprising observation that replacement of the ortho-carboxylic acid of the A-ring in compound 1 with hydrogen as in 3h did not result in great loss of potency prompted us to further probe SAR at this position. Therefore library 6 (Scheme 2) was synthesized to improve our understanding of the binding modes of Aurora A to bisanilinopyrimidines with different groups in the ortho-position of the A-ring. To this end we synthesized analogs of 1 by replacing the carboxylic acid in the ortho-position of the A-ring by fluoro, chloro, bromo, iodo, trifluoromethyl, trifluoromethoxy, methoxy, cyano and phenyl as ortho-substituents (6a-6q, Scheme 2). Compounds with halogens (Cl, F, Br) at ortho-position of the A-ring and para-COOH moiety in the B-ring [compounds 3l (Table 1), 6a and 6i (Table 2) respectively] improved potency by 1.5 to more than 3-fold, whereas compounds with bulky halogenated groups such as OCF3 and CF3 (Entries 27, 31 compounds 6d and 6h respectively in Table 2) were much less active at inhibiting Aurora A. Compound 6q (Entry 40, Table 2) with an ortho-phenyl group was 24-fold less potent than the compound 1 further indicating bulky groups are not tolerated in this region. Compound 6n (IC50 = 43 nM, Entry 37, Table 2) that possess an ortho-CN group is more than 17-fold less active than 3l. The in vitro activity of 3l (IC50 = 2.5 ± 0.3 nM, Entry 13, Table 1) with ortho-Cl and a para-COOH in B-ring was further improved when R1 is fluorine in compound 3o (IC50 = 0.8 ± 0.16 nM, Entry 16, Table 1). The exceptional potency of this compound is thought to derive its novel binding mode, recently described by us.53 Compounds 3o binds to Aurora A in an inactive DFG-out conformation with the activation loop is folded back over the ATP binding site, effectively encapsulating the inhibitor. This binding mode is a consequence of the presence of the ortho-chloro group, which we postulate induces a dipole in the N-flanking alanine adjacent to the DFG motif.53 The contrasting binding modes of 1 and 3o is striking given their high structural similarity and provides chemical tools for the studying the phenotypical consequences of these two binding modes. The para-COOH moiety in the B-ring was important to maintain the Aurora A inhibitory activity in library 6 (Scheme 2). The removal of COOH moiety resulted in loss of in vitro potency as observed with 6b, 6f, 6g, and 6l (Entries 25, 29, 30 and 35, Table 2). The loss of in vitro inhibitory activity in compounds lacking a para-COOH moiety was a general trend observed with libraries 3 and 6 and highlights the importance of H-bond interactions associated with Arg137 and Arg220. Switching the position of B-ring COOH moiety from para- to meta-position as in compound 6o (Entry 38, Table 2) reduced the inhibitory activity compared 3o (Entry 16, Table 1). Modification of B-ring para-COOH to para-CONH2 in compound 6p reduced the in vitro activity by 5-fold (Entry 39, Table 2). Compounds 6r and 6s (Entries 41 and 42, Table 2) with N-alkylated moieties retained the activity; N-methyl derivative was highly potent (IC50 = 8.5 ± 1.2 nM) while the N-ethyl derivative was less active (IC50 = 50.2 ± 2.7 nM). Compound 6t (Entry 43, Table 2) with meta-COOH and fluorine as R1 was 4-fold more potent compared to compound 6o where R1 is hydrogen. Overall our SAR indicated fluorine or chlorine as R1 in this series is beneficial and improve in vitro and in vivo Aurora A activity (see Table 3).
Table 2.
Structure activity relationship studies and in vitro inhibitory activities of library 6 against Aurora A.

| |||||||
|---|---|---|---|---|---|---|---|
| Entry | Compound | R1 | R2 | R3 | R4 | R5 | IC50 (nM)a |
| 24 | 6a | H | H | ortho-F | para-CO2H | H | 3.7 ± 0.7 nM |
| 25 | 6b | H | H | ortho-CF3 | All H | H | 1773 ± 142 nM |
| 26 | 6c | H | H | 2-Cl-4-F | para-CO2H | H | 2.0 ± 0.2 nM |
| 27 | 6d | H | H | ortho-OCF3 | para-CO2H | H | 28 ± 4.8 nM |
| 28 | 6e | H | H | ortho-OMe | para-CO2H | H | 4.0 ± 0.2 nM |
| 29 | 6f | H | H | ortho-OMe | All H | H | 46.6 ± 8.4 nM |
| 30 | 6g | H | H | ortho-CN | All H | H | 560 ± 70.3 nM |
| 31 | 6h | H | H | ortho-CF3 | para-CO2H | H | 35.1 ± 4.0 nM |
| 32 | 6i | H | H | ortho-Br | para-CO2H | H | 2.1 ± 0.4 nM |
| 33 | 6j | H | H | ortho-Cl | para-CH2-CO2H | H | 3.3 ± 1.5 nM |
| 34 | 6k | H | H | ortho-Cl | para-CO2H, meta-OH | H | 6.6 ± 0.6 nM |
| 35 | 61 | H | H | ortho-F | All H | H | 284 ± 11.3 nM |
| 36 | 6m | H | H | ortho-I | para-CO2H | H | 35 ± 3.3 nM |
| 37 | 6n | H | H | ortho-CN | para-CO2H | H | 43 ± 8 nM |
| 38 | 6o | H | H | ortho-Cl | meta-CO2H | H | 18.7 ± 1.5 nM |
| 39 | 6p | H | H | ortho-Cl | para-CONH2 | H | 30.2 ± 1.4 nM |
| 40 | 6q | H | H | ortho-phenyl | para-CO2H | H | 149 ± 23 nM |
| 41 | 6r | H | H | ortho-Cl | para-CO2H | CH3 | 8.5 ± 1.2 nM |
| 42 | 6s | H | H | ortho-Cl | para-CO2H | CH3-CH2 | 50.2 ± 2.7 nM |
| 43 | 6t | F | H | ortho-Cl | meta-CO2H | H | 4.5 ± 1.1 nM |
Determined in-house using the DiscoveRx format.
Table 3.
In-vivo and in vitro Aurora A activities of bisanilinopyrimidines with water-solubilizing moieties
| Entry | Compound ID | Aurora A in vitro IC50 (nM)a | Aurora A in vivo IC50 (μM) [Inhibition of Aurora A T288] | Aurora B in vitro IC50 (nM)b | Aurora B in vivo IC50 (μM) [Inhibition of H3-Ser10] |
|---|---|---|---|---|---|
| 44 |

