Abstract
NR4A family orphan nuclear receptors are an important class of transcription factors for development and homeostasis of dopaminergic neurons that also inhibit expression of inflammatory genes in glial cells. The identification of NR4A2 (Nurr1) as a suppressor of nuclear factor κB (NF-κB)–related neuroinflammatory genes in microglia and astrocytes suggests that this receptor could be a target for pharmacologic intervention in neurologic disease, but compounds that promote this activity are lacking. Selected diindolylmethane compounds (C-DIMs) have been shown to activate or inactivate nuclear receptors, including Nurr1, in cancer cells and also suppress astrocyte inflammatory signaling in vitro. Based upon these data, we postulated that C-DIM12 [1,1-bis(3′-indolyl)-1-(p-chlorophenyl) methane] would suppress inflammatory signaling in microglia by a Nurr1-dependent mechanism. C-DIM12 inhibited lipopolysaccharide (LPS)–induced expression of NF-κB–regulated genes in BV-2 microglia including nitric oxide synthase (NOS2), interleukin-6 (IL-6), and chemokine (C-C motif) ligand 2 (CCL2), and the effects were attenuated by Nurr1-RNA interference. Additionally, C-DIM12 decreased NF-κB activation in NF-κB–GFP (green fluorescent protein) reporter cells and enhanced nuclear translocation of Nurr1 primary microglia. Chromatin immunoprecipitation assays indicated that C-DIM12 decreased lipopolysaccharide-induced p65 binding to the NOS2 promoter and concurrently enhanced binding of Nurr1 to the p65-binding site. Consistent with these findings, C-DIM12 also stabilized binding of the Corepressor for Repressor Element 1 Silencing Transcription Factor (CoREST) and the Nuclear Receptor Corepressor 2 (NCOR2). Collectively, these data identify C-DIM12 as a modulator of Nurr1 activity that results in inhibition of NF-κB–dependent gene expression in glial cells by stabilizing nuclear corepressor proteins, which reduces binding of p65 to inflammatory gene promoters.
Introduction
The signaling pathways regulating expression of inflammatory genes in glial cells have been intensely scrutinized for potential therapeutic targets in Parkinson’s disease (PD) and related neurodegenerative disorders. Among the signaling factors regulating inflammatory gene expression, the nuclear factor-κB (NF-κB) pathway is of considerable interest because it coordinately regulates expression of many proinflammatory genes in glia as well as genes responsible for homeostatic resolution of inflammatory responses (Maguire-Zeiss and Federoff, 2010; Tansey and Goldberg, 2010). Despite increasing attention on the role of NF-κB in glial inflammatory signaling, there are no approved that drugs that successfully target this pathway in patients or in modifying the course of PD or related diseases (Glass et al., 2010; Maguire-Zeiss and Federoff, 2010). Recent studies have demonstrated that cell-specific deletion of NF-κB from microglia or astrocytes is protective in models of inflammatory neurodegeneration (Brambilla et al., 2005; Cho et al., 2008). The results of these knockout studies suggest that selective inhibition of NF-κB could be clinically useful, but current pharmacologic approaches that globally inhibit NF-κB are likely to have undesirable side effects that outweigh any potential neuroprotective benefits due to the importance of this pathway in cellular function and homeostasis (Ghosh et al., 1998; Gilmore, 2006).
Within the central nervous system, there is a high level of regulatory control over inflammatory gene expression in microglia to mitigate neuronal injury during responses to stress and infection (Polazzi and Monti, 2010). One possible approach to selectively inhibit NF-κB signaling in microglia without disrupting its homeostatic regulatory functions is to directly target proteins that modulate p50/p65 transcription factors. Such an approach was suggested in studies reporting that transcriptional regulation of NF-κB–inducible genes in astrocytes and microglia is modulated by NR4A2/Nurr1, a member of the nerve growth factor 1-B family of nuclear receptors (Saijo et al., 2009). In addition to transactivation effects, Nurr1 can also negatively regulate gene expression via protein-protein interactions with transcription factors bound to inflammatory promoters via a mechanism termed “transrepression” (Bensinger and Tontonoz, 2009). Nurr1 is necessary both for development and homeostasis of dopaminergic neurons and has been identified as a genetic mutation involved in familial, late-onset PD (Saucedo-Cardenas et al., 1998; Chu et al., 2006). The discovery that Nurr1 also functions as a transrepressor of NF-κB signaling in glial cells through the active removal of p50/p65 from proinflammatory promoters suggests that it could be a possible therapeutic target for limiting glial inflammation (Saijo et al., 2009).
In the present study we evaluated one of a novel series of para-phenyl substituted diindolylmethane compounds (C-DIMs) shown to be transcriptional modulators of NR4A family nuclear receptors (Inamoto et al., 2008; Lee et al., 2011; Yoon et al., 2011). Selected C-DIMs, such as para-phenyl diindolylmethane (C-DIM12) [1,1-bis(3′-indolyl)-1-(p-chlorophenyl) methane], activates Nurr1 in urothelial carcinoma cells (Inamoto et al., 2008) and in pancreatic cells (Li et al., 2012). We previously demonstrated that C-DIM4 [1,1-bis(3′-indolyl)-1-(p-methoxyphenyl) methane] suppressed NF-κB activation and inhibited inflammatory gene expression in primary astrocytes, leading to a decrease of expressed nitric oxide synthase (NOS2) and tumor necrosis factor α (TNFα) (Carbone et al., 2009). Based on these observations, C-DIM12 was selected for in vivo efficacy studies in a mouse model of PD using subacute dosing with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in combination with probenecid (MPTPp) to induce progressive loss of dopaminergic neurons in the substantia nigra (De Miranda et al., 2013). When delivered orally after 1 week of dosing with MPTPp, C-DIM12 protected against loss of dopamine neurons in the substantia nigra as well as dopamine terminals in the striatum.
It remains unclear whether C-DIM12 can directly inhibit inflammatory gene transcription in microglial cells, despite the evidence that it can reduce dopamine neuron loss in vivo. We therefore postulated that C-DIM12 inhibits inflammatory signaling in BV-2 microglia cells by enhancing Nurr1-dependent transrepression of NF-κB at proinflammatory gene promoters. We present data here that C-DIM12 is able to suppress proinflammatory gene transcription in BV-2 microglia after stimulation with lipopolysaccharide (LPS). Additionally, we investigated whether the transcriptional inhibitory activity of C-DIM12 requires expression of Nurr1 and how modulation of Nurr1 by C-DIM12 affects the activity of nuclear corepressor proteins at p65 DNA-binding sites.
Materials and Methods
Reagents.
C-DIM compounds were synthesized by Dr. Stephen Safe and characterized as described by Qin et al. (2004). LPS from Escherichia coli 0111:b4, NF-κB inhibitor Bay 11-7082 (Bay-11) [(E)-3-(4-methylphenyl)sulfonylprop-2-enenitrile], and all general chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless stated elsewhere. TNFα was purchased from R&D Systems (Minneapolis, MN). The antibodies for p65, Nurr1, and CD11b used in immunofluorescence experiments were purchased from Cell Signaling Technology (Danvers, MA), Santa Cruz Biotechnology (Dallas, TX), and BD Biosciences (San Jose, CA), respectively.
Cell Culture Experiments.
BV-2 microglia were obtained as a generous gift from Dr. Alan Schenkel, Professor of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, Colorado. BV-2 cells were maintained at or below passage 10, cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with l-glutamine (GIBCO/Life Technologies, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA) and a cocktail of 0.001 mg/ml penicillin, 0.002 mg/ml streptomycin, 0.001 mg/ml neomycin; (1× PSN; HyClone Laboratories, Logan, UT). BV-2 cells were grown until 60 to 70% confluent before treatments. Primary mixed glia were obtained through cortical isolations from C57/Bl6 mouse pups (P0, unknown genders). Mixed glial cultures were maintained in minimum Eagle’s medium (supplemented with l-glutamine; Hyclone), 10% FBS, and 1× PSN as described previously. Primary glial cultures were grown until 80% confluent before treatments. Glial purification by our laboratory was performed as described in Carbone et al. (2009). All cell cultures were maintained at 37°C and 5% CO2. All procedures involving animals were approved by the Colorado State University Institutional Animal Care and Use Committee and were conducted in accordance with current National Institutes of Health guidelines.
Gene Knockdown Assays.
RNA interference (siRNA, small interfering RNA) sequences were obtained through Integrated DNA Technologies (IDT DNA, Coralville, IA) and Ambion (Life Technologies, Grand Island, NY). Nurr1 RNAi duplexes were designed against splice common variants of the target gene, and were validated using a dose-response assay with increasing concentrations of the suspended oligo (150–450 ng/μl) using a standard scrambled dicer-substrate RNA (DsiRNA) as control. RNAi oligos were transfected using the TransIT-X2 delivery system (Mirus Bio, Madison, WI) 24 hours before C-DIM12 or LPS treatment. Separate siRNA systems were used to ensure specific knockdown of Nurr1 mRNA, while limiting off-target effects on other nuclear receptor family members (Nur77, Nor1) or corepressor proteins. The Nurr1 siRNA sense sequence was (5′- > 3′) GCAUCGCAGUUGCUUGACAtt, antisense, UGUCAAGCAACUGCGAUGCgt (Ambion Silencer Select siRNA; denoted RNAi-2); and DsiRNA duplex sequences (5′- > 3′) CUAGGUUGAAGAUGUUAUAGGCACT; AGUGCCUAUAACAUCUUCAACCUAGAA (IDT DsiRNA; denoted RNAi-1).
