Abstract
Huntington disease (HD) is a dominant, genetic neurodegenerative disease characterized by progressive loss of voluntary motor control, psychiatric disturbance, and cognitive decline, for which there is currently no disease-modifying therapy. HD is caused by the expansion of a CAG tract in the huntingtin (HTT) gene. The mutant HTT protein (muHTT) acquires toxic functions, and there is significant evidence that muHTT lowering would be therapeutically efficacious. However, the wild-type HTT protein (wtHTT) serves vital functions, making allele-specific muHTT lowering strategies potentially safer than nonselective strategies. CAG tract expansion is associated with single nucleotide polymorphisms (SNPs) that can be targeted by gene silencing reagents such as antisense oligonucleotides (ASOs) to accomplish allele-specific muHTT lowering. Here we evaluate ASOs targeted to HD-associated SNPs in acute in vivo studies including screening, distribution, duration of action and dosing, using a humanized mouse model of HD, Hu97/18, that is heterozygous for the targeted SNPs. We have identified four well-tolerated lead ASOs that potently and selectively silence muHTT at a broad range of doses throughout the central nervous system for 16 weeks or more after a single intracerebroventricular (ICV) injection. With further validation, these ASOs could provide a therapeutic option for individuals afflicted with HD.
Introduction
Huntington disease (HD) is a dominant, genetic neurodegenerative disorder that usually manifests in middle age and is characterized by psychiatric disturbance, chorea, cognitive decline, and death usually 15–20 years later.1 HD is caused by the expansion of a polyglutamine encoding CAG repeat in exon 1 of the huntingtin gene (HTT) to 36 or more,2 resulting in early and selective atrophy of the putamen and caudate nucleus of the striatum, as well as cortical thinning due to degeneration of susceptible neuronal populations.3 Despite the ability to identify HD mutation carriers decades before onset, there is currently no available therapy that can delay onset or slow progression of the disease. Although some symptomatic treatments are available, most are not effective and are often associated with significant side effects.4 Thus, the development of safe and efficacious HD modifying therapies represents a significant unmet medical need.
Mutant huntingtin (muHTT) fails to adequately perform the vital roles of the HTT protein and also acquires toxic functions,3 creating a complex network of cellular dysfunction. As the sole cause of HD, muHTT is an obvious therapeutic target. There is significant evidence from murine studies that reducing muHTT will provide benefit. HTT lowering in HD patient monocytes reverses transcriptional dysregulation and normalizes cytokine release responses.5 Moreover, in a conditional model of HD, turning off muHTT transgene expression postsymptomatically results in significant functional recovery concomitant with clearance of accumulated muHTT,6 indicating that HTT lowering therapies could act not only to halt or slow disease progression, but to reverse pathology. Several HTT lowering preclinical trials in rodents have shown significant benefit to a broad range of HD-like phenotypes even when administered postsymptomatically.7,8,9,10,11,12,13,14,15 One very encouraging study even found that the benefit of HTT lowering much outlasted the duration of HTT reduction,15 suggesting that intermittent therapy may be sufficient to achieve benefit beyond the drug action period.
While these studies are promising, in humans treated with these agents, both mutant and wild-type HTT (wtHTT) would be suppressed, and an ideal therapy would target only muHTT. The wtHTT protein is important for neuronal health throughout life; mice with the murine HTT homolog Hdh inactivated in the neonatal forebrain and testes develop progressive motor impairment and neuropathology.16 Additionally, previous preclinical nonselective HTT lowering studies and wtHTT lowering safety studies have been performed over periods of 6–9 months, and though overt behavioral changes were not observed from nonselective or wtHTT reduction,17,18 transcriptional changes have been reported.13,14 Human therapy will likely be required for decades, and for these reasons, an allele-specific muHTT gene silencing approach that does not disrupt wtHTT expression will likely be preferable for therapeutic applications.
Potential allele-specific muHTT gene silencing approaches include, targeting the expanded CAG tract directly for gene silencing, but this could result in nonselective HTT reduction, as the normal allele does contain a CAG tract, or nonspecific reduction of other CAG tract-containing genes.19 Alternatively, some of the > 3,000 single-nucleotide polymorphisms (SNPs) that have been identified in the HTT gene region are tightly linked to CAG expansion, and population genetics studies have shown that a panel of silencing reagents targeting as few as three to five of these HD-associated SNPs could provide an allele-specific therapeutic option for up to 85% of HD patients.20,21,22 We are developing antisense oligonucleotides (ASOs) targeted to HD-associated SNPs as novel muHTT lowering HD therapeutics.
ASOs are short, synthetic DNA molecules that bind to target RNA and catalyze downstream actions, including RNase H-mediated degradation, resulting in gene product reduction.23 We have previously generated a large number of ASOs targeting six different HD-associated SNPs and counter screened them in cellular systems homozygous for either the targeted or nontargeted allele. This allowed for independent assessments of ASO potency by targeted allele knockdown (KD), or ASO selectivity by nontargeted allele KD,24 and demonstrated the feasibility of a SNP targeted ASO-mediated selective muHTT gene silencing approach.
We have used Hu97/18 mice to evaluate ASOs targeted to HD-associated SNPs in acute in vivo studies. The Hu97/18 mice lack Hdh, but express both human wt and muHTT genes, including the coding, noncoding and associated SNPs in the region.25 This precisely models the human genetic condition of HD, and allows us to identify ASOs with the greatest therapeutic potential. This includes screening of both previously reported and novel ASOs, scoring KD potency and SNP discrimination in the same cells for the first time, as well as assessments of multiple measures of tolerability. Additionally, we have evaluated the drug-like properties of our ASOs, including investigation of ASO persistence in tissue, duration of silencing action in the brain, ASO distribution, KD efficacy regionally throughout the CNS, and dosing of leads (Figure 1).
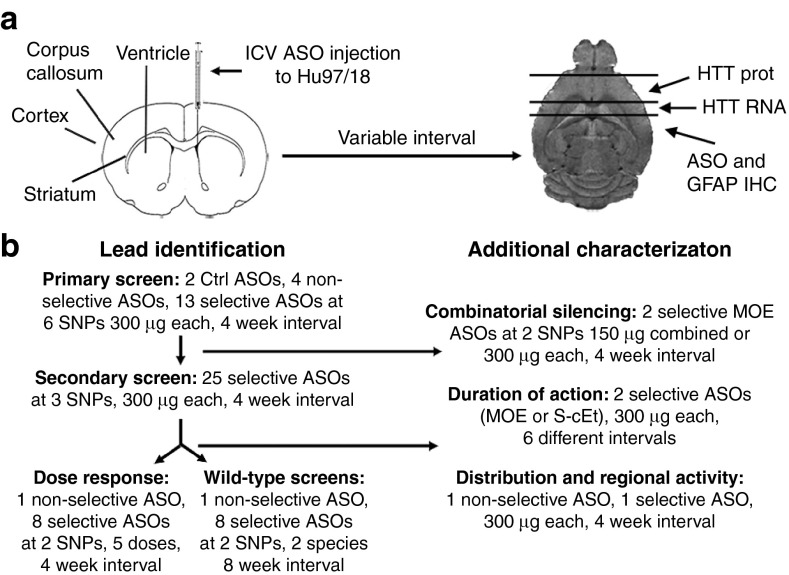
Figure 1.
Study overview. (a) Diagram of intracerebroventricular (ICV) antisense oligonucleotides injection and tissue processing method to allow assessment of HTT mRNA and protein and immunohistochemical analyses from the same brains. (b) Study flow chart.
Results
Primary screen
To determine which SNP sites would be amenable for targeting with ASOs, ASOs targeting six different HD-associated SNPs (Figure 2) previously identified as having significant patient population coverage and being sites of ASO silencing activity24 were delivered by unilateral intracerebroventricular (ICV) injection at a single dose of 300 µg into the right lateral ventricle of Hu97/18 mice. ASOs targeted to all HTT (pan-HTT, PH) or human HTT (H-HTT, HH), which should induce nonselective HTT lowering in Hu97/18 brains, or ASOs that do not match any genes in the mouse genome or lack catalytic silencing activity (control, CTRL), which should not alter HTT levels in Hu97/18 brains, were also evaluated as controls. Four weeks after injection, brains were collected and ASO delivery and distribution were verified by immunohistochemistry (IHC) using an antibody that recognizes the common phosphorothioate (PS) backbone24 (Supplementary Figure S1). WT and muHTT protein levels were assessed by quantitative allelic separation immunoblotting (Figure 3a). HTT levels were quantified by normalizing the density of HTT bands to the density of calnexin loading control and then to the density of the same allele from phosphate-buffered saline (PBS)-treated animals and expressed as % HTT remaining (Figure 3b). An N of four animals per treatment was used and each hemisphere of the brain was analyzed independently, giving eight data points per condition.