|
216 ± 10.3 | 1-3 | 232 | 1-10 |
| 45 |

|
28.1 ± 5.5 | <1 | 93.9 | 1-10 |
| 46 |

|
18 ± 2.8 | 1-3 | 123 | 1-10 |
| 47 |

|
44.9 ± 4.7 | <1 | 180 | > 10 |
| 48 |

|
71.2 ± 9.0 | 1-3 | 174 | >10 |
| 49 |

|
316 ± 44.2 | 3-10 | 361 | 1-10 |
| 50 |

|
116.5 ± 10.6 | 3-10 | 1510 | >10 |
| 51 |

|
27 ± 7.6 | <1 | 194 | <1 |
| 52 |

|
30.2 ± 1.4 | <1 | 50.2 | <1 |
| 53 |

|
253 ± 41.6 | <1 | 43.3 | <1 |
| 54 |

|
21.4 ± 2.5 | <1 | 25.6 | <1 |
| 55 |

|
23.2 ± 1.6 | <1 | 30.0 | <1 |
| 56 |

|
12.3 ± 1.1 | <1 | 40.4 | <1 |
| 57 |

|
14.4 ± 1.7 | <1 | 10.7 | <1 |
| 58 |

|
21.2 ± 2.2 | <1 | 56.6 | >10 |
| 59 |

|
3.1 ± 0.16 | > 10 | 226 | > 10 |
| 60 |

|
2.9 ± 0.30 | 3-10 | 83.7 | >10 |
| 61 |

|
17.3 ± 2.3 | >10 | n.d. | >10 |
| 62 |

|
3.0 ± 0.53 | 3-10 | 106 | 1-10 |
Determined in-house using the DiscoveRx format
measured by Reaction Biology using a 33P assay.
Alignment of the Aurora A-ADP complex with compound 1 indicated R1 and R2 in compound 1 as potential sites for synthetic modifications (Figure 3a, 3d and 3e). The R1 position (Figure 2b) of the pyrimidine ring is in close proximity (~ 4 Å) to the gatekeeper residue Leu210 and R2 is close to Glu211 (~ 3.5 Å) (Figure 3a and 3b). To exploit this narrow space, we first introduced small groups such as methyl (Entry 11, 3j, Table 1), amine (Entry 22, 4a, Table 1), chlorine (Entries 20 and 21 3s and 3t respectively, Table 1) and fluorine (Entries 15 and 17, 3n-3p) as R1. The methyl and NH2 derivatives; compounds 3j and 4a respectively (Entries 11 and 22, Table 1) did not contribute to increased activity most probably due to the steric effect of these groups unable to establish desired interactions. In contrast, compounds with fluorine 3n,3o (Entries 15-16, Table 1) and chlorine 4d (Entry 21, Table 1), were among the most potent inhibitors with IC50 values between 0.8 - 4 nM (Entries 15, 16 and 21, Table 1). It is likely that fluorine and chlorine as R1 (Figure 2b) undergo van der Waals interactions with the side chains of the hydrophobic pocket of Ala160, Leu194 and Leu210 (Figure 4d) as observed in the structures of fluoro-bisanilinopyrimidine inhibitors from Genentech.42 Substitution of the R2 position (Figure 2b) of the pyrimidine moiety with methyl, chloro, and amine as shown in compounds 3k, 4c and 4b (Entries 12, 20 and 23 respectively, Table 1) was detrimental for inhibitory activity probably due to steric clash with the backbone carbonyl of Glu211. By contrast, as mentioned above, synthetic modifications at R3 (Figure 3a and 3b) are largely tolerated since the region around the DFG is less confined than that opposite hinge region.
We examined the isoform selectivity of the most potent compound 3o. The inhibition of Aurora B activity was determined using the Reaction Biology 33P kinase Hotspot℠ assay. The IC50 of 3o for Aurora A and Aurora B, using this assay format was 0.2 and 42.9 nM respectively. Similarly the IC50 of 1 for Aurora A and Aurora B was 5.1 and 31 nM respectively. Significant levels of Aurora A selectivity was also observed for the potent compound 3h (IC50 Aurora A 8.9 nM and Aurora B 193 nM).
Having several potent analogs of bisanilinopyrimidines in hand, we next focused on our attention to obtaining potent Aurora inhibitors that are cell permeable and able to inhibit Aurora A kinase in intact cells. The potent compounds 3l, 3n, 3o, 4d, 6a, 6j, 6k and 6r that showed low nanomolar activities in the enzymatic assay however showed poor aqueous solubility and poor activity in intact cells. Introduction of water-solubilizing groups at the para-position of the B-ring was explored to improve both the solubility and cell permeability. Therefore substitution of the para-carboxylic acid in the B-ring with groups that contain a variety of neutral polar moieties (Scheme 4) were employed to exploit H-bond interactions with Arg137 and Arg220. Direct replacement of the para-COOH by the carboxylic acid isostere tetrazole moiety63 provided compounds 12a-12d, that retained the in vitro potency similar 3l (Entries 59-62, Table 3 and Scheme 5) with good aqueous solubility (49 μg/ml in DMEM buffer at pH 7.4). We were able to obtain the X-ray structure of compound 12a (Scheme 5) bound to Aurora A, and compound 12a adopted a binding conformation similar to compound 3l.53 Our primary aim with B-ring modifications described in Scheme 4 and 5 was to obtain a compound with chlorine at the ortho-position of the A-ring and a water-solubilizing moiety at the para-position of the B-ring.
Determination of the effects of Aurora kinase inhibitors on autophosphorylation of Aurora A on Thr288 and phospho-histone H3 (Ser10) levels in breast cancer cells
We next determined the activity of the most potent Aurora kinase inhibitors in intact cells by assessing the inhibition of phosphorylation of Ser10 on Histone H3 and Aurora A Thr288 as surrogates for Aurora B32 and A24, 30 respectively. MDA-MB-468 breast cancer cells were treated with the inhibitors at various concentrations for 2 hours and processed for immunoblotting as described in the Experimental Section. The hit 1 (Aurora B IC50 31 nM) which contains a carboxylic acid moiety on each of the A-ring and the B-ring as well as the most potent compound 3o (in vitro Aurora B IC50 = 42.9 nM), had little effect on Histone H3 Ser10 phosphorylation levels (Supporting Information, Figure S3). This suggested that the carboxylic acid moiety may hinder the ability of these series of compounds to be taken up by cells. We therefore, set out to improve the ability of these compounds to inhibit Aurora activity in intact cells by varying the substituents on the B-ring. The inhibition of Aurora A and B in vitro and cellular inhibition of Aurora A Thr288 and Histone H3 phosphorylation levels is summarized in Figure 5. Replacement of the B-ring carboxylate with the acid bioisostere63 tetrazole such as in (12a), (12b), (12c) and (12d) did not improve cellular activity. The tetrazole series retained the greater than 30-fold Aurora A:B selectivity in vitro selectivity observed for carboxylic acids 3o and 3h. In contrast, compounds with chlorine on the A-ring, substituting the B-ring carboxylate with a carboxamide (6p, Table 3), sulfonamide (9i, Table 3 ) or morpholino (9h, Table 3) greatly improved their activity and resulted in suppression of Histone H3 and Aurora A Thr288 phosphorylation with concentrations as low as 1 μM. This is consistent with the observed lower selectivity of these compounds which significantly inhibit Aurora B in the in vitro assay. Similarly, compounds with ortho-chlorine on the A-ring and fluorine as R1 on the pyrimidine ring, substitutions of the carboxylic acid moiety with carboxamide (9m, Table 3), morpholino (9j, Table 3) and methylene morpholino (9k, Table 3), sulfonamide (9n, Table 3), also greatly improved the ability to suppress Aurora A autophosphorylation. Other substitutions of the B-ring para carboxylic acid that resulted in moderate improvements in cellular activity included 9a, 9b, 9c, 9l and 9f (Table 3). Taken together, these results demonstrate that in this series of in vitro Aurora A inhibitors which contain carboxylic groups are inactive in intact cells but that those where the B-ring carboxylic moiety was replaced by carboxamide, sulfonamide or morpholino groups are highly potent at inhibiting in intact cells the phosphorylation of the Aurora A kinase substrate.
Figure 5.
Inhibition of phosphorylation of Thr288 on Aurora A in MDA-MB-468 cells by bisanilinopyrimidines with water-solubilizing moieties. Inhibition at three concentrations (1, 3 and 10 μM) for each compound is shown. Alisertib (0.3 μM) was used as a positive control.
Compound 9m was subjected to limited profiling using the Reaction Biology 33P kinase Hotspot℠ kinase profiling service (Table 4).64 At a test concentration of 1 μM, 9m was most potent against Aurora A (99% inhibition). CDK1, CDK2, FLT3 and JAK2 were also inhibited by >85%. JAK2 and FLT3 are often cross-inhibited by Aurora inhibitors, including VX-680 which was included as a control. The Aurora kinases whilst not strictly members of the AGC kinase class65, 66 are structurally-related67 to this group. The AGC kinases ROCK1, AKT1 were not significantly inhibited by 9m. The IC50 of the most potent compounds in the in vitro assay, 3l and 3o were also tested in the Hotspot assay (see supporting information) and had IC50s of 0.74 and 0.2 nM respectively (VX-680 had an IC50 of 0.24 nM)(see Figure S1, supporting information. The IC50 of 3o against JAK2 in this assay was 91 nM, showing the high potency against Aurora A is matched by high selectivity (over 450-fold vs JAK2)(see Figure S2, supporting information). The selectivity of 3o was high when tested against the University of Dundee panel of 131 kinases (see Table S2, supporting information). Aurora A showed the highest inhibition (97%) when tested at 0.5 μM. Only 7 of the 131 kinases (including Aurora A and B) were inhibited by greater than 80%. These include the tyrosine kinase JAK2, in agreement with the Reaction Biology data, and the serine/threonine kinases EIF2AK3, GSK3β, NUAK1 and PIM3. The 12 members of the AGC kinase class tested showed no greater than 35% inhibition.
Table 4.
Selectivity profile of compound 9m against a panel of kinases at 1 μM.
| % Inhibition at 1 μM | ||
|---|---|---|
| Kinase: | 9m | VX-680 |
| ABL2/ARG | 75 | 98 |
| AKT1 | 8 | 3 |
| Aurora A | 99 | 100 |
| Aurora B | 93 | 98 |
| Aurora C | 84 | 86 |
| CDK1/cyclin B | 90 | 5 |
| CDK2/cyclin A | 94 | 7 |
| CHK1 | 8 | 16 |
| CHK2 | 30 | 4 |
| FLT3 | 95 | 102 |
| JAK2 | 94 | 81 |
| NEK11 | 21 | 11 |
| NEK2 | 3 | −3 |
| NEK6 | 11 | −5 |
| PLK1 | 64 | 0 |
| RET | 70 | 96 |
| ROCK1 | −5 | −2 |
| TRKA | 39 | 95 |
| WEE1 | 16 | 2 |
Conclusions
The screening hit 1 has provided an excellent starting point for the design and synthesis of highly potent cell permeable Aurora A inhibitors. The X-ray structure of 1 bound to Aurora A revealed key interactions that were exploited for focused library synthesis and rational design. As expected of a bisanilinopyrimidine, 1 is a type 1 kinase inhibitor that targets the ATP binding site, and binds in a DFG-in conformation. In the course of our study, several X-ray structures were obtained and a novel DFG-out binding mode was discovered with compounds, such as 3o, that contain a halogen at the ortho-position of the A-ring.53 Through SAR, X-ray structures and focused library synthesis, we found that fluorine on the 5-position of the pyrimidine ring in combination with chlorine at the ortho-position of the A-ring greatly increases the in vitro potency. The carboxylic acid in the para-position of the B-ring is important for in vitro potency as exemplified by the DFG-out inhibitor 3o; several compounds 9g-9m with water-solubilizing moieties at the para-position of the B-ring inhibited the phosphorylation of the Aurora A substrate histone H3 in MDA-MB-468 breast cancer cells. The carboxamide 9m displayed selectivity against a small panel of kinases. This selectivity was also observed for the carboxylic acid 3o when screened against a larger panel of kinases.
Experimental Section
Protein expression and purification
Aurora A kinase (123-390) was subcloned into a modified pET28 plasmid, overexpressed in E. coli, and purified through a combination of affinity, ion-exchange, and size exclusion chromatography as described previously.53
In vitro enzyme activity assay
The synthetic peptide LRRASLG served as a substrate for Aurora A. Formation of ADP from ATP was quantified using a coupled enzyme assay (DiscoveRx, Fremont, California) as described previously.53 Inhibitor was added to the assay solution, and the reaction was initiated through the addition of 75 μM ATP and 2 mM peptide substrate. IC50 values for were obtained by fitting the data to equation (1),
| (1) |
where A is the remaining activity, [I] is the concentration of the inhibitor, and n is the Hill slope coefficient. The VX-680 compound was used as a standard for IC50 determinations.23
Protein crystallography
Aurora A (123-390) was crystallized in the presence of 2 mM inhibitor compound using the precipitant PEG3350 as described previously.53 X-ray diffraction data were recorded at −180°C using CuKα radiation generated by a Rigaku Micro-Max 007-HF rotating anode (MSC, The Woodlands, TX) using a CCD Saturn 944+ in the Moffitt Structural Biology Core facility. Data were reduced with XDS.68 The structures were solved by molecular replacement using the MolRep program from CCP469 with the PDB entry 3FDN as the search model. CNS70 was employed for refinement, and model building was performed using Coot.71 Figures were prepared using PYMOL.72 The data and refinement statistics for the co-crystal structures are provided in the supporting information (Table S1). The atomic coordinates and structure factors for Aurora A in complex with compounds 1, 3i, 3g, 13 and ATP have been deposited under accession numbers 3UP7, 4DEA, 4DED, 4DEB, and 4DEE respectively.
Determination of the effects of Aurora kinase inhibitors on autophosphorylation of Aurora A on Thr288 and phospho-histone H3 (Ser10) levels in breast cancer cells
MDA-MB-468 cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS) (Invitrogen, US) at 37 °C and 5% CO2. The cells were plated in 6 cm dishes at a density of 2 × 105 cells/dish. Aurora A activity was determined by measuring autophosphorylation of Aurora A on Thr288 in cells that were synchronized by treatment with nocodazole (100 ng/ml) (Sigma) for 20 h prior to the treatment with Aurora inhibitors, whereas Aurora B activity was determined by measuring phosphorylation of histone H3 on Ser10 (pHisH3-Ser10). Cells were then treated with the compounds (0-10 μM); DMSO was used as a negative control and MLN8237 (Alisertib) (0.3 μM) or VX-680 (Tozasertib) (0.5 μM) (Selleck Chemical LLC) were used as a positive control. Cells were harvested after 2 h of treatment and processed for SDS-PAGE and Western blotting as described previously.73 Briefly, following electro-transfer, the membranes were blocked at room temperature for 1 h with TBS containing 5% (w/v) milk and then washed with a mixture of TBS containing 0.2% Tween 20 (Sigma). The membranes were then gently shook at 4 °C overnight with anti-phospho Aurora A (Thr288) antibody (3079, Cell Signaling), anti-phospho-histone H3 (Ser-10) antibody (9701, Cell Signaling), anti-Aurora A antibody (4718, Cell Signaling) or anti-GAPDH monoclonal antibody (E10086CF, Covance) diluted in TBS containing 5% BSA. The membranes were then incubated with HRP conjugated anti-rabbit or anti-mouse IgG antibody (Jackson ImmunoResearch Lab.) at room temperature for 1 h followed by washing with Tween 20-PBS. The membranes were washed again with PBS and developed with the ECL system (Perkin Elmer) as described previously.74
Chemistry
General
