Abstract
COH29 [N-(4-(3,4-dihydroxyphenyl)-5-phenylthiazol-2-yl)-3,4-dihydroxybenzamide], a novel antimetabolite drug developed at City of Hope Cancer Center, has anticancer activity that stems primarily from the inhibition of human ribonucleotide reductase (RNR). This key enzyme in deoxyribonucleotide biosynthesis is the target of established clinical agents such as hydroxyurea and gemcitabine because of its critical role in DNA replication and repair. Herein we report that BRCA-1–defective human breast cancer cells are more sensitive than wild-type BRCA-1 counterparts to COH29 in vitro and in vivo. Microarray gene expression profiling showed that COH29 reduces the expression of DNA repair pathway genes, suggesting that COH29 interferes with these pathways. It is well established that BRCA1 plays a role in DNA damage repair, especially homologous recombination (HR) repair, to maintain genome integrity. In BRCA1-defective HCC1937 breast cancer cells, COH29 induced more double-strand breaks (DSBs) and DNA-damage response than in HCC1937 + BRCA1 cells. By EJ5– and DR–green fluorescent protein (GFP) reporter assay, we found that COH29 could inhibit nonhomologous end joining (NHEJ) efficiency and that no HR activity was detected in HCC1937 cells, suggesting that repression of the NHEJ repair pathway may be involved in COH29-induced DSBs in BRCA1-deficient HCC1937 cells. Furthermore, we observed an accumulation of nuclear Rad51 foci in COH29-treated HCC1937 + BRCA1 cells, suggesting that BRCA1 plays a crucial role in repairing and recovering drug-induced DNA damage by recruiting Rad51 to damage sites. In summary, we describe here additional biologic effects of the RNR inhibitor COH29 that potentially strengthen its use as an anticancer agent.
Introduction
The prototypic antimetabolite drug hydroxyurea (HU) has been used to treat a variety of human cancers, including chronic myelogenous leukemia, head and neck cancer, and others (Hehlmann, 2003; Shewach and Lawrence, 2007). Its primary anticancer and cellular target is ribonucleotide reductase (RNR), which reduces ribonucleotides to their corresponding deoxy forms to supply dNTPs for DNA replication and repair (Reichard and Ehrenberg, 1983; Xue et al., 2003). The human RNR is composed of the hRRM1 and hRRM2 subunits (Reichard and Ehrenberg, 1983; Xue et al., 2003). After a genotoxic stimulus, an alternate RNR enzyme, which is composed of hRRM1 and p53R2 (a homolog of hRRM2 transactivated by the tumor suppressor protein p53), is induced to supply dNTPs for DNA repair (Shao et al., 2004). Within cells, HU inhibits both types of RNR (Shao et al., 2004) by generating free radicals via oxidative transformation (Young and Hodas, 1964) that quenches free radical–mediated catalysis (Reichard and Ehrenberg, 1983). Blocking this signaling can arrest DNA replication and reduce cell growth (Shewach and Lawrence, 2007); however, therapeutically, HU is limited by its short half-life and problematic side effects, most notably myelosuppression and gastrointestinal and dermatologic effects (Platt, 2008).
COH29 [N-(4-(3,4-dihydroxyphenyl)-5-phenylthiazol-2-yl)-3,4-dihydroxybenzamide] is an RNR inhibitor that demonstrates promise as an anticancer agent and is currently in preclinical development at City of Hope Cancer Center (Duarte, CA). COH29 is an aromatically substituted thiazole compound that occupies a structurally conserved ligand-binding pocket on the hRRM2 subunit located at the hRRM1/hRRM2 interface, thereby inhibiting hRRM1/hRRM2 assembly and effectively inhibiting RNR activity (Zhou et al., 2013). In vitro COH29 inhibits the proliferation of multiple human cancer cell lines with an IC50 less than 10 µM in most cases. Treatment of cancer cells with COH29 led to a dose-dependent S-phase arrest, induction of apoptosis, and cell death (Zhou et al., 2013). One major advantage of COH29 over other RNR inhibitors in development, such as 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (Triapine, 3-AP), is that it does not appear to be an iron chelator, thus reducing the potential side effects.
In response to DNA damage, numerous DNA-repair pathway proteins collectively act to restore DNA continuity and genomic integrity (Helleday et al., 2008). Among these different repair pathways, base-excision repair and nucleotide-excision repair are both involved in the removal of lesions and their replacement with short stretches of DNA. The continuing presence of single-strand breaks during DNA replication will lead to stalled replication forks, whose resolution requires recombination (HR) or nonhomologous end joining (NHEJ) repair, which is responsible for DNA double-strand breaks (DSBs) (Moeller et al., 2009; Gottipati et al., 2010). The biochemical processes of HR are mediated by multiple conserved factors, including the essential recombinase RAD51 and tumor suppressors BRCA1 and BRCA2 (Curtin 2012).
The efficacy of DNA-damaging drugs is highly influenced and is modulated by cellular DNA repair capacity (Helleday et al., 2008). Indeed, small-molecule inhibitors of DNA repair have been combined with conventional chemotherapy drugs in preclinical studies (Miknyoczki et al., 2003), indicating that the DNA repair machinery is a promising target for novel cancer treatments.
Herein we report that COH29 exhibits enhanced cytotoxicity in BRCA1-deficient HCC1937 cells compared with HCC1937 + BRCA1 cells accompanied by significant DNA DSB marker (γH2AX) accumulation in the BRCA1-defective cells, suggesting that BRCA1 prevents the prolonged presence of DSBs. In addition, we also found that COH29 reduced NHEJ repair efficiency in a concentration-dependent manner. In the setting of BRCA1-defective HCC1937 cells, which we also show are HR-deficient, this inhibition of NHEJ by COH29 dramatically reduced repair of DNA lesions. Indicative of this result is that after COH29 treatment, fewer Rad51 nuclear foci were observed in HCC1937 than in HCC1937 + BRCA1 cells. In addition, our microarray results revealed that COH29 downregulated various DNA repair genes. These data suggest that defective HR and NHEJ DNA repair pathways may contribute to the cytotoxicity of COH29 in BRCA1-deficient cells and, as a corollary, that BRCA1 status plays a central role in determining the cytotoxicity of COH29 in cancer cells.