Gene Expression Assays.
Quantitative reverse-transcription polymerase chain reaction (PCR) primers were synthesized by Integrated DNA Technologies (Supplemental Table 1). BV-2 cells were pretreated with 10 μM C-DIM12 (0.1, 1.0, or 10 μM in dose-response assays) for 1 hour in serum and antibiotic free OptiMEM (GIBCO/Life Technologies) before treatment with 1 μg/ml LPS (Sigma-Aldrich). RNA was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA), and purity and concentration were determined using a NanoVue Spectrophotometer (GE Healthcare, Piscataway, NJ). After purification, 1 μg of RNA was used as a template for reverse-transcription reactions using iScript (Bio-Rad Laboratories). The cDNA produced from this reaction was measured for gene expression levels of proinflammatory genes using an Eco Real-Time PCR System (Illumina, San Diego, CA). Relative changes from control conditions (expressed as fold change) in gene expression were calculated using the 2−ΔΔCT method and normalized using the β-actin and HPRT (hypoxanthine-guanine phosphoribosyltransferase) housekeeping genes (Livak and Schmittgen, 2001). Samples were run in duplicate, and biologic replicates were repeated from a minimum of three separate experiments.
NF-κB Reporter Assays.
The NF-κB-293T-GFP-Luc reporter cell line was purchased from System Biosciences (Mountain View, CA). For green fluorescent protein/4′,6-diamidino-2-phenylindole (GFP/DAPI) expression assays, cells were grown in DMEM (Life Technologies) supplemented with 10% FBS and 1× PSN (as described earlier) on 96-well black-walled plates (Thermo Scientific, Waltham, MA). Cells were plated 24 hours before treatment with 30 ng/ml TNFα in the presence or absence of 100 μM of C-DIM12 or 50 μM of Bay-11 (positive control) for 24 hours, or at specific times/dosages as described in the figure legends. Reporter cells were washed with 1× phosphate-buffered saline (PBS) and stained with Hoechst 33342 (Molecular Probes/Life Technologies, Eugene, OR) in Fluorobrite DMEM (Life Technologies) incubated at 37°C, 5% CO2 for 5 minutes, then washed again with 1× PBS. The medium was replaced with fresh Fluorobrite DMEM before reading the plate at 488/519 nm for GFP fluorescence expression and 345/478 nm for DAPI fluorescence expression on a Cytation3 Cell Imaging Multi-Mode Reader (BioTek Instruments, Winooski, VT). The GFP expression intensity values were divided over the DAPI expression intensity values for quantitative analysis. TNFα concentrations were based on a dose–response assay (0–100 ng/ml; Supplemental Fig. 1) for optimal induction of NF-κB–GFP expression.
Chromatin Immunoprecipitation.
BV-2 cells were grown to confluence in 10-cm tissue culture plates (approximately 2 × 107 cells) and treated for the indicated time with LPS (1 μg/ml), with or without C-DIM12 (10 μM) or a dimethylsulfoxide (DMSO) vehicle control before cross-linking with 1% formaldehyde (Thermo Scientific) for 10 minutes. The remaining steps were adapted from the Chromatrap ChIP (chromatin immunoprecipitation) protocol, which accompanies the Chromatrap Pro-A Premium ChIP Kit (Chromatrap, Wrexham, United Kingdom). DNA was sheared into approximately 500 bp fragments before removing 10% (200 ng) for input controls, and 2 μg of chromatin was loaded into the immunoprecipitation reaction with 2 μg precipitating antibody (as suggested by Chromatrap), anticorepressor for repressor element 1 silencing transcription factor [anti-CoREST], anti–nuclear receptor corepressor 2 [anti-NCOR2], and anti-histone deacetylase 3 [anti-HDAC3] from Abcam (Cambridge, United Kingdom); anti-Nurr1 (N-20; 991), and anti-p65 (372) from Santa Cruz Biotechnology. Immunopurified DNA was isolated via the QIAquick PCR Purification Kit (Qiagen; suggested by Chromatrap). A 149-bp region of the NOS2 promoter proximal to the NF-κB site of transcription was amplified via quantitative PCR and performed with SYBR-Green Supermix (Bio-Rad Laboratories, Hercules, CA) and analyzed using the percentage of input method according to Life Technologies.
Immunoblotting.
BV-2 cells were lysed using radioimmunoprecipitation assay buffer with protease inhibitors. Protein was quantified using a BCA protein assay (Thermo Scientific Pierce, Rockford, IL), and equal protein (20 μg) was loaded in a polyacrylamide 12% gel with 4% stacking gradient. Protein quantitation was imaged using a Biorad ChemiDoc XRS System and compared with β-actin control using ImageJ analysis software (National Institutes of Health; http://imagej.nih.gov/ij/). ChIP-grade antibodies, as described, were used at a 1:1000 concentration for Western blot analysis.
Immunofluorescence Microscopy in BV-2 Cells.
BV-2 microglia were plated on cover-glass coated with FBS and pretreated for 1 hour with 10 μM of C-DIM12 followed by 1 μg/ml LPS. After 30 minutes the cells were rinsed with PBS buffer and then paraformaldehyde fixed for 15 minutes at 4°C. Cells were blocked using bovine serum albumin (Sigma-Aldrich) for 1 hour until incubation with primary antibodies overnight at 4°C. Secondary antibody incubation was at room temperature for 3 hours with Alexa Fluor 555, Alexa Fluor 647 (Life Technologies), DAPI counterstain (Vector Laboratories, Burlingame, CA). Slides were imaged using a Zeiss Axiovert 200M inverted fluorescence microscope equipped with a Hammamatsu ORCA-ER–cooled charge-coupled device camera (Hammamatsu Photonics, Hamamatsu City, Japan) using Slidebook software (version 5.5; Intelligent Imaging Innovations, Denver, CO). Quantification of nuclear protein was determined by measuring the fluorescence intensity of p65 or Nurr1 within the boundary of the nucleus of each cell, defined by DAPI counterstain, and segmented using Slidebook 5.0 software function for fluorescence intensity minus background (F/Fo).
Confocal Microscopy in Primary Microglia.
Primary mouse microglia were set on 12-mm round microscopic cover glass slides coated with fetal bovine serum and allowed attach and grow for 48 hours before experimentation. Cells were treated for 1 hour with saline, 1 μg/ml LPS, or 1 μg/ml LPS + 10 μM C-DIM12, fixed in ice-cold methanol and probed with anti-CD11b-fluorescein isothiocyanate (1:250) and anti-Nurr1 (1:500). Nurr1 was visualized with an AlexaFluor-555 conjugated secondary antibody. Microglia were imaged on an Olympus Fluoview 1000 laser-scanning confocal microscope (Olympus America, Center Valley, PA) using a 100× Plan Apochromat oil immersion objective. Each field of view was imaged as a z-stack (8–10 planes, 1-μm step size) transformed into a single maximum projection image using the Stack Focuser plug-in within Fiji open-source image processing software (National Institutes of Health, http://fiji.sc/Fiji).
Statistical Analysis.
Statistical analyses were performed using Prism (version 6.0; Graph Pad Software, San Diego, CA). Data are presented as mean ± S.E.M. Experimental group analyses were performed using a one-way analysis of variance with a Tukey post hoc test. *P < 0.05, **P < 0.001, ***P < 0.001, ****P < 0.0001 were considered statistically significant.
Results
Structure-Activity of C-DIMs in Suppression of Proinflammatory Genes.