Figure 2.

Antisense oligonucleotides (ASOs) in the HTT locus. Diagram showing the location of each targeted single-nucleotide polymorphism (SNP) in the HTT locus and how each evaluated ASO aligns at the SNP. Black-PS, Orange-MOE, Blue-cEt, Green-FHNA, Red-3′ Phosphonate linkage, Target SNP is underlined. Nonunderlined ASOs are from primary screen, underlined ASOs are from secondary screen. Sequence and chemistry of control (CTRL), pan-huntingtin (PH) and human-huntingtin (HH) specific ASOs is also shown.
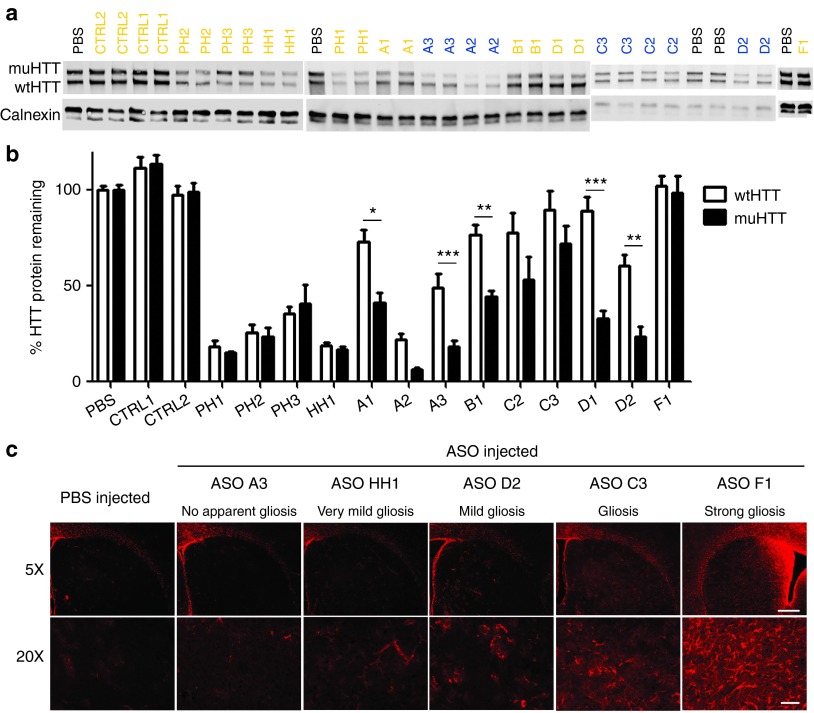
Figure 3.
Single dose screen of 5-9-5 2′-O-methoxyethyl (MOE) and cEt antisense oligonucleotides (ASOs). 300 µg of ASO was delivered by intracerebroventricular (ICV) bolus injection to the right lateral ventricle of 2–3-month-old Hu97/18 mice. Four weeks later, brains were collected and sectioned in a 2 mm coronal rodent brain matrix. The first section containing mostly olfactory bulb was discarded. The second section, containing anterior cortex and striatum, was used for HTT quantitation by allelic separation immunoblotting. The remaining posterior portion of the brain was used for immunohistochemical evaluation of ASO distribution and tolerability. (a) Example western blots showing wt and muHTT protein. ASOs in orange contain only MOE modifications. ASOs in blue contain cEt modifications. (b) Quantitation of HTT protein in both hemispheres of 4 animals. Density of HTT bands was normalized to calnexin loading control and then expressed as a percentage of the same allele (either wtHTT or muHTT) from brain lysates of PBS injected animals on the same membrane. Error bars are SEM. *P < 0.05, **P < 0.01, ***P < 0.001 difference between wt and muHTT by Bonferroni post hoc analysis following two-way analysis of variance. (c) Example immunohistochemistry demonstrating the range of astrogliosis (GFAP reactivity) observed with screened ASOs, as a measure of tolerability. Scale bars are 250 µm for 5× image and 100 µm for 20× image.
Activity of ASOs was assessed by the percentage of muHTT KD as compared to PBS-injected animals (Supplementary Table S1). No reduction of HTT protein was observed with either control ASO; however, a slight induction of both alleles was observed with ASO CTRL1, and thus it is not a good candidate for extended in vivo studies. Between 76 and 85% KD of muHTT protein was observed after treatment with pan-HTT or H-HTT ASOs. Treatment with allele-specific ASOs resulted in muHTT KD between 28 and 94%. Consistent with previous counter screen data,24 S-constrained ethyl (cEt)26-modified ASOs showed enhanced potency compared to 5-9-5 2′-O-methoxyethyl (MOE)27-modified ASOs. Selectivity of ASOs was assessed by the ratio of muHTT KD to wtHTT KD (Supplementary Table S1). Selectivity of pan-HTT and H-HTT ASOs was ~1 as expected, indicating no allelic preference. Selectivity of allele-specific ASOs ranged from a ratio of 1.2 to 6.3, demonstrating preferential KD of muHTT. Tolerability of ASOs was assessed by adverse events at the time of surgery and/or during the 4-week interval before sacrifice (Supplementary Table S1), as well as by qualitative loss of DARPP-32 and NeuN reactivity as a measure of general neuronal health (only observed with ASO F1) or induction of GFAP reactivity as a measure of gliosis (Figure 3c; Supplementary Figure S1 and Supplementary Table S1). GFAP induction was found to be the most sensitive of these measures, and in some cases was the only indicator of reduced tolerability of an ASO. Control ASOs did not induce any adverse effects by these measures. The H-HTT ASO HH1 was the most tolerated nonselective ASO, causing no adverse effects at the time of surgery or over the 4-week interval, and only resulting in very mild gliosis, which is sometimes observed after PBS injection. While no conclusions regarding the long-term tolerability of nonselective HTT reduction can be drawn from these results as the experiments lasted only 4 weeks, this ASO was selected for further study. Allele-specific ASOs that induced adverse effects in any of the tolerability measures were not studied further. ASO D1 was identified from this screen as having moderate activity, excellent selectivity, and excellent tolerability. ASO A3 was identified as having excellent activity, moderate selectivity, and excellent tolerability.
Combinatorial silencing
We next sought to determine if targeting multiple SNPs in the HTT gene could simultaneously induce synergistic rather than additive silencing. Synergistic silencing could potentially increase KD activity while preserving selectivity and limiting adverse effects by reducing the required dose of each independent molecule. For this evaluation, ASOs A1 and D1 were selected because they share common chemistry and design. They are both 5-9-5 MOE gapmers, meaning that the only difference between them is their target sequence. Hu97/18 mice received ICV injections of 150 µg ASO A1 or D1, 300 µg ASO A1 or D1, or a combination of 150 µg each of ASOs A1 and D1. Four weeks later, brains were processed for wt and muHTT levels by immunoblotting (Supplementary Figure S2). The 150 µg doses of ASOs A1 and D1 alone induced 21 and 45% muHTT reduction, respectively. Delivering the combined 150 µg dose of both ASOs together resulted in 56% muHTT reduction. muHTT reduction was found to not be synergistic, and was in fact less than that achieved by delivering the higher dose of the more active molecule, ASO D1, alone (69%). This data suggest that rather than treating a patient simultaneously with multiple ASOs targeted to different SNPs, which could increase potential side effects, it would be most beneficial to treat each patient with the best available single ASO.