All reagents were purchased from commercial suppliers and used without further purification. Melting points were determined using a Barnstead international melting point apparatus and remain uncorrected. Proton NMR spectra were recorded on an Agilent-Varian Mercury 400 MHz spectrometer with CDCl3 or DMSO-d6 as the solvent. Carbon (13C) NMR spectra are recorded at 100 MHz. The 13C spectrum of 1 was recorded at 150 MHz, using an Agilent VNMRS 600 spectrometer with cold probe (University of South Florida Center for Drug Discovery and Innovation). All coupling constants are measured in Hertz (Hz) and the chemical shifts (δH and δC) are quoted in parts per million (ppm) relative to TMS (δ 0), which was used as the internal standard. High resolution mass spectroscopy was carried out on an Agilent 6210 LC/MS (ESI-TOF). Low resolution mass spectroscopy (LRMS) was performed on an Agilent single quad G1956A (Chemistry Department, University of South Florida). Microwave reactions were performed in CEM 908005 model and Biotage initiator 8 machines. All final compounds were purified to ≥95% purity as determined HPLC analysis using a JASCO HPLC system equipped with a PU-2089 Plus quaternary gradient pump and a UV-2075 Plus UV-VIS detector, using an Alltech Kromasil C-18 column (150 × 4.6 mm, 5 μm) and Agilent Eclipse XDB-C18 (150 × 4.6 mm, 5 μm). Melting points were recorded on an Optimelt automated melting point system (Stanford Research Systems). Thin layer chromatography was performed using silica gel 60 F254 plates (Fisher), with observation under UV when necessary. Anhydrous solvents (acetonitrile, dimethylformamide, ethanol, isopropanol, methanol and tetrahydrofuran) were used as purchased from Aldrich. Burdick and Jackson HPLC grade solvents (methanol, acetonitrile and water) were purchased from VWR for HPLC and high resolution mass analysis. HPLC grade TFA was purchased from Fisher. Synthetic protocols and analytical data for compounds 2a to 2o, 5a to 5k, 7a to 7c, 8a to 8c, 10a, 10b, 11a and 11b are reported in the supporting information.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (1, method h in Scheme 1)
A suspension of 3w (0.040 mg, 0.100 mmol) in NaOH (0.2 mL, 4 M) and THF (0.5 mL) was refluxed in seal tube at 85 °C for 2 h. Upon cooling, THF was removed and water (2 mL) was added to the mixture, followed by adding HCl (1M) to acidify (pH = 2) the mixture. The solid obtained was filtered and washed with water (3 mL × 2) and MeOH (3 mL × 2) and dried under high vacuum to afford 1 as a white solid (0.033 mg, 85%). m.p. 256-258 °C. HPLC 98% (Rt = 7.80 min., 45% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.57 (br s, 1H), 10.68 (s, 1H), 9.93 (s, 1H), 8.44 (appt, J = 5.6 Hz, 1H), 8.15 (d, J = 6.0 Hz, 1H), 7.98 (dd, J = 8.0 Hz, 1H), 7.84-7.79 (m, 4 H), 7.60 (t, J = 7.6 Hz, 1H), 7.17 (t, J = 7.6 Hz, 1H), 6.46 (d, J = 5.6 Hz, 1H); 13C NMR (150 MHz, DMSO-d6) δ 169.85, 167.77, 160.93, 158.28, 155.08, 144.94, 141.45, 134.18, 131.90, 130.81, 123.92, 123.22, 122.95, 119.22, 118.83, 101.18; LRMS (ESI−) m/z 351.11 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H15N4O4 (M-Cl)+ 351.1088, found 351.1092.
N4-(2-Carboxyphenyl)-N2-(4-carbamoylphenyl)pyrimidine-2,4-diamine hydrochloride (3a, method d in Scheme 1)
A mixture of 2a (0.050 g, 0.175 mmol) and 2-aminobenzamide (0.027 g, 0.199 mmol) in HCl (1.0 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate obtained upon cooling the mixture was filtered and washed with water (5 mL) and acetone (5 mL × 2) to obtain pure compound 3a (0.036 g, 53%) as a light yellow solid. m.p. 226 °C (dec.). HPLC 96% (Rt = 3.55 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 8.82 (d, J = 7.6 Hz, 1H), 8.63 (appd, J = 7.6 Hz, 1H), 8.17 (d, J = 8.0 Hz, 1H), 8.03 (dd, J = 8.0, 1.4 Hz, 1H), 7.84 (td, J = 7.6, 1.2 Hz, 1H), 7.72-7.69 (m, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.40 (t, J = 7.6 Hz, 1H), 7.30 (t, J = 7.6 Hz, 1H), 6.83 (d, J = 8.0 Hz, 1H); LRMS (ESI+) m/z 333.1 (M-NH2-HCl)+; HRMS (ESI+) m/z calculated for C18H15N5O2 (M-NH2-HCl)+ 333.0982, found 333.1002.
N4-(2-Carboxyphenyl)-N2-(phenyl)pyrimidine-2,4-diamine hydrochloride (3b, method d in Scheme 1)
A mixture of 2a (0.100 g, 0.349 mmol) and aniline (0.038 g, 0.409 mmol) in HCl (1.0 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate formed upon cooling the mixture was filtered and washed with water (5 mL) and hot methanol (5 mL × 2) to obtain 3b (0.065 g, 54%) as a white solid. m.p. 233 °C (dec.). HPLC 91% (Rt = 17.27 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.67 (s, 1H), 9.34 (s, 1H), 8.65 (d, J = 8.0 Hz, 1H), 8.11 (d, J = 5.6 Hz, 1H), 7.97 (d, J = 7.6 Hz, 1H), 7.70 (d, J = 8.4 Hz, 2H), 7.55 (t, J = 6.8 Hz, 1H), 7.25 (t, J = 8.0 Hz, 2H), 7.08 (t, J = 7.6 Hz, 1H), 6.93 (t, J = 6.8 Hz, 1H), 6.35 (d, J = 5.6 Hz, 1H); LRMS (ESI−) m/z 305.1 (M-H-HCl)−; HRMS (ESI+) m/z calculated for C17H15N4O2 (M-Cl)+ 307.1189, found 307.1187.
N4-(2-Carboxyphenyl)-N2-(4-morpholinophenyl)pyrimidine-2,4-diamine hydrochloride (3c, method e in Scheme 1)
A mixture of 2a (0.125 g, 0.437 mmol) and 4-morpholinoaniline (0.089 g, 0.500 mmol) in a solution of EtOH /1 M HCl (1:1, 2 mL) was heated in a microwave reactor at 160 °C for 20 min. Addition of EtOAc (0.5 mL) gave a precipitate. The precipitate was filtered and dried under vacuum to afford the desired compound 3c (0.060 g, 32%) as a light yellow solid. m.p. 162 °C (dec.). HPLC 97% (Rt = 2.99 min., 50% MeOH in 0.1% DEA in water, 20 min.); 1H NMR (400 MHz, CD3OD) δ 8.22 (brs, 1H), 8.14 (dd, J = 8.0, 1.6 Hz, 1H), 7.88 (d, J = 7.2 Hz, 1H), 7.64-7.54 (m, 5H), 7.35 (t, J = 7.6 Hz, 1H), 6.56 (d, J = 7.2 Hz, 1H), 4.06 (appt, J = 4.6 Hz, 4H), 3.14 (appt, J = 4.7 Hz, 4H); 13C NMR (100 MHz, DMSO-d6) δ 168.36, 162.30, 152.53, 144.03, 137.78, 133.70, 131.71, 126.68, 126.62, 124.49, 123.42, 119.53, 100.29, 65.36, 52.16; LC-MS (ESI−)m/z 390.15 (M-HCl)−; HRMS (ESI−) m/z calculated for C21H20N5O3 (M-H-Cl)− 390.1572, found 390.1577.
N4-(2-Carboxyphenyl)-N2-(2-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (3d, method d in Scheme 1)
A suspension of 2,4-dichloropyrimidine (0.149 g, 1.00 mmol) and 2-aminobenzoic acid (0.274 g, 2.00 mmol) in HCl (3.0 ml, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate obtained was filtered and washed with water (10 mL), followed by washing with acetone (5 mL × 2) to obtain the desired compound 3d (0.232 g, 60%) as a white solid. m.p. 254 °C (dec.). HPLC 99% (Rt = 3.53 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 11.80 (s, 1H), 8.91 (d, J = 8.0 Hz, 1H), 8.22 (dd, J = 8.0, 1.2 Hz, 1H), 8.04 (dd, J = 8.0, 1.2 Hz, 1H), 7.97-7.93 (m, 2H), 7.76 (td, J = 7.6, 1.6 Hz, 1H), 7.60 (d, J = 8.0 Hz, 1H), 7.54-7.47 (m, 2H), 7.11 (appd, J = 8.0 Hz, 1H); LRMS (ESI−) m/z 331.0 (M-OH-HCl)-; HRMS (ESI+) m/z calculated for C18H13N4O3 (M-HO-HCl)+ 333.0982, found 333.0997.
N4-(2-Carboxymethylphenyl)-N2-(4-carboxymethylphenyl)pyrimidine-2,4-diamine hydrochloride (3e, method d in Scheme 1)
To a mixture of methyl 4-aminobenzoate (0.166 g, 1.099 mmol) and 2e (0.263 g, 0.876 mmol) was added HCl (3.0 mL, 0.1 M). The reaction mixture was heated in a microwave reactor at 160 °C for 15 min. The crude product that was precipitated was filtered, dried under vacuum and purified using SiO2 chromatography (gradient elution 0-20% EtOAc in hexane) to obtain the desired product 3e (0.116 g, 32%) as a white solid. m.p. 196-198 °C. HPLC 99% (Rt = 5.82 min., 60% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.08 (s, 1H), 9.73 (s, 1H), 8.37 (d, J = 8.4 Hz, 1H), 8.16 (d, J = 6.0 Hz, 1H), 7.94 (dd, J = 8.0, 1.6 Hz, 1H), 7.84 (d, J = 9.2 Hz, 2H), 7.80 (d, J = 9.2 Hz, 2H), 7.64-7.60 (m, 1H), 7.18 (appt, J = 7.2 Hz, 1H), 6.45 (d, J = 6.0 Hz, 1H), 3.81 (s, 3H), 3.79 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 168.25, 166.69, 160.74, 159.58, 157.54, 146.04, 141.41, 134.30, 131.39, 130.62, 123.02, 122.94, 121.99, 118.58, 118.39, 101.12, 52.97, 52.35; LRMS (ESI+) m/z 379.1 (M-Cl)+; HRMS (ESI+) m/z calculated for C20H19N4O4 (M-Cl)+ 379.1401, found 379.1402.
N4,N2-Diphenylpyrimidine-2,4-diamine hydrochloride (3f, method d in Scheme 1)
A mixture of 2,4-dichloropyrimidine (0.050 g, 0.336 mmol) and aniline (0.063 g, 0.677 mmol) in HCl (1.0 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The product obtained was purified using SiO2 chromatography (gradient elution 0-20% EtOAc in hexane) to afford the desired product 3f (0.048 g, 49%) as a white solid. m.p. 144 °C (dec.). HPLC 96% (Rt = 5.33 min., 60% MeOH in 0.1% TFA water, 40 min.); 1H NMR (400 MHz, CD3OD) δ 7.85 (d, J = 6.0 Hz, 1H), 7.59 (d, J = 7.6 Hz, 2H), 7.55 (dd, J = 7.6 Hz, 2H), 7.27 (d, J = 7.6 Hz, 2H), 7.23 (d, J = 7.6 Hz, 2H), 7.05-6.98 (m, 2H), 6.20 (d, J =6.0 Hz, 1H); LRMS (ESI+) m/z 263.1 (M-Cl)+; HRMS (ESI+) m/z calculated for C16H15N4 (M-Cl)+ 263.1291, found 263.1293.
N4-(2-Carbamoylphenyl)-N2-(4-carbamoylphenyl)pyrimidine-2,4-diamine hydrochloride (3g, method d in Scheme 1)
A mixture of 2b (0.248 g, 0.871 mmol) and 4-aminobenzamide (0.136 g, 1.00 mmol) in HCl (3.0 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate was filtered and washed with water (10 mL), hot MeOH (10 mL), hot THF (10 mL), dioxane (5 mL), DMF (5 mL) and MeOH (10 mL) sequentially to obtain pure 3g (0.160 g, 48%) as a light yellow solid. m.p. 257-260 °C. HPLC 97% (Rt = 3.32 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ10.63 (s, 1H disappeared on D2O shake), 9.65 (s, 1H disappeared on D2O shake), 8.57 (d, J = 8.4 Hz, 1H), 8.15 (d, J = 5.2 Hz, 1H), 7.98 (dd, J = 7.6, 1.6 Hz, 1H), 7.83-7.73 (m, 5H becomes 4H on D2O shake), 7.63-7.58 (m, 1H), 7.17 (brs, 1H disappeared on D2O shake), 7.12 (t, J = 8.4 Hz, 1H), 6.42 (d, J = 6.0 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 170.31, 168.30, 160.55, 159.67, 157.36, 144.07, 142.41, 134.35, 131.96, 128.81, 127.22, 122.25, 121.83, 118.50, 117.57, 101.05; LRMS (ESI+) m/z 349.1 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H17N6O2 (MCl)+ 349.1408, found 349.1407.
N4-(Phenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (3h, method d in Scheme 1)
A mixture of 2c (0.020 g, 0.082 mmol) and 4-aminobenzoic acid (0.013 g, 0.097 mmol) in HCl (1.5 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The solid obtained upon cooling the mixture was filtered, washed with water (5 mL) and dried under vacuum to obtain the desired compound 3h (0.020 g, 71%) as a white solid. m.p. 215 °C (dec.). HPLC 99% (Rt = 7.28 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, CD3OD) δ 8.00 (d, J = 8.8 Hz, 2H), 7.86 (d, J = 7.2 Hz, 1H), 7.63 (d, J = 8.4 Hz, 2H), 7.57 (d, J = 7.6 Hz, 2H), 7.41 (t, J = 7.6 Hz, 2H), 7.28 (t, J = 7.6 Hz, 1H), 6.46 (d, J = 7.2 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 167.44, 161.79, 152.70, 144.17, 141.82, 137.81, 130.87, 129.50, 126.80, 126.15, 123.30, 121.17, 100.58. LRMS (ESI−) m/z 305.0 (M-Cl)−; HRMS (ESI+) m/z calculated for C17H15N4O2 (M-Cl)+ 307.1190, found 307.1187.
N4,N2-Di(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (3i, method d in Scheme 1)
A mixture of 2d (0.100 g, 0.350 mmol) and 4-aminobenzoic acid (0.055 g, 0.401 mmol) in HCl (1.5 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate obtained upon cooling the mixture was filtered and washed with water (10 mL), followed by quick wash with acetone (5 mL × 2) to give pure 3i (0.136 g, 99%) as a white solid. m.p. 281-283 °C. HPLC 98% (Rt = 4.97 min., 50% MeOH in 0.1% TFA in water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.71 (brs, 1H), 10.46 (brs, 1H), 8.09 (d, J = 6.8 Hz), 7.90 (d, J = 6.0 Hz, 2H partially overlapping) 7.88 (d, J = 6.0 Hz, 2H partially overlapping), 7.79 (d, J = 8.8 Hz, 2H), 7.71 (d, J = 8.4 Hz, 2H), 6.50 (d, J = 6.4 Hz, 1H); LRMS (ESI−) m/z 349.0 (M-H-HCl)−; HRMS (ESI+) m/z calculated for C18H15N4O4 (M-Cl)+ 351.1088, found 351.1082.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)-5-methylpyrimidine-2,4-diamine hydrochloride (3j, method g in Scheme 1)
A mixture of 2j (0.115 g, 0.383 mmol) and 4-aminobenzoic acid (0.179 g, 1.307 mmol) in HCl (1 M) /THF (2:1, 3.0 mL) was heated in a microwave reactor at 160 °C for 15 min. The precipitate obtained upon cooling the mixture was filtered and washed with water (5 mL) and MeOH (5 mL × 2). The solid obtained was slurried in DMF (2 mL), filtered and washed with MeOH (5 mL), acetone (5 mL) and DCM (5 mL) sequentially to obtain the desired compound 3j (0.055 g, 36%) as a white solid. m.p. 213 °C (dec.). HPLC 97% (Rt = 5.17 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.50 (brs, 1H, disappeared on D2O shake), 11.00 (s, 1H disappear on D2O shake), 9.68 (s, 1H disappeared on D2O shake), 9.01 (d, J = 8.4 Hz, 1H), 8.08 (s, 1H), 8.03 (d, J = 7.9 Hz, 1H), 7.85-7.80 (m, 4H), 7.60 (t, J = 7.9 Hz, 1H), 7.11 (t, J = 7.6 Hz, 1H), 2.16 (s, 3H); LC-MS (ESI+) m/z 365.13 (M-Cl)+; HRMS (ESI+) m/z calculated for C19H17N4O4 (M-Cl)+ 365.1244, found 365.1240.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)-6-methylpyrimidine-2,4-diamine hydrochloride (3k, method d in Scheme 1)
A mixture of 2f (0.263 g, 0.877 mmol) and 4-aminobenzoic acid (0.137 g, 1.00 mmol) in HCl (3.0 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The precipitate obtained was filtered and washed with water (10 mL), MeOH (10 mL × 2), DMSO (1 mL, quick wash), and acetone (5 ml) to obtain the desired compound 3k (0.187 g, 53%) as a white solid. m.p. 243 °C (dec.). HPLC 90% (Rt = 5.11 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 1H), 9.73 (s, 1H), 8.55 (d, J = 8.4 Hz, 1H), 7.98 (d, J = 6.8 Hz, 1H), 7.86 (d, J = 8.8 Hz, 2H), 7.81 (d, J = 8.8 Hz, 2H), 7.58 (t, J = 7.2 Hz, 1H), 7.11 (t, J = 7.2 Hz, 1H), 6.33 (s, 1H), 2.28 (s, 3H); LC-MS (ESI−) m/z 363.11 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C19H15N4O4 (M-H-HCl)+ 363.1099, found 363.1107.
N4-(2-Chlorophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (3l, method e in Scheme 1)
A mixture of 2l (0.094 g, 0.340 mmol) and 4-aminobenzoic acid (0.107 g, 0.781 mmol) in EtOH /1M HCl (2.0 mL, 1:1) was heated in a microwave reactor at 160 °C for 15 min. The mixture was cooled to r.t. The precipitate obtained was filtered, washed with water (2 mL), and MeOH (2 mL) sequentially to afford the desired compound 3l (0.055 g, 43%) as a white solid. m.p. 234 °C (dec.). HPLC 99% (Rt = 4.32 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.59 (s, 1H), 10.00 (s, 1H), 9.76 (s, 1H), 8.06 (d, J = 6.0 Hz, 1H), 7.70-7.68 (m, 3H), 7.63-7.58 (m, 3H), 7.43 (t, J = 7.8 Hz, 1H), 7.33 (t, J = 7.4 Hz, 1H), 6.40 (d, J = 6.1 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 167.36, 163.18, 152.30, 145.07, 141.91, 134.61, 130.72, 130.64, 130.47, 129.67, 129.52, 128.56, 126.30, 120.01, 95.58; LC-MS (ESI+) m/z 341.09 (MCl)+; HRMS (ESI+) m/z calculated for C17H14ClN4O2 (M-Cl)+ 341.0800, found 341.0810.
N4-(2-Carboxyphenyl)-N2-phenyl-5-fluoropyrimidine-2,4-diamine hydrochloride (3m, method e in Scheme 1)
A mixture of 2g (0.134 g, 0.441 mmol) and aniline (0.140 g, 1.50 mmol) in 1:1 ratio of EtOH /1M HCl (2.0 mL) was heated in a microwave reactor at 160 °C for 30 min. The precipitate formed upon cooling the mixture was filtered and washed with MeOH (5 mL), acetone (5 mL) and dried under vacuum to afford the desired compound 3m (0.150 g, 94%) as a white solid. m.p. 240 °C (dec.). HPLC 99% (Rt = 4.21 min., 70% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 11.50 (s, 1H), 9.41 (s, 1H), 8.96 (d, J = 8.4 Hz, 1H), 8.21 (d, J = 3.1 Hz, 1H), 8.04 (dd, J = 7.9, 1.5 Hz, 1H), 7.67 (d, J = 8.4 Hz, 2H), 7.60 (t, J = 8.4 Hz, 1H), 7.27 (t, J = 8.4 Hz, 2H), 7.12 (t, J = 7.6 Hz, 1H), 6.94 (t, J = 7.3 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ -166.03 (s); 13C NMR (100 MHz, DMSO-d6) δ 170.83, 156.07 (d, J = 3.0 Hz), 149.60 (d, J = 10.0 Hz), 142.26, 141.68 (d, J = 245 Hz), 141.52, 141.33, 141.25, 134.72, 132.02, 129.05, 122.15 (d, J = 21.0 Hz), 120.74, 119.80, 116.24; LC-MS (ESI+) m/z 325.12 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H14FN4O2 (M-Cl)+ 325.1095, found 325.1093.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)-5-fluoropyrimidine-2,4-diamine acid (3n, method e in Scheme 1)
A mixture of 2g (0.134 g, 0.44 mmol) and 4-aminobenzoic acid (0.206 g, 1.50 mmol) in 1:1 ratio of EtOH/HCl (2.0 mL, 1 M) was heated in a microwave reactor at 160 °C for 30 min. The precipitate formed upon cooling the mixture was filtered and washed with sat. NaHCO3 (3 mL) and water (5 mL). The solid obtained was slurried in hot DMF (3 mL), filtered and washed with MeOH (5 mL) and dried under vacuum to afford the desired compound 3n (0.096 g, 59%) as a white solid. m.p. 287-290 °C. HPLC 99% (Rt = 14.73 min., 55% MeOH in 0.1% TFA, water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 11.42 (s, 1H disappeared on D2O shake), 9.83 (s, 1H disappeared on D2O shake), 8.94 (d, J = 8.6 Hz, 1H), 8.29 (appd, J = 2.7 Hz, 1H), 8.05 (d, J = 7.9 Hz, 1H), 7.83 (d, J = 8.4 Hz, 2H), 7.83 (d, J = 8.4 Hz, 2H), 7.63 (t, J = 7.2 Hz, 1H), 7.16 (t, J = 7.5 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ -164.42 (s); 13C NMR (100 MHz, DMSO-d6) δ 170.76, 167.82, 155.42 (d, J = 3.5 Hz), 149.76 (d, J = 10 Hz), 145.58, 142.09 (d, J = 247 Hz), 142.00, 141.25 (d, J = 18 Hz), 134.70, 132.03, 130.87, 123.39, 122.57, 120.93, 118.13, 116.65; LC-MS (ESI+) m/z 369.10 (M+H)+; HRMS (ESI+) m/z calculated for C18H14FN4O4 (M+H)+ 369.0994, found 369.0994.