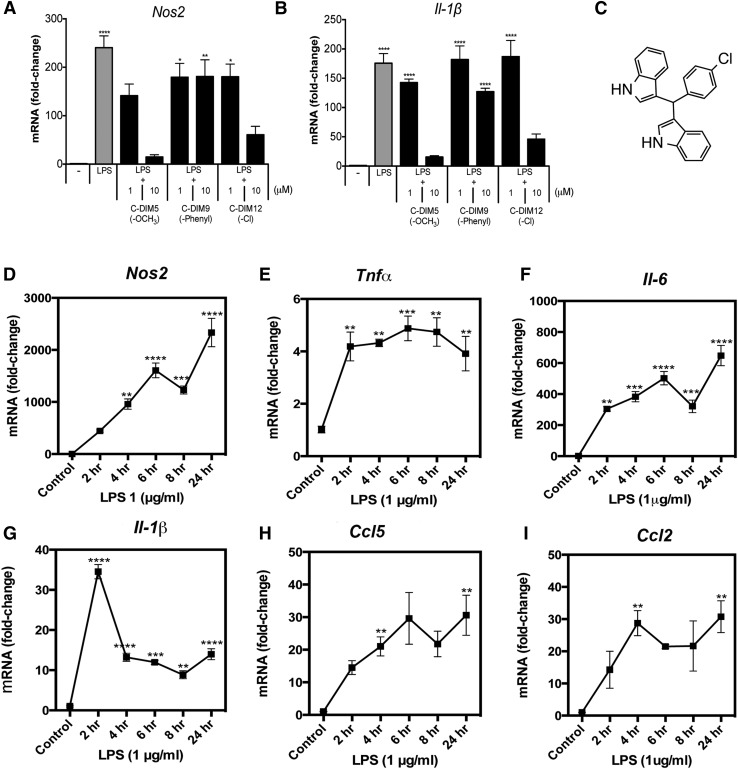
An initial analysis of a series of C-DIM analogs (C-DIM1 through C-DIM14) was performed to determine their effect on inflammatory gene expression in primary mixed cultures of astrocytes and microglia (Li et al., 2012). To assess the efficacy and structure-activity relationships of C-DIM compounds on the expression of representative NF-κB–regulated neuroinflammatory genes, primary mixed glia were treated with 1 μg/ml LPS and either 1 or 10 μM of each C-DIM compound (Table 1). Compared with LPS treatment alone, several of the C-DIM compounds (C-DIM 1, 3, 5, 7, 8, 10, 11, 12, and 13) significantly decreased NOS2 and interleukin-1β (IL-1β) expression at a concentration of 10 μM. The substituted -R position of the para-phenyl group appeared to change the anti-inflammatory activity of specific C-DIMs, consistent with a structure-activity relationship indicating that C-DIMs with smaller, more polar para substituents on the phenyl group are even more potent as suppressors of inflammatory gene expression in primary mixed glial cells. This structure-activity relationship study showed that C-DIM5, and C-DIM12, which contain para-methoxy- (C-DIM5) and para-chloro-phenyl- substituents (C-DIM12; NR4A2/Nurr1 activity), suppressed NOS2 and IL-1β, whereas phenyl-substituted C-DIM9 did not decrease the LPS-induced expression of these inflammatory genes (Fig. 1, A and B). Based on these results and on previous pharmacokinetic and efficacy screening of C-DIMs in mice (De Miranda et al., 2013), we selected C-DIM12 (Fig. 1C) for further in vitro analysis of its mechanism of inhibition of inflammatory cytokine expression in microglia.
TABLE 1.
Structure-dependent suppression of LPS-induced inflammation by C-DIMs
Primary murine mixed glial cultures were treated with saline or 1 μg/ml LPS and 1 or 10 μM doses of one of 14 C-DIM analogs. Structural differences displayed as R-Group, and significant suppression of NOS2 and IL-1β mRNA levels from LPS control are indicated. Data are expressed as mean ± S.D.; mRNA fold induction over LPS (β-actin internal control).
| C-DIM | R-Group | NOS2 mRNA (Fold-Induction) |
IL-1β mRNA (Fold-Induction) |
||
|---|---|---|---|---|---|
| Saline 1.0 ± 0.2 | LPS 240.5 ± 24.4a | Saline 1.0 ± 0.2 | LPS 175.7 ± 16.5a | ||
| LPS + |
LPS + |
||||
| C-DIM (1 μM) | C-DIM (10 μM) | C-DIM (1 μM) | C-DIM (10 μM) | ||
| 1 | -CF3 | 160.6 ± 13.1 | 50.8 ± 8.7b | 167.6 ± 18.2a | 34.1 ± 3.1b |
| 2 | -Br | 175.5 ± 34.9 | 73.1 ± 23.1 | 129.4 ± 43.0 | 40.6 ± 3.7b |
| 3 | -F | 133.8 ± 8.6 | 3.3 ± 1.5b | 156.7 ± 10.6a | 7.1 ± 1.8b |
| 4 | -tert-Butyl | 107.8 ± 20.5 | 84.8 ± 8.0 | 125.3 ± 8.0 | 93.9 ± 10.2 |
| 5 | -OCH3 | 141.4 ± 24.0 | 14.7 ± 4.7b | 142.6 ± 6.1a | 15.4 ± 2.3b |
| 6 | -N(CH3)2 | 224.2 ± 68.0a | 99.5 ± 21.7 | 164.9 ± 30.0a | 65.1 ± 9.1 |
| 7 | -H | 184.9 ± 36.1a | 12.4 ± 4.4b | 142.9 ± 44.5a | 12.6 ± 2.3b |
| 8 | -OH | 189.3 ± 61.8a | 0.7 ± 0.4b | 136.2 ± 48.3a | 9.3 ± 1.4b |
| 9 | -Phenyl | 136.2 ± 47.7 | 180.9 ± 34.5a | 137.4 ± 47.4a | 127.2 ± 5.8 |
| 10 | -CN | 96.0 ± 21.6 | 34.5 ± 5.8b | 140.1 ± 7.5a | 23.8 ± 1.8b |
| 11 | -CH3 | 140.0 ± 5.5 | 32.0 ± 6.8b | 152.2 ± 15.7a | 36.4 ± 3.5b |
| 12 | -Cl | 180.4 ± 26.2 | 60.6 ± 17.5b | 186.9 ± 27.6a | 45.9 ± 8.9b |
| 13 | -COOCH3 | 255.5 ± 78.8a | 90.9 ± 7.8b | 185.2 ± 11.5a | 44.0 ± 15.2b |
| 14 | -I | 242.9 ± 83.9a | 122.7 ± 33.5b | 161.4 ± 43.6a | 72.4 ± 15.0 |
Different from control group (P < 0.05).
Different from LPS group (P < 0.05).
Fig. 1.
C-DIM inflammatory gene suppression in glia is structure dependent. Primary murine mixed glial cultures were treated with saline or 1 μg/ml LPS and 1 or 10 μM of DIM-C-pPhOCH3 (C-DIM5), DIM-C-pPhPh (C-DIM9), or DIM-C-pPhCl (C-DIM12) for 8 hours, and assessed for (A) NOS2 or (B) IL-1β expression with real-time PCR. (C) The structure of chloro-substituted C-DIM12. (D–I) BV-2 microglia were treated with saline or 1 μg/ml LPS over a 24-hour time point, and RNA was collected for real-time PCR analysis of cytokine mRNA expression. Data are expressed as mean ± S.E.M. (n = 4); mRNA fold change; internal control (β-actin). Statistical significance is expressed as mean compared with saline control. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
C-DIM12 Decreases Inflammatory Gene Expression in BV-2 Microglia.
Cytokine expression in glia is well documented both in vivo and in vitro after stimulation of microglial Toll-like receptor (TLR) and subsequent IκB kinase (IKK)–dependent activation of NF-κB (Saijo et al., 2013). BV-2 microglia were challenged with 1 μg/ml LPS and assayed for the time course of expression of the NF-κB–regulated genes NOS2, TNFα, IL-6, IL-1β, CCL5 [chemokine (C-C motif) ligand 5], and CCL2 over a period of 24 hours (Fig. 1, D–I). All genes assayed were maximally induced at 24 hours after LPS treatment with the exception of IL-1β, which peaked after 2 hours followed by a decline in expression that remained above the control levels.
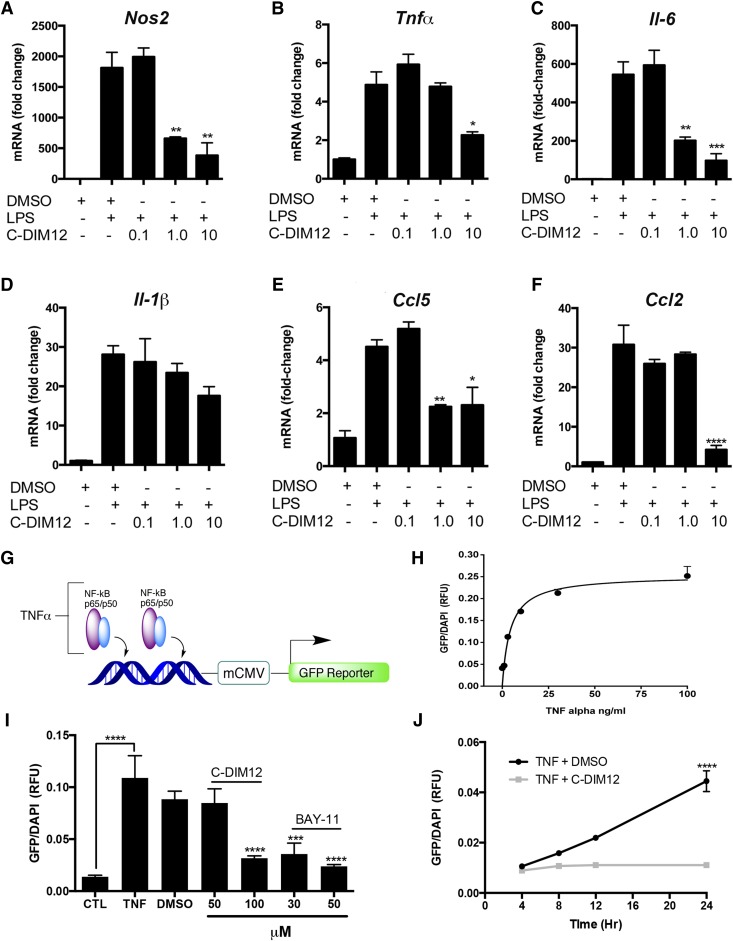
We next examined the potency of C-DIM12 in suppressing the expression of selected neuroinflammatory genes (Fig. 2, A–F) by treating BV-2 cells with LPS (10 μg/ml) for 24 hours in the presence of increasing concentrations of C-DIM12 (0, 0.1, 1, or 10 μM). Inflammatory gene expression of NOS2, IL-6, and CCL5 was significantly decreased over LPS and vehicle control (DMSO) expression levels with 1 μM of C-DIM12, whereas TNFα and CCL2 were significantly suppressed with 10 μM C-DIM12. A trend toward suppression of IL-1β is evident, and a higher concentration of C-DIM12 may be required to elicit a significantly decreased response (Fig. 2D).