Duration of action
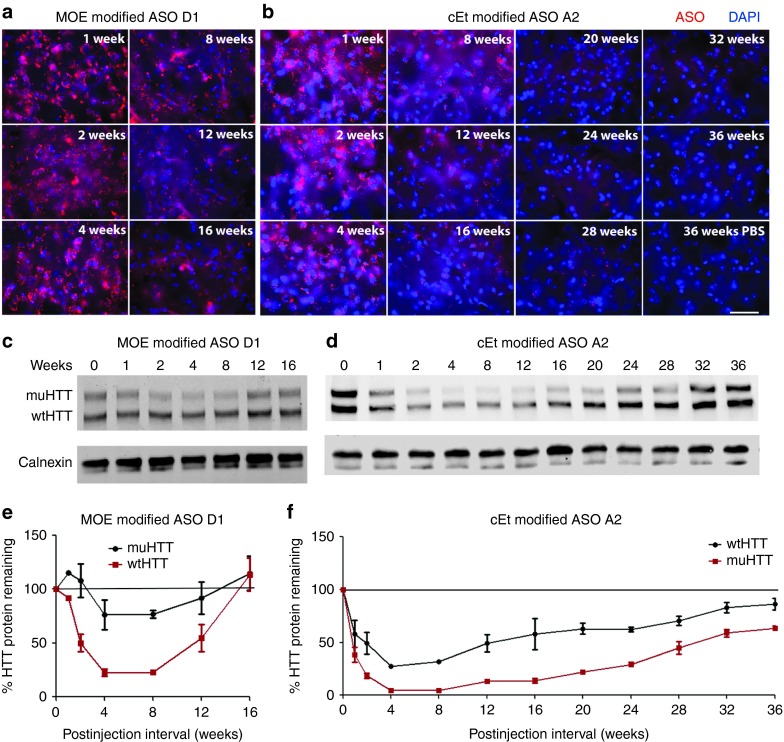
We have previously shown that the incorporation of cEt-modified nucleotides increases the potency of ASO-mediated muHTT KD beyond that of molecules containing only MOE-modified nucleotides.24 To compare the kinetics of ASO-mediated muHTT silencing between MOE-modified and cEt-modified ASOs, 2-month-old Hu97/18 mice were injected ICV with 300 µg of MOE-modified ASO D1, cEt-modified ASO A2, or PBS vehicle. ASOs D1 and A2 were selected for this evaluation because they were, respectively, the most active MOE-modified and cEt-modified molecules identified through primary screening. Mice were sacrificed 1, 2, 4, 8, 12, or 16 weeks later and brains were processed for ASO distribution (Figure 4a,b) and GFAP induction (Supplementary Figure S3a–c) by IHC, wt and muHTT mRNA by RT-qPCR (Supplementary Figure S3d), wt and muHTT protein by immunoblotting (Figure 4c–f), and muHTT protein by time resolved fluorescence resonance energy transfer (TR-FRET) (Supplementary Figure S3e). For both ASOs, maximal silencing was observed 4 weeks postinjection (Figure 4 and Supplementary Figure S3). For MOE-modified ASO D1, muHTT protein returned to approximately basal levels by 16 weeks postinjection (Figure 4c,e and Supplementary Figure S3e), which is consistent with previous reports of duration of MOE ASO-induced HTT lowering.15 However, for cEt-modified ASO A2, which exhibits increased silencing activity compared to MOE-modified ASOs, 75–85% muHTT reduction was observed at 16 weeks postinjection (Figure 4d,f and Supplementary Figure S3d,e). In an effort to measure the complete duration of action of ASO A2, a second group of mice was treated with PBS or ASO A2 and sacrificed 20, 24, 28, 32, or 36 weeks postinjection. Over these time points, muHTT protein slowly increases toward baseline. However, at the longest postsurgical interval evaluated, 36 weeks, 25–35% muHTT reduction was still observed in ASO A2-treated brains (Figure 4d,f). IHC of ASO in the brain at all time points revealed similar levels of ASO for MOE- and cEt-modified oligos (Figure 4a,b), indicating that this increased duration of action for ASO A2 is not the result of increased tissue persistence, but is a function of increased silencing potency. At the longest time point evaluated, 36 weeks postinjection, ASO puncta are still apparent in ASO A2 treated, but not in PBS-treated brain (Figure 4b).
Figure 4.
Duration of antisense oligonucleotides (ASO) action. Hu97/18 mice received a single 300 µg intracerebroventricular (ICV) injection of ASO. At the indicated intervals brains were harvested and assayed for HTT protein and for ASO by immunohistochemistry (IHC). (a,b) IHC from the central portion of the striatum showing (a) MOE modified ASO D1 or (b) cEt modified ASO A2 in red with DAPI nuclear stain in blue. Scale bar is 25 µm. (c,d) Example western blots showing wt and muHTT protein. (e,f) Quantitation of HTT protein in both hemispheres of two animals. Density of HTT bands was normalized to calnexin loading control and then expressed as a percentage of the same allele (either wtHTT or muHTT) from brain lysates of phosphate-buffered saline–injected animals from the same interval and on the same membrane. Errors bars are SEM.
Secondary screen of enhanced selectivity ASOs
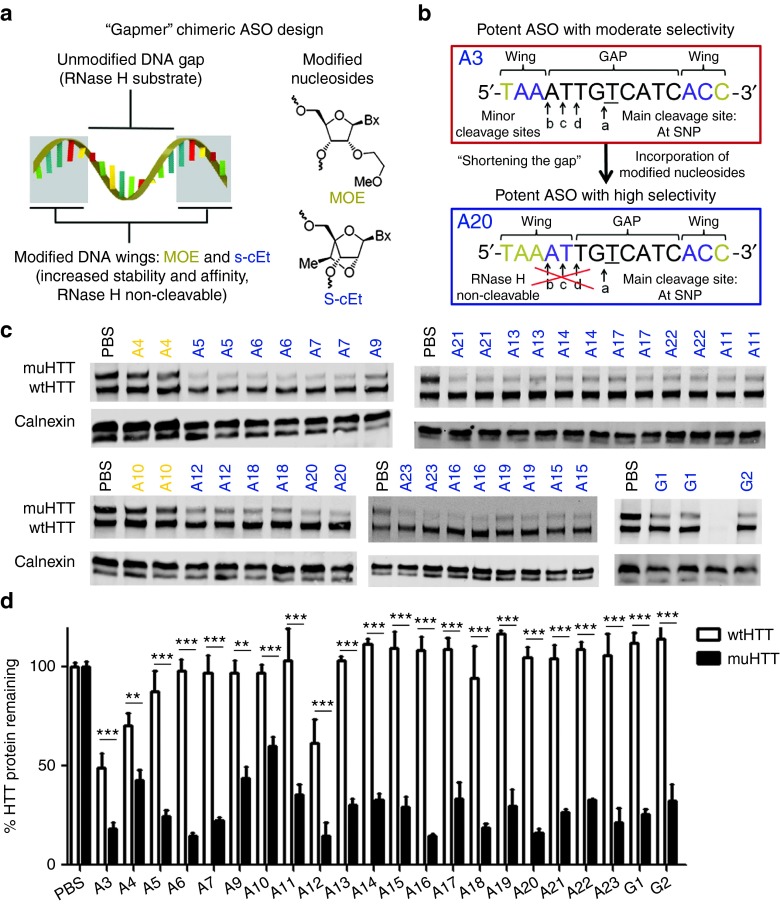
Primary screening identified two well-tolerated lead allele-specific ASOs. ASO A3, which exhibited excellent activity but only moderate selectivity, and ASO D1, which exhibited excellent selectivity but only moderate activity. Based on our hypothesis that enhancing selectivity through chemical and design modification would be more tractable than enhancing activity, ASO A3 was chosen for further structure activity relationship studies. In an effort to improve the selectivity of ASO A3 while maintaining the potency and tolerability, several strategies were employed (Figure 2), including microwalking the sequence around the target SNP, changing the number and position of high-affinity MOE- and cEt-modified nucleotides in the wings (Figure 5a), incorporation of other modifications such as FHNA and methyl phosphonate linkage, and limiting minor RNase H cleavage sites by shortening the gap region (Figure 5b).28 We found that ASOs with a shorter gap region retain the potency of ASO A3 with enhanced selectivity. To expand upon this strategy, gap shortening was then applied to ASOs targeting two other SNPs: rs363088 (D) and rs6446723 (G). Secondary screening of enhanced selectivity ASOs was performed as above using HTT quantitation by immunoblotting (Figure 5c,d). KD of muHTT from 40 to 85% was observed (Supplementary Table S2), and ASOs that induced less than 50% KD were excluded from further study. Selectivity ratios from 1.94 to greater than 85 were observed (Supplementary Table S2), and ASOs that were less than 10-fold selective were excluded from further study. In many cases, KD of wtHTT was not observed, making this ratio impossible to accurately calculate. For these data, 1% KD of wtHTT was used to calculate selectivity. These values are proceeded by a > sign, indicating that the selectivity ratio is greater than the given value. Tolerability was assessed by adverse events at the time of surgery or during the four week interval, as well as by GFAP IHC (Supplementary Table S2 and Supplementary Figure S4). Interestingly, all ASOs from this enhanced selectivity screen that were targeted to rs363088 (ASOs D4, D5, and D6) were toxic. As a result, this SNP target was not pursued further. ASOs targeting the other two SNPs, A or G, that caused adverse time of surgery events or induced anything more than very mild gliosis, which is sometimes seen with PBS injection, were excluded from further study. Eight ASOs, six targeting rs7685686 (ASOs A9, A16, A18, A20, A21, and A23) and two targeting rs6446723 (ASOs G1 and G2), emerged from this secondary screen as lead ASOs having high activity, high selectivity, and good tolerability.
