Abstract
Enzalutamide is a potent second-generation androgen receptor (AR) antagonist with activity in metastatic castrate-resistant prostate cancer (CRPC). Although enzalutamide is initially effective, disease progression inevitably ensues with the emergence of resistance. Intratumoral hypoxia is also associated with CRPC progression and treatment resistance. Given that both AR and hypoxia inducible factor-1 α (HIF-1α) are key regulators of these processes, dual targeting of both signaling axes represents an attractive therapeutic approach. Crosstalk of the AR and HIF-1α signaling pathways were examined in prostate cancer cell lines (LNCaP, 22Rv1) with assays measuring the effect of androgen and hypoxia on AR-dependent and hypoxia-inducible gene transcription, protein expression, cell proliferation, and apoptosis. HIF-1α inhibition was achieved by siRNA silencing HIF-1α or via chetomin, a disruptor of HIF-1α-p300 interactions. In prostate cancer cells, the gene expression of AR targets (KLK3, FKBP5, TMPRSS2) was repressed by HIF-signaling; conversely, specific HIF-1α target expression was induced by dihydrotestosterone-mediated AR signaling. Treatment of CRPC cells with enzalutamide or HIF-1α inhibition attenuated AR-regulated and HIF-1α–mediated gene transcription. The combination of enzalutamide and HIF-1α inhibition was more effective than either treatment alone. Similarly, the combination also reduced vascular endothelial growth factor protein levels. HIF-1α siRNA synergistically enhanced the inhibitory effect of enzalutamide on cell growth in LNCaP and enzalutamide-resistant 22Rv1 cells via increased enzalutamide-induced apoptosis. In conclusion, the combination of enzalutamide with HIF-1α inhibition resulted in synergistic inhibition of AR-dependent and gene-specific HIF-dependent expression and prostate cancer cell growth.
Introduction
The androgen receptor (AR), a member of the nuclear hormone receptor superfamily of proteins, is the transcription factor responsible for mediating the effects of androgens on target tissues. AR plays a critical role in the development and progression of prostate cancer. Mechanisms that underlie the transition from hormone-dependent prostate cancer to that of castration resistance involve regulation at the AR level (Chen et al., 2004; Debes and Tindall, 2004). Despite androgen deprivation therapy, low levels of circulating androgens persist (Pienta and Bradley, 2006) and the AR can become transactivated in this low-androgen environment through various pathways and genetic aberrations involving molecular alterations to the receptor that lead to castration resistance (Scher and Sawyers, 2005).
Therapy targeting persistent AR-mediated signaling led to the development of enzalatumide (MDV3100), a small-molecule, pure AR antagonist, second generation antiandrogen that improves upon the effects of current first generation antiandrogens (e.g., bicalutamide). The drug was selected as a result of its activity in metastatic castrate-resistant prostate cancer (CRPC) models of AR overexpression, in which it showed selective, potent affinity for AR while being devoid of any agonist AR activity compared with bicalutamide (Tran et al., 2009). Preclinical studies have shown that enzalutamide effectively inhibits AR nuclear translocation and DNA binding to androgen response elements, induces apoptotic effects on prostate cancer tumors, and prevents coactivator recruitment. Although enzalutamide has provided benefit to many, not all patients respond to therapy, and among those who do, the duration of response is often limited with the emergence of resistance (Nelson and Yegnasubramanian, 2013).
Androgen deprivation as a result of castration results in hypoxia in prostate cancer cells (Shabsigh et al., 2001; Halin et al., 2007) and subsequently enhances the transcriptional activity of AR. Thus, understanding the molecular mechanisms of aberrant AR signaling under hypoxic conditions is important. Intratumoral hypoxia is emerging as a common feature of prostate cancers that are associated with poor prognosis (Stewart et al., 2010). In prostate cancer progression, tumor cells acquire the ability to adapt to hypoxic environments by co-opting blood vessel formation while also migrating and invading toward vessels. The transcription factor hypoxia-inducible factor (HIF)-1α mediates key hypoxia-associated genes involved in angiogenesis, metabolism, survival, immunity, and invasion (reviewed in Semenza, 2010, 2012; Greer et al., 2012).
The results of several studies (Mabjeesh et al., 2003; Horii et al., 2007; Mitani et al., 2011; Park et al., 2012) suggest that there is crosstalk between the AR and HIF-1α pathways that may converge on AR, HIF-1α, and β-catenin, forming a ternary complex on androgen response elements of AR target genes (Mitani et al., 2012). Thus HIF-1α might be involved in AR-mediated gene expression in prostate cancer and implicated in tumor growth and progression. Given that both AR and HIF-1α are key regulators of multiple cellular processes in prostate cancer, dual targeting of both axes represents an attractive therapeutic approach. The goal of our study was to evaluate the combination treatment of the novel AR antagonist enzalutamide with HIF inhibition in preclinical models of prostate cancer and to define the molecular mechanism by which HIF-1α inhibition potentiates anti-AR therapy in CRPC.
Materials and Methods
Cell Culture.
LNCaP and 22Rv1 (AR-positive human prostate cancer) cells (American Type Culture Collection, Manassas, VA) were maintained in phenol red–free RPMI 1640 medium supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 mg/ml streptomycin unless otherwise indicated. For hypoxic conditions, cells were treated with hypoxia mimetic cobalt chloride (150 μM CoCl2) (Sigma-Aldrich, St. Louis, MO).
Antibodies and Reagents.