N4-(2-Chlorophenyl)-N2-(4-carboxyphenyl)-5-fluoropyrimidine-2,4-diamine hydrochloride (3o, method d in Scheme 1)
A mixture of 2k (0.300 g, 1.12 mmol) and 4-aminobenzoic acid (0.153 g, 1.12 mmol) in ethanol (1.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The reaction mixture was cooled, and stirred at room temperature for 48 hours. The white precipitate was isolated by filtration and washed with ethyl acetate (5 mL). The product was suspended in ethyl acetate (5 mL) and sonicated for 5 min. and filtered to provide 3o (0.232 g, 52%) as a white powder. m.p. 308 °C (dec.). HPLC 99% (Rt = 7.57 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.44 (s, 1H), 9.59 (s, 1H), 9.38 (s, 1H), 8.15 (d, J = 3.4 Hz, 1H), 7.62-7.52 (m, 6H), 7.45-7.36 (m, 2H); 19F NMR (376 MHz, DMSO-d6) δ -164.92; 13C NMR (100 MHz, DMSO-d6) δ 167.77, 155.67 (d, J = 3.0 Hz), 151.48 (d, J = 12.0 Hz), 145.80, 141.60 (d, J = 20.0 Hz), 141.46 (d, J = 245.0 Hz), 136.04, 131.63, 130.55, 130.40, 130.19, 128.44, 128.38, 122.66, 117.37; LC-MS (ESI+) m/z 359.07 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H13ClFN4O2 (M-Cl)+ 359.0706, found 359.0709.
N4-(2-Chlorophenyl)-N2-phenyl-5-fluoropyrimidine-2,4-diamine hydrochloride (3p, method m in Scheme 2)
A mixture of 2k (0.061 g, 0.207 mmol) and aniline (0.020 g, 0.207 mmol) in EtOH (2.0 mL) was heated in a microwave reactor at 150 °C for 20 min. The precipitate formed upon cooling the mixture was filtered and quickly washed with MeOH (1 mL) to afford the desired product 3p (0.037 g, 51%) as a white solid. m.p. 145-148 °C. HPLC 99% (Rt = 9.43 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.18 (s, 1H disappeared on D2O shake), 9.12 (s, 1H disappeared on D2O shake), 8.08 (d, J = 4.0 Hz, 1H), 7.59-7.56 (m, 2H), 7.44 (d, J = 8.4 Hz, 2H), 7.39 (td, J = 7.6, 1.6 Hz, 1H), 7.31 (td, J = 7.6, 1.6 Hz, 1H), 7.02 (t, J = 7.2 Hz, 2H), 6.77 (t, J = 7.6 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ -166.49; 13C NMR (100 MHz, DMSO-d6) δ 155.97, 151.42 (d, J = 12 Hz), 141.40, 141.24 (d, J = 20 Hz), 141.11 (d, J = 244 Hz), 136.05, 131.18, 130.35, 129.80, 128.76, 128.29, 128.13, 121.33, 118.80; LC-MS (ESI+) m/z 315.08 (M-Cl)+; HRMS (ESI+) m/z calculated for C16H13ClFN4 (M-Cl)+ 315.0807, found 315.0812.
N4 -(2-Carboxyphenyl)-N2-(3-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (3q method d in Scheme 1)
A mixture of 2a (0.050 g, 0.175 mmol) and 3-aminobenzoic acid (0.024 g, 0.175 mmol) in HCl (0.1 mL, 0.1 M) was heated in a microwave reactor at 160 °C for 15 min. The solution was cooled and the precipitate obtained was filtered and washed quickly with hot methanol (3 ml) to obtain the desired product 3q (0.056 g, 72%) as a white solid. m.p. 275 °C (dec). HPLC 99% (Rt = 6.31 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.85 (brs, 1H), 10.73 (s, 1H), 9.52 (s, 1H), 8.67 (d, J = 8.4 Hz, 1H), 8.27 (s, 1H), 8.12 (d, J = 5.7 Hz, 1H), 7.96 (d, J = 7.9 Hz, 2H), 7.54-7.47 (m, 2H), 7.51 (t, J = 7.7 Hz, 2H), 7.05 (t, J = 7.6 Hz, 1H), 6.36 (d, J = 5.7 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 170.49, 168.16, 160.41, 159.99, 157.52, 142.75, 141.52, 134.46, 131.93, 131.72, 129.22, 124.08, 122.79, 121.79, 121.25, 120.71, 116.79, 100.98. LC-MS (ESI−) m/z 349.10 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C18H14N4O4 (M-H-HCl)− 349.0942, found 397.0940.
N4-(2-Carboxyphenyl)-N2-(3-carboxyphenyl)-5-fluoropyrimidine-2,4-diamine hydrochloride (3r method m Scheme 2)
A mixture of 2g (0.100 g, 0.329 mmol) and 3-aminobenzoic acid (0.048 g, 0.350 mmol) in HCl (1.0 mL, 0.1 M) was heated in a microwave reactor at 150 °C for 20 min. The mixture was cooled to r.t. and the precipitate obtained was filtered and washed quickly with ethanol (3 ml) to obtain the desired product 3r (0.089 g, 66%) as a white solid. m.p. 272 °C (dec.). HPLC 99% (Rt = 14.34 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 13.07 (brs, 1H), 11.48 (s, 1H), 9.61 (s, 1H), 8.97 (d, J = 8.4 Hz, 1H), 8.31 – 8.23 (m, 2H), 8.03 (d, J = 7.9 Hz, 1H), 7.60–7.48 (m, 2H), 7.39 (t, J = 7.9 Hz, 1H), 7.12 (t, J = 7.6 Hz, 1H); 13C NMR (101 MHz, DMSO-d6) δ 170.87, 168.29, 155.90, 149.69 (d, J = 9.7 Hz), 151.37 (d, J = 11.5 Hz), 141.91 (d, J = 235.3 Hz), 142.22, 141.54, 134.89, 132.04, 131.77, 129.32, 123.78, 122.87, 122.29, 120.62, 120.37, 116.19; 19F NMR (376 MHz, DMSO) δ -165.34; Elemental analysis: calculated for C18H14ClFN4O4 C, 53.41; H, 3.49; N, 13.84. Found: C, 53.22; H, 3.37; N, 13.57.
N4-(2-Carboxyphenyl)-N2-(4-ethoxycarbonylphenyl)-5-chloropyrimidine-2,4-diamine hydrochloride (3s, method f (ii) in Scheme 1)
A mixture of 2m (0.063 g, 0.193 mmol) and ethyl 4-aminobenzoate (0.043 g, 0.260 mmol) in ethanol (0.8 mL), was heated in a sealed tube at 120 °C (oil bath temperature) overnight. The resulting precipitate was filtered and washed with ethanol (1 mL × 2), Et2O (3 mL), hexane (2 mL) sequentially and dried under vacuum to afford the title compound 3s (0.079 g, 90%), as a white solid. m.p. 221 °C (dec.). 1H NMR (400 MHz, DMSO-d6) δ 13.77 (s, 1H), 11.46 (s, 1H), 9.97 (s, 1H), 8.93 (d, J = 8.3 Hz, 1H), 8.34 (s, 1H), 8.04 (dd, J = 1.5, 7.9 Hz, 1H), 7.86 (d, J = 9.1 Hz, 2H), 7.83 (d, J = 9.1 Hz, 2H), 7.64–7.60 (m, 1H), 7.19–7.15 (m, 1H), 4.26 (q, J = 7.1 Hz, 2H), 1.29 (t, J = 7.1 Hz, 3H). LC-MS (ESI−) m/z 412.09 (M+H)+; HRMS (ESI−) m/z calculated for C20H18ClN4O4 (M+H)+ 413.1011, found 413.0989.
N4-(2-Carboxyphenyl)-N2-(4-ethoxycarbonylphenyl)-5,6-dichloropyrimidine-2,4-diamine hydrochloride (3t, method f (ii) in Scheme 1)
A mixture of 2n (0.091 g, 0.257 mmol) and ethyl 4-aminobenzoate (0.044 g, 0.266 mmol) in ethanol (1.0 mL), was heated in a sealed tube at 110 °C (oil bath temperture) for 4 days. The resulting precipitate was filtered and washed with ethanol (1 mL × 2) and suspended in methanol (1 mL). The mixture was sonicated and the solid was filtered and dried under vacuum to afford the title compound 3t (0.022 g, 17%) as a white solid. m.p. 206 °C (dec.). 1H NMR (400 MHz, DMSO-d6) δ 11.52 (s, 1H), 10.25 (s, 1H), 8.74 (s, 1H), 8.04 (dd, J = 1.5, 7.9 Hz, 1H,), 7.86 (d, J = 8.8 Hz, 2H), 7.75 (d, J = 8.9 Hz, 2H), 7.64–7.60 (m, 1H,), 7.23-7.20 (m, 1H), 4.27 (q, J = 7.1 Hz, 2H,), 1.29 (t, J = 7.1 Hz, 3H). LC-MS (ESI−) m/z 447.07 (M-Cl)+; HRMS (ESI−) m/z calculated for C20H17Cl2N4O4 (M-Cl)+ 447.0621, found 447.00598.
5-Amino-N4-(2-carboxyphenyl)-N2-(4-ethoxycarbonylphenyl)pyrimidine-2,4-diamine hydrochloride (3u, method f (i) in Scheme 1)
This was prepared by using a method described Gray and co-workers.47 A mixture of 2h (0.080 g, 0.266 mmol), ethyl 4-aminobenzoate (0.099 g, 0.600 mmol) and HCl (0.15 mL, 4M in dioxane) in 2-butanol (1.0 mL) was heated in a sealed tube at 120 °C (oil bath temperature) for 24 h. After cooling the mixture to r.t., the suspension obtained was filtered and washed with water (5 mL), MeOH (3 mL) to afford the desired compound 3u (0.082 g, 72%) as a yellow solid. m.p. 262 °C (dec.). HPLC 99% (Rt = 15.48 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.88 (s, 1H), 9.87 (s, 1H), 9.18 (s, 1H), 7.94-7.84 (m, 5H), 7.80 (d, J = 8.0 Hz, 1H), 7.42 (t, J = 8.4 Hz, 1H), 7.19 (d, J = 8.0 Hz, 1H), 7.04 (t, J = 7.5 Hz, 1H), 4.26 (q, J = 7.0 Hz, 2H), 1.30 (t, J = 7.0 Hz, 3H); LC-MS (ESI+) m/z 376.15 (M-OH-HCl)+; HRMS (ESI+) m/z calculated for C20H18N5O3 (M-OH-HCl)+ 376.1404, found 376.1405.
6-Amino-N4-(2-carboxyphenyl)-N2-(4-ethoxycarbonylphenyl)pyrimidine-2,4-diamine hydrochloride (3v, method e in Scheme 1)
A mixture of 2i (0.265 g, 0.880 mmol) and ethyl 4-aminobenzoate (1.65 g, 10.00 mmol) in EtOH /1 M HCl (1:1, 12 mL) was heated in a microwave reactor at 160 °C for 1 h. The mixture was cooled to r.t., and the solid obtained was filtered, washed with MeOH (5 mL) and slurried in acetone (10 × 5 mL) until no impurity was shown by NMR to afford the desired compound 3v (0.125 g, 33%) as a beige color solid. m.p. 280 °C (dec.). 1H NMR (400 MHz, DMSO-d6) δ 10.65 (s, 1H disappeared on D2O shake), 9.36 (s, 1H disappeared on D2O shake), 8.85 (appd, J = 7.6 Hz, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.83 (d, J = 8.0 Hz, 2H), 7.65 (d, J = 8.4 Hz, 2H), 7.47 (t, J = 8.4Hz, 1H), 6.94 (t, J = 8.0 Hz, 1H), 6.54 (brs, 2H disappeared on D2O shake), 5.54 (s, 1H), 4.26 (q, J = 7.0 Hz, 2H), 1.29 (t, J = 7.0 Hz, 3H); LC-MS (ESI+) m/z 394.15 (M-Cl)+; HRMS (ESI+) m/z calculated for C20H20N5O4 (M-Cl)+ 394.1510, found 394.1509.
N4-(2-Carboxyphenyl)-N2-(4-methoxycarbonylphenyl)pyrimidine-2,4-diamine hydrochloride (3w, method m in Scheme 2)
A suspension of 2a (0.060 g, 0.210 mmol) and methyl 4-aminobenzoate (0.032 g, 0.210 mmol) in MeOH ( 1 mL) was heated in a microwave reactor at 150 °C for 20 min. Upon cooling, the resulting precipitate was filtered and washed with MeOH (2 mL) to afford 3w (0.064 mg, 76%) as a yellow solid which was used without further purification. 1H NMR (400 MHz, DMSO-d6) δ 13.40 (br s, 1H), 10.74 (s, 1H), 10.11 (s, 1H), 8.33 (br d, J = 5.6 Hz, 1H), 8.14 (d, J = 6.0 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.84-7.76 (m, 4 H), 7.63 (t, J = 7.6 Hz, 1H), 7.22 (t, J = 7.6 Hz, 1H), 6.50 (d, J = 6.4 Hz, 1H), 3.80 (s, 3H); LC-MS (ESI+) m/z 365.13 (M-Cl)+; HRMS (ESI+) m/z calculated for C19H17N4O4 (M-Cl)+ 365.1244, found 365.1243.
5-Amino-N4-(2-carboxyphenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine (4a, method h in Scheme 1)
A suspension of 3u (0.069 g, 0.161 mmol) in NaOH/THF (2 M, 0.44 mL/0.2 mL) was heated at 100 °C (oil bath temperature) in a sealed tube for 30 min. A solution of HCl (1 M) was added to acidify the solution to pH = 1-2 after removing THF. The solid obtained was filtered and washed with water (5 mL), sat. NaHCO3 (3 mL), water (5 mL), acetone (3 mL), and MeOH (3 mL) sequentially to afford the desired compound 4a (0.038 g, 65%) as a brown solid. m.p. 160 °C (dec.). HPLC 96% (Rt = 4.19 min., 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.33 (brs, 1H disappeared on D2O shake), 8.86 (brs, 1H), 7.99 (brs, 1H), 7.88-7.78 (m, 5H), 7.50 (brs, 1H), 7.01 (brs, 1H); LC-MS (ESI+) m/z 366.12 (M+H)+; HRMS (ESI+) m/z calculated for C18H16N5O4 (M+H)+ 366.1197, found 366.1198.
6-Amino-N4-(2-carboxyphenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine (4b, method h in Scheme 1)
A suspension of 3v (0.070 g, 0.163 mmol) in NaOH (0.45 mL, 2 M) and THF (0.25 mL) was heated at 100 °C (oil bath temperature) in a sealed tube for 16 h. The THF in the mixture was evaporated and HCl (1 M) was added to acidify (pH = 1-2) the mixture. The solid obtained was filtered and washed with water (3 mL), sat. aq. NaHCO3 (3 mL) and water (3 mL) sequentially, and dried to afford the desired compound 4b (0.047 g, 79%) as a yellow solid. m.p. 220 °C (dec.). HPLC 95% (Rt = 5.20 min., 55% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.65 (s, 1H disappeared on D2O shake), 9.78 (s, 1H disappeared on D2O shake), 8.54 (brs, 1H), 7.96 (appd, J = 5.2 Hz, 1H), 7.82-7.80 (m, 2H), 7.59-7.53 (m, 3H), 7.34-7.10 (m, 3H), 5.60 (s, 1H); LC-MS (ESI+) 366.13 m/z (M+H)+; HRMS (ESI+) m/z calculated for C18H16N5O4 (M+H)+ 366.1197, found 366.1194.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)-5,6-dichloropyrimidine-2,4-diamine hydrochloride (4c, method h in Scheme 1)
A mixture of 3t (0.018 g, 0.034 mmol) in THF (0.3 mL) and NaOH (0.1 mL, 2 M) was heated in a sealed tube at 110 °C (oil bath temperature) overnight. The THF was then removed under reduced pressure and HCl (1 M aq., 0.5 mL) was added to the residue. The resulting precipitate was filtered and washed with water (2 mL) and dried under vacuum. The solid obtained was then slurried in methanol (1 mL), filtered and dried under vacuum to afford the title compound 4c (0.005 g, 32%) as a white solid. m.p. 230 °C (dec.). HPLC 84% [Rt = 10.84 min., 20% MeOH, 80% water (with 0.1% DEA), 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 12.62 (s, 1H), 11.47 (s, 1H), 10.21 (s, 1H), 8.72 (s, 1H), 8.04 (dd, J = 7.9, 1.4 Hz, 1H), 7.84 (d, J = 8.8 Hz, 2H), 7.72 (d, J = 8.7 Hz, 2H), 7.65–7.54 (m, 1H), 7.23-7.20 (m, 1H); LC-MS (ESI−) m/z 417.02 (M-HHCl)−; HRMS (ESI−) m/z calculated for C18H13Cl2N4O4 (M-H-HCl)− 417.0163, found 417.0160.
N4-(2-Carboxyphenyl)-N2-(4-carboxyphenyl)-5-chloropyrimidine-2,4-diamine hydrochloride (4d, method h in Scheme 1)
A mixture of 3s (0.058 g, 0.119 mmol) in THF (0.4 mL) and NaOH (2 M aq. 0.2 mL) was heated in a sealed tube at 110 °C (oil bath temperature) overnight. The THF was then removed under reduced pressure and HCl (1 M aq., 0.6 mL) was added to the residue. The resulting precipitate was filtered, washed with water (2 mL × 3) and dried under vacuum. The solid obtained was then slurried in DMF (2 mL), filtered, washed with DMF (5 mL), methanol (1 mL) , Et2O (3 mL) sequentially and dried under vacuum to afford the title compound 4d (0.036 g, 72%) as a white solid. m.p. 261 °C (dec.). 1H NMR (400 MHz, DMSO-d6) δ 12.59 (s, 1H), 11.45 (s, 1H), 9.90 (s, 1H), 8.87 (s, 1H), 8.30 (s, 1H), 8.01-7.55 (m, 6H), 7.13 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 170.57 167.72, 157.79, 155.85, 155.36, 145.16, 141.90, 134.51, 131.90, 130.84, 123.84, 122.82, 121.54, 118.76, 117.26, 107.11; HPLC 97% [Rt = 8.74 min., 15% MeOH, 85% water (with 0.1% DEA) 20 min.]; LC-MS (ESI−) m/z 383.04 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C18H14ClN4O4 (M-H-HCl)− 383.0553, found 383.0552.
N4-(2-Fluorophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6a, method m in Scheme 2)
A mixture of 2-chloro-N-(2-fluorophenyl)pyrimidin-4-amine (5a) (0.096 g, 0.428 mmol) and 4-aminobenzoic acid (0.069 g, 0.503 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The mixture was filtered and the resulting precipitate was washed with EtOH (0.5 mL × 2) to provide the title compound 6a (0.107 g, 69%) as a white solid. m.p. 264 °C (dec). HPLC 98.7% (Rt = 3.8 min.,, 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, d6-DMSO) δ 12.79 (s, 1H), 11.04 (s, 1H), 10.87 (s, 1H), 8.13 (d, J = 7.0 Hz, 1H), 7.77 (d, J = 8.7 Hz, 2H), 7.66 (t, J = 7.9 Hz, 1H), 7.56 (d, J = 8.5 Hz, 2H), 7.46–7.36 (m, 2H), 7.33–7.25 (m, 1H), 6.58 (d, J = 6.7 Hz, 1H); 19F NMR (400 MHz, DMSO-d6) δ 121.12; 13C NMR (100 MHz, DMSO-d6) δ 167.40, 163.00, 156.59 (d, J = 245 Hz), 152.55, 145.08, 141.92, 130.79, 129.14, (d, J = 9.27 Hz), 128.30, 126.43, 125.39, 125.04 (d, J = 12.15 Hz), 120.29, 116.88(d, J = 18.72 Hz), 99.84, LC-MS (ESI−) m/z 323.10 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C17H12FN4O2 (M-H-HCl)− 323.0950, found 323.0974.
N4-[2-(Trifluoromethyl)phenyl]-N2-phenylpyrimidine-2,4-diamine hydrochloride (6b, method m in Scheme 2)
A mixture of chloropyrimidine 5f (0.076 g, 0.277 mmol) and aniline (0.03 mL, 0.328 mmol) in EtOH (0.4 mL) was heated in a microwave reactor at 150 °C for 20 min. The solvent was removed under reduced pressure to provide an off-white solid. Ethyl acetate (3 mL) was added to the reaction mixture. The mixture was left at r.t. for 30 min and sonicated occasionally. The resulting precipitate was isolated by filtration and washed with ethyl acetate (1 mL × 5) and hexane (3 mL) to afford 6b (0.085 g, 84%) as a white solid. m.p. 207 °C (dec). HPLC 100% (Rt = 9.1 min., 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.62 (s, 1H), 10.43 (s, 1H), 8.01 (apparent s, 1H), 7.86–7.80 (m, 2H), 7.71–7.48 (m, 2H), 7.27 (d, J = 7.2 Hz, 2H), 7.14 (t, J = 7.5 Hz, 2H), 7.03 (t, J = 7.3 Hz, 1H), 6.46 (s, 1H); LC-MS (ESI+) m/z 331.13 (MCl)+; HRMS (ESI+) m/z calculated for C17H14F3N4 (M-Cl)+ 331.1165, found 331.1170.
N4-(2-Chloro-4-fluorophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6c, method m in Scheme 2)