Figure 5.
Secondary screen of enhanced selectivity antisense oligonucleotides (ASO). (a) Typical design of a “gapmer” ASO with high affinity, non-RHaseH cleavable wings surrounding a PS, RNaseH cleavable gap. (b) One strategy for increasing selectivity of a potent ASO by shortening the gap: replacing PS nucleosides at minor cleavage sites with RNAseH noncleavable nucleosides thus restricting cleavage to the main site at the single-nucleotide polymorphism (SNP) of interest.28 Modified ASOs were then screened in Hu97/18 mice. (c) Example western blots showing wt and muHTT protein. ASOs in orange contain only 5-9-5 2′-O-methoxyethyl modifications. ASOs in blue contain cEt modifications. (d) Quantitation of HTT protein in both hemispheres of four animals. Errors bars are SEM. **P < 0.01, ***P < 0.001 difference between wt and muHTT by Bonferroni post hoc analysis following two-way analysis of variance. Data from the primary screen for the parent molecule, A3, are included on the graph to allow direct comparison.
Distribution and regional silencing
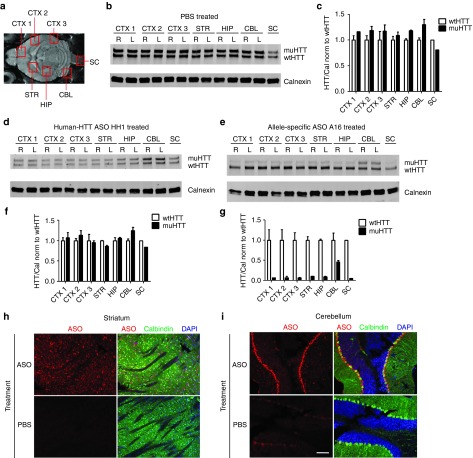
To determine the full CNS distribution of ASOs and the regional effects of HTT silencing, 300 µg of lead ASO A16, the most active and selective lead identified through secondary screening, H-HTT ASO HH1, or PBS vehicle was injected into the right lateral ventricle of 2 month old Hu97/18 mice. Four weeks later, brains were removed and bilaterally dissected into three separate regions of cortex (an anterior portion similar to the region used for assessing HTT protein levels in our screening experiments-CTX1, a medial portion adjacent to the striatum-CTX2, and a posterior portion-CTX3), striatum (STR), hippocampus (HIP), the most posterior portion of cerebellum (CBL), in addition to a piece of spinal cord (SC) (Figure 6a). Each of these regions were processed for wt and muHTT protein levels by immunoblotting and densitometry (Figure 6b–g). PBS-injected mice showed uniform HTT protein levels throughout the brain with lower basal levels in the spinal cord (Figure 6b). In all areas evaluated, wt and muHTT levels were approximately equivalent (Figure 6c). HH1 treatment resulted in reduced levels of both wt and muHTT protein with maintenance of the basal wt to muHTT ratio in all areas of the CNS (Figure 6d,f) indicating nonselective HTT lowering, while ASO A16 induced allele-specific muHTT reduction resulting in increased wt to muHTT ratios throughout the CNS (Figure 6e,g). ASO silencing was found to be least robust in the cerebellum (Figure 6d,e). Because ASO effect was greater in the spinal cord than the cerebellum, which is further from the injection site, the reduced action in the cerebellum is not likely the result of less ASO reaching this tissue. Comparison of ASO distribution by IHC in the striatum (Figure 6h) versus the cerebellum (Figure 6i) revealed that ASOs distribute evenly in the striatum while concentrating in the Purkinje cell layer of the cerebellum, which likely accounts for the reduced activity observed when this tissue is processed as a whole.
Figure 6.
A single intracerebroventricular (ICV) injection of antisense oligonucleotides (ASO) induces HTT silencing throughout the central nervous system (CNS). Hu97/18 mice received a 300 µg ICV injection of ASO or phosphate-buffered saline (PBS) vehicle. Four weeks later, brains were microdissected into the indicated areas and wt and muHTT were quantified by immunoblot. (a) Diagram of brain areas evaluated. (b) Immunoblot of PBS treated brain demonstrating approximately equivalent wt and muHTT levels in all evaluated regions. (c,f,g) Density of HTT bands is normalized first to calnexin and then to wtHTT density of the same lane. Error bars for bilateral structures are SEM between the two hemispheres. (c) quantitation of HTT in PBS-treated brain. (d) Immunoblot of human-HTT ASO HH1 treated brain showing nonselective reduction of HTT throughout the CNS. (e) Immunoblot of allele-specific ASO A16 treated brain showing selective reduction of muHTT protein throughout the CNS. (f,g) Quantitation of wt and muHTT protein in HH1 and A16 treated brains, respectively. (h,i) immunohistochemistry (IHC) of sagitally sectioned ASO A16 or PBS-injected brain stained for ASO in red, calbindin in green, and 4',6-diamidino-2-phenylindole (DAPI) in blue. (h) In the striatum, ASO distributes evenly, while (i) in the cerebellum it is concentrated in the Purkinje cell layer. Scale bar is 100 µm.