Materials and Methods
Cell Lines.
All cell lines were acquired from the American Type Culture Collection (Manassas, VA), and were maintained in RPMI 1640 medium (Mediatech, Inc., Manassas, VA) with 10% fetal bovine serum, 2 mM glutamine, and 100 U of penicillin and 100 µg of streptomycin per milliliter of medium (Sigma-Aldrich, St. Louis, MO) at 37°C in 5% CO2. To isolate HCC1937 + BRCA1 cells, parental HCC1937 cells were transfected with pcDNA3.1 plasmid expressing full-length BRCA1 cDNA. Stable transfectant clones were selected and used for drug sensitivity assays. For stable transfection, cells at 30–40% confluence were incubated overnight with 2 μg of plasmid DNA, using FuGENE 6 transfectin reagent (Roche Molecular Biochemical, Monza, Italy) according to the manufacturer’s instructions. Cells were then selected in puromycin (1 μg/ml) (Invitrogen Life Technologies, La Jolla, CA). After 20–30 days, viable puromycin-resistant colonies from HCC1937 transfections were expanded and screened. The clones that stably expressed puromycin and retained growth potential were assayed for BRCA1 expression by Western blot analysis. By Western blot analysis, we evaluated the restoration of BRCA1 expression in the puromycin-resistant cDNA/transfectant cells. These transfected cells showed an increased expression of BRCA1 protein, suggesting effective restoration of protein expression.
Reagents.
COH29 was synthesized and purified at City of Hope. All other recombinant proteins and antibodies were obtained from commercial sources: γH2AX, Cell Signaling Technology (Danvers, MA); Rad51, Novus (Littleton, CO); β-actin, Millipore (Billerica, MA); antibodies specific to FOXO3 (H-144 and N-16, 1:1000), phospho-H2AX serine-139 (γ-H2AX, 1:1,000), Rad51 (1:1000), β-tubulin (1:1000), Lamin A/C (1:2000 dilution) PARP, and anti-mouse, and anti-rabbit IgGs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against FOXO3 (1:1000) and phospho–ataxia-telangiectasia-mutated (ATM) serine-1981 (ATM-pS1981; 1:1000 dilution) were obtained from Epitomics (Burlingame, CA) and Millipore, respectively. An antibody against p53-pS15 was purchased from Cell Signaling Technology. An anti-p27Kip1 antibody was purchased from BD PharMingen (San Diego, CA). Alexa 488 (green)– and Alexa 594 (red)–conjugated secondary antibodies were obtained from Molecular Probes (Eugene, OR). Anti-rabbit IgG (whole molecule)–fluorescein isothiocyanate antibody was purchased from Sigma. Rhodamine Red-X goat anti-mouse IgG was purchased from Invitrogen (Carlsbad, CA).
Immunofluorescence.
Immunofluorescence experiments on HCC1937 and HCC1937 + BRCA1 cells were conducted as described previously (Chung et al., 2012; Hu et al., 2014). Briefly, cells were grown on glass coverslips. After treatment with COH29 (10 μM) for 24 hours, cells were fixed with 4% paraformaldehyde for 10 minutes and permeabilized with Triton X-100 (0.5%). The coverslips were washed with phosphate-buffered saline (PBS) and blocked with PBS-containing 2% bovine serum albumin, incubated with an antibody specific to FOXO3 or ATM-pS1981 or γ-H2AX or Rad51 (1:50–1:200 dilution), followed by Alexa 488–conjugated anti-rabbit or anti-mouse (1:200), Alexa 594–conjugated anti-goat (1:100) secondary antibodies (Molecular Probes). Cells were incubated with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) to stain the nuclei. Specific staining was visualized, and images were captured with a Leica SP2 AOBS confocal laser scanning microscope (Leica Microsystems, Buffalo Grove, IL). To measure foci-positive cells, we used ∼300 cells randomly captured by confocal microscopy. The percentages of foci-positive cells were calculated from cells containing at least five foci. Each error bar presented is the mean of standard deviation.
Subcellular Fractionation and Immunoblotting.
For details of the subcellular fractionation, cells were trypsinized and washed with cold PBS solution twice. After centrifugation at 1200g for 5 minutes, cells were incubated in buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing 0.2% NP40, supplemented with protease inhibitors (5 μg/ml each of pepstatin, leupeptin, and aprotinin), and phosphatase inhibitors on ice for 5 minutes. After centrifugation at 1000g for 5 minutes, the supernatant was collected (i.e., cytoplasmic fraction), and pellets were washed with the same buffer twice. The washed samples were extracted for 40 minutes on ice with fractionation buffer containing 0.5% NP40 for nuclear fraction. All the samples were sonicated and clarified by centrifugation at 16,000g for 15 minutes. Protein concentrations of all fractions were determined using Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA). Immunoblotting was performed as described previously (Chung et al., 2012; Hu et al., 2014).
Cytotoxicity and Viability Assays.
Cells were seeded into 96-well plates in 100 µl of complete medium at 2000 to 5000 cells per well, depending on the cell line’s growth rate. After overnight incubation, test compound was added to each well at various concentrations in 50 µl of culture medium. After a further incubation for 96 hours at 37°C, fluorescein diacetate (final concentration: 10 mg/ml) and eosin Y [final concentration: 0.1% (w/v)] were added to each well, and the cells were incubated for an additional 20 minutes at 37°C. Cytotoxicity was assessed by Digital Imaging Microscopy System detection (Keshelava et al., 2005).
Viability was assessed using MTS [(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)] as previously described (Zhou et al., 2013).
Orthotopic Tumor Model.