A mixture of chloropyrimidine 5b (0.096 g, 0.372 mmol) and 4-aminobenzoic acid (0.058 g, 0.422 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The resulting precipitate was isolated by filtration and washed with EtOH (0.5 mL × 2), diethyl ether (2 mL) and hexane (2 mL) sequentially to provide the title compound 6c (0.102 g, 69%) as a white solid. m.p. 268 °C (dec). HPLC 97.7% (Rt = 4.8 min., 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.89 (s, 1H), 10.72 (s, 1H), 8.12 (d, J = 7.0 Hz, 1H), 7.75 (d, J = 8.7 Hz, 2H), 7.72–7.63 (m, 2H), 7.50 (d, J = 8.6 Hz, 2H), 7.40 (td, J = 8.5, 2.9 Hz, 1H), 6.53 (d, J = 7.0 Hz, 1H); 19F NMR (400 MHz, DMSO-d6) δ 112.66; 13C NMR (100 MHz, DMSO-d6) δ 167.37 163.39, 161.22 (d, J = 246.3 Hz), 152.39, 145.37, 141.91, 131.87 (d, J = 11.12 Hz); LC-MS (ESI−) m/z 357.06 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C17H11ClFN4O2 (M-HHCl)− 357.0560, found 357.0521.
N4-[2-(Trifluoromethoxy)phenyl]-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6d, method m in Scheme 2)
A mixture of chloropyrimidine 5c (0.074 g, 0.255 mmol) and 4-aminobenzoic acid (0.042 g, 0.306 mmol) in EtOH (0.5 mL) was heated with a microwave reactor at 150 °C for 20 min. The resulting precipitate was isolated by filtration and washed with EtOH (0.5 mL × 2), diethyl ether (2 mL) and hexane (2 mL) sequentially to provide the title compound 6d (0.63 g, 58%) as a white solid. m.p. 228 °C (dec). HPLC 96.4% (Rt = 5.8 min.,, 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.93 (s, 1H), 10.75 (s, 1H), 8.12 (d, J = 7.0 Hz, 1H), 7.77–7.73 (m, 3H), 7.63–7.39 (m, 5H), 6.58 (d, J = 6.7 Hz, 1H); 19F NMR (400 MHz, d6-DMSO) δ 57.09; 13C NMR (100 MHz, DMSO-d6) δ 167.40, 163.23, 152.77, 145.96, 143.26, 142.04, 130.77, 130.16, 129.31, 129.10, 128.65, 126.30, 122.51, 120.22, 118.10 (q, J = 256 Hz), 99.76. LC-MS (ESI−) m/z 389.07 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C18H12F3N4O3 (M-H-HCl)− 389.0867, found 389.0818.
N4-(2-Methoxyphenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6e, method m in Scheme 2)
A mixture of chloropyrimidine 5d (0.075 g, 0.317 mmol) and 4-aminobenzoic acid (0.046 g, 0.328 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. Ethanol (0.5 mL) was added to the reaction mixture. The resulting precipitate was isolated by filtration and washed with EtOH (0.5 mL) and hexane (5 mL) to provide the title compound 6e (0.8 g, 68%) as a white solid. m.p. 247.5-249.3 °C. HPLC 99.6% (Rt = 6.0 min., 50% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.75 (s, 1H), 10.85 (s, 1H), 10.41 (s, 1H), 8.03 (d, J = 7.1 Hz, 1H), 7.77 (d, J = 8.6 Hz, 2H), 7.59-7.56 (m, 3H), 7.33 (t, J = 7.9 Hz, 1H), 7.18 (d, J = 8.3 Hz, 1H), 7.02 (t, J = 7.5 Hz, 1H), 6.50 (apparent s, 1H), 3.80 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 167.42, 162.78, 153.64, 152.19, 144.09, 142.02, 130.81, 128.73, 127.42, 126.30, 125.47, 120.89, 120.17, 112.66, 99.85, 56.32; LC-MS (ESI−) m/z 335.11 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C18H15N4O3 (M-H-HCl)− 335.1150, found 335.1153.
N4-(2-Methoxyphenyl)-N2-(phenyl)pyrimidine-2,4-diamine (6f, method m in Scheme 2)
A mixture of chloropyrimidine 5d (0.105 g, 0.444 mmol) and aniline (0.05 mL, 0.548 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The solvent was removed under reduced pressure. Aqueous saturated NaHCO3 (10 mL) was added to the residue and extracted with ethyl acetate (10 mL × 2). The combined organic phase was dried (Na2SO4), filtered and concentrated to dryness. The residue was purified by flash chromatography (10 g silica gel, Hex/EtOAc) to afford the title compound 6f (0.100 g, 76%) as an off-white solid. m.p. 142.3-144.5 °C. HPLC 99.4% (Rt = 4.90 min, 60% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.06 (s, 1H), 8.54 (s, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.96 (d, J = 5.7 Hz, 1H), 7.69 (d, J = 7.6 Hz, 2H), 7.18–7.14 (m, 2H), 7.11–7.02 (m, 2H), 6.97–6.89 (m, 1H), 6.85 (t, J = 7.3 Hz, 1H), 6.32 (d, J = 5.8 Hz, 1H), 3.82 (s, 3H); 13C NMR (100 MHz, CDCl3) 161.00, 160.08, 156.52, 149.55, 140.01, 128.97, 128.31, 123.69, 122.60, 121.50, 120.91, 120.33, 110.62, 98.24; LC-MS (ESI+) m/z 293.15 (M+H)+; HRMS (ESI+) m/z calculated for C17H17N4O (M+H)+ 293.1397, found 293.1393.
N4-(2-Cyanophenyl)-N2-(phenyl)pyrimidine-2,4-diamine (6g, method m in Scheme 2)
A mixture of chloropyrimidine 5e (0.092 g, 0.398 mmol) and aniline (0.037 ml, 0.398 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The solvent was removed under reduced pressure. The solid obtained was slurried in ethyl acetate (3 mL), filtered, and washed with ethyl acetate (5 mL), and hexane (5 mL). The dried solid was dissolved in methanol (3 mL). Diisopropylethylamine (1 ml) was added and the solvent was removed under reduced pressure. The solid obtained was slurried with water (5 ml), filtered, washed with water (10 ml × 2), ether (5 ml), slurried with methanol (1 ml), filtered and dried under vacuum to provide the title compound 6g (0.027 g, 24%) as an off-white solid. m.p. 170.0.2-172.7 °C. HPLC 100% [Rt = 5.1 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 9.52 (s, 1H), 9.15 (s, 1H), 8.07 (d, J = 5.7 Hz, 1H), 7.82 (d, J = 8.8 Hz, 1H), 7.77–7.66 (m, 2H), 7.59 (d, J = 8.8 Hz, 2H), 7.32 (t, J = 8.0 Hz, 1H), 7.11 (t, J = 7.9 Hz, 2H), 6.83 (t, J = 7.3 Hz, 1H), 6.28 (d, J = 5.7 Hz, 1H); LC-MS (ESI+) m/z 288.13 (M+H)+; HRMS (ESI+) m/z calculated for C17H14N5 (M+H)+ 288.1244, found 288.1241.
N4-[2-(Trifluoromethyl)phenyl]-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6h, method m in Scheme 2)
A mixture of chloropyrimidine 5f (0.055 g, 0.2 mmol) and 4-aminobenzoic acid (0.03 g, 0.218 mmol) in EtOH (0.3 mL) was heated in a microwave reactor at 150 °C for 20 min. Ethanol (0.5 mL) was added to the reaction mixture and sonicated at room temperature for 5 min. The resulting precipitate was isolated by filtration and washed with EtOH (0.5 mL × 2), and hexane (3 mL × 2) to afford the title compound 6h (0.067 g, 82%) as a white solid. m.p. 240.0-242.6 °C. HPLC 99.6% (Rt = 7.7 min., 50% MeOH in 0.1% TFA water 30 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.91 (s, 1H), 10.76 (s, 1H), 8.11 (d, J = 7.0 Hz, 1H), 7.90–7.85 (m, 2H), 7.69–7.64 (m, 4H), 7.38 (d, J = 8.5 Hz, 2H), 6.53 (d, J = 6.7 Hz, 1H); 19F NMR (400 MHz, DMSO-d6) δ 59.99 (s); 13C NMR (100 MHz, d6-DMSO) δ 167.35, 164.21, 152.47, 145.71, 142.05, 135.37, 134.40, 131.77, 130.65, 129.09, 127. (q, J = 4.6 Hz), 126.62 (q, J = 29.5 Hz), 126.10, 124.03 (q, J = 271.5 Hz), 119.77; LC-MS (ESI−) m/z 373.08 (M+H)+; LC-MS (ESI+) m/z 375.11 (M+H)+; HRMS (ESI+) m/z calculated for C18H14F3N4O2 (M+H)+ 375.1063, found 375.1068.
N4-(2-Bromophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6i, method m in Scheme 2)
A mixture of chloropyrimidine 5g (0.100 g, 0.350 mmol) and 4-aminobenzoic acid (0.048 g, 0.350 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The resulting precipitate was isolated by filtration and washed with EtOH (0.5 mL × 2) and dried under vacuum to provide the title compound 6i (0.069 g, 47%) as a white solid. m.p. 240 °C (dec.). HPLC 98% [Rt = 7.40 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 10.46 (s, 2H), 8.06 (d, J = 6.8 Hz, 1H), 7.78 (d, J = 8.3 Hz, 1H), 7.68 (d, J = 8.5 Hz, 2H), 7.59 (d, J = 8.3 Hz, 1H), 7.51–7.47 (m, 3H), 7.32 (t, J = 8.3 Hz, 1H), 6.45 (s, 1H); 13C NMR (100 MHz, d6-DMSO) 167.43, 163,07, 153.17, 146.60, 142.37, 136.36, 133.80, 130.71, 129.95, 129.71, 129.19, 125.91, 121.27, 119.83 , 99.45; LC-MS (ESI+) m/z 385.02 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H14BrN4O2 (M-Cl)+ 385.0295, found 385.0292.
N4-(2-Chlorophenyl)-N2-(4-methylcarboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6j, method l in Scheme 2)
A mixture of chloropyrimidine 2l (0.120 g, 0.500 mmol) and 4-aminophenylacetic acid (0.076 g, 0.5 mmol) in EtOH (2 mL with 1 drop of 1 M hydrochloric acid) was heated in a microwave reactor at 160 °C for 15 min. A clear solution was obtained. Ethanol was removed from the mixture under vacuum, and the analysis of the crude NMR showed formation of 25% ethyl ester of 6j. The crude material was stirred in THF (0.5 mL) and NaOH solution (1 mL, 2 M) at r.t. for 16 h. The THF was evaporated from the mixture and HCl (1 M) was added to acidify the mixture (pH = 1-2). The precipitate obtained was filtered, washed with water (5 mL) and dried under high vacuum to afford 6j (0.137 g, 70%) as a beige solid. m.p. 132 °C (dec). HPLC 97% (Rt = 3.77 min., 50% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.19 (s, 1H disappeared on D2O shake), 9.30 (s, 1H disappeared on D2O shake), 9.21 (s, 1H disappeared on D2O shake), 8.01 (d, J = 6.0 Hz, 1H), 7.79 (d, J = 8.0 Hz, 1H), 7.54 (dd, J = 8.0, 1.4 Hz, 1H partially overlapping), 7.51 (d, J = 8.4 Hz, 2H partially overlapping with dd), 7.38 (apptd, J = 8.0, 1.4 Hz, 1H), 7.24 (apptd, J = 8.0, 1.4 Hz, 1H), 7.03 (d, J = 8.4 Hz, 2H), 6.30 (d, J = 6.0 Hz, 1H), 3.45 (s, 2H); 13C NMR (100 MHz, DMSO-d6) δ 173.65, 162.04, 158.90, 155.15, 139.43, 136.41, 130.31, 129.87, 129.85, 128.56, 128.44, 128.10, 126.90, 119.66, 119.63, 98.71; LC-MS (ESI+) m/z 355.11 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H16ClN4O2(M-Cl)+ 355.0956, found 355.0971.
N4-(2-Chloromethyl)phenyl]-N2-(4-carboxy-3-hydroxyphenyl)pyrimidine-2,4-diamine hydrochloride (6k, methods n and o in Scheme 2)
A mixture of chloropyrimidine 2l (0.100 g, 0.416 mmol) and methyl 5-aminosalicylate (0.070 g, 0.416 mmol) in THF (10 mL) was stirred at room temperature for 10 min. Two drops of concentrated HCl were added and the mixture heated at reflux for 14 hours. The solvent was removed under reduced pressure to provide a gray solid which was suspended in sodium bicarbonate (sat. aq. 50 mL), sonicated for 10 min, filtered, and washed with water (10 mL) to provide a white powder. The crude material was washed with methanol (2 × 10 ml) and finally with ethyl acetate (20 ml) to give methyl 4-(4-(2-chlorophenylamino)pyrimidin-2-ylamino)-2-hydroxybenzoate (6u) (0.081 g, 56%). 1H NMR (400 MHz, d6-DMSO) δ 10.61 (s, 1H), 9.63 (s, 1H), 9.15 (s, 1H), 8.07 (s, 1H), 7.76 (s, 1H), 7.59–7.33 (3 × broad s, 3H), 7.25– 7.10 (2 × broad s, 2H), 6.35 (s, 1H), 3.83 (s, 3H). LC-MS (ESI+) 371.09, m/z calculated for C18H16ClN4O3 (M+H)+ 371.0905, found 371.0914. To a stirred solution of ester 6u (200 mg, 0.540 mmol) in THF (15 ml), was added sodium hydroxide (108 mg, 2.70 mmol) in water (1.5 ml). The reaction was heated under reflux for 14 hours. The solvent was removed under reduced pressure to provide a white solid which was then dissolved in water (20 ml) and acidified to pH ~6-7 by addition of HCl (1 M). The colorless precipitate was filtered, washed with water and dried under vacuum. The crude solid was suspended in methanol (20 ml), sonicated for 10 min., filtered, washed with methanol (5 ml) and dried under vacuum to provide the title compound 6k (118 mg, 62%) as an off-white powder. m.p. 202-204 °C. HPLC 100% [Rt = 8.3 min, 45% MeOH, 65% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 13.55 (brs, 1H), 11.29 (brs, 1H), 10.14 (brs, 1H), 9.93 (s, 1H), 8.07 (d, J = 6.7 Hz, 1H), 7.70 (d, J = 7.8 Hz, 1H), 7.57 (d, J = 7.6 Hz, 1H), 7.53 (d, J = 8.5 Hz, 1H), 7.41 (t, J = 7.8 Hz, 1H), 7.30 (t, J = 8.0 Hz, 1H), 7.25 (s, 1H), 7.02 (d, J = 8.5 Hz, 1H), 6.43 (d, J = 5.7 Hz, 1H); 13C NMR (100 MHz, d6-DMSO), δ 172.34, 162.82, 162.58, 155.69, 146.06, 135.35, 131.18, 130.58, 129.32, 128.64, 128.53, 128.25, 111.11, 107.27, 106.54, 99.78; LC-MS (ESI+) m/z 357.08 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H14ClN4O3 (M-Cl)+ 357.0749, found 357.0751.
N4-(2-Fluorophenyl)-N2-4-phenylpyrimidine-2,4-diamine (6l, method m in Scheme 2)
A mixture of chloropyrimidine 5a (0.073 g, 0.325 mmol) and aniline (0.04 mL, 0.438 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The solvent was removed under reduced pressure. Aqueous saturated NaHCO3 (10 mL) was added to the residue and extracted with ethyl acetate (10 mL × 2). The combined organic phase was dried (Na2SO4), filtered and concentrated to dryness. The residue was purified by flash chromatography (10 g silica gel, hex/EtOAc) to afford the title compound 6l (0.050 g, 55%) as an off-white solid. m.p. 133.2-134.6 °C. HPLC 99.5% (Rt = 4.4 min, 60% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.11 (s, 1H), 9.07 (s, 1H), 8.02 (d, J = 5.7 Hz, 1H), 7.97 (t, J = 7.9 Hz, 1H), 7.65 (d, J = 8.6 Hz, 2H), 7.32–7.23 (m, 1H), 7.22–7.07 (m, 4H), 6.86 (t, J = 7.3 Hz, 1H), 6.30 (d, J = 5.7 Hz, 1H); LC-MS (ESI+) m/z 281.11 (M-Cl)+; HRMS (ESI+) m/z calculated for C16H14FN4 (M-Cl)+ 281.1197, found 281.1209.
N4-(2-Iodophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6m, method m in Scheme 2)
A mixture of chloropyrimidine 5h (0.119 g, 0.358 mmol) and 4-aminobenzoic acid (0.048 g, 0.350 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The solvent was then removed under reduced pressure to provide an off-white solid. The solid was slurried in methanol (2 mL), filtered, washed with methanol (2 mL), and dried under vacuum to provide the title compound 6m (0.053 g, 32%) as a white solid. m.p. 253 °C (dec). HPLC 99.7% [Rt = 7.80 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 12.76 (bs, 1H), 10.72 (s, 1H), 10.62 (s, 1H), 8.07 (d, J = 7.0 Hz, 1H), 8.01 (dd, J = 1.2, 7.9 Hz, 1H), 7.67 (d, J = 8.5 Hz, 2H), 7.54–7.45 (m, 4H), 7.19-7.15 (m, 1H), 6.44 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 167.36, 163.09, 152.32, 145.44, 142.04, 139.99, 139.67, 130.68, 130.04, 129.94, 129.45, 126.12, 119.90, 99.24; LC-MS (ESI+) m/z 433.02 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H14IN4O2 (M-Cl)+ 433.0156, found 433.0150.
N4-(2-Cyanophenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6n, method m in Scheme 2)
A mixture of chloropyrimidine 5e (0.094 g, 0.406 mmol) and 4-aminobenzoic acid (0.037 mg, 0.408 mmol) in EtOH (0.5 mL) was heated with in a microwave reactor at 150 °C for 20 min. The solvent was removed under reduced pressure. The compound was purified by reverse phase C-18 preparative HPLC [Eclipse XDB-C18 PrepHT 21.2 × 250 mm, 7 μm; 40% MeOH, 60% water (with 0.1% TFA) 20 min, 20mL/min.] to provide the title compound as its TFA salt. The solid obtained was suspended in methanol (9 mL) followed by the addition of HCl (4 M in dioxane, 1 mL). The solvent was then removed in a Genevac evaporator. The dried solid was slurried with acetonitrile (1 mL), filtered, washed with acetonitrile (1 mL) and dried under vacuum to provide the title compound 6n (0.022 g, 15%) as an off-white solid. m.p. 222 °C (dec). HPLC 97.6% [Rt = 5.5 min., 40% MeOH, 60% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 10.67 (s, 1H), 10.43 (s, 1H), 8.14 (d, J = 6.8 Hz, 1H,), 7.93 (d, J = 7.6 Hz, 1H), 7.80 (t, J = 7.6 Hz, 1H), 7.72–7.68 (m, 3H), 7.55 (d, J = 7.3 Hz, 2H), 7.50 (t, J = 7.6 Hz, 1H), 6.50 (d, J = 6.8 Hz, 1H), LC-MS (ESI+) m/z 332.11 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H14N5O2 (M-Cl)+ 332.1142, found 332.1139.
N4-(2-Chlorophenyl)-N2-(3-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6o, methodm in Scheme 2)
A mixture of 2l (0.100 g, 0.417 mmol) and 3-aminobenzoic acid (0.057 g, 0.417 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The precipitate obtained upon cooling the reaction mixture was filtered and washed quickly with ethanol (3 ml) to obtain the 6o (0.097 g, 62%) as a white solid. m.p. 290-296 °C (dec). HPLC 100% (Rt = 7.58 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 13.03 (s, 1H), 10.49 (brs, 1H), 8.04 (d, J = 6.4 Hz, 1H), 7.88 (brs, 1H), 7.73 (appd, J = 7.6 Hz, 1H), 7.65 (d, J = 8.0 Hz, 1H), 7.60 (d, J = 7.6 Hz, 1H), 7.57 (dd, J = 7.9, 1.5 Hz, 1H), 7.37 (td, J = 7.6, 0.8 Hz, 1H), 7.32-7.27 (m, 2H), 6.49 (s, 1H); 13C NMR (101 MHz, DMSO) δ 167.5, 162.95, 153.3, 145.9, 138.08, 134.6, 132.1, 130.5, 129.6, 129.5, 129.1, 128.89, 128.36, 125.9, 125.5, 122.6, 99.5; elemental analysis calculated for C17H14Cl2N4O2: C, 54.13; H, 3.74; N, 14.85, Found: C, 53.78; H, 3.64; N, 14.66.