Dose response of lead ASOs
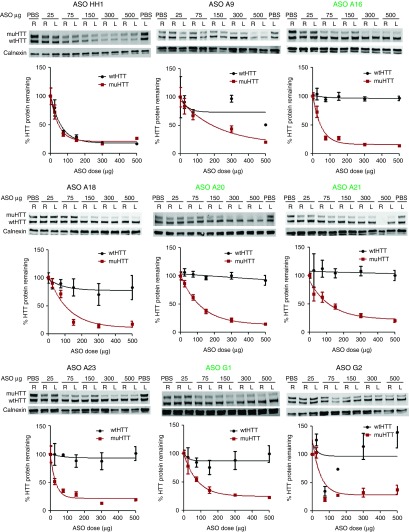
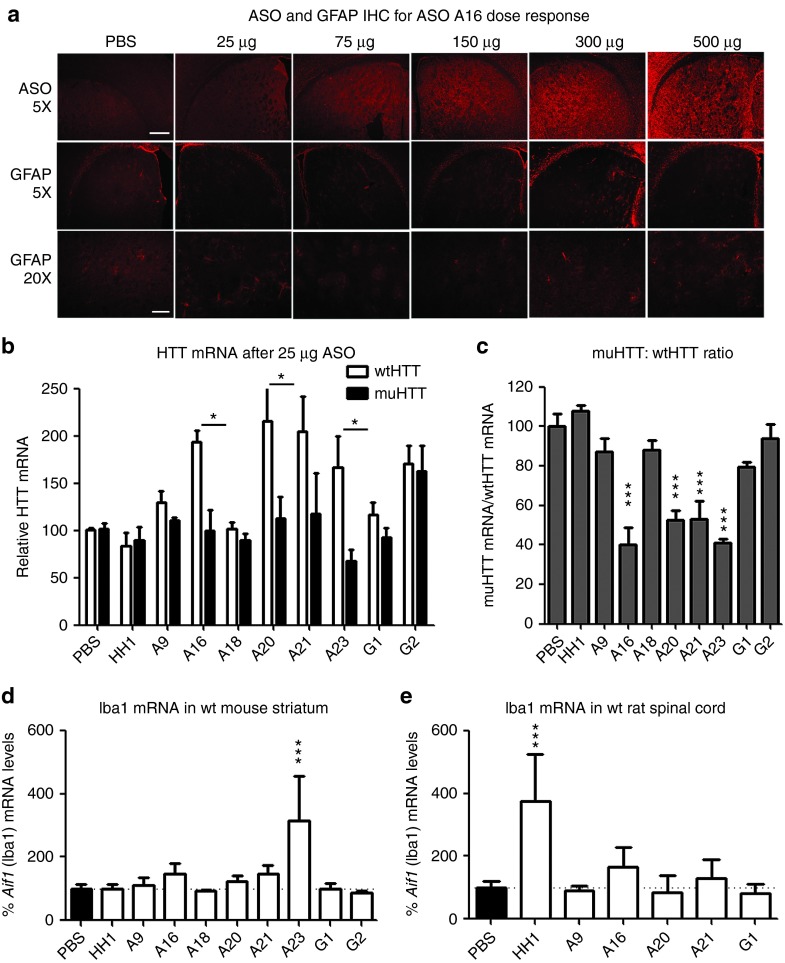
In parallel with the wt tolerability screens, ASOs that passed secondary screening in Hu97/18 mice were also evaluated at several doses to determine optimal therapeutic dosing and to further evaluate candidates (Supplementary Table S3). ASO was delivered to Hu97/18 mice by unilateral ICV injection at 25, 75, 150, 300, or 500 µg to the right lateral ventricle. Four weeks later, brains were collected and processed for wt and muHTT protein levels (Figure 7), ASO distribution, and general CNS tolerability (Figure 8a and Supplementary Figure S5) as above. Additionally, wt and muHTT mRNA was quantified at the lowest dose evaluated, 25 µg (Figure 8b,c) to identify the ASOs with the broadest therapeutic window. ASOs that did not induce sufficient muHTT silencing (<75% max KD, ED50 >100 µg), or induced significant wtHTT silencing at any dose (>10%), were discarded due to lack of potency or selectivity, respectively. Additionally, any ASO that induced adverse events greater than sedation at the time of surgery or very mild gliosis by GFAP IHC were discarded due to lack of tolerability. ASO A9, the only 9-base gap ASO included in the dosing study, was discarded due to low potency (muHTT ED50 216 µg), low selectivity (max wtHTT KD 49%), and poor tolerability (weight loss and gliosis observed at highest dose). ASO G2 was discarded due to lack of potency (max muHTT KD 67%), and ASOs A18 and A23 were discarded due to poor tolerability (Hind limb ataxia and gliosis observed at highest dose). This is consistent with the wild-type animal screens where ASOs A9, A18, A23, and G2 failed due to poor tolerability. Three lead ASOs targeting rs7685686, A16, A20, A21, and one lead ASO targeting rs6446723, G1, met all of our criteria and were determined to be excellent candidate therapeutic ASOs for further preclinical validation.
Figure 7.
Lead antisense oligonucleotides (ASO) potently and selectively lower muHTT at a broad range of doses. ASO was delivered by intracerebroventricular (ICV) injection at the indicated doses and brains were processed as in screening. Quantitation of HTT protein was performed in both hemispheres of four animals. Density of HTT bands was normalized to calnexin loading control and then expressed as a percentage of the same allele (either wtHTT or muHTT) from brain lysates of phosphate-buffered saline–injected animals on the same membrane. Error bars are SEM. Lead ASOs that passed all screening and dosing criteria are in green.
Figure 8.
Lead antisense oligonucleotides (ASO) identified in secondary screen are tolerated at a broad range of doses and efficacious at very low doses. (a–c) ASO was delivered by intracerebroventricular (ICV) injection into Hu97/18 at the indicated doses and brains were processed as in screening. (a) ASO and GFAP immunohistochemistry (IHC) for lead ASO A16 dose response. Induction of gliosis was not observed even at the highest dose. Scale bars are 250 µm for 5× image and 100 µm for 20× image. (b,c) Quantitation of HTT mRNA after ICV delivery of 25 µg of the indicated ASO. (b) relative levels of allelic HTT mRNA in both hemispheres of four animals. *P < 0.05 difference between wt and muHTT by Bonferroni post hoc analysis following two-way analysis of variance (ANOVA). Error bars are SEM. (b) muHTT:wtHTT mRNA ratio. ***P < 0.001 different from phosphate-buffered saline (PBS)–injected animals by Bonferroni post hoc analysis following two-way ANOVA. Error bars are SEM. (d) ASO was delivered to wild-type mice and assessed for tolerability. Aif1 mRNA levels in striatum. PBS compiled from four studies. Error bars are SD (e) ASO was delivered by repeated IT bolus into rats and assessed for tolerability. Aif1 mRNA levels in lumbar spinal cord. PBS compiled from three studies. Error bars are SD. ***P < 0.001 different from PBS-injected animals by Bonferroni post hoc analysis following one-way ANOVA. Data from A20 reported previously.28
Tolerability of leads in wild-type animals
To further investigate the tolerability of the ASOs identified in the secondary screen, we performed assessments in wt mice and rats (Supplementary Table S4). As these ASOs do not target rodent Htt, assessment in wt animals allows for analysis of ASO tolerability without any potential influence from lowering Htt levels. The eight lead ASOs identified through secondary Hu97/18 screening were first taken into an 8-week tolerability screen in wt mice. Mice were assessed for acute tolerability and monitored weekly for 8 weeks for body weight and adverse events. Following the 8-week screen, tissue was collected for quantification of microgliosis by qRT-PCR of Aif1 (Iba1) mRNA (Figure 8d). The data between the Hu97/18 mice and the wt mice were remarkably consistent and five SNP ASOs (A9, A16, A20, A21, and G1) and the nonallele selective ASO (HH1) were sufficiently tolerated to warrant further screening in rats. Three of the ASOs (A18, A23, and G2) that passed the Hu97/18 screen failed the longer more rigorous screen in the wt mice, as one or more of the animals scored positive for an adverse event during their weekly neurological examination. Despite failing the wt screen, these ASOs were sufficiently tolerated to include in subsequent dose response studies to further understand their activity profiles, but are not sufficiently tolerated to be considered lead ASOs.
In human patients, ASOs are delivered to the CSF via lumbar puncture into the intrathecal (IT) space. To mimic this in an animal model, rats were implanted with IT catheters and subcutaneous ports and given 300 µg of ASO every 2 weeks for 8 weeks (total of 1.2 mg). Rats were assessed weekly for 8 weeks and at the end of 8 weeks, tissue was collected for quantification of microgliosis and astrocytosis by qRT-PCR of Iba1 (Figure 8b) or GFAP, respectively. Four of the SNP ASOs were well tolerated (A16, A20, A21, and G1), as they did not cause an acute response at the time of dosing, weight loss or induce microgliosis or astrocytosis. A9 was not sufficiently tolerated, as one of the six animals exhibited neurological dysfunction after the third dose. The nonallele specific ASO (HH1), which does not alter rodent Htt levels, did increase Iba1, a marker for microglia, and is not sufficiently tolerated either (Figure 8e).
Lessons learned for ASO design
Each ASO at each SNP has different tolerability, toxicity and efficacy of KD and, therefore, must be evaluated independently. Through these studies, we have attempted to derive a better understanding of the determinants of these properties, and build some general guidelines that can be employed during ASO design to limit the number of molecules that must be evaluated. We found that ASO activity can vary by SNP target. In previous studies we have found that ASOs targeted to some SNPs do not display any silencing activity.24 In this study, we have found that ASOs that are identical in chemistry and design and only vary by target sequence, such as ASOs A1 and D1, can have different levels of activity. These differences in target site activity may be the result of differences in the tertiary structure of the transcript, aptameric, or protein binding effects, or in the case of intronic SNP targets, the kinetics of transcript splicing. For our lead SNP target, A, we found that SNP discrimination is most effective with the SNP centered in the gap region. For this SNP, moving the SNP toward to 5′ end of the gap resulted in diminished tolerability (Figure 2; Supplementary Table S2 and Supplementary Figure S4), while moving it toward the 3′ end resulted in diminished activity (data not shown). However, this is not expected to be the case for all SNP targets. Incorporation of cEt nucleosides in the wings can increase activity over molecules with entirely MOE modified wings. For SNP target A, ASOs with entirely cEt-modified wings were not as well tolerated as those containing both MOE and cEt nucleosides (Figure 2; Supplementary Table S2 and Supplementary Figure S4). Both total number of cEt modifications and arrangement of MOE and cEt modifications appear to impact activity and tolerability, though we were unable to determine specific guidelines for these effects. Additionally, we found that ASO length affects tolerability; in some cases, removing a single base, such as from ASO A19 (a 16-mer) to ASO A20 (a 15-mer), can change tolerability from poor (induction of hind limb ataxia and gliosis) to excellent (no adverse events). In other cases, removing a single base, such as from ASO A15 (a 17-mer) to ASO A16 (a 16-mer) can change tolerability from excellent (no adverse events) to poor (decreasing survival and inducing gliosis) (Figure 2; Supplementary Table S2 and Supplementary Figure S4). For each of these properties: SNP position, wing modification, and oligo length, we were unable to define governing principles of ASO design and recommend evaluation of multiple molecules to identify optimal ASO candidate drugs. However, we have observed for multiple molecules and SNP targets, that minimizing secondary cleavage sites with shorter gap regions yields more selective molecules without negatively impacting activity or tolerability (Figure 2; Supplementary Table S2 and Supplementary Figure S4). This strategy can be applied to new ASO target sites, facilitating ASO design and shortening the time required for generation of potential ASO drugs for human use.