Monoclonal HIF-1α antibody was purchased from BD Biosciences (San Diego, CA), and monoclonal Actin (C-2) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 680 goat anti-mouse IgG was obtained from Molecular Probes (Eugene, OR). Odyssey blocking buffer was from LI-COR (Lincoln, NE). Dihydrotestosterone (DHT) was purchased from Steraloids, Inc. (Wilton, NH), Chetomin was purchased from Sigma-Aldrich, and enzalutamide was purchased from Selleck Chemicals (Houston, TX). The ARE3-TK-luc plasmid was kindly provided by Dr. Nima Sharifi (Sharifi et al., 2008). The pRL-TK control and HRE-luc (pGL4.42) plasmids were obtained from Promega (Fitchburg, WI).
siRNA Transfection.
Specific knockdown was achieved using siRNAs against HIF-1α (Qiagen, Valencia, CA) or Allstars negative control siRNA (Qiagen). 22Rv1 or LNCaP cells were transiently transfected with control or HIF-1α siRNA at 20 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions for 24 or 48 hours.
Western Blot Analysis.
Cells were lysed with 400 μl ice-cold lysis buffer [2.5% (v/v) Tris pH 8, 3% (v/v) 5 M NaCl, 0.4% (v/v) 0.5 M EDTA, 1% (v/v) Triton, and complete protease inhibitors]. After 30 minutes of incubation on ice, cell lysates were centrifuged at 7500 rpm for 10 minutes. Supernatants were collected and total protein concentration was determined using the BCA assay (Thermo Scientific, Waltham, MA) according to the manufacturer's protocol. Cell lysates were subjected to SDS-PAGE and analyzed by Western blotting with anti–HIF-1α monoclonal antibody or anti-actin monoclonal antibody. Primary antibody was immunoreacted with fluorophore-conjugated goat anti-mouse IgG. Bound antibodies were visualized, and densitometry was completed via the Odyssey Infrared Imaging System and Odyssey software (LI-COR).
Cell Viability Assays.
LNCaP cells were seeded in 96-well plates in 100 μl 10% fetal bovine serum supplemented phenol red–free RPMI 1640 medium. After overnight incubation at 37°C, cells were transiently transfected with siRNA. Forty-eight hours after siRNA transfection, cells were treated with charcoal-stripped phenol red–free RPMI 1640 containing 150 μM CoCl2 and 1 nM DHT with or without 1 μM enzalutamide (day 0). Cell viability was measured on days 0, 1, 2, 3, and 4 using the Cell Counting Kit-8 cell viability assay according to the manufacturer’s instructions (Dojindo, Rockville, MD), and absorbance was read at 450 nm using a SpectraMax M2 fluorescence plate reader (Molecular Devices, Sunnyvale, CA).
Semiquantitative Real-Time Polymerase Chain Reaction.
LNCaP cells were plated at a density of 400,000 cells per well in a 6-well dish in phenol red–free RPMI 1640 charcoal-stripped media for 48 hours and then treated for 18 hours with combinations of DHT, chetomin, and enzalutamide in normoxia or hypoxia (150 µM CoCl2). Treatments were completed in duplicate, and total RNA extraction was performed using the QIAshredder and RNAeasy mini kit (Qiagen) according to the manufacturer’s protocol. RNA concentration was determined using a NanoDrop spectrophotometer (Molecular Devices). Purified RNA (0.24–0.32 μg) from LNCaP cells was reverse transcribed per 30 μl cDNA synthesis reaction using The Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s protocol.
Two microliters of cDNA reaction products (30 μl total) were amplified with forward and reverse primers: VEGFA (Hs00900055_m1, Applied Biosystems, Foster City, CA), ENO1 (Hs00361415_m1, Applied Biosystems), LDHA (Hs00855332_g1, Applied Biosystems), KLK3 (Hs02576345_m1, Applied Biosystems), FKBP5 (Hs01561006_m1, Applied Biosystems), TMPRSS2 (Hs01120965_m1, Applied Biosystems), HIF-1α (Hs00153153_m1, Applied Biosystems), and ACTB (Hs99999903_m1, Applied Biosystems). For each sample, 2 µl cDNA was mixed with 18 µl reaction mixture containing 1 µl forward and reverse primers, 7 µl water, and 10 µl Taqman Gene Expression Master Mix (Applied Biosystems) for a total volume of 20 µl. Semiquantitative real-time polymerase chain reaction (qPCR) was performed using an Applied Biosystems StepOnePlus Real-Time PCR system with StepOne Software. All qPCR reactions were run in triplicate. β-Actin (ACTB) was used as a reference housekeeping gene. Fold-change in RNA levels was calculated using the ΔΔCt method (SABiosciences 2009 RT2 Profiler PCR Array System User Manual; SABiosciences, Frederick, MD).
Enzyme-Linked Immunosorbent Assay.
LNCaP cells were plated at a density of 10,000 cells per well in a 96-well dish in phenol red–free RPMI 1640 charcoal stripped media for 48 hours at 37°C and then treated for 18 hours at 37°C with combinations of DHT, chetomin, enzalutamide, and hypoxic conditions (treatment with 150 µM CoCl2). Conditioned media were collected and assayed with a vascular endothelial growth factor (VEGF) enzyme-linked immunosorbent assay (ELISA; R&D systems, Minneapolis, MN).
Luciferase Reporter Assay.
LNCaP cells were grown in 96-well plates in phenol red–free RPMI 1640 with 10% fetal bovine serum, and cells were transiently transfected with siRNA. After 48 hours of siRNA treatment, cells were transiently transfected with reporter plasmids using Lipofectamine 2000 according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were then treated in phenol red–free, charcoal-stripped media containing combinations of DHT and enzalutamide under normoxia or hypoxia (150 µM CoCl2) for 18 hours. Transfection efficiency was normalized using pRL-TK (control reporter vector). Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase measurements were taken on a GloMax Multi+ Microplate reader (Promega) and normalized using the Renilla luciferase measurements. All measurements were done in quadruplicate.