Fig. 2.
Dose-dependent suppression of inflammatory genes in BV-2 microglia and NF-κB transactivation in reporter cells. (A–F) BV-2 microglia were pretreated for 1 hour with 0.1, 1.0, or 10 μM C-DIM12 followed by 24 hours of LPS (1 μg/ml). The mRNA expression was assessed for the fold change of cytokine expression (β-actin internal control). Data are expressed as mean ± S.E.M. (n = 4); mRNA fold change. (G) NF-κB/GFP/Luc HEK. (H) Dose-response of NF-κB–GFP expression per cell (DAPI counterstain) in NF-κB/GFP/Luc HEK cells with increasing TNFα concentrations. (I) NF-κB/GFP/Luc HEK cells were treated with C-DIM12 (25, 50, 100 μM) or Bay-11 (30, 50 μM) as a positive control (CTL) for NF-κB suppression. (J) Time course data from NF-κB/GFP/Luc HEK cells that received TNFα and were assayed over 24 hours with (gray line) or without (black line) 100 μM C-DIM12. The mRNA were assessed for fold change expression, and data are expressed as mean ± S.E.M. (n = 16). **P < 0.01; ***P < 0.001; ****P < 0.0001.
C-DIM12 Decreases Expression of NF-κB–Enhanced GFP Expression in Human Embryonic Kidney 293 Reporter Cells.
To further examine the effect C-DIM12 on NF-κB transcriptional activity, we exposed NF-κB–GFP human embryonic kidney 293 (HEK293) cells (NF-κB/293/GFP-Luc Transcriptional Reporter Cells; System Biosciences, Mountain View, CA) to TNFα in the presence of C-DIM12 (Fig. 2, G–J). These cells stably express an NF-κB reporter construct consisting of high affinity p65-binding sites driving coexpression of GFP and luciferase in response to stimulation with TNFα (Fig. 2G). Exposing NF-κB–GFP HEK cells to increasing concentrations of TNFα (1–100 ng/ml) for 24 hours resulted in a dose-dependent increase in total GFP fluorescence per cell (Fig. 2H). Cotreatment with C-DIM12 (100 μM) significantly reduced TNFα (30 ng/ml)-induced NF-κB–GFP expression, comparable to that observed for Bay-11 (50 μM), a positive control for inhibition of NF-κB that blocks IKK activity (Krishnan et al., 2013). C-DIM12 (100 μM) and Bay-11 (50 μM) were equally efficient at blocking NF-κB–GFP expression in the NF-κB–GFP HEK cells after TNFα treatment, displaying a statistically significant reduction in total GFP fluorescence per cell (Fig. 2I). The time course for C-DIM12–dependent suppression of TNFα-induced NF-κB–GFP is shown in Fig. 2J, where no detectable increase in NF-κB–GFP fluorescence was observed up to 24 hours in cells exposed to 30 ng/ml of TNFα in the presence of 100 μM C-DIM12 (P < 0.0001 compared with TNFα plus DMSO vehicle control).
Nurr1 Is Required for Suppression of Inflammatory Genes by C-DIM12.
Previous data indicated that lentiviral-mediated RNAi knockdown of Nurr1 increases both basal and inducible levels of inflammatory cytokines in BV-2 microglial cells and enhances loss of dopamine neurons in the SN after LPS treatment (Saijo et al., 2009). We therefore used RNAi to examine the capacity of C-DIM12 to function as a pharmacologic modulator of Nurr1 though its effects on transrepression of NF-κB–regulated inflammatory genes.
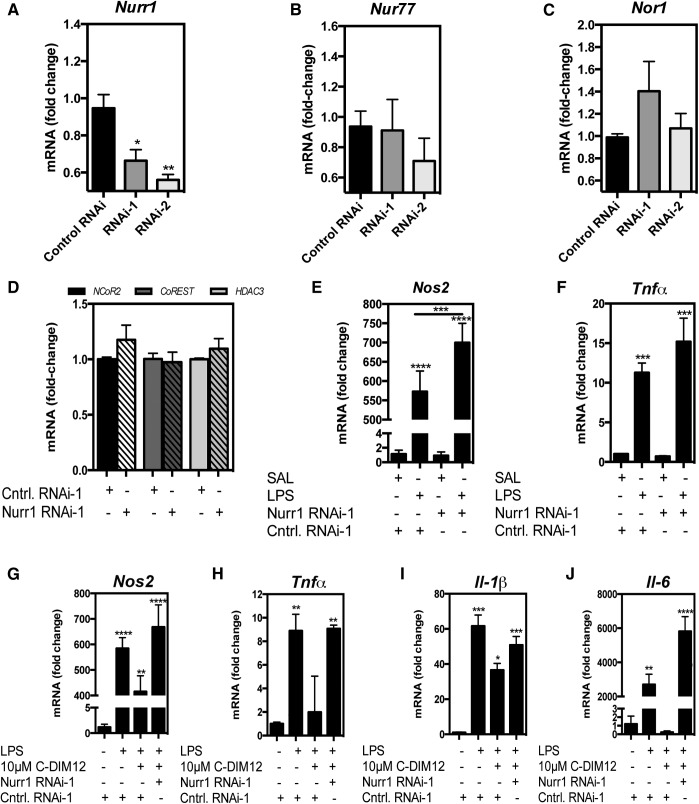
The data shown in Fig. 3A indicated that treatment of BV-2 microglia with two different sequences of siRNA against Nurr1 (RNAi-1 or RNAi-2) resulted in significant knockdown of Nurr1 mRNA expression. To address whether knockdown of Nurr1 affected other NR4A family members (NR4A1/Nur77, NR4A3/Nor1), mRNA expression was assayed after Nurr1 RNAi treatment, which did not result in significantly altered gene expression, though Nur77 mRNA showed a trend toward decrease compared with control in RNAi-2–treated cells, and Nor1 showed a trend toward increasing in RNA-1–treated cells, respectively (Fig. 3, B and C). Expression of corepressor proteins involved in Nurr-1–mediated transrepression (NCOR2, CoREST, and HDAC3) were unaffected by RNAi interference, (Fig. 3D). Nurr1 RNAi enhanced expression of NOS2 after 24 hours of stimulation with LPS (Fig. 3E). This same trend was observed with TNFα, though it was not statistically significant (Fig. 3F). After RNAi knockdown of Nurr1, BV-2 cells were also treated with LPS in both the presence and absence of C-DIM12 (10 μM) for 24 hours and then assayed for expression of the representative NF-κB–regulated inflammatory genes NOS2, TNFα, IL-1β, and IL-6 (Fig. 3, G–J). In the treatment groups receiving Nurr1 RNAi, C-DIM12 no longer suppressed the LPS-induced expression NOS2, TNFα, IL-1β, or IL-6 (Fig. 3, G–J). These data indicate that C-DIM12 requires expression of Nurr1 to fully suppress proinflammatory gene expression in BV-2 cells stimulated by LPS.
Fig. 3.
C-DIM12–dependent inhibition of inflammatory gene expression requires Nurr1. (A–C) BV-2 microglia were treated with two different sequences of siRNA (denoted RNAi-1, RNAi-2) or scrambled RNAi (control RNAi). (D) NCor2, CoREST, HDAC3 mRNA were measured using real-time PCR after treatment with RNAi-1 (β-actin internal control). (E–F) BV-2 microglia were treated with Nurr1-RNAi-1 or control RNAi-1 for 24 hours followed by saline or 1 μg/ml LPS for 24 hours and assessed for mRNA expression of NOS2 and TNFα. (G–J) BV-2 cells were treated with RNAi-1 or control RNAi-1 for 24 hours followed by 10 μM C-DIM12 (1 hour pretreatment) and 1 μg/ml LPS treatment and assessed for mRNA expression of NOS2, TNFα, IL-1β, and IL-6. Data are expressed as mean ± S.E.M. (n = 4); mRNA fold change. Statistical significance is compared with saline control. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
NF-κB-p65 Translocation Is Not Inhibited by C-DIM12.