Discussion
It has been over 30 years since identification of the first DNA marker for HD29 and the development of genetic predictive testing, and over 20 years since discovery of the causative mutation,2 yet there is still no effective treatment available for HD sufferers. One difficulty with development of effective and/or disease-modifying treatments for HD is the complex pathogenesis. The HTT protein is a large, promiscuous protein that associates with many molecular partners and cellular components and plays roles in numerous cellular processes including transcriptional regulation, apoptosis suppression, ER stress signaling, calcium homeostasis, axonal transport, endocytosis, and synaptic transmission.3 Without a central pathologic process to target, small molecule drugs are unlikely to be effective, necessitating development of genetic interventions. Though HD may not result from a simple pathologic process, it does result from a single cause: CAG tract expansion in the HTT gene, which provides an obvious gene therapy target. Reducing expression of the muHTT gene and thus lowering levels of muHTT protein should ameliorate all aspects of HD pathogenesis and prevent or delay onset or slow progression of the disease. There is even evidence from animal studies that lowering muHTT postsymptomatically could result in functional recovery,15,16 rather than just halting progression.
Because the HTT protein plays so many important cellular roles, allele-specific muHTT lowering strategies will likely exhibit greater long-term safety and tolerability than nonselective HTT lowering strategies. Allele-specific muHTT silencing can be achieved either by directly targeting the expanded CAG tract or by targeting genetic elements in linkage with the expanded CAG tract. While targeting the expanded CAG tract is the simplest approach and has the potential for multidisease therapy, it is associated with certain caveats, principally the potential for off-target silencing of normal CAG tract containing genes19 including, but not limited to, the wtHTT gene. Selectivity for muHTT has been demonstrated with CAG tract-targeting ASOs or ss-siRNAs.30,31,32,33,34,35,36 However, these reagents become less selective as the upper and lower CAG tract lengths approach one another.30,31,32,34,36 The mean normal CAG tract length is 17, but high normal (20–26) and intermediate allele (27–35) tract lengths are common as well,37 making it unlikely that a CAG targeting reagent could easily distinguish all of the potential upper and lower allelic combinations.
Alternatively, allele-specific muHTT silencing can be achieved by targeting genetic polymorphisms that are linked to, but distinct from the CAG tract expansion. Toward this end, previous studies have identified SNPs that are enriched in muHTT genes and heterozygous in significant portions of the HD population.20,24 Gene silencing reagents targeting 3–5 HD-associated SNPs with minimally overlapping populations could provide a therapy for >85% of the HD population.20,21 It is important to note that this SNP-targeted approach would require additional patient genotyping to determine which ASO(s) could be used, and that some patients are expected to be homozygous at all target SNPs; unfortunately, these patients would not be candidates for SNP-targeted allele-specific treatment. Because neither the CAG-targeting nor SNP-targeting approaches can provide an allele-specific option for all HD patients, both strategies should be pursued.
We selected ASOs to target HD-associated SNPs for several reasons, including targeting, delivery, and clinical accessibility. ASOs are especially well-suited for SNP targeting in HD because >90% of the SNPs identified in the HTT gene are intronic,38 meaning they cannot be targeted for degradation by the RNAi machinery, which works in the cytosol on mature mRNA. Conversely, RNase H-mediated degradation can take place in the cytosol, or in the nucleus on pre-mRNA,23 greatly increasing the number of potential silencing targets for an ASO-mediated approach. Moreover, ASOs are freely taken up by neurons, glia, and ependymal cells, thus do not require viral or lipid carriers, and distribute throughout the CNS when delivered to the CSF.15,39 Due to the broad CNS distribution, the intrathecal delivery capabilities, and the potential for treatment termination in the case of adverse events, ASOs are currently in development for the treatment of a number of neurodegenerative disorders.38,40 In fact, two phase 1 clinical trials of intrathecally delivered ASOs for the treatment of neurodegenerative diseases have been completed (clinicaltrials.gov identifiers: NCT01041222, NCT01494701), and adverse events related to treatment have not been reported.41 Several similar clinical trials are planned, which will further increase clinical accessibility of CNS ASO drugs in man.
We have previously generated and screened a large number of ASOs targeted to 6 different HD-associated SNPs, each heterozygous in ~50% of the sequenced HD population.24 These SNPs are in high linkage disequilibrium with one another, meaning that they each target the same or similar portions of the HD population and could not be used to develop a panel of complimentary ASO therapeutics. Our strategy is instead to use these SNP targets to develop the best single candidate ASO therapeutic, after which complimentary molecules targeting other SNPs could be developed in subsequent studies. ASOs were first evaluated for HTT mRNA reduction in HD patient derived fibroblast lines homozygous for either the targeted (on-target) or nontargeted (off-target) allele. ASOs that showed sufficient activity (on-target reduction) and selectivity (lack of off-target reduction) were then counter screened for HTT protein lowering in HD mouse primary neuronal cultures. Cultures from either BACHD mice,42 which express human muHTT with HD-associated SNPs, or YAC18 mice,43 which express human wtHTT without HD-associated SNPs, were used to provide independent assessments of potency and selectivity. KD of muHTT between 39 and 68% and negligible wtHTT KD was observed. Finally, one lead ASO was injected intrastriatally into the brains of either BACHD or YAC18 mice resulting in 55% reduction of muHTT and negligible reduction of wtHTT.24
In an effort to provide a more relevant model system for evaluation of ASOs with both the targeted and nontargeted alleles expressed in the same brains and cells, we interbred BACHD and YAC18 mice on the Hdh-/- background to generated a fully humanized mouse model of HD that is genetically similar to HD patients, the Hu97/18 mouse.25 Hu97/18 mice express transgenic full-length human muHTT with 97Q and HD-associated SNPs, transgenic full-length human wtHTT with 18Q without HD-associated SNPs, and lack murine endogenous Htt. Hu97/18 mice develop a progressive HD-like phenotype and allow direct evaluation of human HTT single nucleotide discrimination in the living brain.
In the Hu97/18 mouse model, we began screening the ASOs that had demonstrated potency and selectivity in our previous cultured neuron counter screen: a 5-9-5 MOE gapmer and a 15-mer cEt-modified ASO at each of 6 SNPs (ASOs 1 and 3 at SNP targets A-F), a pan-HTT ASO, PH1, and a control ASO CTRL1. Additionally, we screened novel ASOs including a 17-mer cEt modified ASO at each SNP site (ASOs A2, B2, C2, D2, E2, and F2) as well as two new pan-HTT ASOs, PH2, and PH3, two human HTT specific ASOs, HH1 and HH2, and an additional control ASO, CTRL2. Our primary screen consisted of ICV injection of ASO at a single 300 µg dose followed by assessment of wt and muHTT protein KD, and multiple assessments of tolerability. ICV injection of ASO A1, the one lead ASO previously counter-screened in vivo in BACHD and YAC18 mice, resulted in 58% muHTT KD, a value similar to previous findings.24 However, in Hu97/18 brain, 27% KD of wtHTT was observed. In counter-screened mice, this ASO did not induce any wtHTT KD, indicating that the Hu97/18 system is more sensitive for evaluating SNP discrimination, and highlighting the necessity of using appropriate model systems that contain both the targeted and nontargeted alleles in the same cells. Two lead ASOs emerged from our primary screen as candidates for further study: ASO A3, which demonstrated excellent activity, moderate selectivity, and excellent tolerability, and ASO D1, which demonstrated moderate activity, excellent selectivity, and excellent tolerability. Because we believed it would be more feasible to enhance selectivity than activity, ASO A3 was selected for further structure activity relationship studies.