Experiments in mice were conducted under a protocol approved by the Institutional Animal Care and Use Committee of City of Hope. Female NOD scid γ mice with HCC1937 and HCC1937 + BRCA1 cells implanted into the mammary fat pads around the inguinal area were administered 400 mg/kg COH29 in 30% solutol (BASF North America, Florham Park, NJ) or vehicle by daily gavage for 28 days. Because HCC1937 and HCC1937 + BRCA1 cells form slowly growing tumors, they were implanted using Matrigel (Becton-Dickinson Biosciences, San Jose, CA). To establish tumors, 4 × 106 cells in 200 µl serum-free medium containing 50% Matrigel were injected into the mammary fat pads around the inguinal area of a pair of 8-week-old female NOD scid γ mice. Once the initial tumors reached 13 mm in diameter, they were dissected out, minced into 3-mm pieces, and implanted into the inguinal area of the mammary fat pads of the experimental mice. When the transplanted tumors reached approximately 50 mm2, drug treatment was initiated. Tumor diameters were measured by digital calipers over a 28-day period, and the tumor volume was calculated using the formula 0.5 × width2 × length for each time point. Mice were euthanized once the tumors reached approximately 500 mm3, in compliance with City of Hope’s Institutional Animal Care and Use Committee stopping rules. Student’s t test was used to determine the statistical significance between COH29 treatment and corresponding vehicle control. P value less than 0.05 (two sides) was considered to indicate statistical significance.
DNA Repair Assays.
Reporter cell lines for GFP-based DNA damage repair assays were established by stable transfection of HCC1937 and HCC1937 + BRCA1 cells with the pimEJ5GFP reporter plasmid for NHEJ (Bennardo et al., 2008) and the pHPRT-DRGFP reporter plasmid for HR (Pierce et al., 2001), respectively, and selected with 0.3 μg/ml puromycin. The resultant HCC1937-EJ5GFP and HCC1937 + BRCA1-DRGFP cells were first pretreated with COH29 for 24 hours and then transiently transfected with a predetermined mixture of pCBA-Scel plasmid to express I-Scel endonuclease and a plasmid to express DsRed (red fluorescent protein) protein, which served as the control for transfection efficiency. After incubation with COH29 for another 48 hours, 5 × 105 cells per transfection were analyzed by fluorescence-activated cell sorting to count total GFP and DsRed protein positive cells. Each assay was performed three times, and data were presented as the ratio of GFP-positive to DsRed-positive cells among whole cells.
Small Interfering RNA Interference Assay.
The construction of the anti-human BRCA1 small interfering RNA (siRNA)–expressing plasmid was performed as described (Un et al., 2006) using previously published anti-human BRCA1 siRNA sequences (5′-UCACAGUGUCCUUUAUGUA-3′ and 5′-UACAUAAAGGACACUGUGA-3′). In each case, the annealed oligonucleotide duplex encoding the siRNA was subcloned into the expression vector psiRNA-hH1zeo (InvivoGen, San Diego, CA) to express under the control of the RNA polymerase III–dependent H1 RNA promoter. Cells were transfected with the indicated plasmid at equimolar concentrations via electroporation.
Zebrafish Genotoxicity Assay.
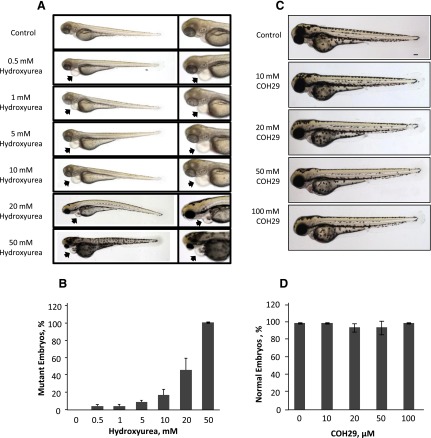
Zebrafish (Danio rerio) were obtained from Zebrafish Core Facility of Taipei Medical University and maintained at 28°C on a 14 hours light/10 hours dark cycle. Embryos were incubated at 28°C, and different developmental stages were determined as described (Westerfield, 1993). Fifteen wild-type embryos each were treated with concentrations of HU (0, 5, 10, 20, 50 mM) or COH29 (0, 10, 20, 50, 100 μM) at 20 hours of postfertilization to evaluate the mutagenic effect. Treated embryos were observed at 2, 3, 4, 5, and 6 days postfertilization (dpf). At 6 dpf, the percentage exhibiting developmental abnormalities and the survival rate were determined. Embryos were observed using an Olympus IX70-FL inverted fluorescence microscope (Scientific Solutions Americas Corp., Waltham, MA). Images were taken using SPOT digital camera system (Diagnostic Instruments, Sterling Heights, MI) and assembled with ImageJ Software (Schneider et al., 2012).
Microarray Analysis.
For microarray analysis, HCC1937 and HCC1937 + BRCA1 cells were treated with 10 µM COH29 or vehicle for 24 hours. Total RNA was extracted using TRIzol reagent (Invitrogen). Synthesis and labeling of complementary RNA (cRNA) targets, hybridization of GeneChips, and signal detection were carried out by the Integrated Genomics Core Facility at City of Hope. The Affymetrix Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA) was used for microarray gene expression profiles. The microarray was carried out using Ambion’s WT Expression kit (Life Technologies) and Affymetrix’s GeneChip Terminal labeling system. Briefly, 100 ng of total RNA was used to start the first-strand cDNA synthesis using an engineered random primer plus polyT7 promoter. After the second strand cDNA synthesis, the antisense cRNA (in vitro transcription) was generated using T7 RNA polymerase. Then 10 μg of cRNA was used to start the second cycle of cDNA synthesis using random primers plus dUTP and dNTP. The single-strand cDNA was fragmented and then end-labeled with biotinylated nucleotides in the presence of terminal deoxynucleotidyl transferase using Affymetrix’s WT Terminal Labeling kit. Labeled single-stranded cDNA (5 µg) was hybridized with an Affymetrix Human Gene 1.0 ST array, and the array was scanned using an Affymetrix GeneChip Scanner 3000 7G. Data have been deposited into NCBI Gene Expression Omnibus [GSE55004].