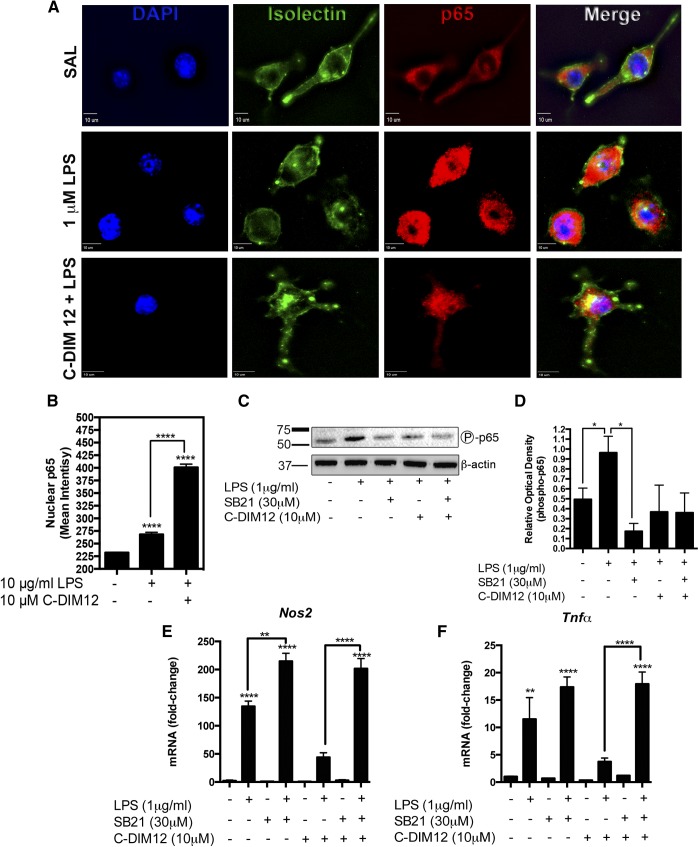
Binding of LPS to TLR4 receptors in microglia activates NF-κB through IKK-dependent phosphorylation and degradation of IκBα, leading to translocation of p50/p65 heterodimers into the nucleus where they activate inflammatory gene promoters at cis-acting promoter elements (He et al., 2002; Saijo et al., 2013). To determine whether C-DIM12 inhibited inflammatory gene expression by preventing translocation of p65 to the nucleus, we treated BV-2 microglial cells with 1 μg/ml LPS and measured the nuclear translocation of p65 using immunofluorescence. After 30 minutes, the p65 levels in the nucleus of BV-2 cells were markedly increased over the saline control (Fig. 4A), indicated by the disappearance of nuclear voids evident before LPS treatment. C-DIM12 did not inhibit the nuclear translocation of p65 in BV-2 microglia treated with 1 μg/ml LPS but rather enhanced the nuclear levels of p65 (Fig. 4B).
Fig. 4.
C-DIM12 does not prevent p65 translocation and blocks NF-κB–transactivation in reporter cells. (A) BV-2 cells were treated with 10 μM C-DIM12 for 1 hour followed by saline or 1 μg/ml LPS for 30 minutes and fixed for immunofluorescence to examine p65 translocation with DAPI (blue), isolectin (green), p65 (red). (B) The p65 nuclear expression was quantified by mean fluorescence intensity encompassing the nuclei (DAPI boundary; background subtracted). (C) Representative protein expression of phosphorylated p65 after LPS and LPS + C-DIM12 treatment. (D) Relative optical density of phospho-p65 immunoblot, mean of three replicate experiments. Protein levels were normalized to β-actin (± S.E.M.). (E and F) BV-2 cells were treated with 30 μM SB216763 (SB21) to inhibit GSK3β for 1 hour, followed by 1 μg/ml LPS and cotreatment with 10 μM C-DIM12 for 24 hours. Data are expressed as mean ± S.E.M. (n = 3). **P < 0.01; ***P < 0.001; ****P < 0.0001. SAL, saline.
It was recently reported that glycogen synthase kinase 3β (GSK3β) is required for the binding of Nurr1 to p65 at inflammatory gene promoters (Saijo et al., 2009). Phosphorylation of p65 at serine provides the docking site necessary to recruit Nurr1 to p65 and thereby block the removal of transcriptional inhibitory protein complexes at inflammatory gene promoters (Saijo et al., 2009). Protein expression of phosphorylated p65 after treatment with SB216763 [3-(2,4-dichlorophenyl)-4-(1-methylindol-3-yl)pyrrole-2,5-dione], an inhibitor of GSK3β, indicated that LPS-induced p65 phosphorylation of serine was decreased after GSK3β inhibition (Fig. 4, C–D). C-DIM12 treatment did not significantly alter the amount of phosphorylated p65 though a trend of decreased phosphorylation was visible, which correlates with the global NF-κB suppression that was observed after C-DIM12 treatment.
To determine whether C-DIM12 requires GSK3β to block LPS-induced expression of neuroinflammatory genes, we treated BV-2 cells with SB216763 and challenged them with 10 μg/ml LPS for 24 hours with and without the cotreatment of C-DIM12. When GSK3β was inhibited, expression of NOS2 and TNFα were both significantly induced in BV-2 cells in both the presence and absence of C-DIM12 (Fig. 4, E and F).
C-DIM12 Modulates Cellular Localization of Nurr1.
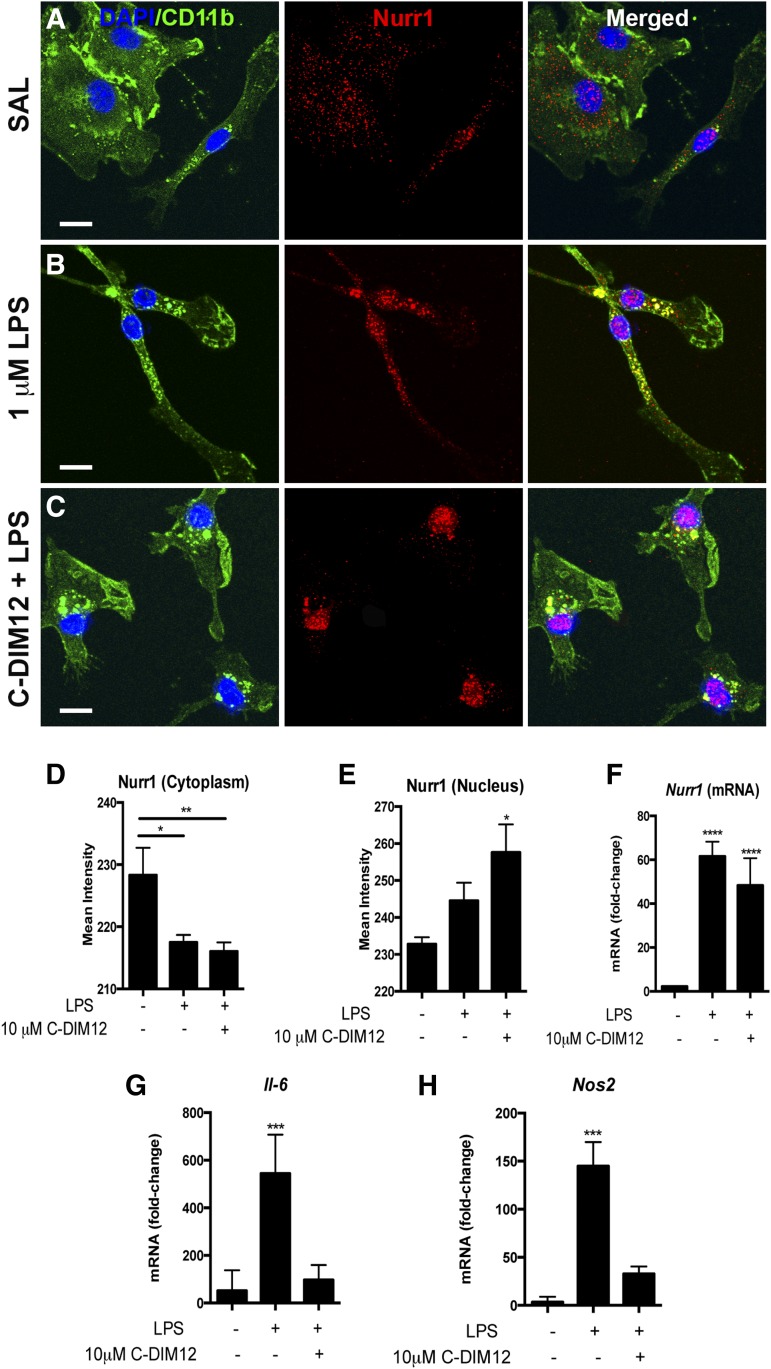
Because suppression of LPS-induced expression of inflammatory genes by C-DIM12 required functional levels of Nurr1, we sought to determine whether C-DIM12 altered the expression or subcellular localization of Nurr1 in microglia. Highly enriched cultures of primary mouse microglia were treated with LPS (10 ng/ml) or with LPS + C-DIM12 (10 μM), and Nurr1 protein expression and subcellular localization was examined by confocal microscopy (Fig. 5, A–C; Nurr1, red; CD11b, green; DAPI, blue). Nurr1 expression was visibly increased over saline in microglia treated with LPS; pronounced nuclear translocation of Nurr1 was evident with colocalization of Nurr1 and DAPI in merged images. Quantitation of the fluorescence intensity of Nurr1 protein colocalizing with the cytoplasm or the nucleus resulted in a significant increase of nuclear Nurr1 in microglia treated with LPS + C-DIM12 (Fig. 5, D and E). Additionally, LPS and LPS + C-DIM12 treatments significantly increased Nurr1 mRNA after 24 hours (Fig. 5C). Despite the observation that Nurr1 expression and nuclear localization were equivalent between the LPS and LPS + C-DIM12 groups, LPS-induced expression of IL-6 and NOS2 was significantly suppressed only in the LPS + C-DIM12 treatment groups (Fig. 5, G and H), indicating that C-DIM12 may not further increase Nurr1 above LPS-induced levels in BV-2 cells but may activate or enhance Nurr1-dependent function in microglia.