In an effort to improve the selectivity of ASO A3 while maintaining the activity, many analogs were generated with either the SNP position moved within the gap region, an altered number and position of high affinity MOE- and cEt-modified nucleotides, or a shortened gap region. These A3 analogs were then screened for HTT mRNA suppression in HD patient derived fibroblasts to identify those suited for in vivo characterization (ASOs A20, A21, A22, and A23).28 Secondary screening with these ASOs and novel ASOs designed by similar strategies (ASOs A4-A19) was then performed in Hu97/18 brain. Six active and well-tolerated molecules with enhanced selectivity were identified (ASOs A9, A16, A18, A20, A21, and A23). All but one of these ASOs is a short gap ASO, and the other, ASO A9, has a phosphonate modification within the gap that may act in a similar way to disrupt secondary cleavage sites. Interestingly, ASO A9 was the only one of these six leads targeting SNP A to induce any reduction of wtHTT protein at this dose. Gap shortening was then applied to two other SNP targets, D and G. All of the shorter gap ASOs tested for target D (ASOs D4, D5, and D6) were not well tolerated. The same was true for the cEt modified ASOs targeting SNP D from our primary screen (ASOs D2 and D3), indicating that this SNP sequence may be in some way toxic. Conversely, both of the shorter gap ASOs for target G (ASOs G1 and G2) were reasonably active, very selective, and well-tolerated. While many ASOs against SNP A were generated and screened to identify leads that met our criteria, only these two were required for target G. This acts as validation of our ASO designs and demonstrates that as long as SNP targets are active (i.e., available for ASO binding) and nontoxic (i.e., tolerant of high potency molecules), the design principles we employed can be applied to new targets, greatly shortening the required time to develop safe and efficacious candidate therapeutics. Although there are guiding principles, it is important to keep in mind that each ASO is a distinct molecule, with specific tolerability, efficacy, and dosing properties, and just like other small and large molecules drugs, each must be evaluated independently.
With this in mind, it is important to employ rigorous screening paradigms to ensure that the best candidates are chosen. To this end, our lead compounds were tested in both wt and HD mouse lines, in a repeated dosing rat study, and were assessed independently in two laboratories. The compounds identified here were well tolerated in both the Hu97/18 mice and nontransgenic animals, and were selected because they did not induce any significant changes in any of the endpoints assessed. Taken together, these data consistently affirm the tolerability of the lead compounds and support their consideration for further development.
In addition to screening, we have investigated the drug-like properties of our lead ASOs, including duration of action, distribution, and dosing. First, we compared the duration of action of a moderately potent MOE-modified ASO, D1, and a very potent cEt-modified ASO, A2. Both ASOs reached maximal muHTT lowering at 4 weeks postinjection and were detectable for long periods in the brain as assessed by IHC. By 16 weeks postinjection, muHTT protein levels in the MOE ASO-injected brain had returned to nearly basal levels, consistent with previous reports of ASO duration of action.15 However, at this time point, the more potent ASO still induced greater than 75% muHTT reduction, and muHTT levels had still not returned to basal levels at 36 weeks postinjection. Though we were unable to evaluate the full duration of action of ASO A2 since muHTT had not returned to basal levels after our longest evaluated interval, at greater than 36 weeks, the duration of action of our enhanced ASOs far exceeds the 16-week duration of action previously reported for therapeutic HTT ASOs.15 Combined with previous findings that the period of therapeutic benefit from HTT-lowering far outreaches the actual period of HTT protein reduction,15 these data indicate that treatment with the maximum tolerated ASO dose could prolong duration of action and provide long lasting therapeutic benefit from intermittent treatment. Considering these data, we envision that intrathecal ASO treatments for HD could be performed intermittently or infrequently while still providing a therapeutic benefit.
Next we sought to explore ASO distribution and activity throughout the CNS. We have found that a single unilateral ICV injection of ASO is sufficient to silence muHTT expression in the entire brain and spinal cord of a mouse. Silencing is less robust in the cerebellum due to the focused concentration of ASO in the Purkinje cells, which would be expected to result in maximal HTT lowering in Purkinje cells and minimal HTT lowering in other cell types leading to moderation of effect when cerebellar tissue is homogenized. However, we feel this is acceptable and even potentially beneficial because the cerebellum is relatively spared from HD pathology,44 with the greatest effect being early and dramatic loss of Purkinje cells,45 where silencing and therapeutic benefit is expected to be highest.
Distribution of ASOs delivered into the CSF, though still broad and reaching all areas, is less uniform in larger brains, such as in nonhuman primates, resulting in a gradient with high ASO concentrations in more superficial areas such as cortex, and lower concentrations in deeper structures such as the caudate.15,46 For this reason, it is necessary to identify ASOs with a large therapeutic window of safe and efficacious doses. To investigate this, we performed dose response studies using the six ASOs targeting SNP A and the two ASOs targeting SNP G that passed our secondary screening criteria. Four of these ASOs, ASOs A16, A20, A21, and G1, maintained activity at low doses and selectivity and tolerability at high doses, indicating that they would be efficacious across the concentration gradient that is expected after delivery to a larger brain. Three of these ASOs, ASOs A16, A20, and A21, induced significant suppression of muHTT mRNA and protein after delivery of only 25 µg, an extremely low dose. The allele-selectivity of our leads surpasses that of previously reported therapeutic HTT ASOs. In fact, two of these ASOs, ASOs A21 and G1, did not induce any reduction of wtHTT protein even at the highest dose evaluated, 500 µg. Moreover after high doses in mice and repeated dosing in rats, these ASOs did not induce any adverse events or neuroinflammatory signatures, indicating excellent tolerability.
These four lead ASOs are promising potential HD therapeutics suitable for further pre-clinical validation including evaluation of therapeutic efficacy in Hu97/18 mice and tolerability in nonhuman primates. Contingent on findings from those studies and using delivery and dosing information gained from ongoing CNS ASO clinical trials, a primary SNP-targeted ASO could be fairly rapidly translated for human applications.
Materials and Methods
Mice. Experiments were performed using Hu97/18 HD model mice,25 wt C57Bl6 mice, and Sprague Dawley rats. Animals were maintained under a 12-hour light:12-hour dark cycle in a clean facility and given free access to food and water. Experiments were performed with the approval of the animal care committee at the University of British Columbia and Isis Pharmaceuticals. All studies in Hu97/18 mice were performed at University of British Columbia and tolerability studies in wt mice and rats were conducted independently at Isis.
ASO injection surgery. ASO in a final volume of 10 µl in sterile PBS was delivered to mice by ICV injection to the right lateral ventricle. Mice were anesthetized with isoflurane and placed into a stereotaxic frame. The scalp was prepared and an incision made along the mid-line. The skull was then dried to enhance visibility of sutures and landmarks. A Hamilton syringe with 26 gauge needle (Hamilton, Reno, NV) was used to punch through the skull at 0.3 mm anterior and 1 mm lateral to Bregma. The needle was then lowered to 3 mm below the surface of the skull, and the ASO was injected over 10 seconds. The needle was left in place for 2 minutes and then withdrawn slowly. Tolerability studies in wild-type mice and rats were performed as described previously.28
Tissue collection and processing. Hu97/18 mice were anesthetized with an overdose of Avertin and brains removed. Brains were sectioned using a 1 mm coronal brain matrix (ASI Instruments, Warren, MI). A section containing the first 2 mm, predominately olfactory bulb, was discarded. A section containing the second 2 mm was divided into right and left hemispheres and snap frozen in liquid nitrogen for protein isolation. For animals used for dose and duration studies, an additional 1 mm coronal section was divided into right and left hemispheres and snap frozen in liquid nitrogen for RNA isolation. The remaining, posterior portion of the brain was postfixed overnight in 4% paraformaldehyde, cryoprotected in 30% sucrose, and cut by cryostat into 25 µm free-floating coronal sections for IHC. For regional ASO distribution, whole brains were sectioned into 25 µm free-floating sagital sections for IHC. Wild-type mice and rats in tolerability screens were euthanized with CO2, cortex, striatum, and lumbar spinal cord was dissected and immediately frozen on dry ice and stored at −80 °C until processing.