Statistical Processing of Microarray Data.
Microarray samples were Robust Multi-Array Analysis normalized (Irizarry et al., 2003) using Partek Genomics Suite (Version 6.6; Partek, Inc.), and genes were defined as differentially expressed if they showed at least a 1.2-fold change and a false discovery rate of <0.05. False discovery rate values were calculated using the method of Benjamini and Hochberg (1995) from the distribution of analysis of variance with Linear Contrast P values. Gene ontology (GO) (Ashburner et al., 2000) enrichment analysis was performed within Partek Genomics Suite, and GO categories were defined significant by Fisher’s exact test P < 0.05.
Results
COH29 Targets BRCA1-Defective Human Cancer Cells.
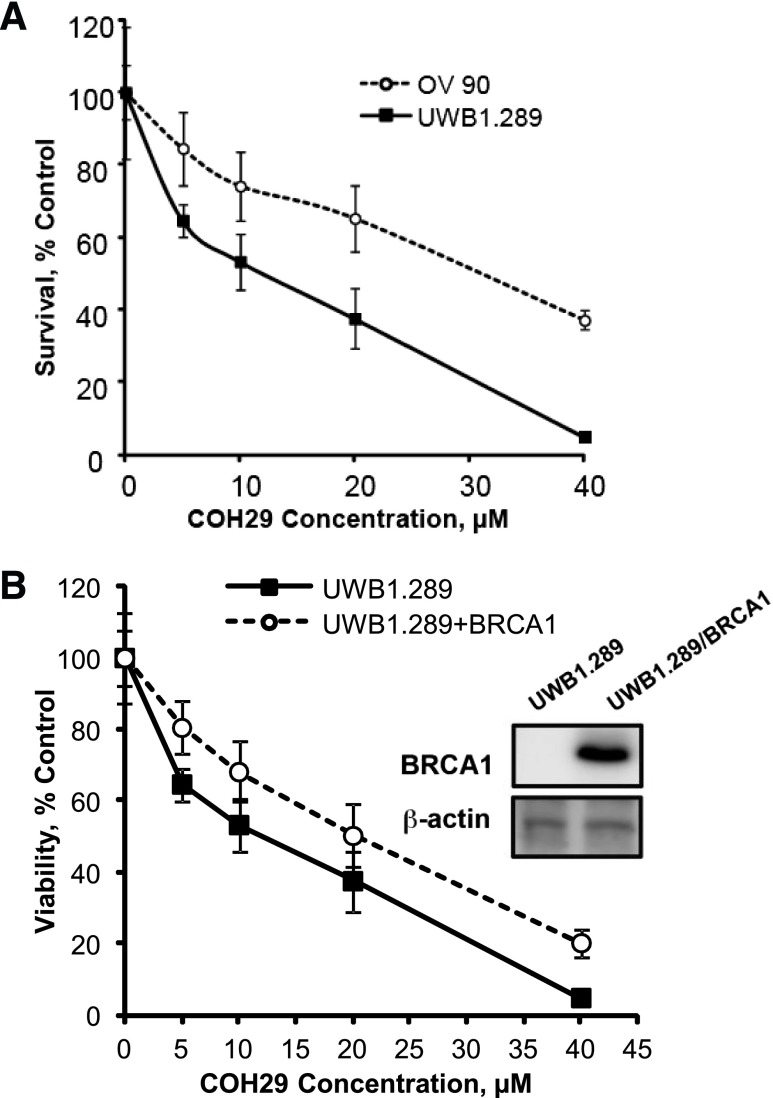
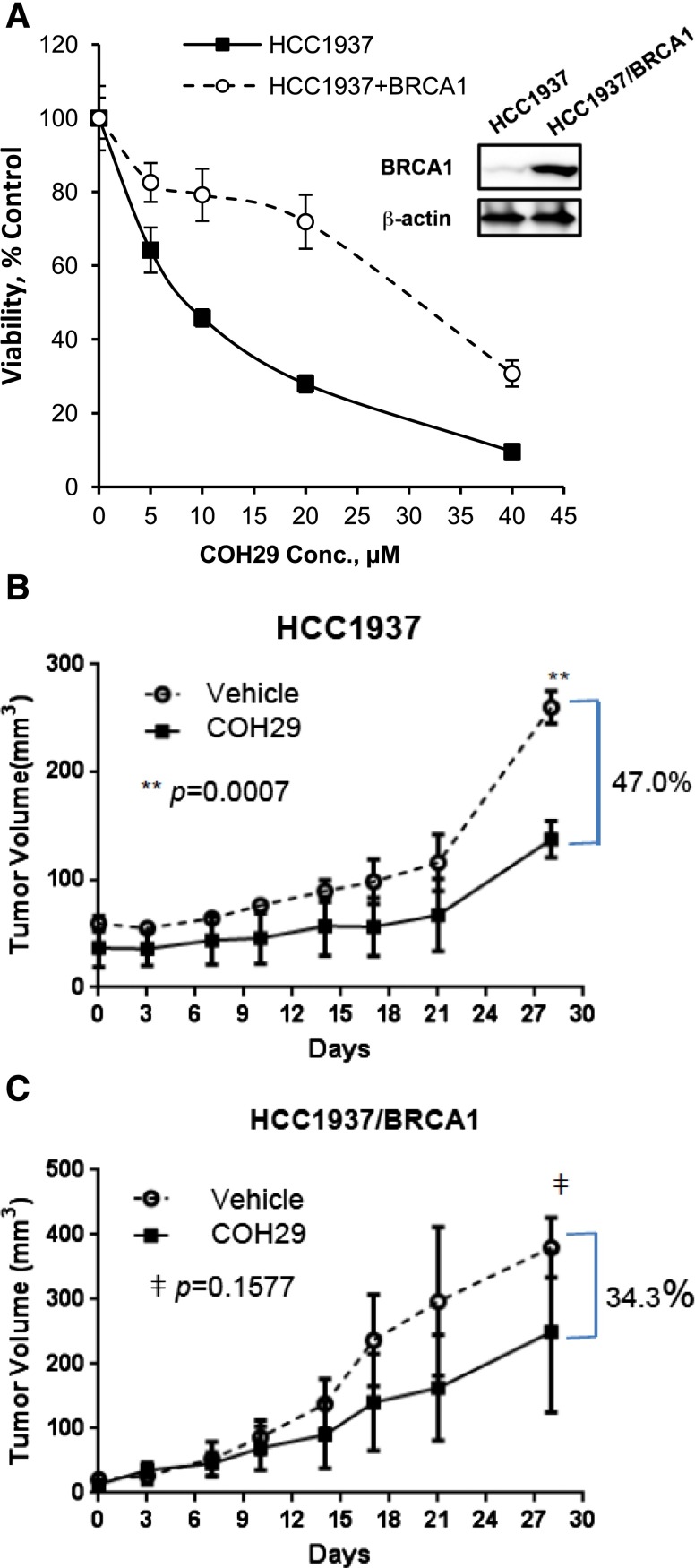
Our previous data showed the broad antitumor activity of COH29 in the NCI-60 cell line panel and that multiple human breast cancer cell lines, as well as human ovarian cancer cell lines, are sensitive to COH29 (Zhou et al., 2013). Breast and ovarian cancers occur with a greater frequency in carriers of a mutant BRCA1 gene than in the general population (Wooster and Weber, 2003). We therefore investigated the activity of COH29 in several cell lines with differing BRCA1 status, including OV90 (BRCA1 wild-type), UWB1.289 (BRCA1-mutant), HCC1937 (BRCA1-mutant), and HCC1937 + BRCA1 cells. As shown in Fig. 1A, the UWB1.289 ovarian cancer cell line, which expresses truncated BRCA1 protein as a result of the homozygous 2594delC mutation (DelloRusso et al., 2007), was more sensitive to COH29 (IC50: 12.30 ± 1.15 µM) than the OV90 human ovarian cancer cell line that expresses wild-type BRCA1 (IC50: 31.57 ± 3.35 µM). We further stably expressed BRCA1 in UWB1.289 ovarian cancer cells, which resulted in a more resistant phenotype in response to COH29 treatment compared with the parental UWB1.289 cells (Fig. 1B). Likewise, similar results were observed in an isogenic pair of human breast cancer cell lines. HCC1937 cells were more sensitive to COH29 than their BRCA1 wild type–expressing counterpart (HCC1937 + BRCA1) (Fig. 2A). The sensitivity of BRCA1-deficient cells to COH29 was further tested in an orthotopic tumor explant model. The growth of HCC1937 tumors implanted into mouse mammary fat pads was significantly (47.0%; P = 0.0007) suppressed by daily oral dosing with 