Fig. 5.
Nurr1 translocation and expression following LPS and C-DIM12 treatment. (A–C) Primary mouse microglia were plated onto cover glass and treated with 10 μM C-DIM12 for 1 hour followed by 1 μg/ml LPS for 24 hours. Cells were fixed and imaged (100×) by confocal microscopy for immunofluorescence of Nurr1 (red), CD11b (green) and DAPI (blue). (D and E) Quantification of Nurr1 protein translocation from the cytoplasm to the nucleus. (F) Nurr1 mRNA levels in BV-2 microglia treated with 1 μg/ml LPS, or 1 μg/ml LPS + 10 μM C-DIM12 for 24 hours. (G and H) IL-6 and Nos2 mRNA levels from the same samples corresponding to Nurr1 mRNA. Data are expressed as mean ± S.E.M.; mRNA fold change (n = 3); *P<0.05; ***P < 0.001; ****P < 0.0001. Scale bar = 10 μm. SAL, saline.
Nurr1 Transcriptional Repression Is Enhanced by C-DIM12.
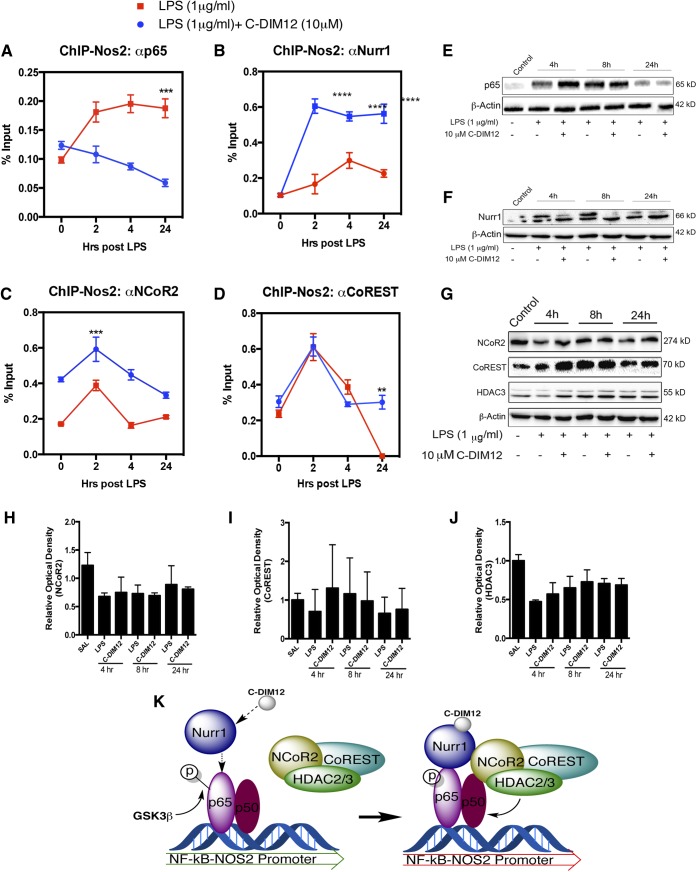
Recruitment of Nurr1 monomers to p65 involves several proteins with kinase action (GSK3β, Nemo-like kinase) as well as proteins that modify Nurr1 to recruit corepressors such as histone deacetylases (HDAC2/3), nuclear receptor corepressor 2 (NCOR2), and the CoREST complex (Saijo et al., 2009). We performed ChIP assays to determine the capacity of C-DIM12 to modulate protein-DNA interactions at NF-κB binding sites in the NOS2 promoter (Fig. 6), postulating that C-DIM12–dependent activation of Nurr1 would stabilize corepressor protein complexes at NF-κB cis-acting promoter elements. Time course data indicate that in BV-2 microglia treated with 10 μg/ml LPS, p65 binding to the NOS2 promoter increased and remained elevated for up to 24 hours (Fig. 6A), whereas cotreatment with C-DIM12 (10 μM) significantly reduced the amount of p65 binding (P < 0.001 relative to LPS + C-DIM12). Simultaneously, Nurr1 was strongly recruited to the p65 binding site by 24 hours in cells treated with LPS + C-DIM12 (Fig. 6B; P < 0.0001, relative to LPS + DMSO vehicle control). Overall, BV-2 cells treated with LPS + C-DIM12 had a significantly lower amount of p65 bound to the NOS2 promoter throughout the 24-hour time course.
Fig. 6.
C-DIM12 enhances Nurr1 recruitment to NOS2 promoter, decreases p65 binding, and stabilizes binding of nuclear corepressors. BV-2 cells were treated with 10 μM C-DIM12 for 1 hour followed by 1 μg/ml LPS over a 24-hour time point and assessed at the NOS2 promoter using ChIP. (A) The amount of p65 bound to the NOS2 promoter was measured in LPS or LPS + C-DIM treatments. (B) The level of Nurr1 bound to the NOS2 promoter with or without C-DIM12 over a 24-hour time course with LPS. (C and D) ChIP assessment of nuclear corepressor NCOR2 and corepressor complex CoREST bound to the NOS2 promoter. (E and F) Representative protein quantitation of total Nurr1 and p65 after LPS and LPS + C-DIM12 treatment. (G) NCOR2, HDAC3, and CoREST protein from 24-hour time course treatment with saline or LPS + 10 μM C-DIM12. (H–J) Relative optical density of total cellular corepressor protein expression, mean of three replicate experiments. Protein levels were normalized to β-actin (±S.E.M.). (K) Hypothesized mechanism by which C-DIM12 promotes Nurr1-dependent transrepression of p65 at the NOS2 promoter in BV-2 microglia. ChIP data are expressed as the percentage of input ± S.E.M. over control IgG (n = 3). **P < 0.01; ***P < 0.001; ****P < 0.0001. SAL, saline.
DNA binding and expression of the corepressor proteins NCOR2 and CoREST were also examined under these conditions. In cells treated with LPS + C-DIM12, cyclic increases in NCOR2 bound to the NOS2 promoter occurred 2 and 8 hours after LPS treatment (Fig. 6C; P < 0.001 and P < 0.0001 compound to LPS + DMSO vehicle control). In contrast, CoREST binding to the NOS2 promoter increased 3-fold by 2 hours in both the LPS and LPS + C-DIM12 groups, but remained elevated at 24 hours in the LPS + C-DIM12 group (Fig. 6D; P < 0.01 compared with LPS + DMSO vehicle control). In summary, cells treated with C-DIM12 had sustained levels of both Nurr1 and CoREST bound to the NOS2 promoter 24 hours after exposure to LPS in the presence of C-DIM12, which correlated with decreased binding of p65.
Representative protein expression of total p65 shows an apparent decrease from LPS treatment with the addition of C-DIM12 (Fig. 6E); however, consistent with mRNA expression, Nurr1 protein levels are not significantly altered by C-DIM12 treatment (Fig. 6F). The protein expression of NCOR2, HDAC3, and CoREST was determined in BV-2 cells over 24 hours of exposure to LPS and LPS + C-DIM12 (Fig. 6G). Immunoblot data from these experiments indicated that NCOR2, CoREST, and HDAC3 protein levels decreased slightly after LPS treatment (4 hours), but no significant differences were detected at the level of total protein expression (Fig. 6, H–J).