Statistical Considerations.
The Student’s t test was used for comparisons between treatment and expression of gene transcripts, luciferase reporter assays, and ELISAs. Linear regression was used to test for the relationship between luciferase induction and increasing concentrations of HIF-1α inhibitors or DHT. One-way analysis of variance followed by Dunnett’s multiple comparisons test was performed to test for differences in treatments with percent cell count versus time. All comparisons were conducted using GraphPad Prism software (GraphPad Prism version 6.00, GraphPad Software, La Jolla CA). Significance was assigned to any observation in which P ≤ 0.05.
Results
DHT- and Cobalt Chloride-Induced Transcription of AR and HIF-1α Target Genes.
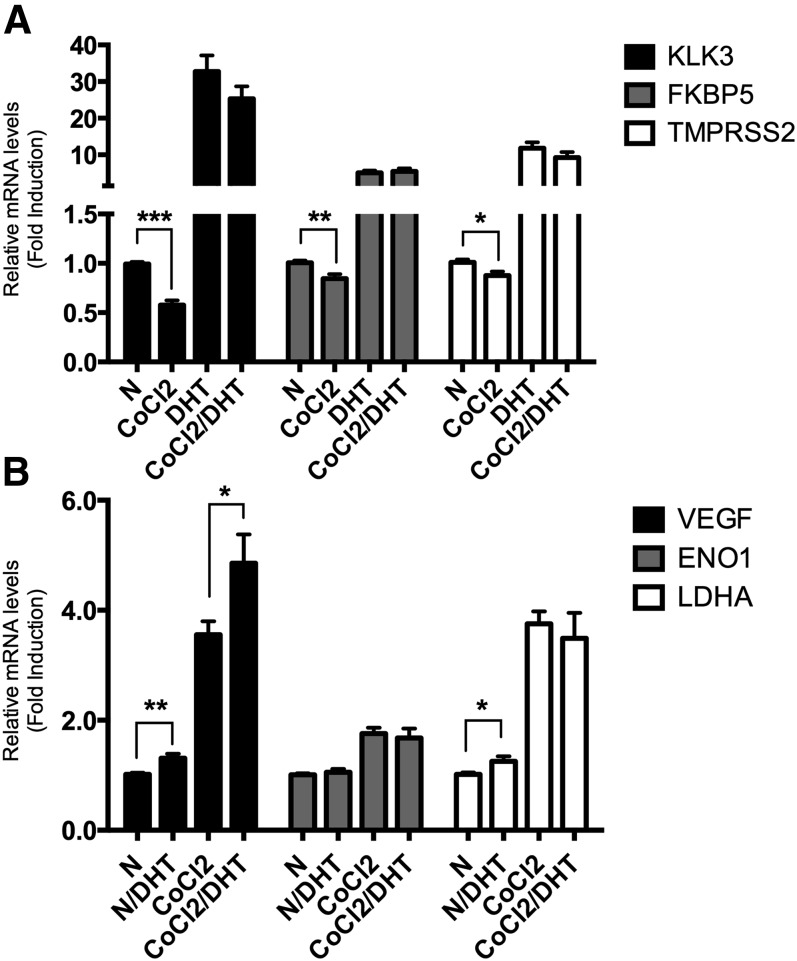
To delineate the crosstalk between HIF-1α and AR pathways, prostate cancer hormone-sensitive LNCaP cells were treated with 1 nM dihydrotestosterone (DHT) and 150 μM of the hypoxia mimetic cobalt chloride (CoCl2) and assessed for the expression of androgen-responsive genes (KLK3, FKBP5, TMPRSS2) and hypoxia-induced genes (VEGF, ENO1, LDHA). Although DHT increased the expression of the AR-regulated genes by ≥5.1-fold (P < 0.0001; Fig. 1A), CoCl2 treatment alone reduced expression of all AR targets by ≤0.875-fold (P ≤ 0.011). The addition of CoCl2 did not change DHT-induced transcription (FKPB5 and TMPRSS2, P ≥ 0.19). CoCl2 treatment increased the transcription of three HIF-1α targets by ≥1.8-fold (P < 0.0001; Fig. 1B), whereas DHT treatment alone under normoxia caused increased transcription of two HIF-1α targets, VEGF and LDHA, by ≥1.3-fold (P ≤ 0.03); ENO1 expression was not affected (P = 0.55). The addition of DHT either increased CoCl2-induced transcription (VEGF 3.6-fold versus 4.9-fold over untreated control; P = 0.021) or did not change gene expression (ENO1 and LDHA; P ≥ 0.60). Therefore, gene expression of all studied AR targets was repressed by hypoxia; conversely, specific HIF-1α target genes were induced by DHT-mediated AR signaling. This suggests that the crosstalk between the two pathways is complex and gene specific; simultaneous treatment with both DHT and CoCl2 typically minimized the effects of such crosstalk on transcription, producing no synergistic effect.
Fig. 1.
Effect of DHT and CoCl2 on AR-dependent and hypoxia-inducible gene transcription. (A) LNCaP cells were cultured in the presence or absence of DHT (1 nM) under normoxic (N) or hypoxic (150 μM CoCl2) conditions for 18 hours. Total RNA was extracted and qPCR analyses were performed for AR-dependent target genes (KLK3, FKBP5, TMPRSS2) (A); hypoxia-dependent target genes (VEGF, ENO1, LDHA) and β-actin mRNA expression (B). Relative mRNA levels of each target gene expression are normalized by β-actin expression, and the results are indicated as fold change from those in the absence of DHT in normoxia. The result is representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Repression of AR-Responsive Gene Transcription by Enzalutamide and HIF-1α Inhibition.