N4-(2-Chlorophenyl)-N2-(4-carbamoyl)pyrimidine-2,4-diamine hydrochloride (6p, method l in Scheme 2)
A mixture of chloropyrimidine 2l (0.100 g, 0.417 mmol) and 4-aminobenzamide (0.057 g, 0.417 mmol) in EtOH (2.0 mL with 1 drop of 1 M hydrochloric acid) was heated with a microwave reactor at 160 °C for 15 min. The mixture was filtered and the solid obtained was washed with MeOH (1 mL) and dried to afford the desired compound 6p (0.110 g, 70%) as a white solid. m.p. 227 °C (dec.). HPLC 99% (Rt = 3.70 min., 50% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.75 (brs, 2H disappeared on D2O shake), 8.09 (dd, J = 6.1, 3.5 Hz, 1H), 7.91 (brs, 1H disappeared on D2O shake), 7.71–7.64 (m, 4H), 7.50–7.42 (m, 4H), 7.30 (brs, 1H disappeared on D2O shake), 6.52 (brs, 1H); 13C NMR (100 MHz, DMSO-d6) δ 167.76, 163.15, 152.33, 144.97, 140.39, 134.55, 130.62, 130.29, 130.02, 129.54, 129.49, 128.90, 128.55, 119.98, 99.43; LC-MS (ESI+) m/z 340.10 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H15ClN5O (M-Cl)+ 340.0960, found 340.0971.
N4-(2-Biphenyl)-N2-(4-carboxyphenyl)pyrimidine-2,4-diamine hydrochloride (6q, method m in Scheme 2)
A mixture of chloropyrimidine 5i (0.096 g, 0.34 mmol) and 4-aminobenzoic acid (0.054 g, 0.393 mmol) in EtOH (0.4 mL) was heated in a microwave reactor at 150 °C for 20 min. The resulting precipitate was filtered and washed with EtOH (0.5 mL × 3) to provide the title compound 6q (0.098 g, 67%), as a white solid. m.p. 259 °C (dec). HPLC 100% (Rt = 8.7 min., 55% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.83 (s, 1H), 10.96 (s, 1H), 10.71 (s, 1H), 7.96 (d, J = 7.2 Hz, 1H), 7.73 (d, J = 8.3 Hz, 2H), 7.61–7.42 (m, 6H), 7.42–7.17 (m, 5H), 6.31 (apparent s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 167.41, 163.26, 152.00, 144.30, 142.07, 138.96, 134.37, 131.35, 130.75, 129.35, 129.09, 128.74, 128.17, 126.18, 119.89, 99.22; LC-MS (ESI−) m/z 381.13 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C23H17N4O2 (M-HHCl)− 381.1357, found 381.11366.
N4-(2-Chlorophenyl)-N2-(4-carboxyphenyl)-N4-methylpyrimidine-2,4-diamine hydrochloride (6r, method m in Scheme 2)
This was obtained as a white solid (0.127 g, 0.36 mmol, 80%) from 5j (0.115 g, 0.452 mmol) and 4-aminobenzoic acid (0.062 g, 0.452 mmol) in the same manner as described for 6s. m.p. 288 °C (dec.). HPLC 98% [Rt = 8.39 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]. 1H NMR (400 MHz, CD3OD) δ 8.17-8.04 (m, 2.57 H), 7.77-7.74 (m, 3H), 7.70-7.67 (m, 2H), 7.58-7.54 (m, 4H), 7.49-7..41 (m, 0.42 H), 7.17 (d, J = 8.8 Hz, 1H, major), 6.83 (d, J = 7.5 Hz, 0.41 H, minor), 5.80 (d, J = 7.4 Hz, 1H, major ), 3.55 (s, 4H); LC-MS (ESI+) m/z 355.10 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H16ClN4O2 (M-Cl)+ 355.0956, found 355.0962; Elemental Analysis: Found C, 54.98%; H, 4.12; N, 14.18. C18H16Cl2N4O2 requires C, 55.26%; H, 4.12; N, 14.32.
N4-(2-Chlorophenyl)-N2-(4-carboxyphenyl)-N4-ethylpyrimidine-2,4-diamine hydrochloride (6s, method m in Scheme 2)
A solution of 5k (0.151 g, 0.673 mmol) and 4-aminobenzoic acid (0.092 g, 0.674 mmol) in anhydrous ethanol (0.673 mL) was heated in a microwave reactor at 150 °C for 20 min. The resulting precipitate was isolated by filtration and washed with EtOH (1 mL × 2) and dried under vacuum to provide the title compound 6s (0.172 g, 71%), as a white solid. m.p. 284 °C (dec). HPLC 100% [Rt = 13.96 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]. Two rotamers are present: 1H NMR (400 MHz, CD3OD) δ 8.11 (d, J = 8.8 Hz, 2H, major), 8.07 (d, J = 7.6 Hz, 0.4 H, minor), 7.75-7.66 (m, 5H), 7.63-7.54 (m, 5H), 7.15 (d, J = 8.8 Hz, 1H, major), 6.87 (d, J = 7.5 Hz, 0.5 H, minor), 5.74 (d, J = 7.4 Hz, 1H, major),4.30- 4.21 (m, 1H), 4.09-4.02 (m, 0.6 H, minor), 3.93-3.83 (m, 2H), 1.35 (t, J = 7.2 Hz, 1.5 H, minor), 1.29 (t, J = 7.2 Hz, 3H, major); LC-MS (ESI+) m/z 369.12 (M-Cl)+; HRMS (ESI+) m/z calculated for C19H18Cl2N4O (M-Cl)+ 369.1113, found 369.1123; Elemental Analysis: Found C, 56.30%; H, 4.45; N, 13.76. C19H18Cl2N4O2 requires C, 56.31%; H, 4.48; N, 13.82.
N4-(2-Chlorophenyl)-N2-(3-carboxyphenyl)-5-fluoropyrimidine-2,4-diamine hydrochloride (6t, method m in Scheme 2)
A mixture of 2k (0.100 g, 0.350 mmol) and 3-aminobenzoic acid (0.048 g, 0.350 mmol) in EtOH (0.5 mL) was heated in a microwave reactor at 150 °C for 20 min. The reaction mixture was cooled and the precipitate obtained was filtered and washed quickly with ethanol (3 ml) to provide 6t (0.080 g, 57%) as a white solid. m.p. 257 °C (dec.). HPLC 98% (Rt = 9.45 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.74 (brs,1H), 9.32 (s, 1H), 9.18 (s, 1H), 8.12 (d, J = 3.5 Hz, 1H), 8.02 (s, 1H), 7.80 (d, J = 8.2 Hz, 1H), 7.61 (d, J = 7.8 Hz, 1H), 7.56 (d, J = 7.9 Hz, 1H), 7.40-7.36 (m, 2H), 7.29 (t, J = 7.6 Hz, 1H), 7.11 (t, J = 7.9 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 168.14, 156.0 (d, J = 2.9 Hz), 151.37 (d, J = 11.5 Hz), 141.75, 141.43 (d, J = 246.44 Hz), 141.39 (d, J = 18.4 Hz), 135.92, 131.54, 130.88, 130.38, 129.55, 128.90, 128.32, 128.03, 122.84, 122.18, 119.87; 19F NMR (376 MHz, DMSO) δ -165.76; LC-MS (ESI−) m/z 357.06 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C17H12ClFN4O2 (M-H-HCl)− 357.0560 , found 357.0554.
N4-(2-Chlorophenyl)-N2-[4-N-(2-morpholinoethyl)carbamoylphenyl]pyrimidine-2,4-diamine dihydrochloride (9a, method r in Scheme 4)
A solution of 2l (0.100 g, 0.42 mmol) and 8a (0.105 g, 0.42 mmol) in isopropyl alcohol (8 mL) and concentrated HCl (0.315 mL) was heated in a microwave reactor at 170 °C for 20 min. The solvent was then removed under reduced pressure. The resulting solid was dissolved in methanol (2 mL) followed by the addition of diethyl ether (10 mL). The precipitate was filtered and dried under vacuum to afford the title compound 9a (0.054 g, 29%) as white crystals. m.p. 173.0-174.6 °C. HPLC 99.0% [Rt = 5.53 min., 50% MeOH, 40% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 11.37–10.62 (m, 1H), 8.88 (s, 1H), 8.59–8.43 (m, 1H), 8.12 (s, 1H), 7.78 (s, 2H), 7.69–7.61 (m, 2H), 7.50 (s, 2H), 7.43 (s, 1H), 6.76–6.41 (m, 2H), 4.03–2.95 (m, 12H); 13C NMR (100 MHz, DMSO-d6) δ 166.39, 163.08, 152.86, 145.97, 140.91, 134.67, 130.67, 130.15, 129.42, 129.24, 128.82, 128.60, 119.81, 99.46, 63.80, 56.32, 51.87, 34.28. LCMS (ESI+) m/z 453.17 (M-HCl-Cl)+; HRMS (ESI+) m/z calculated for C23H25ClN6O2 (M-HCl-Cl)+ 453.1800, found 453.1797.
N4-(2-Chlorophenyl)-N2-[4-N-(2-dimethylaminoethyl)carbamoylphenyl]pyrimidine-2,4-diamine dihydrochloride (9b, method r in Scheme 4)
This was obtained as a white solid (0.02 g, 0.04 mmol, 8%) from 2l (0.100 g, 0.48 mmol) and 8b (0.116 g, 0.48 mmol) in a similar manner as described for 9a. m.p. 143.1-145.8 °C. HPLC 98% [Rt = 5.10 min., 60% MeOH, 40% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 10.65 (s, 1H), 9.96 (s, 1H), 8.75 (s, 1H), 8.09 (s, 1H), 7.73 (s, 2H), 7.71–7.61 (m, 2H), 7.58–7.45 (m, 3H), 7.45–7.35 (m, 1H), 6.50 (s, 1H), 3.62–3.55 (m, 2H), 3.23 (d, J = 5.7 Hz, 2H), 2.80 (d, J = 4.0 Hz, 6H); 13C NMR (100 MHz, DMSO-d6) δ 166.50, 163.10, 152.72, 145.80, 140.87, 134.73, 130.67, 130.19, 129.45, 129.31, 128.61, 119.80, 99.50, 56.76, 43.01, 35.16; LC-MS (ESI+) m/z 411.18 (M-HCl-Cl)+; HRMS (ESI+) m/z calculated for C21H24ClN6O (M-HCl-Cl)+ 411.1695, found 411.1693.
N4-(2-Chlorophenyl)-N2-[4-N-(2-methoxyethyl)carbamoylphenyl]pyrimidine-2,4-diamine hydrochloride (9c, method s in Scheme 4)
This was obtained as a white solid (0.047 g, 0.12 mmol) from 2l (0.092 g, 0.47 mmol) and 8c (0.060 g, 0.47mmol) in a similar manner as described for compound 6s m.p. 210.4-213.0 °C. HPLC 97% [Rt = 5.61 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 10.71 (s, 2H), 8.44 (t, J = 5.6 Hz, 1H), 8.08 (d, J = 7.3 Hz, 1H), 7.68–7.62 (m, 4H), 7.49–7.38 (m, 3H), 6.52 (s, 1H), 3.41–3.37 (m, 4H), 3.24 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 166.06, 163.12, 152.41, 145.17, 140.32, 134.55, 130.60, 130.23, 130.08, 129.50, 129.42, 128.56, 119.98, 99.48, 71.50, 58.58, 58.55 LCMS (ESI+) m/z 398.14 (M-Cl)+. HRMS (ESI+) m/z calculated for C20H21ClN5O2 (M-Cl)+ 398.1378, found 398.1370.
N4-(2-Chlorophenyl)-N2-[4-(methylsulfonyl)phenyl]pyrimidine-2,4-diamine (9d, method t in Scheme 4)
A solution of 2l (0.098 g, 0.41 mmol) and 4-(methylsulfonyl)aniline (0.070 g, 0.41 mmol) in ethanol (1 mL) and 1M HCl (aq., 1.0 mL) was heated in a microwave reactor at 180 °C for 15 min. The solvent was removed under reduced pressure. The solid obtained was slurred in a saturated solution of sodium bicarbonate filtered, slurried with methanol (1 mL), then filtered to yield the title compound 9d (0.077 g, 44%) as a white solid. m.p. 169.3-171.3 °C. HPLC 99% [Rt = 4.15 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 9.72 (s, 1H), 9.17 (s, 1H), 8.08 (d, J = 5.7 Hz, 1H), 7.84 (d, J = 8.7 Hz, 2H), 7.74 (d, J = 7.8, 1H), 7.65–7.51 (m, 3H), 7.41 (t, J= 7.6, 1H), 7.26 (t, J = 7.6 Hz, 1H), 6.35 (d, J = 5.7 Hz, 1H), 3.09 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 162.03, 159.47, 156.92, 146.25, 136.52, 132.20, 130.40, 129.03, 128.38, 128.34, 128.21, 127.12, 118.38, 99.77, 44.70; LC-MS (ESI+) m/z 375.06 (M+H)+; HRMS (ESI+) m/z calculated for C17H16ClN4O2S (M+H)+ 375.0677, found 375.0670.
N4-(2-Chlorophenyl)-N2-(3-acetamidophenyl)pyrimidine-2,4-diamine hydrochloride (9e, method s in Scheme 4)
This compound was prepared from 2l (0.105 g, 0.44 mmol) and N-(3-aminophenyl)acetamide (0.066 g, 0.44 mmol) in a similar manner as described for 6s. The solvent was then removed under reduced pressure. The resulting solid was dissolved in methanol (2 ml), followed by the addition of ethyl acetate (10 ml). A precipitate formed and the mixture was sonicated for 30 min at room temperature. The precipitate was filtered and washed with ethyl acetate (10 mL) and hexane (20 mL), affording the pure product 9e (0.166 g, 95%), as an off-white solid. m.p. 150 °C (dec). HPLC 100% [Rt = 5.30 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 10.63 (s, 1H), 10.45 (s, 1H), 10.00 (s, 1H), 8.00 (d, J = 5.0 Hz, 1H), 7.70–7.52 (m, 3H), 7.4–7.28 (m, 2H), 7.24–7.13 (m, 2H), 7.13–7.07 (m, 1H), 2.02 (s, 3H); LC-MS (ESI+) m/z 354.11 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H17ClN5O (M-Cl)+ 354.1116, found 354.1119.
N4-(2-Chlorophenyl)-N2-[6-(1H-benzo[d]imidazol-2(3H)-one)]pyrimidine-2,4-diamine hydrochloride (9f, method s in Scheme 4)
A solution of 2l (0.105g, 0.437 mmol) and 5-amino-1H-benzo[d]imidazol-2(3H)-one (0.065 g, 0.437 mmol) and ethanol (0.437 mL) was heated in a microwave reactor at 150 °C for 40 min. The resulting precipitate was filtered, dried under vacuum and suspended in ethanol. The suspension was sonicated for thirty minutes, filtered and dried under vacuum to provide the title compound 9f (0.112 g, 66%) as an off-white solid. m.p. 263 °C (dec). HPLC 98% [Rt = 3.21 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, CD3OD) δ 7.74 (s, 2H), 7.65-7.63 (m, 1H), 7.54-7.52 (m, 1H), 7.35-7.28 (m, 2H), 7.04 (d, J = 7.4 Hz, 3H), 6.42 (s, 1H); LC-MS (ESI+) m/z 353.10 (M-Cl)+; HRMS (ESI+) m/z calculated for C17H14ClN6O (M-Cl)+ 353.0912, found 353.0913.
N4-(2-Chlorophenyl)-N2-(4-carboxyl-3-methoxyphenyl)pyrimidine-2,4-diamine hydrochloride (9g, method s in Scheme 4)
A mixture of chloropyrimidine 2l (0.100 g, 0.416 mmol) and 4-amino-2-methoxybenzoic acid (0.070 g, 0.418 mmol) in EtOH (0.5 mL) was heated with a microwave reactor at 150 °C for 20 min. The solvent was evaporated from the resulting thick mass under reduced pressure. The residue was suspended in ethyl acetate (8 mL) and sonicated for 10 min. The mixture was filtered and the solid was washed with ethyl acetate (8 mL) and ethyl acetate:methanol (1:1, 1 mL) to provide the title compound 9g (0.068 g, 57%) as a gray solid. m.p. 160-162 °C. HPLC 95% [Rt = 4.5 min, 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6), δ 12.69 (s, 1H), 10.42 (s, 1H), 10.24 (s, 1H), 7.95 (d, J = 6.2 Hz, 1H), 7.64–7.51 (m, 3H), 7.48 (d, J = 8.4 Hz, 1H), 7.36 (t, J = 7.7 Hz, 1H), 7.29 (t, J = 7.4 Hz, 1H), 6.98 (d, J = 8.8 Hz, 1H), 6.45 (s, 1H), 3.76 (s, 3H); 13C NMR (100 MHz, DMSO-d6), δ 167.37, 162.97, 155.67, 152.95, 144.61, 134.39, 130.51, 129.61, 129.00, 128.35, 127.11, 124.90, 122.22, 113.36, 99.26, 56.70; LC-MS (ESI+) m/z 371.09 (M-Cl)+; HRMS (ESI+) m/z calculated for C18H16ClN4O3 (M-Cl)+ 371.0905, found 371.0909.
N4-(2-Chlorophenyl)-N2-[4-(N -morpholino)phenyl]pyrimidine-2,4-diamine hydrochloride (9h, method t in Scheme 4)
A mixture of chloropyrimidine 2l (0.120 g, 0.500 mmol) and 4-morpholinoaniline (0.089 g, 0.500 mmol) in EtOH (2 mL with 1 drop of 1 M of hydrochloric acid) was heated in a microwave reactor at 160 °C for 15 min. The reaction mixture was cooled to r.t. and the product precipitated. The mixture was filtered, and the product obtained was quickly washed with MeOH (1 mL), DCM (1 mL) and dried to afford 9h (0.14 g, 67%) as a green solid. m.p. 136-138 °C. HPLC 99% (Rt = 7.43 min., 50% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 12.11 (s, 1H disappeared on D2O shake), 10.63 (s, 1H disappeared on D2O shake), 10.37 (s, 1H disappeared on D2O shake), 7.94 (brs, 1H), 7.65–.60 (m, 2H), 7.43 (appt, J = 7.6 Hz, 1H), 7.36 (appt, J = 7.6 Hz, 1H), 7.22 (d, J = 8.1 Hz, 2H), 6.84 (brs, 2H), 6.45 (brs, 1H), 3.73 (appt, J = 4.4 Hz, 4H), 3.05 (brt, 4H); 13C NMR (100 MHz, DMSO-d6) δ 163.03, 152.56, 148.63, 144.36, 134.58, 130.53, 129.99, 129.37, 129.12, 128.43, 122.90, 116.05, 98.78, 66.65, 49.38; LC-MS (ESI+) m/z 382.15 (M-Cl)+; HRMS (ESI+) m/z calculated for C20H21ClN5O (M-Cl)+ 382.1429, found 382.1434.
N4-(2-Chlorophenyl)-N2-[4-(aminosulfonyl)phenyl]pyrimidine-2,4-diamine hydrochloride (9i, method t in Scheme 4)
A mixture of chloropyrimidine 2l (0.120 g, 0.500 mmol) and 4-aminobenzenesulfonamide (0.086 g, 0.500 mmol) in EtOH (2 mL with 1 drop of 1 M HCl) was heated in a microwave reactor at 160 °C for 15 min. The precipitate formed upon cooling filtered and washed with MeOH (2 mL), then slurried and sonicated in DCM (5 mL) and filtered. The product obtained was dried to afford 9i (0.155 g, 75%) as a white solid. m.p. 215 °C (dec.). HPLC 96% (Rt = 3.23 min., 50% MeOH in 0.1% TFA water 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.81 (s, 1H), 10.67 (s, 1H), 8.10 (d, J = 6.9 Hz, 1H), 7.67–7.63 (m, 2H), 7.61–7.56 (m, 4H), 7.47 (td, J = 7.7, 1.6 Hz, 1H), 7.41 (td, J = 7.7, 1.6 Hz, 1H), 7.28 (s, 2H), 6.53 (appd, J = 5.2 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 163.15, 152.43, 145.28, 140.86, 139.52, 134.61, 130.64, 130.38, 129.60, 129.50, 128.57, 127.10, 120.32, 99.66; LC-MS (ESI+) m/z 376.06 (M-Cl)+; HRMS (ESI+) m/z calculated for C16H15ClN5O2S (M-Cl)+ 376.0629, found 376.0634.
N4-(2-Chlorophenyl)-N2-[4-(N -morpholino)phenyl]-5-fluoropyrimidine-2,4-diamine (9j, method s in Scheme 4)
A mixture of chloropyrimidine 2k (0.100 g, 0.387 mmol) and 4-morpholinoaniline (0.069 g, 0.387 mmol) in ethanol (1 mL) was heated in a microwave reactor at 150 °C for 20 min. The reaction mixture was cooled, and stirred at room temperature for 30 min. The solvent was removed in vacuo. The residual solid was suspended in ethyl acetate (10 mL) and sonicated for 10 min, filtered and washed with ethyl acetate (5 mL). The solid was suspended in sodium bicarbonate (sat. aq. 50 mL), filtered, and washed with water (10 mL) to provide the title compound 9k (0.92 g, 60%) as an off-white solid. m.p. 209-211 °C. HPLC 99% [Rt = 7.2 min., 50% MeOH, 50% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, DMSO-d6) δ 9.63 (s, 1H), 9.36 (s, 1H), 8.11 (d, JH-F = 4.2 Hz, 1H), 7.61–7.54 (m, 2H), 7.44–7.37 (m, 1H), 7.36–7.25 (m, 3H), 6.79 (s, 2H), 3.73 (s, 4H), 3.03 (s, 4H). 1H NMR (400 MHz, DMSO-d6+D2O) δ 8.06 (d, J = 4.1 Hz, 1H), 7.59–7.51 (m, 2H), 7.39 (td, J = 7.6, 1.5 Hz, 1H), 7.32 (td, J = 7.7, 1.7 Hz, 1H), 7.27 (d, J = 9.0 Hz, 2H), 6.79 (d, J = 8.9 Hz, 2H), 3.71 (t, J = 4.0 Hz, 4H), 3.03 (t, J = 4.4 Hz, 4H); 19F NMR (376 MHz, DMSO-d6), δ -165.59; 13C NMR (100 MHz, DMSO-d6) δ 154.38, 152.31 (d, J = 12.0 Hz), 141.63, 139.04, 139.19, 135.42, 134.20, 130.43, 129.76, 120.74, 117.11, 66.31, 50.71; LC-MS (ESI+) m/z 400.13 (M-Cl)+; HRMS (ESI+) m/z calculated for C20H20ClFN5O (MCl)+ 400.1335, found 400.1344.