400 mg/kg COH29 compared with vehicle by day 28 (Fig. 2B). In contrast, the growth of tumors established with the isogenic HCC1937 + BRCA1 cells in COH29-treated mice was not significantly different from that in vehicle controls at the same time point (34.3%; P = 0.1577) (Fig. 2C). As the HCC1937 + BRCA1–bearing animals were sacrificed per institutional guidelines at this time, no further comparisons between the effect of COH29 on the growth of the HCC1937 deficient xenografts and HCC1937 + BRCA xenografts could be made. The HCC1937 xenografts were continued for a total of 60 days, however, in which the suppression of tumor growth by COH29 continued (data not shown). The in vitro data indicated COH29 is more potent in BRCA1-defective cells. Among the IC50 values shown in Table 1, COH29 showed 4.8 times more potency in HCC1937 cells compared with HCC1937 + BRCA1. Therefore, we used these two cell lines in the subsequent experiments to investigate the cause of the differential sensitivity to COH29.
Fig. 1.
BRCA1 status affects COH29 cytotoxicity in ovarian cancer cells. (A) Dose-response curves for ovarian cancer cells expressing wild-type BRCA1 (OV90) or mutant BRCA1 (UWB1.289) incubated with COH29 for 72 hours. (B) Dose-dependent effects of COH29 on cell viability in UWB1.289 and UWB1.289 + BRCA1 cells. Cell viability was assessed by MTS assay. UWB1.289 + BRCA1 is a stable cell line derived from BRCA1-null UWB1.289 described in Materials and Methods. The points depicted represent an average of three independent experiments with error bars indicated.
Fig. 2.
BRCA1 status affects COH29 antitumor activity in breast cancer cells. (A) Cell viability of COH29 in HCC1937 and HCC1937 + BRCA1 cells assessed by MTS assay, and growth of tumor explants was established with HCC1937 (B) and HCC1937 + BRCA1 (C) cells in the mammary fat pads of female NOD scid γ (NSG) mice. Mice were treated daily with 400 mg/kg COH29 or vehicle as indicated. Results are the mean ± standard error of tumor measurements from four mice per group.
TABLE 1.
Comparison of the effect of COH29 in several cell lines
| BRCA1 | Cell Line | IC50 of COH29 |
|---|---|---|
| µM | ||
| Wild-type | OV90 | 31.57 ± 3.35 |
| Mutant | UWB1.289 | 12.30 ± 1.25 |
| UWB1.289/BRCA1 | 23.52 ± 2.38 | |
| Mutant | HCC1937 | 7.25 ± 0.64 |
| HCC1937/BRCA1 | 35.01 ± 3.63 |
Effect of COH29 on DNA Damage Checkpoints.