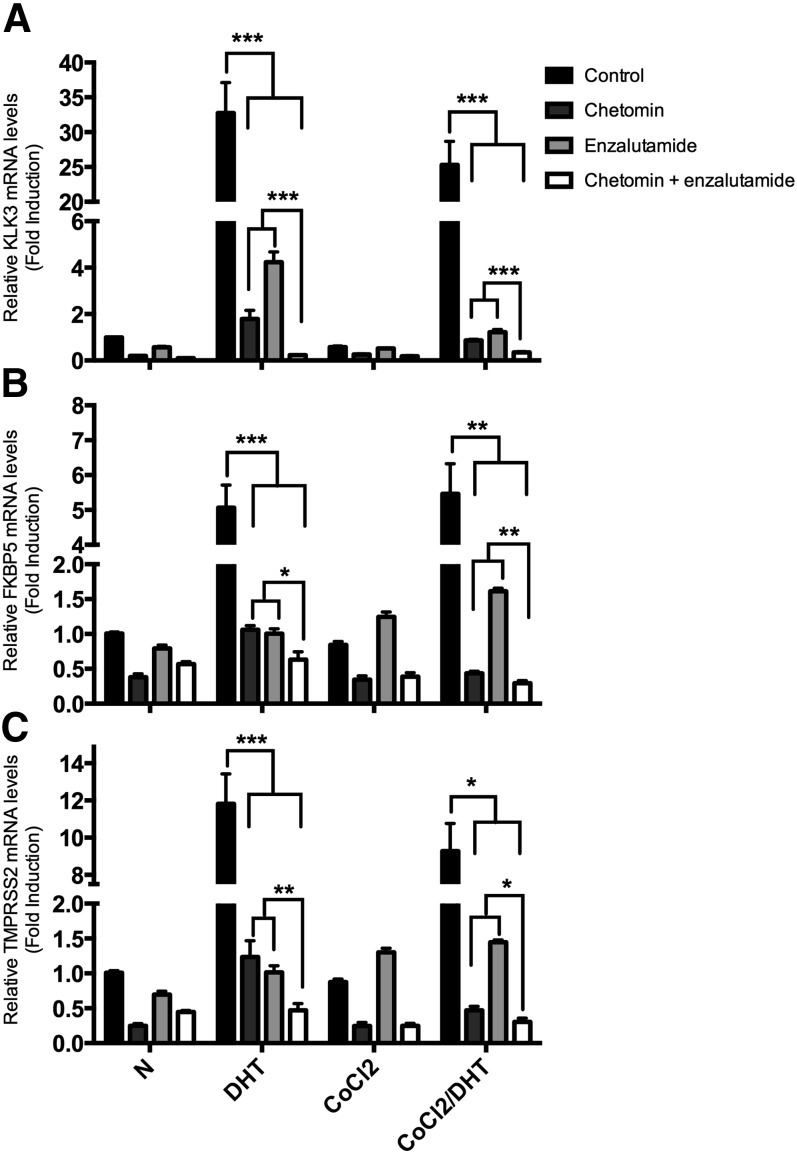
We next ascertained whether repression of AR signaling was potentiated by repression of HIF-1α signaling. LNCaP cells were exposed to the AR inhibitor enzalutamide in the presence or absence of chetomin, a known HIF-1α inhibitor that disrupts the binding of p300 to both HIF-1α and HIF-2α (Kung et al., 2004). Without CoCl2 stimulation, chetomin or enzalutamide mono-treatment similarly repressed transcription of FKBP5 (5.1-fold versus 1.1- and 1.0-fold, respectively) and TMPRSS2 (11.8-fold versus 1.6- and 1.0-fold, respectively) in the presence of DHT (see Fig. 2). Chetomin was actually a better inhibitor of KLK3 transcription than enzalutamide (38.2-fold versus 1.8- and 4.2-fold, respectively; P < 0.001). Cotreatment with chetomin and enzalutamide reduced transcription of all AR target genes to an even greater extent than either chetomin or enzalutamide alone (P ≤ 0.012). During simultaneous DHT and CoCl2 treatment, chetomin repressed the transcription of all AR targets to an even greater extent than enzalutamide (P ≤ 0.0080), and cotreatment with chetomin and enzalutamide repressed transcription even further (P ≤ 0.047). Similar results were observed in another AR-positive prostate cancer 22Rv1 cell line (see Supplemental Fig. 1).
Fig. 2.
Effect of the AR antagonist enzalutamide and the HIF-1α inhibitor chetomin on AR-dependent gene transcription. LNCaP cells were cultured in the presence or absence of DHT (1 nM) under normoxic (N) or hypoxic (150 μM CoCl2) conditions for 18 hours. Cells were treated with 1 μM enzalutamide or 100 nM chetomin. Total RNA was extracted and qPCR analyses were performed for AR-dependent target genes (KLK3, FKBP5, TMPRSS2) and β-actin mRNA expression. Relative mRNA levels of each target gene expression are normalized by β-actin expression, and the results are indicated as fold change from those in the absence of DHT in normoxia. The result is representative of three independent experiments: KLK3 (A), FKBP5 (B), TMPRSS2 (C). *P < 0.05; **P < 0.01; ***P < 0.001.
Although chetomin treatment resulted in suppression of AR target gene expression in LNCaPs, it counterintuitively caused a greater dose-dependent increase in AR transcriptional activity (as determined by androgen-responsive luciferase reporter assay) than even DHT (see Supplemental Fig. 2, A and B). Shikonin, another HIF-1α inhibitor, also increased DHT-induced AR transactivation, albeit to a lesser extent in the concentrations tested (Supplemental Fig. 2C). A natural product-based screen using a HIF-1α-p300 assay developed in our laboratory led to the identification of shikonin being able to reduce the tight interaction between a HIF-1α fragment and the CH1 domain of p300 (unpublished observations). This effect is likely due to the pleiotropic effects of chetomin and shikonin, which inhibit transcriptional complexes containing the rather promiscuous p300.