4-(2-Chlorophenylamino)-2-(4-(morpholinomethyl)phenyl)-5-fluoropyrimidine (9k, method u in Scheme 4)
A mixture of chloropyrimidine 2k (0.100 g, 0.387 mmol), 4-(morpholinomethyl)aniline (0.087 g, 0.452 mmol), X-Phos (2-dicyclohexylphosphino-2′,4′,6′-triisopropylbiphenyl) (0.019 g, 0.0387 mmol), bis(dibenzylideneacetone) palladium(0) (0.022 g, 0.0387 mmol), potassium carbonate (0.117 g, 0.851 mmol) in tert-BuOH (3.0 mL) was heated under reflux for 18 hours. The solvent was evaporated from the resulting dark solution. The dark residue was purified by chromatography (SiO2, ethyl acetate and hexanes) to provide the title compound 9k (0.062 g, 39%) as an off-white solid. m.p. 142-144 °C. HPLC 98% [Rt = 23.4 min., 50% MeOH, 50% water (with 0.1% TFA) 36 min.]; 1H NMR (400 MHz, DMSO-d6), δ 9.16 (s, 1H), 9.10 (s, 1H), 8.07 (d, JH-F = 2.6 Hz, 1H), 7.58 (t, J = 6.8 Hz, 2H), 7.42–7.34 (m, 3H), 7.31 (t, J = 8.2 Hz, 1H), 6.94 (d, J = 7.7 Hz, 2H), 3.52 (s, 4H), 2.26 (s, 4H); 13C NMR (100 MHz, DMSO-d6), δ 156.22 (d, JCF = 2.8 Hz), 151.27 (d, JCF = 11.4 Hz), 141.15 (d, JCF = 245 Hz), 141.62 (d, JCF = 18.9 Hz), 140.44, 136.11, 131.08, 130.33, 130.26, 129.73, 129.52, 128.26, 128.01, 118.57, 66.84, 62.79, 53.74; 19F NMR (376 MHz, DMSO-d6), δ -166.63 (s); LC-MS (ESI+) m/z 327.08 (M-morpholine); HRMS (ESI+) m/z calculated for C21H22ClFN5O (M+H)+ 414.1491, found 414.1497 (M+H)+, 327.0819 (M - morpholine).
N4-(2-Chlorophenyl)-N2-[4-N -(2-hydroxyethyl)carbamoylphenyl]pyrimidine-2,4-diamine (9l, method t in Scheme 4)
A mixture of chloropyrimidine 2l (0.100 g, 0.416 mmol) and 4-amino-N-(2-hydroxyethyl)benzamide (0.075 g, 0.416 mmol) in HCl (1 mL of 0.1 N aq.) was heated in a microwave reactor at 140 °C for 30 min. The contents were then stirred in the microwave vial at room temperature for 30 min. The mixture was filtered and the obtained solid was washed with water (20 mL) and sodium bicarbonate (sat. aq. 50 mL). The crude product was purified by chromatography (silica gel, hexane: ethyl acetate) to provide 9l as an off-white solid (0.087 g, 54%). m.p. 171-173 °C, HPLC 100% [Rt = 8.8 min, 40% MeOH, 60% water (with 0.1% TFA) 20 min.]; 1H NMR (400 MHz, MeOH-d4), δ 8.28 (t, J = 6.2 Hz, 1H, peak disappeared upon D2O shake), 8.00 (d, J = 5.9 Hz, 1H), 7.84 (dd, J = 8.0, 1.5 Hz, 1H), 7.71–7.64 (m, 4H), 7.50 (dd, J = 8.0, 1.5 Hz, 1H), 7.37 (td, J = 8.0, 1.5 Hz, 1H), 7.22 (td, J = 8.0, 1.5 Hz, 1H), 6.30 (d, J = 5.9 Hz, 1H), 3.70 (t, J = 5.5 Hz, 2H), 3.49 (q, J = 5.5 Hz, 2H); 13C NMR (100 MHz, DMSO-d6), δ 172.68, 166.63, 161.90, 159.62, 156.84, 144.10, 136.56, 130.31, 128.61, 128.27, 128.07, 126.92, 126.82, 117.93, 99.18, 60.56, 42.68; LC-MS (ESI+) m/z 384.12 (M+H)+; HRMS (ESI+) m/z calculated for C19H19ClN5O2 (M+H)+ 384.1222, found 384.1219.
N4-(2-Chlorophenyl)-N2-(4-carbamoyl)-5-fluoropyrimidine-2,4-diamine (9m, method v in Scheme 4)
A mixture of chloropyrimidine 2k (0.500 g, 1.935 mmol), 4-aminobenzamide (0.265 g, 1.935 mmol) in methanol (3 mL), was heated in 5 ml sealed pressure tube at 100 °C for 6 hrs. The white precipitate was isolated by filtration and washed with methanol (2 mL). The white solid was sonicated for 5 min. in sodium bicarbonate (aq. sat., 10 mL). The mixture was filtered and the solid washed with water (10 mL) and finally with methanol (2 mL) to provide 9m (0.439 g, 63%) as an off-white powder. m.p. 250-252 °C. HPLC 100% [Rt = 7.8 min, 50% MeOH, 50% water (with 0.1% formic acid) 20 min.]; 1H NMR (400 MHz, DMSO-d6), δ 9.42 (s, 1H), 9.31 (s, 2H), 8.12 (d, JH-F = 3.6 Hz, 1H), 7.70 (s, 1H), 7.59 (dd, J = 8.6, 1.3 Hz, 1H), 7.56 (dd, J = 8.6, 1.3 Hz) overlapping 7.54 (d, J = 8.7 Hz, 2H), 7.48 (d, J = 8.7 Hz, 2H), 7.42 (td, J = 7.7, 1.5 Hz, 1H), 7.35 (td, J = 7.6, 1.6 Hz, 1H), 7.08 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 168.57, 155.66 (d, JC-F = 3.0 Hz), 151.35 (d, JC-F = 11.7 Hz), 144.27, 142.54, 141.66 (d, JC-F = 19.0 Hz), 140.09, 135.77, 131.27, 130.41, 129.79, 128.67, 128.41, 126.18, 117.31; 19F NMR (376 MHz, DMSO-d6), δ -165.45 (bd, JH-F = 3.4 Hz) LC-MS (ESI+) m/z 358.09 (M+H)+; HRMS (ESI+) m/z calculated for C17H14ClFN5O (M+H)+ 358.0865, found 358.0868.
N4-(2-Chlorophenyl)-N2-(4-aminosulfonyl)-5-fluoropyrimidine-2,4-diamine (9n, method u in Scheme 4)
A mixture of chloropyrimidine 2k (free base) (0.100 g, 0.387 mmol), 4-aminobenzenesulfonamide (0.066 g, 0.0.387 mmol), X-Phos (2-dicyclohexylphosphino-2′,4′,6′-triisopropylbiphenyl) (0.019 g, 0.0387 mmol), bis(dibenzylideneacetone) palladium(0) (0.022 g, 0.0387 mmol), potassium carbonate (0117 g, 0.851 mmol) in tert-BuOH (2.0 mL) was heated in 5 ml sealed pressure tube at 100 °C for 64 hours under argon. The solid was isolated by filtration and suspended in ethyl acetate (5 mL) and sonicated for 5 minutes. The mixture was filtered and the solid washed with ethyl acetate (5 mL). The crude product was dissolved in DMSO (2 mL) and filtered to remove undissolved material. The DMSO was removed in vacuo at 50 °C to provide the title sulfonamide 9n (0.080 g, 53%) as a brown solid. m.p. 253-255 °C. HPLC 99% [Rt = 11.2 min, 40% MeOH, 60% water (with 0.1% TFA) 66 min.]; 1H NMR (400 MHz, DMSO-d6), δ 9.57 (s, 1H), 9.39 (s, 1H), 8.13 (d, JH-F = 3.7 Hz, 1H), 7.61–7.52 (m, 4H), 7.46–7.38 (m, 3H), 7.35 (t, J = 8.0 Hz, 1H), 7.07 (s, 1H). 1H NMR (400 MHz, DMSO-d6+D2O), δ 8.09 (d, JH-F = 3.5 Hz, 1H), 7.58 (dd, J = 7.9, 1.5 Hz, 1H), 7.54–7.48 (m, 3H), 7.44–7.39 (m, 3H), 7.35 (dt, J = 7.4, 1.7 Hz, 1H); 19F NMR (376 MHz, DMSOd6-+D2O), δ -164.56 (d, J = 3.1 Hz); 13C NMR (100 MHz, DMSO-d6) 155.67 (d, JC-F = 2.8 Hz), 151.57, 151.46, 144.66, 142.73, 141.79, 141.60, 140.27, 136.06, 135.87, 131.65, 130.42, 130.19, 128.42, 126.85, 117.46. LC-MS (ESI+) m/z 394.05 (M+H)+; HRMS (ESI+) m/z calculated for C16H14ClFN5O2S (M+H)+ 394.0535, found 394.0537.
N4-(2-Chlorophenyl)-N2-[4-(2H -tetrazol-5-yl)phenyl]pyrimidine-2,4-diamine hydrochloride (12a, method a in Scheme 5)
A mixture of 2l (0.072 g, 0.299 mmol) and 4-(2H-tetrazol-5-yl)phenylamine (11a) (0.050 g, 0.310 mmol) in EtOH (2.0 mL) was heated in a microwave reactor at 150 °C for 40 min. The precipitate obtained upon cooling the reaction mixture was filtered and washed with EtOH (5 mL) to provide 12a (0.095 g, 79%) as a yellow solid (1H NMR analysis indicated the presence of trace impurities). The product was further purified by washing with methanol (5 mL) to give pure 12a (0.045 g, 37%) as a yellow solid. m.p. 250 °C (dec.). HPLC 100% (Rt = 6.21 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.76 (s, 1H), 10.64 (s, 1H), 8.09 (d, J = 6.9 Hz, 1H), 7.86 (d, J = 8.4 Hz, 2H), 7.69-7.59 (m, 4H), 7.48 (t, J = 7.4 Hz, 1H), 7.41 (t, J = 7.5 Hz, 1H), 6.52 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 163.07, 153.08, 146.47, 140.76, 134.74, 130.64, 130.17, 129.48, 129.32, 129.31, 128.57, 128.21, 121.13, 119.58; elemental analysis calculated for C17H14Cl2N8 : C, 50.89; H, 3.52; N, 27.93. Found: C, 51.22; H, 3.50; N, 27.54; LC-MS (ESI−) m/z 363.08 (M-H-HCl)−; HRMS (ESI−) m/z calculated for C17H12ClN8 (M-H-HCl)− 363.0879, found 363.0871.
N4-(2-Chlorophenyl)-N2-[4-(2H -tetrazol-5-yl)phenyl]-5-fluoropyrimidine-2,4-diamine hydrochloride (12b, method a in Scheme 5)
A mixture of 2k (free base) (0.076 g, 0.298 mmol) and 4-(2H-tetrazol-5-yl)phenylamine (11a) (0.050 g, 0.310 mmol) in EtOH (2.0 mL) was heated in a microwave reactor at 170 °C for 40 min. The precipitate obtained upon cooling the reaction mixture was filtered and washed with EtOH (5.0 mL) to get 12b (0.090 g, 72%) as a brown-yellow solid. The 1H NMR spectrum showed the presence of a baseline impurity and the product was further purified by washing with methanol (5 mL) to provide pure 12b (0.041 g, 33%) as a brown-yellow solid. m.p.: 223 °C (dec.). HPLC 99% (Rt = 10.37 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.98 (s, 1H), 9.91 (s, 1H), 8.25 (d, J = 4.0 Hz, 1H), 7.74 (d, J = 8.7 Hz, 2H), 7.64 (d, J = 7.8 Hz, 1H), 7.62 – 7.56 (m, 3H), 7.50-7.38 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 155.74 (d, J = 3.0 Hz), 151.50 (d, J = 11.7 Hz), 144.37, 141.45 (d, J = 247.45 Hz), 141.68 (d, J = 17.6 Hz), 136.06, 131.58, 130.43, 130.15, 128.41, 127.85, 118.47. 19F NMR (376 MHz, DMSO) δ -165.12 (s); elemental analysis calculated for C17H13Cl2FN8: C, 48.70; H, 3.13; N, 26.73. Found: C, 49.09; H, 3.13; N, 26.48. LC-MS (ESI−) m/z 381.1 (M-H)−; HRMS (ESI−) m/z calculated for C17H11ClFN8 (M-H)− 381.0785, found 381.0784.
N4-(2-Chlorophenyl)-N2-[3-hydroxy-4-(2H -tetrazol-5-yl)phenyl]-5-fluoropyrimidine-2,4-diamine hydrochloride (12c, method a Scheme 5)
A mixture of 2k (freebase) (0.072 g, 0.279 mmol) and tetrazole 11b (0.050 g, 0.282 mmol) in EtOH:HCl (1:1, 2.0 mL, 0.1 M HCl) was heated in a microwave reactor at 160 °C for 40 min. The precipitate obtained upon cooling was filtered and rinsed with EtOH (5 mL) to obtain the product with base-line impurity (0.096 g, 77%) as a yellow solid. This solid was further purified by washing with methanol (5 mL) to provide pure 12c (0.044 g, 36%) as a yellow solid. m.p. 214 °C (dec.). HPLC 93% (Rt = 12.99 min., 50% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 10.66 (s, 1H), 9.44 (s, 1H), 9.24 (s, 1H), 8.13 (d, J = 3.3 Hz, 1H), 7.66 – 7.54 (m, 4H), 7.41 (t, J = 7.5 Hz, 1H), 7.33 (t, J = 7.4 Hz, 1H), 7.29 – 7.23 (m, 2H); 13C NMR (100 MHz, DMSO-d6) 156.34, 155.80 (d, J = 3.1 Hz), 151.46 (d, J = 11.5 Hz), 145.58, 141.61 (d, J = 247.2 Hz), 141.23 (d, J = 19.1 Hz), 135.90, 131.04, 130.42, 129.71, 129.25, 128.38, 128.20, 110.49, 105.32, 103.39; 19F NMR (376 MHz, DMSO-d6) δ -165.94 (s); LC-MS (ESI−) m/z 397.07 (M-H)−; HRMS (ESI−) m/z calculated for C17H11ClFN8O (M-H)− 397.0734 , found 397.0730.
N4-(2-Chlorophenyl)-N2-[4-(2H -tetrazol-5-yl)phenyl]-5-chloropyrimidine-2,4-diamine hydrochloride (12d, method a Scheme 5)
A mixture of chloropyrimidine 2o (0.093 g, 0.3 mmol) and 4-(2H-tetrazol-5-yl)phenylamine (11a) (50 mg, 0.3 mmol) in EtOH (2 mL) was heated in a microwave reactor at 150 °C for 40 min. The precipitate obtained upon cooling was filtered and washed with EtOH (5 mL) to provide impure 12d (105 mg, 81%) as a yellow color solid. Analysis of the 1H NMR spectrum indicated the presence of trace impurities. The product was further purified by washing with hot ethyl acetate (5 mL) and hot methanol (5 mL) to provide pure 12d (0.032 g, 25%) as a yellow solid. m.p. 238 °C (dec.). HPLC 91% (Rt = 5.71 min., 70% MeOH in 0.1% TFA water, 20 min.); 1H NMR (400 MHz, DMSO-d6) δ 9.72 (s, 1H), 9.05 (s, 1H), 8.19 (s, 1H), 7.71–7.56 (m, 6H), 7.52–7.38 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 157.73, 157.53, 154.65, 143.69, 136.39, 131.71, 130.38, 130.32, 128.69, 128.49, 127.87, 119.04, 104.93; LC-MS (ESI−) m/z 397.05 (M-H)−; HRMS (ESI−) m/z calculated for C17H11Cl2N8 (M-H)− 397.0489, found 397.0472.
Acknowledgments
We thank the Chemical Biology Core at the Moffitt Cancer Center and Research Institute for assistance with the Z-lyte kinase assay (Dr Richard Yip, HTS Unit), mass spectrometry and HPLC analysis (Chemistry Unit). We also thank the Moffitt Structural Biology Core for use of the X-ray crystallography facility. We thank the Drs Gary Daughdrill Hongwei Wu for access to the 600 MHz NMR facility and acknowledge that this work has been supported in part by the Florida Center of Excellence for Drug Discovery & Innovation at the University of South Florida.
Abbreviations
- CML
Chronic Myeloid Leukemia
- DCM
Dichloromethane
- DEA
Diethylamine
- DMEM
Dulbecco's Modified Eagle's Medium
- EI
Electrospray Ionization
- ECL
Enhanced Chemiluminescence
- EIF2AK3
Eukaryotic translation Initiation Factor 2 Alpha Kinase 3
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GSK3β
Glycogen Synthase Kinase 3 beta
- HPLC
High Performance Liquid Chromatography
- HTS
High Throughput Screening
- HRP
horseradish peroxidase
- JAK2
Janus kinase 2
- LCMS
Liquid Chromatography Mass Spectroscopy
- PBS
Phosphate Buffered Saline
- PIM3
Provirus Integration site for Moloney murine leukaemia virus
- NUAK1
SnF1-like Kinase
- SAR
Structure Activity Relationship
- TLC
Thin Layer Chromatography
- TFA
Trifluoroacetic acid
Footnotes
Associated Content
Supporting Information
(i) Synthetic protocols and characterization data for 2a-o, 5a-k, 7a-b, 8a-b, 10a-b and 11a-b, (ii) scanned NMR spectra, HPLC and LC-MS reports for 1, 3l, 3n, 3o, 6c, 6i, 9h, 9m, 9n and 12a, (iii) determination of cocrystal structures of 1, 3i, 3g, 13 and ADP with Aurora A, (iv) kinase profiling of 3o, (v) IC50 data (Reaction Biology) for 9h, 3l, 3o against Aurora A and JAK2, (vi) Aurora A IC50 data of compounds 13-24. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Green MR, Woolery JE, Mahadevan D. Update on aurora kinase targeted therapeutics in oncology. Expert Opin. Drug Discovery. 2011;6:291–307. doi: 10.1517/17460441.2011.555395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marumoto T, Zhang D, Saya H. Aurora-A - A guardian of poles. Nat. Rev. Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 3.Asteriti IA, Rensen WM, Lindon C, Lavia P, Guarguaglini G. The Aurora-A/TPX2 complex: A novel oncogenic holoenzyme? Biochim. Biophys. Acta, Rev. Cancer. 2010;1806:230–239. doi: 10.1016/j.bbcan.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Saeki T, Ouchi M, Ouchi T. Physiological and oncogenic Aurora-A pathway. Int. J. Biol. Sci. 2009;5:758–762. doi: 10.7150/ijbs.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 6.Libertini S, Abagnale A, Passaro C, Botta G, Portella G. Aurora A and B kinases - targets of novel anticancer drugs. Recent Pat. Anti-Cancer Drug Discovery. 2010;5:219–241. doi: 10.2174/157489210791760517. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura T, Hasako S, Nakatsuru Y, Mita T, Ichikawa K, Kodera T, Sakai T, Nambu T, Miyamoto M, Takahashi I, Miki S, Kawanishi N, Ohkubo M, Kotani H, Iwasawa Y. MK-5108, a Highly Selective Aurora-A Kinase Inhibitor, Shows Antitumor Activity Alone and in Combination with Docetaxel. Mol. Cancer Ther. 2010;9:157–166. doi: 10.1158/1535-7163.MCT-09-0609. [DOI] [PubMed] [Google Scholar]
- 8.Ochi T, Fujiwara H, Yasukawa M. Aurora-A kinase: a novel target both for cellular immunotherapy and molecular target therapy against human leukemia. Expert Opin. Ther. Targets. 2009;13:1399–1410. doi: 10.1517/14728220903307483. [DOI] [PubMed] [Google Scholar]
- 9.Shang X, Burlingame SM, Okcu MF, Ge N, Russell HV, Egler RA, David RD, Vasudevan SA, Yang J, Nuchtern JG. Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Mol. Cancer Ther. 2009;8:2461–2469. doi: 10.1158/1535-7163.MCT-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bearss DJ. Shining the Light on Aurora-A Kinase as a Drug Target in Pancreatic Cancer. Mol. Cancer Ther. 2011;10:2012. doi: 10.1158/1535-7163.MCT-11-0720. [DOI] [PubMed] [Google Scholar]
- 11.Keen N, Taylor S. Mitotic drivers-inhibitors of the Aurora B Kinase. Cancer Metastasis Rev. 2009;28:185–195. doi: 10.1007/s10555-009-9184-9. [DOI] [PubMed] [Google Scholar]
- 12.Girdler F, Gascoigne KE, Eyers PA, Hartmuth S, Crafter C, Foote KM, Keen NJ, Taylor SS. Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 2006;119:3664–3675. doi: 10.1242/jcs.03145. [DOI] [PubMed] [Google Scholar]
- 13.Portella G, Passaro C, Cheiffi P. Aurora B: a new prognostic marker and therapeutic target in cancer. Curr. Med. Chem. 2011;18:482–496. doi: 10.2174/092986711794480203. [DOI] [PubMed] [Google Scholar]
- 14.Yeung S-CJ, Gully C, Lee M-H. Aurora-B kinase inhibitors for cancer chemotherapy. Mini-Rev. Med. Chem. 2008;8:1514–1525. doi: 10.2174/138955708786786480. [DOI] [PubMed] [Google Scholar]
- 15.Garuti L, Roberti M, Bottegoni G. Small molecule aurora kinases inhibitors. Curr. Med. Chem. 2009;16:1949–1963. doi: 10.2174/092986709788682227. [DOI] [PubMed] [Google Scholar]