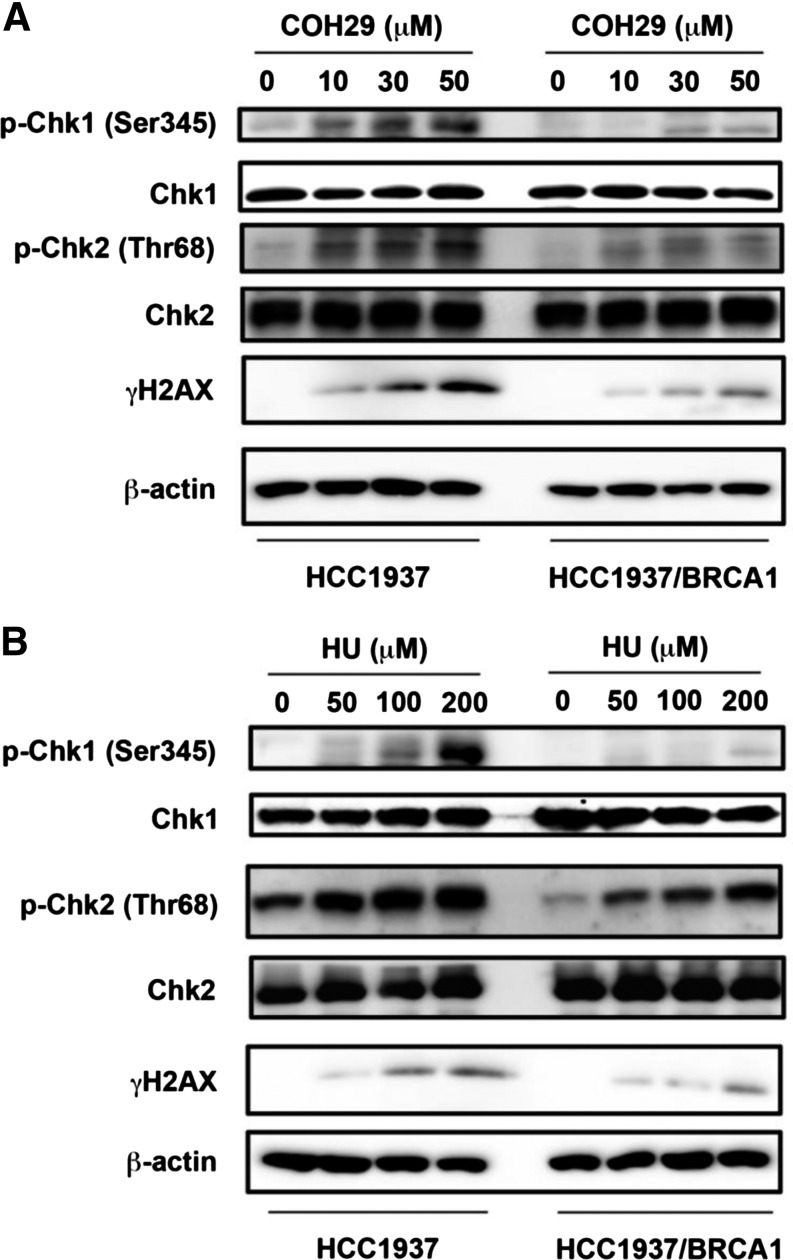
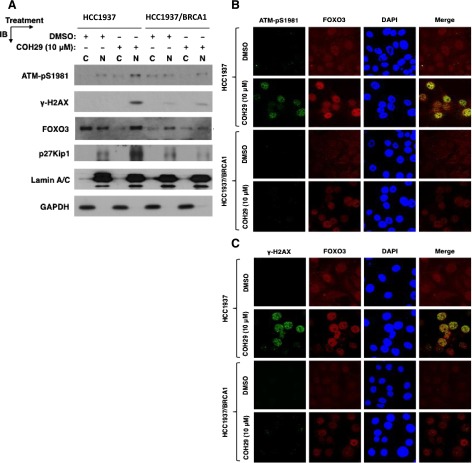
Next, we evaluated the effect of COH29 on DNA damage signaling in HCC1937 and HCC1937 + BRCA1 cells. COH29 induced significant phosphorylation of checkpoint kinase proteins Chk1 and Chk2 and increased the level of the DSB marker γ-H2AX in both cell lines (Fig. 3A). Notably, COH29 triggered more obvious signaling in HCC1937 cells compared with HCC1937 + BRCA1 cells in the same concentration range. A similar effect was also detected with HU treatment (Fig. 3B). It has been reported that foxo3 is necessary for ATM-mediated apoptotic signaling after DNA damage (Chung et al., 2012). As shown in Fig. 4A, induction of accumulation of p-ATM, γH2AX, foxo3, and its target protein p27 in the nucleus in response to COH29 was also observed. Furthermore, we found that γH2AX and p-ATM colocalize with foxo3 in the nucleus by confocal immunofluorescence microscopy (Fig. 4, B and C). These data suggest COH29 activates DNA damage signaling and recruitment of activated-ATM and foxo3 at DNA damage sites.
Fig. 3.
COH-29 treatment activates DNA damage checkpoint. (A) The effect of COH29 treatment on DNA damage checkpoint proteins in HCC1937 and HCC1937 + BRCA1 human breast cancer cells assessed by Western blot. (B) The effect of HU on DNA-damage response (DDR)–associated proteins was assessed by Western blot analysis. Cells were treated with COH29 at the indicated doses for 24 hours, and cell lysates were subjected to immunoblotting using the indicated antibodies.
Fig. 4.
Effect of COH29 on colocalization of DNA-damage response (DDR)–related proteins. (A) The effect of COH29 on DDR-associated proteins was assessed in cytoplasm and nucleus by Western blot analysis. Cells were treated with COH29 at the indicated doses for 24 hours, and cell lysates were subjected to immunoblotting (IB) using the indicated antibodies. FOXO3 activity is indicated by the levels of its downstream target p27Kip1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lamin A/C represent the fractionation and loading controls of cytosolic (C) and nuclear (N) extracts. (B) Phospho-ATM, γH2AX, together with foxo3 in the nucleus, were assessed by indirect immunofluorescence assay. DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethylsulfoxide.
Differential Gene Expression in COH29-Treated BRCA1-Defective Human Breast Cancer Cells.