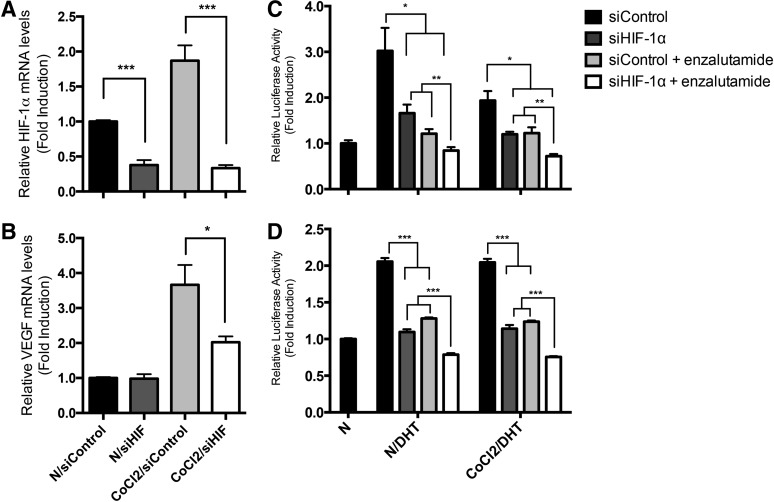
Therefore, we tested the hypothesis that specific HIF-1α knockdown potentiates the AR-repressive effect of enzalutamide. LNCaP cells transfected with HIF-1α siRNA specifically knocked down the mRNA expression of both HIF-1α (1.0- versus 0.38-fold and 1.9- versus 0.22-fold; P ≤ 0.001; Fig. 3A) and VEGFA (during CoCl2 treatment, 3.6- versus 2.0-fold; P = 0.019; Fig. 3B) as well as other transcriptional targets of HIF-1α (ENO1, LDHA; see Supplemental Fig. 3), indicating the specificity and effectiveness of the HIF-1α siRNA tested. We next assessed the effect of HIF-1α siRNA on DHT- and CoCl2-enhanced AR transactivation in the presence of enzalutamide. As expected, enzalutamide decreased AR transactivation (P ≤ 0.0071; Fig. 3C). HIF-1α siRNA had similar effects as chetomin in reducing DHT- and CoCl2-enhanced AR transactivation (P ≤ 0.034) and had a synergistic AR-repressive effect in the presence of enzalutamide (P ≤ 0.010; Fig. 3C). Similar observations were noted in 22Rv1 cells with the combination of HIF-1α siRNA and enzalutamide being more effective in reducing AR transactivation than either treatment alone (P < 0.001; Fig. 3D). The similarity between the results presented in Figs. 2 and 3 suggest that chetomin’s HIF/p300 repressive effect on specific gene expression in prostate cancer cells supersedes its ability to promote androgen response element–mediated transactivation on the global transcription machinery.
Fig. 3.
Effects of HIF siRNA and enzalutamide on AR transcriptional activation. LNCaP cells were treated with 20 nM HIF-1α siRNA (siHIF-1α) or control siRNA (siControl) for 48 hours. Cells were incubated in normoxia (N) or CoCl2 for an additional 18 hours. HIF-1α siRNA repressed HIF (A) and VEGF (B) transcripts. (C) After siRNA treatment, cells were transfected with pARE-Luc and pRL-TK for 24 hours followed by treatment with enzalutamide in the presence and absence of 1 nM DHT in normoxia or hypoxia for an additional 18 hours. Luciferase activities were determined, and data are expressed as means ± S.E.M. (n = 3). HIF-1α siRNA and enzalutamide repressed ARE luciferase activation (P ≤ 0.034), whereas combined HIF-1α siRNA and enzalutamide treatment was more effective than either treatment alone (P ≤ 0.010). (D) HIF siRNA and enzalutamide repressed ARE-luc activation in 22Rv1 cells (P < 0.001), and enzalutamide treatment was more effective than either treatment alone (P < 0.001). *P < 0.05; **P ≤ 0.01; ***P < 0.001.
Repression of Hypoxia-Mediated Gene Expression by Enzalutamide and HIF-1α Inhibition.
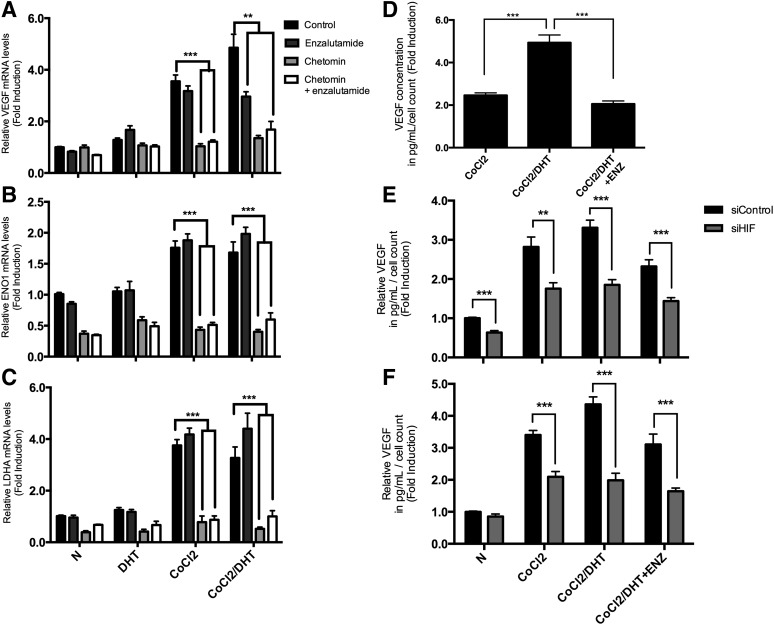
We next determined the effect of AR and HIF-1α inhibition on hypoxia-induced gene expression. LNCaP cells were cultured in the presence or absence of DHT (1 nM) under normoxic or hypoxic (150 μM CoCl2) conditions for 18 hours and treated with 1 μM enzalutamide or 100 nM chetomin or both drugs followed by qPCR analysis. Under CoCl2 treatment, chetomin significantly reduced all HIF-1α targets to near or below basal levels (P < 0.001) (Fig. 4, A–C). During CoCl2 and DHT cotreatment, enzalutamide decreased VEGF transcription (3.0- versus 4.9-fold; P < 0.001; Fig. 4A), whereas other HIF targets were not affected (P > 0.25; see Fig. 4, B and C). We further measured VEGF at the protein level in the conditioned medium by ELISA. Enzalutamide treatment resulted in reduced VEGF release in the medium, and the inhibitory effect was synergistic with HIF-1α silencing using HIF-1α siRNA (P < 0.0001; Fig. 4, D and E). Similar results were obtained in the 22Rv1 prostate cancer cell line (Fig. 4F).