A schematic depicting the proposed activation of Nurr1 by C-DIM12 is illustrated in Fig. 6K, represented as an adaptation from the mechanism described by Saijo et al. (2009). The hypothesized mechanism includes the enhanced binding of Nurr1 to the NF-κB response element region of the promoter, and the recruitment of corepressor complexes responsible for the decrease in p65 bound to the NOS2 promoter in microglial cells.
Discussion
The nuclear receptor NR4A2/Nurr1 is described as a potential therapeutic target for mitigating the damaging effects of glial activation, but compounds that selectively modulate this receptor to inhibit NF-κB–regulated neuroinflammatory genes have not been reported (Bensinger and Tontonoz, 2009; Nolan et al., 2013). We selected C-DIM12 for examination following several lines of evidence for its use as a potential neuroprotective compound. C-DIM12 displays the ability to suppress inflammatory gene expression in astrocytes in vitro (Table 1), and it has significant distribution to the brain after oral dosing (De Miranda et al., 2013). In an in vivo MPTPp mouse model of PD, postlesion treatment with C-DIM12 attenuated dopamine neuron loss and reduced glial activation and cytokine induction with greater significance than other C-DIM compounds (De Miranda et al., 2015).
The data presented here indicate that C-DIM12 suppresses NF-κB–induced gene expression in microglia through a mechanism involving transcriptional repression via Nurr1 (Saijo et al., 2009). Structure-activity studies indicated that C-DIM12 and related halogenated C-DIM structures had more potent anti-inflammatory action in primary mouse mixed glia than several other C-DIM analogs with bulkier aliphatic or aromatic substituents (Fig. 1; Table 1).
The structure-dependent binding of some C-DIMs to the ligand-binding domain of NR4A1 has been reported (Lee et al., 2014), and these compounds can either activate or inactivate NR4A1- and NR4A2-dependent transactivation in a structure-specific manner (Cho et al., 2010; Yoon et al., 2011; Li et al., 2012). Through this mechanism, these studies indicated that C-DIM12 inhibits growth of urothelial carcinoma cells (KU7, 253J cell lines) and also decreased tumor growth in vivo (Inamoto et al., 2008). The anticancer properties of C-DIM12 were Nurr1-dependent (Inamoto et al., 2008); however, the effects on of this compound on Nurr1 in normal cells have been less well studied and are likely to involve distinct protein complexes of transcription activators and repressors, and other nuclear cofactors.
Dose-dependent inhibition of inflammatory gene expression in BV-2 microglia indicated that C-DIM12 blocks expression of NF-κB–regulated inflammatory cytokines and chemokines (Fig. 2). In addition, C-DIM12 suppressed NF-κB activation in NF-κB–GFP HEK reporter cells stimulated with TNFα (Fig. 2, I and J) but did not prevent the nuclear translocation of p65 in BV-2 cells exposed to LPS (Fig. 4A). These findings indicate that C-DIM12 acts downstream of NF-κB-p50/65 translocation and therefore likely prevents transcriptional activation of NF-κB–regulated genes by a direct effect on the transcriptional activity of nuclear proteins, rather than on prevention of upstream NF-κB activating kinases.
The observed increase in nuclear p65 after LPS + C-DIM12 treatment may be indicative of an increased amount of Nurr1 bound to p65, causing an accumulation in the nucleus. Knockdown of Nurr1 by RNAi-1 markedly decreased the ability of C-DIM12 to suppress mRNA levels for multiple inflammatory genes (Fig. 3, G–J), indicating that the transrepressive activity C-DIM12 likely requires Nurr1. Moreover, knockdown of Nurr1 had no effect on the effect on the expression of corepressor proteins NCOR2, CoREST, or HDAC3, supporting the conclusion that C-DIM12 directly modulates Nurr1 and not its downstream effector proteins. It cannot be completely ruled out that a change in expression of Nur77 or Nor1 (Fig. 3, B and C) may contribute to the alteration of inflammatory gene expression that was observed following Nurr1 RNAi, but such gene interactions are poorly understood in glia and warrant a comprehensive study of their own.
Nurr1-mediated transrepression of Nos2 in BV-2 microglia involves the recruitment of the CoREST corepressor complex to remove p65 (Saijo et al., 2009). The transrepressive activity of Nurr1 toward NF-κB requires phosphorylation of p65 at serine 468 by GSK3β, and inhibition of GSK3β significantly increases the transcription of NF-κB–regulated genes such as TNFα and NOS2 (Buss et al., 2004; Saijo et al., 2009). We noted a similar effect with C-DIM12 (Fig. 4), where the GSK3β inhibitor SB216763 reduced the capacity of C-DIM12 to suppress LPS-induced expression of NOS2 and TNFα. This suggests that C-DIM12–mediated repression also requires phosphorylation of p65 for recruitment of Nurr1 and inhibition of p65 transcriptional activation. The studies in Fig. 3 indicated that C-DIM12 did not suppress LPS-induced expression of IL-1β to the same extent as the other cytokines assayed, likely because expression of IL-1β in microglia is highly regulated by mitogen-activated protein kinase pathways in addition to NF-κB, which appears to support the specificity of C-DIM12 for suppression of NF-κB–regulated proteins (Kim et al., 2004). However, RNAi studies (Fig. 3I) indicated that knockdown of Nurr1 expression significantly attenuated the inhibitory effects of C-DIM12 on LPS-induced expression of IL-1β, supporting the importance of NF-κB/Nurr1 interactions in regulating this pathway. Although cross-talk between inflammatory cascade proteins is known to occur upon stimulation of TLRs on the surface of cells, it is likely that mitogen-activated protein kinase proteins are able to separately upregulate IL-1β expression independently of NF-κB. Therefore, suppression of p65-mediated inflammatory gene transcription likely results in only partial suppression of IL-1β.
The NR4A family of nuclear receptors is described as comprising immediate-early response genes involved in cellular maintenance and development, as well as in regulating inflammation (Zhao and Bruemmer, 2010). Although Nurr1 is primarily located in the nucleus in neurons and cancer cells (Boldingh Debernard et al., 2012; Li et al., 2012; García-Yagüe et al., 2013), our data in primary microglia suggest that Nurr1 is dynamically regulated in glial cells and that its subcellular localization changes after inflammatory stimulation with LPS (Fig. 5). This is supported by previous studies reporting that stimulation of TLR4 with LPS causes nuclear import of Nurr1, although the mechanisms regulating this effect are not well understood (Fan et al., 2009). Additionally, the cellular distribution of Nurr1 appears to vary between cell types. In SH-SY5Y and MN9D neuronal cell lines, changes in the subcellular localization of Nurr1 occurred within 1 hour of sodium arsenite–induced oxidative stress, which caused exposure of a nuclear export signal (García-Yagüe et al., 2013) and thereby decreased its transcriptional activity. Similarly, the intracellular location of NR4A1/Nur77 in cancer cells is variable; for example, treatment with apoptosis-inducing agents induces nuclear export of Nur77 where it forms a complex with Bcl-2 on mitochondria (Zhang et al., 2009), whereas the C-DIMs that bind the receptor activate or inactivate nuclear Nur77 (Lee et al., 2014).
In non-neuronal cells, nuclear import signals on Nurr1 may be alternatively uncovered for import into the nucleus for feedback inhibitory control over inflammatory proteins, although the mechanism is not well established (García-Yagüe et al., 2013). Here, we report that the translocation of Nurr1 from the cytosol to the nucleus in primary microglia treated with LPS occurred in both the presence and absence of C-DIM12 (Fig. 5, A and B). Inflammatory gene expression, however, was only blocked in the presence of C-DIM12, despite increases in Nurr1 mRNA in both groups, indicating that C-DIM12 likely stimulates the anti-inflammatory activity of Nurr1 by direct action on nuclear proteins.
The use of ChIP assays to examine the effects of C-DIM12 on LPS-induced protein-DNA interactions (Fig. 6) revealed that initial increases in p65 binding to the NOS2 promoter were maintained up to 24 hours, consistent with the time course for suppression of inflammatory gene expression (Figs. 2A and 6A). C-DIM12 decreased p65 interaction at the NOS2 promoter, with the greatest reduction seen at 24 hours. The interaction of Nurr1 with the NOS2 promoter was also increased in cells treated with C-DIM12, with a marked increase in recruitment of Nurr1 to the promoter regions encompassing the p65 binding site at 24 hours (Fig. 6B). In addition, the amount of CoREST and NCOR2 bound to the NOS2 promoter was increased over LPS (24 hours) in cells treated with C-DIM12, a mechanism that corresponds with the reduction of inflammatory gene expression in BV-2 microglia (Figs. 2G and 6, C and D). The total protein levels of Nurr1, NCOR2, CoREST, and HDAC3 (Fig. 6, E–G) are additional evidence that C-DIM12 action does not produce sizeable increases in corepressor proteins but instead may enhance Nurr1-mediated recruitment of corepressors that mediate transrepression of inflammatory genes in microglia. Consistent with these findings, there were no significant changes in the expression of either p65 or Nurr1 between the LPS and LPS + C-DIM12 treatment groups (Fig. 6, E and F).