We next performed genome-wide microarray analysis using the Affymetrix GeneChip microarray platform to identify the gene expression profiles and pathways affected by COH29 treatment. The RNA expression profile of COH29 treated HCC1937 breast cancer cells lacking BRCA1 was compared with that of COH29 treated HCC1937 + BRCA1 cells. Both HCC1937 and HCC1937 + BRCA1 cells showed GO enrichment for DNA repair genes (Table 2; P values ranging from 0.0046 to 0.0069) after exposure to 10 µM COH29 for 24 hours. These data suggest COH29 may interfere with several DNA repair pathways. This enrichment remained significant for the various subsets of DNA repair genes, including DNA ligation involved in DNA repair, but not HR repair genes when COH29-treated HCC1937 and HCC1937-BRCA1 cells were compared. This finding is consistent with what we observed in the NHEJ and HR repair reporter assays described as follows.
TABLE 2.
Gene ontology enrichment of genes downregulated by COH29 treatment
| Comparison | COH29 effect in BRCA1-deficient cells | COH29 effect in BRCA1 wild-type cells | BRCA1 wild-type vs. BRCA1 deficient cells |
|---|---|---|---|
| Treatment groups | HCC1937-COH29 vs. HC1937 | HCC1937 + BRCA1-COH29 vs. HC1937 + BRCA1 | HCC1937 + BRCA1-COH29 versus HCC1937-COH29 |
| DNA repair, P | 0.018 | 1.6 × 10−5 | 0.001 |
| DNA ligation involved in DNA repair, P | 0.00065 | 0.0066 | 0.06 |
| Double-strand break repair via nonhomologous end joining, P | 0.049 | 0.041 | 0.04 |
| Double-strand break repair via homologous recombination, P | 0.0069 | 0.0046 | 0.26 |
| Double-strand break repair, P | 0.0015 | 6.9 × 10−5 | 0.3 |
Effect of COH29 on DNA Double-Strand Breaks Repair.
DSBs can be repaired either by the HR or NHEJ pathways. Previous studies have shown that the EJ5- and DR-GFP reporter assays could be used to measure the ability of NHEJ and HR repair respectively (Bennardo et al., 2008). We next sought to elucidate the role of COH29 in DSB DNA repair by using integrated EJ5-GFP and DR-GFP reporters in HCC1937 cells. In this system, transient expression of I-Scel induces DSBs, which if repaired results in the generation of GFP+ cells. GFP signal was hardly detected in HCC1937-DR-GFP cells, whereas red fluorescent protein signal indicated successful transfection (Fig. 5B), consistent with the role of BRCA1 as an essential component of HR repair and indicating HR deficiency in BRCA1-mutant HCC1937 cells. In addition, we found that COH29 suppressed NHEJ repair efficiency in a concentration-dependent manner as shown by reduction in the percentage of GFP+ cells in HCC1937-EJ5-GFP assay (Fig. 5B). These data suggest that the COH29-suppressed NHEJ repair pathway may also contribute to the accumulation of DSBs in HR-deficient HCC1937 cells. Recruitment of Rad51 protein at lesions is a well documented step in the HR repair process to facilitate DNA damage repair (Deng and Brodie, 2000). Indeed, we detected obvious Rad51 foci formation in the nucleus in COH29-treated HCC1937/BRCA1 cells compared with COH29-treated HCC1937 cells (Fig. 5C). Similar effects were also observed in HU-treated cells. Taken together, these data further support our hypothesis that BRCA1 may be the key player in determining the sensitivity of cancer cells to COH29.
Fig. 5.
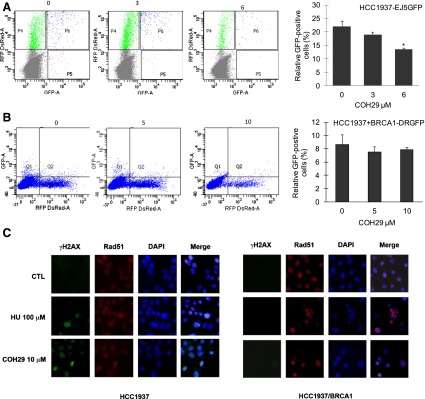
Effect of COH29 on DNA repair, and expression of DSB markers. Flow cytometric analysis of (A) HCC1937-EJ5GFP and (B) HCC1937 + BRCA1-DRGFP cells treated with 0, 0.25, and 0.5 of the IC50 concentration of COH29, with the normalized percentage of GFP-positive cells shown to the right. *P < 0.05, compared with COH-29 untreated cells. (C) The effect of COH29 on RAD51 and γ-H2AX foci was assessed by immunofluorescence assay described in Materials and Methods. CTL, control; DAPI, 4′,6-diamidino-2-phenylindole; RFP, red fluorescent protein.
Genotoxicity of COH29 in Embryos of Zebrafish.
We next assessed the genotoxic effect of COH29 in wild-type zebrafish embryos treated for 1–7 dpf (day postfertilization) with a range of doses of COH29 (0–100 μM). HU (0–50 mM) was included as a positive control because it is known to cause developmental defects. As expected, HU caused defects in the eyes and the heart by 4 dpf (Fig. 6A) and resulted in a dose-dependent increase in the number of mutant embryos (Fig. 6B). It is noteworthy that no developmental defects (Fig. 6C) or decreases in viability (Fig. 6D) were observed in the presence of COH29, indicating that COH29 exhibits antitumor activity without causing genotoxicity.
Fig. 6.
Effect of COH29 compared with HU in zebrafish genotoxicity assay. (A) Wild-type zebrafish embryos at 4 dpf exposed to HU as indicated. Morphologic changes in the eye and heart development are indicated by the arrowheads. (B) Bar graph of the effect (mutant embryos) of a series of different concentrations of HU on zebrafish (0, 0.5, 1, 5, 10, 20, 50 mM; n = 50, performed in triplicate). (C) Wild-type zebrafish embryos at 4 dpf exposed to COH29 as indicated; Bar = 100 µm. (D) Bar graph of the effect (survival embryos) of a series of different concentrations of COH29 on zebrafish (0, 10, 20, 50, 100 μM; n = 46, performed in triplicate).