Fig. 4.
Effect of enzalutamide and HIF inhibition on hypoxia-induced gene expression. (A–C) LNCaP cells were cultured in the presence or absence of DHT (1 nM) under normoxic (N) or hypoxic (150 μM CoCl2) conditions for 18 hours. Cells were treated with 1 μM enzalutamide or 100 nM chetomin. Total RNA was extracted and mRNA levels were analyzed by qPCR for hypoxia-dependent target genes (VEGF, ENO1, LDH1) and β-actin as an internal standard. Relative mRNA levels of each target gene expression are normalized by β-actin expression, and the results are indicated as fold change from those in the absence of DHT in normoxia. The result is representative of three independent experiments: VEGF (A); ENO1 (B); LDHA (C). (D–F) Enzalutamide inhibits VEGF secretion. (D) LNCaP cells treated with 1 μM enzalutamide in the presence or absence of 1 nM DHT under normoxic or hypoxic conditions for 18 hours. Conditioned media were collected and VEGF protein levels were measured by ELISA. The concentrations of VEGF (picogram per milliliter) per the total number of cells in each well were calculated, and results are expressed as the fold induction of VEGF secretion compared with untreated controls. Each point represents the mean ± S.E.M. of three different determinations each performed in triplicate. (E) LNCaP cells were treated with 20 nM HIF-1α siRNA (siHIF) or control siRNA (siControl) for 48 hours followed by treatment as above. (F) 22Rv1 cells were treated with 20 nM HIF-1α siRNA or control siRNA for 24 hours followed by treatment as above. **P ≤ 0.01; ***P < 0.001.
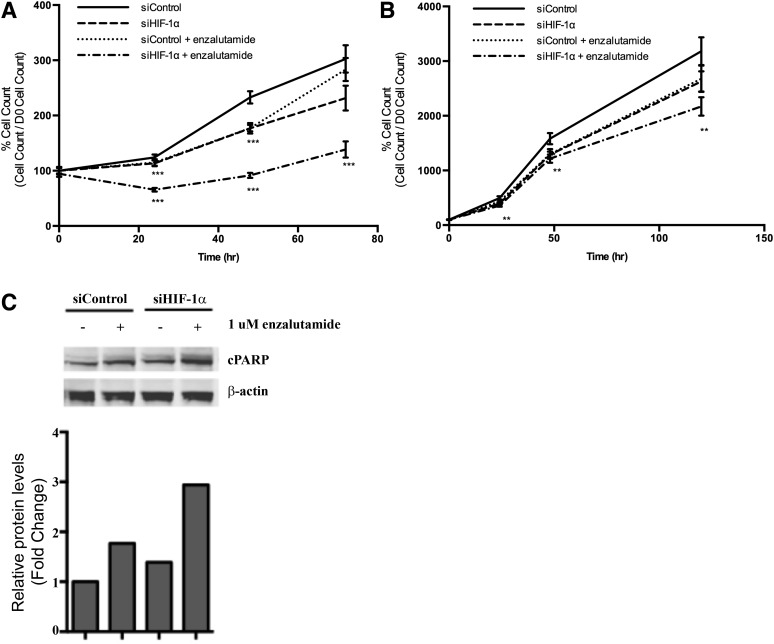
HIF-1α Knockdown Synergistically Enhances Enzalutamide Activity.
Given the significant crosstalk between the HIF and AR pathways, we determined the biologic implications of this interaction by assessing the effects of AR inhibition and HIF-1α inhibition on the growth of LNCaP cells during CoCl2 and DHT treatment. As shown in Fig. 5A, enzalutamide inhibited LNCaP cell growth in cells transfected with scrambled siRNA. Similarly, HIF-1α siRNA also reduced cell growth when treated with CoCl2. Simultaneous treatment with HIF-1α siRNA synergistically enhanced the inhibitory effect of enzalutamide on LNCaP cell viability. We evaluated the effect of HIF-1α inhibition on the enzalutamide-resistant 22Rv1 prostate cancer cell line and found that the treatment combination was also effective at reducing cell growth of the enzalutamide-resistant 22Rv1 prostate cancer cell line, albeit at a delayed response (48 hours) compared with LNCaPs (24 hours) (Fig. 5B). In addition, the combination approach also resulted in HIF-1α inhibition, synergistically enhancing enzalutamide-induced apoptosis, as shown by cleaved poly (ADP-ribose) polymerase (Fig. 5C).