- 16.Pollard JR, Mortimore M. Discovery and Development of Aurora Kinase Inhibitors as Anticancer Agents. J. Med. Chem. 2009;52:2629–2651. doi: 10.1021/jm8012129. [DOI] [PubMed] [Google Scholar]
- 17.Cheung CHA, Coumar MS, Hsieh H-P, Chang J-Y. Aurora kinase inhibitors in preclinical and clinical testing. Expert Opin. Invest. Drugs. 2009;18:379–398. doi: 10.1517/13543780902806392. [DOI] [PubMed] [Google Scholar]
- 18.Lok W, Klein RQ, Saif MW. Aurora kinase inhibitors as anti-cancer therapy. Anti-Cancer Drugs. 2010;21:339–350. doi: 10.1097/CAD.0b013e3283350dd1. [DOI] [PubMed] [Google Scholar]
- 19.Dar AA, Goff LW, Majid S, Berlin J, El-Rifai W. Aurora Kinase Inhibitors - Rising Stars in Cancer Therapeutics? Mol. Cancer Ther. 2010;9:268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez Fidalgo JA, Roda D, Rosello S, Rodriguez-Braun E, Cervantes A. Aurora kinase inhibitors: a new class of drugs targeting the regulatory mitotic system. Clin. Transl. Oncol. 2009;11:787–798. doi: 10.1007/s12094-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 21.Boss DS, Beijnen JH, Schellens JHM. Clinical experience with Aurora kinase inhibitors: a review. Oncologist. 2009;14:780–793. doi: 10.1634/theoncologist.2009-0019. [DOI] [PubMed] [Google Scholar]
- 22.Cheung CHA, Coumar MS, Chang J-Y, Hsieh H-P. Aurora kinase inhibitor patents and agents in clinical testing: an update (2009 - 10). Expert Opin. Ther. Pat. 2011;21:857–884. doi: 10.1517/13543776.2011.574614. [DOI] [PubMed] [Google Scholar]
- 23.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JMC, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 24.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, Ray ET, Sells TB, Stringer B, Stroud SG, Vos TJ, Weatherhead GS, Wysong DR, Zhang M, Bolen JB, Claiborne CF. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodson CA, Kosmopoulou M, Richards MW, Atrash B, Bavetsias V, Blagg J, Bayliss R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem. J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 26.Chakravarty A, Shinde V, Tabernero J, Cervantes A, Cohen RB, Dees EC, Burris H, Infante JR, Macarulla T, Elez E, Andreu J, Rodriguez-Braun E, Rosello S, von Mehren M, Meropol NJ, Langer CJ, Oneil B, Bowman D, Zhang M, Danaee H, Faron-Yowe L, Gray G, Liu H, Pappas J, Silverman L, Simpson C, Stringer B, Tirrell S, Veiby OP, Venkatakrishnan K, Galvin K, Manfredi M, Ecsedy JA. Phase I Assessment of New Mechanism-Based Pharmacodynamic Biomarkers for MLN8054, a Small-Molecule Inhibitor of Aurora A Kinase. Cancer Res. 2011;71:675–685. doi: 10.1158/0008-5472.CAN-10-1030. [DOI] [PubMed] [Google Scholar]
- 27.Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, Santo L, Tai Y-T, Nahar S, Zheng M, Bandi M, Carrasco RD, Raje N, Munshi N, Richardson P, Anderson KC. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomita M, Mori N. Aurora A selective inhibitor MLN8237 suppresses the growth and survival of HTLV-1-infected T-cells in vitro. Cancer Sci. 2010;101:1204–1211. doi: 10.1111/j.1349-7006.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloane DA, Trikic MZ, Chu MLH, Lamers MBAC, Mason CS, Mueller I, Savory WJ, Williams DH, Eyers PA. Drug-Resistant Aurora A Mutants for Cellular Target Validation of the Small Molecule Kinase Inhibitors MLN8054 and MLN8237. ACS Chem. Biol. 2010;5:563–576. doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- 30.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shinde V, Huck JJ, Wysong DR, Janowick DA, Hyer ML, LeRoy PJ, Gershman RE, Silva MD, Germanos MS, Bolen JB, Claiborne CF, Sells TB. Characterization of Alisertib (MLN8237), an Investigational Small-Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin. Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 31.Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann J-JM, Warin N, Renaud F, De Savi C, Roberts NJ, Johnson T, Dousson CB, Hill GB, Perkins D, Hatter G, Wilkinson RW, Wedge SR, Heaton SP, Odedra R, Keen NJ, Crafter C, Brown E, Thompson K, Brightwell S, Khatri L, Brady MC, Kearney S, McKillop D, Rhead S, Parry T, Green S. Discovery, Synthesis, and in Vivo Activity of a New Class of Pyrazolylamino Quinazolines as Selective Inhibitors of Aurora B Kinase. J. Med. Chem. 2007;50:2213–2224. doi: 10.1021/jm061335f. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson RW, Odedra R, Heaton SP, Wedge SR, Keen NJ, Crafter C, Foster JR, Brady MC, Bigley A, Brown E, Byth KF, Barrass NC, Mundt KE, Foote KM, Heron NM, Jung FH, Mortlock AA, Boyle FT, Green S. AZD1152, a Selective Inhibitor of Aurora B Kinase, Inhibits Human Tumor Xenograft Growth by Inducing Apoptosis. Clin. Cancer Res. 2007;13:3682–3688. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- 33.Fancelli D, Berta D, Bindi S, Cameron A, Cappella P, Carpinelli P, Catana C, Forte B, Giordano P, Giorgini ML, Mantegani S, Marsiglio A, Meroni M, Moll J, Pittala V, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Varasi M, Vulpetti A, Vianello P. Potent and selective Aurora inhibitors identified by the expansion of a Novel scaffold for protein kinase inhibition. J. Med. Chem. 2005;48:3080–3084. doi: 10.1021/jm049076m. [DOI] [PubMed] [Google Scholar]
- 34.Fancelli D, Moll J, Varasi M, Bravo R, Artico R, Berta D, Bindi S, Cameron A, Candiani I, Cappella P, Carpinelli P, Croci W, Forte B, Giorgini ML, Klapwijk J, Marsiglio A, Pesenti E, Rocchetti M, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Zugnoni P, Vianello P. 1,4,5,6-Tetrahydropyrrolo[3,4-c]pyrazoles: Identification of a Potent Aurora Kinase Inhibitor with a Favorable Antitumor Kinase Inhibition Profile. J. Med. Chem. 2006;49:7247–7251. doi: 10.1021/jm060897w. [DOI] [PubMed] [Google Scholar]
- 35.Soncini C, Carpinelli P, Gianellini L, Fancelli D, Vianello P, Rusconi L, Storici P, Zugnoni P, Pesenti E, Croci V, Ceruti R, Giorgini ML, Cappella P, Ballinari D, Sola F, Varasi M, Bravo R, Moll J. PHA-680632, a Novel Aurora Kinase Inhibitor with Potent Antitumoral Activity. Clin. Cancer Res. 2006;12:4080–4089. doi: 10.1158/1078-0432.CCR-05-1964. [DOI] [PubMed] [Google Scholar]
- 36.Fancelli D, Moll J. Discovery of PHA-739358. Kinase Inhib. Drugs. 2009:281–307. [Google Scholar]
- 37.Adams ND, Adams JL, Burgess JL, Chaudhari AM, Copeland RA, Donatelli CA, Drewry DH, Fisher KE, Hamajima T, Hardwicke MA, Huffman WF, Koretke-Brown KK, Lai ZV, McDonald OB, Nakamura H, Newlander KA, Oleykowski CA, Parrish CA, Patrick DR, Plant R, Sarpong MA, Sasaki K, Schmidt SJ, Silva DJ, Sutton D, Tang J, Thompson CS, Tummino PJ, Wang JC, Xiang H, Yang J, Dhanak D. Discovery of GSK1070916, a Potent and Selective Inhibitor of Aurora B/C Kinase. J. Med. Chem. 2010;53:3973–4001. doi: 10.1021/jm901870q. [DOI] [PubMed] [Google Scholar]
- 38.Belanger DB, Curran PJ, Hruza A, Voigt J, Meng Z, Mandal AK, Siddiqui MA, Basso AD, Gray K. Discovery of imidazo[1,2-a]pyrazine-based Aurora kinase inhibitors. Bioorg. Med. Chem. Lett. 2010;20:5170–5174. doi: 10.1016/j.bmcl.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Cee VJ, Schenkel LB, Hodous BL, Deak HL, Nguyen HN, Olivieri PR, Romero K, Bak A, Be X, Bellon S, Bush TL, Cheng AC, Chung G, Coats S, Eden PM, Hanestad K, Gallant PL, Gu Y, Huang X, Kendall RL, Lin M-HJ, Morrison MJ, Patel VF, Radinsky R, Rose PE, Ross S, Sun J-R, Tang J, Zhao H, Payton M, Geuns-Meyer SD. Discovery of a Potent, Selective, and Orally Bioavailable Pyridinyl-Pyrimidine Phthalazine Aurora Kinase Inhibitor. J. Med. Chem. 2010;53:6368–6377. doi: 10.1021/jm100394y. [DOI] [PubMed] [Google Scholar]
- 40.Yu T, Tagat JR, Kerekes AD, Doll RJ, Zhang Y, Xiao Y, Esposite S, Belanger DB, Curran PJ, Mandal AK, Siddiqui MA, Shih N-Y, Basso AD, Liu M, Gray K, Tevar S, Jones J, Lee S, Liang L, Ponery S, Smith EB, Hruza A, Voigt J, Ramanathan L, Prosise W, Hu M. Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]- Pyrazine Core. ACS Med. Chem. Lett. 2010;1:214–218. doi: 10.1021/ml100063w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coumar MS, Tsai M-T, Chu C-Y, Uang B-J, Lin W-H, Chang C-Y, Chang T-Y, Leou J-S, Teng C-H, Wu J-S, Fang M-Y, Chen C-H, Hsu JTA, Wu S-Y, Chao Y-S, Hsieh H-P. Identification, SAR Studies, and X-ray Co-crystallographic Analysis of a Novel Furanopyrimidine Aurora Kinase A Inhibitor. ChemMedChem. 2010;5:255–267. doi: 10.1002/cmdc.200900339. [DOI] [PubMed] [Google Scholar]
- 42.Aliagas-Martin I, Burdick D, Corson L, Dotson J, Drummond J, Fields C, Huang OW, Hunsaker T, Kleinheinz T, Krueger E, Liang J, Moffat J, Phillips G, Pulk R, Rawson TE, Ultsch M, Walker L, Wiesmann C, Zhang B, Zhu BY, Cochran AG. A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B. J. Med. Chem. 2009;52:3300–7. doi: 10.1021/jm9000314. [DOI] [PubMed] [Google Scholar]
- 43.Moriarty K,J, Koblish H,K, Garrabrant T, Maisuria J, Khalil E, Ali F, Petrounia I,P, Crysler C,S, Maroney A,C, Johnson D,L, Galemmo RA., Jr. The synthesis and SAR of 2-amino-pyrrolo[2,3-d]pyrimidines: a new class of Aurora-A kinase inhibitors. Bioorg. Med. Chem. Lett. 2006;16:5778–83. doi: 10.1016/j.bmcl.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 44.Tari LW, Hoffman ID, Bensen DC, Hunter MJ, Nix J, Nelson KJ, McRee DE, Swanson RV. Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity disubstituted pyrimidine inhibitors. Bioorg. Med. Chem. Lett. 2007;17:688–691. doi: 10.1016/j.bmcl.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 45.Bebbington D, Binch H, Charrier J-D, Everitt S, Fraysse D, Golec J, Kay D, Knegtel R, Mak C, Mazzei F, Miller A, Mortimore M, O'Donnell M, Patel S, Pierard F, Pinder J, Pollard J, Ramaya S, Robinson D, Rutherford A, Studley J, Westcott J. The discovery of the potent aurora inhibitor MK-0457 (VX-680). Bioorg. Med. Chem. Lett. 2009;19:3586–92. doi: 10.1016/j.bmcl.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 46.Breault GA, Ellston RPA, Green S, James SR, Jewsbury PJ, Midgley CJ, Pauptit RA, Minshull CA, Tucker JA, Pease JE. Cyclin-dependent kinase 4 inhibitors as a treatment for cancer. Part 2: identification and optimization of substituted 2,4-bis anilino pyrimidines. Bioorg. Med. Chem. Lett. 2003;13:2961–2966. doi: 10.1016/s0960-894x(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 47.Gray N, Sim T, Choi HG. Preparation of amino substituted pyrimidine, pyrrolopyridine and pyrazolopyrimidine derivatives useful as kinase inhibitors and in treating proliferative disorders and diseases associated with angiogenesis. 2009 Mar 12;A1:2009. WO 2009/032694. [Google Scholar]
- 48.Barlaam B, Ducray R, Lambert-van der Brempt C, Ple P, Bardelle C, Brooks N, Coleman T, Cross D, Kettle JG, Read J. Inhibitors of the tyrosine kinase EphB4. Part 4: Discovery and optimization of a benzylic alcohol series. Bioorg. Med. Chem. Lett. 2011;21:2207–2211. doi: 10.1016/j.bmcl.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Harrison RJ, Hobson A, Ramsden N. Ortho substituted pyrimidine compounds as JAK inhibitors. 2011 WO 2011/029807 A1, 20100907.
- 50.Liu M, Wang S, Clampit JE, Gum RJ, Haasch DL, Rondinone CM, Trevillyan JM, Abad-Zapatero C, Fry EH, Sham HL, Liu G. Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies. Bioorg. Med. Chem. Lett. 2007;17:668–72. doi: 10.1016/j.bmcl.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 51.Kettle JG, Read J, Leach A, Barlaam BC, Ducray R, Lambert-Van Der Brempt CMP. Pyrimidine derivatives as EphB4 inhibitors and their preparation, pharmaceutical compositions and use in the treatment of cancer. 2007 Aug 2;A2:2007. WO 2007/085833. [Google Scholar]
- 52.Adams JL, Drewry DH, Linn JA. Compounds. 2007 Feb 15;A2:2007. WO 2007/018941. [Google Scholar]
- 53.Martin MP, Zhu J-Y, Lawrence H, Pireddu R, Luo Y, Alam R, Ozcan S, Sebti SM, Lawrence NJ, Schonbrunn E. A novel mechanism by which small molecule inhibitors induce the DFG flip in Aurora A. ACS Chem. Biol. 2012;7:698–706. doi: 10.1021/cb200508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anandan S-K, Gless RD. Exploration of secondary and tertiary pharmacophores in unsymmetrical N,N'-diaryl urea inhibitors of soluble epoxide hydrolase. Bioorg. Med. Chem. Lett. 2010;20:2740–2744. doi: 10.1016/j.bmcl.2010.03.074. [DOI] [PubMed] [Google Scholar]
- 55.Maeda DY, Zebala JA. Pyridine- and pyrimidinecarboxamides as CXCR2 modulators and their preparation and use for the treatment of inflammatory and neoplastic diseases. 2010 Aug 19;A1:2010. US 2010/0210593. [Google Scholar]
- 56.Kang NS, Lee GN, Kim CH, Bae MA, Kim I, Cho YS. Identification of small molecules that inhibit GSK-3[beta] through virtual screening. Bioorg. Med. Chem. Lett. 2009;19:533–537. doi: 10.1016/j.bmcl.2008.10.120. [DOI] [PubMed] [Google Scholar]
- 57.Koresawa M, Okabe T. High-throughput screening with quantitation of ATP consumption: A universal non-radioisotope, homogeneous assay for protein kinase. Assay Drug Dev. Technol. 2004;2:153–160. doi: 10.1089/154065804323056495. [DOI] [PubMed] [Google Scholar]
- 58.Charter NW, Kauffman L, Singh R, Eglen RM. A generic, homogenous method for measuring kinase and inhibitor activity via adenosine 5′-diphosphate accumulation. J. Biomol. Screening. 2006;11:390–399. doi: 10.1177/1087057106286829. [DOI] [PubMed] [Google Scholar]
- 59.Lietha D, Eck MJ. Crystal structures of the FAK kinase in complex with TAE226 and related bis anilino pyrimidine inhibitors reveal a helical DFG conformation. PloS one. 2008;3:e3800. doi: 10.1371/journal.pone.0003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bardelle C, Coleman T, Cross D, Davenport S, Kettle JG, Ko EJ, Leach AG, Mortlock A, Read J, Roberts NJ, Robins P, Williams EJ. Inhibitors of the tyrosine kinase EphB4. Part 2: structure-based discovery and optimisation of 3,5-bis substituted anilinopyrimidines. Bioorg. Med. Chem. Lett. 2008;18:5717–21. doi: 10.1016/j.bmcl.2008.09.087. [DOI] [PubMed] [Google Scholar]
- 61.Beattie JF, Breault GA, Ellston RP, Green S, Jewsbury PJ, Midgley CJ, Naven RT, Minshull CA, Pauptit RA, Tucker JA, Pease JE. Cyclin-dependent kinase 4 inhibitors as a treatment for cancer. Part 1: identification and optimisation of substituted 4,6-bis anilino pyrimidines. Bioorg. Med. Chem. Lett. 2003;13:2955–60. doi: 10.1016/s0960-894x(03)00202-6. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herr RJ. 5-Substituted 1H-tetrazoles as carboxylic acid isosteres: medicinal chemistry and synthetic methods. Bioorg. Med. Chem. 2002;10:3379–3393. doi: 10.1016/s0968-0896(02)00239-0. [DOI] [PubMed] [Google Scholar]
- 64.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 66.Jester BW, Gaj A, Shomin CD, Cox KJ, Ghosh I. Testing the Promiscuity of Commercial Kinase Inhibitors Against the AGC Kinase Group Using a Split-luciferase Screen. J. Med. Chem. 2012;55:1526–1537. doi: 10.1021/jm201265f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vader G, Lens SMA. The Aurora kinase family in cell division and cancer. BBA-Rev. Cancer. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 69.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 70.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 71.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 72.DeLano WL. PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- 73.Gizatullin F, Yao Y, Kung V, Harding MW, Loda M, Shapiro GI. The Aurora kinase inhibitor VX-680 induces endoreduplication and apoptosis preferentially in cells with compromised p53-dependent postmitotic checkpoint function. Cancer Res. 2006;66:7668–7677. doi: 10.1158/0008-5472.CAN-05-3353. [DOI] [PubMed] [Google Scholar]
- 74.Berndt N, Yang H, Trinczek B, Betzi S, Zhang Z, Wu B, Lawrence NJ, Pellecchia M, Schonbrunn E, Cheng JQ, Sebti SM. The Akt activation inhibitor TCN-P inhibits Akt phosphorylation by binding to the PH domain of Akt and blocking its recruitment to the plasma membrane. Cell Death Differ. 2010;17:1795–1804. doi: 10.1038/cdd.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]