A theoretical model for the anti-inflammatory action of C-DIM12 is illustrated in Fig. 6H, where NOS2 promoter transcription activated by NF-κB-p50/p65 is reduced by corepressor complex proteins (HDAC, NCOR2, and CoREST) that are recruited to the NOS2 promoter by Nurr1. Pharmacologic modulation of Nurr1 by C-DIM12 increases the nuclear receptor interactions with corepressor proteins, preventing binding of the RNA polymerase complex and leading to eventual removal p65 from the NOS2 promoter, thereby reducing inflammatory gene transcription.
The discovery of Nurr1 as a transrepressor of NF-κB–regulated genes in glia demonstrated that this receptor is a potential target for development of therapeutic agents for treatment of neuroinflammatory disease (Saijo et al., 2009). Our studies with the para-phenyl substituted diindolylmethane demonstrate that this compound induces anti-inflammatory activity in glial cells through a Nurr1-dependent mechanism. Pharmacologic stimulation of Nurr1-mediated transrepression by C-DIM12 represents a novel mechanism to decrease expression of NF-κB–regulated inflammatory genes in glial cells relevant to neurodegenerative disease.
Supplementary Material
Abbreviations
- Bay-11
Bay 11-7082, (E)-3-(4-methylphenyl)sulfonylprop-2-enenitrile
- CCL
chemokine (C-C motif) ligand
- C-DIM
para-phenyl substituted diindolylmethane compound
- C-DIM4
1,1-bis(3′-indolyl)-1-(p-methoxyphenyl) methane
- C-DIM12
1,1-bis(3′-indolyl)-1-(p-chlorophenyl) methane
- ChIP
chromatin immunoprecipitation
- CoREST
corepressor for repressor element 1 silencing transcription factor
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethylsulfoxide
- DsiRNA
dicer-substrate RNA
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- GSK
glycogen synthase kinase
- HDAC
histone deacetylase
- HEK
human embryonic kidney cells
- IKK
IκB kinase
- LPS
lipopolysaccharide
- MPTPp
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine + probenecid
- NCOR
nuclear receptor corepressor
- NF-κB
nuclear factor κB
- NOS2
nitric oxide synthase
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PD
Parkinson’s disease
- PSN
penicillin-streptomycin-neomycin
- SB216763
3-(2,4-dichlorophenyl)-4-(1-methylindol-3-yl)pyrrole-2,5-dione
- siRNA
small interfering RNA
- TLR
Toll-like receptor
- TNFα
tumor necrosis factor α
Authorship Contributions
Participated in research design: De Miranda, Popichak, Hammond, Safe, Tjalkens.
Conducted experiments: De Miranda, Popichak, Hammond, Jorgensen, Phillips.
Contributed new reagents or analytic tools: Safe.
Performed data analysis: De Miranda, Popichak, Hammond, Jorgensen, Tjalkens.
Wrote or contributed to the writing of the manuscript: De Miranda, Popichak, Hammond, Safe, Tjalkens.
Footnotes
This work was supported by grants from the Michael J. Fox Foundation for Parkinson’s Research and the Consolidated Anti-Aging Foundation, and the National Institutes of Health National Institute of Environmental Health Sciences [Grants ES021656, P30-023512].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Bensinger SJ, Tontonoz P. (2009) A Nurr1 pathway for neuroprotection. Cell 137:26–28. [DOI] [PubMed] [Google Scholar]
- Brambilla R, Bracchi-Ricard V, Hu W-H, Frydel B, Bramwell A, Karmally S, Green EJ, Bethea JR. (2005) Inhibition of astroglial nuclear factor kappaB reduces inflammation and improves functional recovery after spinal cord injury. J Exp Med 202:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss H, Dörrie A, Schmitz ML, Frank R, Livingstone M, Resch K, Kracht M. (2004) Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. J Biol Chem 279:49571–49574. [DOI] [PubMed] [Google Scholar]
- Carbone DL, Popichak KA, Moreno JA, Safe S, Tjalkens RB. (2009) Suppression of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced nitric-oxide synthase 2 expression in astrocytes by a novel diindolylmethane analog protects striatal neurons against apoptosis. Mol Pharmacol 75:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I-H, Hong J, Suh EC, Kim JH, Lee H, Lee JE, Lee S, Kim C-H, Kim DW, Jo E-K, et al. (2008) Role of microglial IKKbeta in kainic acid-induced hippocampal neuronal cell death. Brain 131:3019–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Le W, Kompoliti K, Jankovic J, Mufson EJ, Kordower JH. (2006) Nurr1 in Parkinson’s disease and related disorders. J Comp Neurol 494:495–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SD, Lee SO, Chintharlapalli S, Abdelrahim M, Khan S, Yoon K, Kamat AM, Safe S. (2010) Activation of nerve growth factor-induced B alpha by methylene-substituted diindolylmethanes in bladder cancer cells induces apoptosis and inhibits tumor growth. Mol Pharmacol 77:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Miller JA, Hansen RJ, Lunghofer PJ, Safe S, Gustafson DL, Colagiovanni D, Tjalkens RB. (2013) Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson’s disease. J Pharmacol Exp Ther 345:125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Miller JA, Safe S, Tjalkens RB. (2015) Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol Sci 143:360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldingh Debernard KB, Mathisen GH, Paulsen RE. (2012) Differences in NGFI-B, Nurr1, and NOR-1 expression and nucleocytoplasmic translocation in glutamate-treated neurons. Neurochem Int 61:79–88. [DOI] [PubMed] [Google Scholar]
- Fan X, Luo G, Ming M, Pu P, Li L, Yang D, Le W. (2009) Nurr1 expression and its modulation in microglia. Neuroimmunomodulation 16:162–170. [DOI] [PubMed] [Google Scholar]
- García-Yagüe ÁJ, Rada P, Rojo AI, Lastres-Becker I, Cuadrado A. (2013) Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J Biol Chem 288:5506–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. (1998) NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16:225–260. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. (2006) Introduction to NF-kappaB: players, pathways, perspectives. Oncogene 25:6680–6684. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140:918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BP, Wen W, Strong MJ. (2002) Activated microglia (BV-2) facilitation of TNF-alpha-mediated motor neuron death in vitro. J Neuroimmunol 128:31–38. [DOI] [PubMed] [Google Scholar]
- Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. (2008) 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther 7:3825–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Smith CJ, Van Eldik LJ. (2004) Importance of MAPK pathways for microglial pro-inflammatory cytokine IL-1 beta production. Neurobiol Aging 25:431–439. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Bencze G, Cohen P, Tonks NK. (2013) The anti-inflammatory compound BAY-11-7082 is a potent inhibitor of protein tyrosine phosphatases. FEBS J 280:2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-O, Li X, Hedrick E, Jin U-H, Tjalkens RB, Backos DS, Li L, Zhang Y, Wu Q, Safe S. (2014) Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Mol Endocrinol 28:1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-O, Li X, Khan S, Safe S. (2011) Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin Ther Targets 15:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lee S-O, Safe S. (2012) Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol 83:1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Maguire-Zeiss KA, Federoff HJ. (2010) Future directions for immune modulation in neurodegenerative disorders: focus on Parkinson’s disease. J Neural Transm 117:1019–1025. [DOI] [PubMed] [Google Scholar]
- Nolan YM, Sullivan AM, Toulouse A. (2013) Parkinson’s disease in the nuclear age of neuroinflammation. Trends Mol Med 19:187–196. [DOI] [PubMed] [Google Scholar]
- Polazzi E, Monti B. (2010) Microglia and neuroprotection: from in vitro studies to therapeutic applications. Prog Neurobiol 92:293–315. [DOI] [PubMed] [Google Scholar]
- Qin C, Morrow D, Stewart J, Spencer K, Porter W, Smith R, 3rd, Phillips T, Abdelrahim M, Samudio I, Safe S. (2004) A new class of peroxisome proliferator-activated receptor gamma (PPARgamma) agonists that inhibit growth of breast cancer cells: 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes. Mol Cancer Ther 3:247–260. [PubMed] [Google Scholar]
- Saijo K, Crotti A, Glass CK. (2013) Regulation of microglia activation and deactivation by nuclear receptors. Glia 61:104–111. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. (2009) A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. (1998) Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA 95:4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey MG, Goldberg MS. (2010) Neuroinflammation in Parkinson’s disease: its role in neuronal death and implications for therapeutic intervention. Neurobiol Dis 37:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Lee S-O, Cho SD, Kim K, Khan S, Safe S. (2011) Activation of nuclear TR3 (NR4A1) by a diindolylmethane analog induces apoptosis and proapoptotic genes in pancreatic cancer cells and tumors. Carcinogenesis 32:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wang P, Ren H, Fan J, Wang G. (2009) NGFI-B nuclear orphan receptor Nurr1 interacts with p53 and suppresses its transcriptional activity. Mol Cancer Res 7:1408–1415. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Bruemmer D. (2010) NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol 30:1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.