Fig. 5.
Effect of HIF-1α siRNA and enzalutamide on LNCaP cell growth over 72 hours (A) and 22Rv1 cell growth over 120 hours (B) (**P ≤ 0.01; ***P < 0.001). Cells were treated with HIF-1α siRNA (siHIF-1α) or control siRNA (siControl) for 48 hours followed by 1 μM enzalutamide in the presence of 150 μM CoCl2 and 1 nM DHT. Cell viability was determined using the Cell Counting Kit-8 cell viability assay at the indicated time points. (C) Effect of HIF-1α siRNA and enzalutamide on cleavage of poly (ADP) ribose polymerase (PARP). Cells were treated as above, and whole cell lysates were analyzed by Western blot analysis.
Discussion
Prostate cancer is largely driven by aberrant androgen signaling and tumor hypoxia. Thus, inhibiting hypoxia may be key in increasing the efficacy of AR-targeted therapy. Here, we proposed that inhibiting HIF-1α potentiates enzulatamide treatment, thereby demonstrating that simultaneously targeting both HIF-1α and AR pathways is an effective treatment option for CRPC.
Given the conflicting literature reports concerning AR activity under hypoxic conditions in different experimental systems, we first examined the crosstalk between HIF-1α and AR pathways by evaluating the effect of androgen and hypoxia on multiple AR- and HIF-1α targets. We found that hypoxia repressed all AR target gene expression; this is consistent with findings from Ghafar et al. (2003), which showed that hypoxia-response (up to 24 hours) acutely decreased expression of prostate-specific antigen protein levels. Conversely, specific HIF-1α target genes were induced by liganded AR activation. Previous studies have found that hypoxia activated AR only in the presence of androgen (Horii et al., 2007; Park et al., 2012). Others showed unliganded AR transactivation in cells exposed to hypoxia (Khandrika et al., 2009) or hypoxia only activated the AR at low androgen concentrations mimicking the castration-resistant stage (Mitani et al., 2011), involving AR, HIF-1α, and β-catenin forming a ternary complex on AREs (Mitani et al., 2012). Similarly, the effect of AR activation on HIF-mediated transcription was either induction (Mabjeesh et al., 2003; Horii et al., 2007) or no synergistic effect (Park et al., 2012). Our findings in the current study suggest that the crosstalk between the two pathways is complexly regulated in a gene-specific manner; however, the simultaneous treatment with both DHT and CoCl2 typically minimized the effects of such crosstalk on gene transcription.
We determined the effect of enzalutamide and HIF-1α inhibition on mediating AR-responsive and hypoxia-induced gene expression. We found that HIF-1α repression (using either chetomin or HIF-1α siRNA) potentiated the AR-repressive effect of enzalutamide; however, the combination of enzalutamide and HIF-1α inhibition resulted in specific repression of hypoxia-induced VEGF expression. Simultaneous treatment with HIF-1α siRNA synergistically enhanced the inhibitory effect of enzalutamide on LNCaP cell viability through increased apoptosis. Surprisingly, this treatment combination was also effective at reducing the growth rate of the enzalutamide-resistant 22Rv1 prostate cancer cell line, suggesting that HIF-1α inhibition can restore sensitivity to enzalutamide-resistant cells. Li et al. (2013) demonstrated that C-terminally truncated, constitutively active AR splice variants (AR-Versus) lacking the ligand binding domain limit the efficacy of enzalutamide, because only knockdown of AR-Versus, not of full-length AR, was able to restore the ability of enzalutamide to inhibit cell proliferation of AR/AR-V coexpressing 22Rv1 prostate cancer cells. Exploration of novel avenues to potentiate anti-AR therapy is needed to overcome the effects of AR/AR-V signaling or enzalutamide resistance. Potential pathways include inhibition of cellular signaling networks based on targeting compensatory survival pathways associated with relief of feedback inhibition. Indeed recent studies have proposed that combined targeting of the PI3K/AKT and AR pathways (Carver et al., 2011; Toren et al., 2014) suggest a possible approach to overcome enzalutamide resistance, providing support for further evaluation in the clinic.
In the present study, we provide a strong rationale for the combined targeting of the HIF-1α and AR pathways. Enzalutamide’s effect on HIF-1α–regulated gene expression is specifically targeted to VEGF, thus making VEGF a potential biomarker for assessing enzalutamide response; studies are underway to evaluate this effect in clinical treatment samples. Our results are consistent with a gene expression profiling study in LNCaP cells that indicate enzalutamide opposing agonist-induced changes in genes involved in processes such as angiogenesis (Guerrero et al., 2013). Moreover, a recent clinical study found that nonspecific HIF-1α inhibitors (digoxin, metformin, angiotensin-2 receptor blockers) appear to increase progression-free survival and reduce the risk of developing CRPC and metastases in patients on continuous androgen deprivation therapy (Ranasinghe et al., 2014). Whether combining nonspecific HIF-1α inhibitors with existing second generation antiandrogens might produce an added survival benefit remains to be investigated. Our laboratory is involved in developing several specific small molecule HIF-1α inhibitors, and ongoing preclinical studies evaluating the combination of these HIF-1α inhibitors with enzalutamide are currently underway. Taken together, dual targeting of the AR and HIF-1 axis represents an attractive approach for CRPC treatment with HIF-1α inhibition as a possible mechanism for overcoming enzalutamide resistance and potentiating anti-AR therapy.
Supplementary Material
Abbreviations
- AR
androgen receptor
- CRPC
castrate-resistant prostate cancer
- DHT
dihydrotestosterone
- ELISA
enzyme-linked immunosorbent assay
- HIF-1α
hypoxia-inducible factor-1 α
- qPCR
semiquantitative real-time polymerase chain reaction
- VEGF
vascular endothelial growth factor
Authorship Contributions
Participated in research design: Fernandez, Reece, Troutman, Sissung, Price, Chau, Figg.