mRNA quantitation. Tissue was lysed in guanidinium thiocyanate containing 8% β-mercaptoethanol. RNA was isolated using the RNeasy96 Kit (Qiagen, Germantown, MD). Allelic HTT qPCR was performed as in ref. [28]. HTT mRNA levels were first normalized to total RNA levels using Ribogreen, and then to mean mRNA levels of the same allele from PBS-injected animals. The muHTT:wtHTT mRNA ratio was calculated as an assessment of allele-specificity. For quantification of Iba1 (marker for microgliosis) and GFAP (marker for astrocytosis) qPCR was performed as described previously.28
HTT protein quantitation. Each 2 mm coronal hemisphere was lysed and 40 µg of total protein was resolved on 10% low-BIS acrylamide gels (200:1 acrylamide:BIS) as in ref. [24]. Proteins were transferred to 0.45 µmol/l nitrocellulose membranes that were blotted for HTT (MAB2166, 1:2,000, Millipore, Billerica, MA) and calnexin (1:10,000, Sigma C4731, St Louis, MO). Proteins were detected with IR dye 800CW goat anti-mouse (1:250, Rockland 610-131-007, Gilbertsville, PA) and AlexaFluor 680 goat anti-rabbit (1:250, Molecular Probes A21076, Eugene, OR)-labeled secondary antibodies and the LiCor Odyssey Infrared Imaging system. Mutant and wtHTT intensities were normalized to calnexin loading control and then to the average level of HTT of the same allele from PBS-injected animals.
For the quantitation of muHTT protein by FRET, 10 µg of total protein was mixed with 0.2 ng/ml terbium (FRET donor)-labeled BKP1 anti-HTT antibody47 and 2 ng/ml of D2 (FRET acceptor)-labeled MW1 antiexpanded CAG antibody,48 in a white 384-well plate. After excitation at 340 nm, FRET was measured as the ratio of 655 nm (D2)/615 nm (TB) emission. FRET signal was normalized to the mean signal from PBS-injected animals of the same postinjection interval.
Immunohistochemistry. Sections were blocked for 30 minutes at room temperature (RT) in 3% bovine serum albumin, 10% normal goat serum, and 0.1% Triton X-100 in PBS. Primary antibodies: rabbit anti-ASO (1:7,500),24 rabbit anti-GFAP (1:1,000 Dako, Carpinteria, CA), mouse anti-NeuN (1:1,000, Millipore), rat anti-dopamine- and cyclic AMP-regulated phosphoprotein (DARPP-32) (1:1,000, R&D Systems, Minneapolis, MN), or mouse-anti calbindin (1:200, Abcam, Cambridge, UK) were diluted in blocking solution and incubated on sections overnight at 4 °C. Secondary Alexa-fluro 488-conjugated goat anti-mouse or Alexa-fluor 568 conjugated goat anti-rabbit antibodies (1:250, Invitrogen, Carlsbad, CA) were incubated on sections for 1.5 hours at RT. Sections were then mounted on slides with ProLong Gold antifade reagent with DAPI (Invitrogen). Sections were photographed using a Zeiss Axioplan 2 microscope and Coolsnap HQ Digital CCD camera (Photometrics, Tucson, AZ).
SUPPLEMENTARY MATERIAL Figure S1. IHC analysis from primary screen. Figure S2. Treating with multiple ASOs does not yield synergistic silencing. Figure S3. Duration of action of MOE and cEt modified ASOs Figure S4. IHC assessment of secondary screen. Figure S5. IHC assessment of dose response. Table S1. Summary of primary screen. Table S2. Summary of secondary screen. Table S3. Summary of dose response. Table S4. Summary of tolerability assessment in wildtype animals.
Acknowledgments
The authors thank Mahsa Amirabassi and Mark Wang for excellent animal care; Gene Hung and Curt Mazar for assistance optimizing ASO delivery techniques; and Simon Warby, Chris Kay, Shaun Sanders, and Mahmoud Pouladi for discussion and support. This work was supported by grants from The Canadian Institutes of Health Research (CIHR: MOP-84438) and ISIS Pharmaceuticals and postdoctoral fellowships for A.L.S. and N.H.S. from CIHR and for A.L.S. from The Huntington Society of Canada and The Michael Smith Foundation for health Research. M.R.H. is a Killam University Professor and holds a Canada Research Chair. M.O., H.K., A.T.W, S.M.F, E.S., P.S., and C.F.B. are employees of, with a financial interest in, ISIS Pharmaceuticals. ISIS Pharmaceuticals synthesized and provided the ASOs used in this study, and supported work in the laboratory of M.R.H.
Supplementary Material
References
- Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- Videnovic A. Treatment of Huntington disease. Current Treatment Options in Neurology. 2013. pp. 1–15. [DOI] [PMC free article] [PubMed]
- Träger U, Andre R, Lahiri N, Magnusson-Lind A, Weiss A, Grueninger S, McKinnon C, et al. HTT-lowering reverses Huntington's disease immune dysfunction caused by NFκB pathway dysregulation. Brain. 2014;137 Pt 3:819–833. doi: 10.1093/brain/awt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Franich NR, Fitzsimons HL, Fong DM, Klugmann M, During MJ, Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington's disease. Mol Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB. Sustained therapeutic reversal of Huntington's disease by transient repression of Huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, et al. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin R, Kaytor MD, Ai Y, Nelson PT, Thakker DR, Heisel J, Weatherspoon MR, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135 Pt 4:1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland SL, Devon RS, Huang Y, Mead CL, Meynert AM, Neal SJ, Lee SS, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, Collins JA, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Gillis T, Mysore JS, Ramos EM, Myers RH, Hayden MR, Morrison PJ, et al. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet. 2012;90:434–444. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, Warby SC, Carroll JB, Doty CN, Skotte NH, Zhang W, Villanueva EB, et al. A fully humanized transgenic mouse model of Huntington disease. Hum Mol Genet. 2013;22:18–34. doi: 10.1093/hmg/dds397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, et al. Short antisense oligonucleotides with novel 2'-4' conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J Med Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, et al. Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA. Nat Struct Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- Oestergaard ME. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, Watkins PC, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers MM, Pepers BA, van Deutekom JC, Mulders SA, den Dunnen JT, Aartsma-Rus A, van Ommen GJ, et al. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS One. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Yu D, Chu Y, Corey DR. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res. 2012;40:11270–11280. doi: 10.1093/nar/gks907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszer A, Olejniczak M, Galka-Marciniak P, Mykowska A, Krzyzosiak WJ. Self-duplexing CUG repeats selectively inhibit mutant huntingtin expression. Nucleic Acids Res. 2013;41:10426–10437. doi: 10.1093/nar/gkt825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pendergraff H, Narayanannair KJ, Lackey JG, Kuchimanchi S, Rajeev KG, Manoharan M, et al. RNA duplexes with abasic substitutions are potent and allele-selective inhibitors of huntingtin and ataxin-3 expression. Nucleic Acids Res. 2013;41:8788–8801. doi: 10.1093/nar/gkt594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaka A, Kay C, Doty CN, Collins JA, Tam N, Hayden MR. High frequency of intermediate alleles on Huntington disease-associated haplotypes in British Columbia's general population. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:864–871. doi: 10.1002/ajmg.b.32193. [DOI] [PubMed] [Google Scholar]
- Southwell AL, Skotte NH, Bennett CF, Hayden MR. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Shi F, Gounko NV, Wang X, Ronken E, Hoekstra D. In situ entry of oligonucleotides into brain cells can occur through a nucleic acid channel. Oligonucleotides. 2007;17:122–133. doi: 10.1089/oli.2007.0034. [DOI] [PubMed] [Google Scholar]
- DeVos SL, Miller TM. Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics. 2013;10:486–497. doi: 10.1007/s13311-013-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH, Tao J, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, et al. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Jeste DV, Barban L, Parisi J. Reduced Purkinje cell density in Huntington's disease. Exp Neurol. 1984;85:78–86. doi: 10.1016/0014-4886(84)90162-6. [DOI] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, Graham RK, Loubser O, et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain Res Bull. 2001;56:319–329. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.