Conducted experiments: Fernandez, Reece, Ley, Troutman.
Performed data analysis: Fernandez, Reece, Ley, Troutman, Sissung, Price, Chau, Figg.
Wrote or contributed to the writing of the manuscript: Fernandez, Reece, Ley, Troutman, Sissung, Price, Chau, Figg.
Footnotes
This research was supported in part by the Intramural Research Program of the National Institutes of Health National Cancer Institute, Center for Cancer Research.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
References
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. (2011) Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. (2004) Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39. [DOI] [PubMed] [Google Scholar]
- Debes JD, Tindall DJ. (2004) Mechanisms of androgen-refractory prostate cancer. N Engl J Med 351:1488–1490. [DOI] [PubMed] [Google Scholar]
- Ghafar MA, Anastasiadis AG, Chen MW, Burchardt M, Olsson LE, Xie H, Benson MC, Buttyan R. (2003) Acute hypoxia increases the aggressive characteristics and survival properties of prostate cancer cells. Prostate 54:58–67. [DOI] [PubMed] [Google Scholar]
- Greer SN, Metcalf JL, Wang Y, Ohh M. (2012) The updated biology of hypoxia-inducible factor. EMBO J 31:2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero J, Alfaro IE, Gómez F, Protter AA, Bernales S. (2013) Enzalutamide, an androgen receptor signaling inhibitor, induces tumor regression in a mouse model of castration-resistant prostate cancer. Prostate 73:1291–1305. [DOI] [PubMed] [Google Scholar]
- Halin S, Hammarsten P, Wikström P, Bergh A. (2007) Androgen-insensitive prostate cancer cells transiently respond to castration treatment when growing in an androgen-dependent prostate environment. Prostate 67:370–377. [DOI] [PubMed] [Google Scholar]
- Horii K, Suzuki Y, Kondo Y, Akimoto M, Nishimura T, Yamabe Y, Sakaue M, Sano T, Kitagawa T, Himeno S, et al. (2007) Androgen-dependent gene expression of prostate-specific antigen is enhanced synergistically by hypoxia in human prostate cancer cells. Mol Cancer Res 5:383–391. [DOI] [PubMed] [Google Scholar]
- Khandrika L, Lieberman R, Koul S, Kumar B, Maroni P, Chandhoke R, Meacham RB, Koul HK. (2009) Hypoxia-associated p38 mitogen-activated protein kinase-mediated androgen receptor activation and increased HIF-1alpha levels contribute to emergence of an aggressive phenotype in prostate cancer. Oncogene 28:1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, et al. (2004) Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 6:33–43. [DOI] [PubMed] [Google Scholar]
- Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. (2013) Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 73:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabjeesh NJ, Willard MT, Frederickson CE, Zhong H, Simons JW. (2003) Androgens stimulate hypoxia-inducible factor 1 activation via autocrine loop of tyrosine kinase receptor/phosphatidylinositol 3′-kinase/protein kinase B in prostate cancer cells. Clin Cancer Res 9:2416–2425. [PubMed] [Google Scholar]
- Mitani T, Harada N, Nakano Y, Inui H, Yamaji R. (2012) Coordinated action of hypoxia-inducible factor-1α and β-catenin in androgen receptor signaling. J Biol Chem 287:33594–33606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani T, Yamaji R, Higashimura Y, Harada N, Nakano Y, Inui H. (2011) Hypoxia enhances transcriptional activity of androgen receptor through hypoxia-inducible factor-1α in a low androgen environment. J Steroid Biochem Mol Biol 123:58–64. [DOI] [PubMed] [Google Scholar]
- Nelson WG, Yegnasubramanian S. (2013) Resistance emerges to second-generation antiandrogens in prostate cancer. Cancer Discov 3:971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Kim Y, Shim M, Lee Y. (2012) Hypoxia enhances ligand-occupied androgen receptor activity. Biochem Biophys Res Commun 418:319–323. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Bradley D. (2006) Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res 12:1665–1671. [DOI] [PubMed] [Google Scholar]
- Ranasinghe WK, Sengupta S, Williams S, Chang M, Shulkes A, Bolton DM, Baldwin G, Patel O. (2014) The effects of nonspecific HIF1α inhibitors on development of castrate resistance and metastases in prostate cancer. Cancer Med 3:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Sawyers CL. (2005) Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol 23:8253–8261. [DOI] [PubMed] [Google Scholar]
- Semenza GL. (2010) Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabsigh A, Ghafar MA, de la Taille A, Burchardt M, Kaplan SA, Anastasiadis AG, Buttyan R. (2001) Biomarker analysis demonstrates a hypoxic environment in the castrated rat ventral prostate gland. J Cell Biochem 81:437–444. [PubMed] [Google Scholar]
- Sharifi N, Hurt EM, Thomas SB, Farrar WL. (2008) Effects of manganese superoxide dismutase silencing on androgen receptor function and gene regulation: implications for castration-resistant prostate cancer. Clin Cancer Res 14:6073–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GD, Ross JA, McLaren DB, Parker CC, Habib FK, Riddick AC. (2010) The relevance of a hypoxic tumour microenvironment in prostate cancer. BJU Int 105:8–13. [DOI] [PubMed] [Google Scholar]
- Toren P, Kim S, Cordonnier T, Crafter C, Davies BR, Fazli L, Gleave ME, Zoubeidi A. (2014) Combination AZD5363 with Enzalutamide Significantly Delays Enzalutamide-resistant Prostate Cancer in Preclinical Models. Eur Urol, in press. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. (2009) Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324:787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.