Discussion
In this study, we sought to further define the biologic effects of the novel RNR inhibitor COH29, which effectively inhibits proliferation of various cancer cell lines, especially ovarian cancer and leukemic cells, and overcomes resistance to the RNR inhibitor hydroxyurea (Zhou et al., 2013). Building on our initial observation that the BRCA1-deficient ovarian cancer cell line UWB1.289 was particularly sensitive to COH29, we determined that BRCA1 status itself could account for this effect. Reconstitution of BRCA1 activity in the HCC1937 human breast cancer cell line, which expresses a truncated, inactive BRCA1 protein (Tomlinson et al., 1998) in the stable transfectant clone HCC1937 + BRCA1 blunted response to COH29 in vitro and in vivo. In addition, siRNA knockdown of BRCA1 increased sensitivity to COH29 in A2780 (BRCA1 wild-type) cells. As shown in Supplemental Fig. 1, 72-hour treatment with COH29 resulted in lower survival in A2780 cells transfected with BRCA1 siRNA than in those transfected with control siRNA. These data suggested that BRCA1 deficiency exaggerates the antiproliferative effect of COH29 treatment.
The signaling initiated by DNA damage is initially mediated by ATM and ATM and Rad3-related kinases. The Chk1 and Chk2 DNA-damage response kinases lie downstream of ATM and ATM and Rad3-related kinases (Abbas et al., 2013). We observed that COH29 treatment led to more significant activation of ATM, Chk1, Chk2, and γH2AX in HCC1937 compared with HCC1937 + BRCA1 cells. In addition, γH2AX and phospho-ATM colocalize with foxo3 in the nucleus. These data indicate COH29 triggers ATM-foxo3-γH2AX complexes at sites of DNA damage.
BRCA1 is a critical mediator of cellular response to DNA damage and of HR repair, (Moeller et al., 2009; Curtin, 2012), which suggested that COH29 potency is modulated by the perturbed DNA repair pathways in BRCA1-deficient cells. Our observation that the GFP + cell population was undetectable in HCC1937-DR-GFP cells suggested that HCC1937 cells are HR-deficient. When we investigated the effect of COH29 on the NHEJ repair pathway in HCC1937 cells using the EJ5-GFP reporter system, we found that COH29 suppressed NHEJ repair efficiency (Fig. 5A). Our microarray analysis (Table 2) also provided preliminary data in support of this conclusion, although further experiments covering a time course and validation of identified genes would be needed to draw a strong conclusion. This is the subject of ongoing investigation.
In response to DNA damage, Rad51 translocates from the cytosol to the nucleus to form nucleofilaments on single-stranded DNA, which is an essential step to promote the HR repair pathway (Haaf et al., 1995; Baumann et al., 1996). By confocal microscopy, we observed that COH29 induced significantly more γH2AX foci in HCC1937 cells compared with HCC1937 + BRCA1 cells. In contrast, COH29 induced Rad51 nuclear foci in HCC1937 + BRCA1 cells (Fig. 5C), suggesting that Rad51 has been recruited at damage sites to repair COH29-triggered DNA damage via the HR repair pathway in these BRCA1 wild-type cells. This effect of COH29 on Rad51 is similar to that documented for HU, which is known to stall replication forks (Petermann et al., 2010), with the important distinction that COH29 is 20-fold more potent than HU (Zhou et al., 2013) and is not appreciably genotoxic (Fig. 6). Taken together, these results suggested inhibition of the NHEJ repair pathway by COH29 could also contribute to COH29-induced DSBs in HR-deficient HCC1937 cells.
The NHEJ pathway is reported to be the major pathway for DNA repair of radiation-induced DSBs in mammalian cells (Riballo et al., 2004). Furthermore, a previous report has also proposed that quiescent/slowly cycling cancer stem cells are more likely to use the error prone NHEJ pathway, resulting in offspring with enhanced chemoresistance and metastatic abilities after replication (Maugeri-Sacca et al., 2012). This suggests that targeting the NHEJ pathway may be an effective way to kill cancer stem cells. Therefore, NHEJ inhibition may represent a potential strategy in patients with proficient NHEJ to increase the response to treatment.
The underlying mechanism of the effect of COH29 on DNA repair needs further evaluation, however. For instance, it is unclear whether this is a direct effect on the DNA repair machinery or a consequence of depletion of dNTPs resulting from RNR inhibition. A handful of reports have indicated that other RNR inhibitors also affect DNA repair pathways and or checkpoints. As mentioned, the prototypic RNR inhibitor HU, causes upregulation of Rad51, and formation of both Rad51 and γH2AX foci (Petermann et al., 2010). Gemcitabine, a nucleoside analog that also inhibits the RNR large subunit (Shao et al., 2006), has been shown to be more potent in BRCA1 deficient cells, synergize with cisplatin, and induce Rad51 and γH2AX foci (Alli et al., 2011). Lin and colleagues knocked down RRM2 subunit expression and observed results consistent with what we see for COH29, which is a specific RRM2 inhibitor (Zhou et al., 2013), activation of Chk1, and upregulation of γH2AX (Lin et al., 2011). The same group has recently shown that the RRM1 inhibitor Triapine (3-AP, 3-aminopyridine-2-carboxaldehyde thiosemicarbazone) caused Chk1 activation (Lin et al., 2014).
Collectively, our data provide evidence that COH29 is an RNR inhibitor that can activate DNA damage checkpoints and suppress DNA repair functions without significant genotoxicity. In summary, this report provides initial evidence that COH29 suppresses NHEJ repair, whether directly or indirectly, and that in the setting of HR deficiency, such as in BRCA1 mutants, this is a particularly effective approach in vitro and provides information to guide initial clinical development of this compound.
Supplementary Material
Abbreviations
- ATM
ataxia-telangiectasia-mutated
- COH29
N-(4-(3,4-dihydroxyphenyl)-5-phenylthiazol-2-yl)-3,4-dihydroxybenzamide
- cRNA
complementary RNA
- dpf
days post-fertilization
- DSB
double-strand breaks
- GFP
green fluorescent protein
- GO
gene ontology
- HR
homologous recombination
- HU
hydroxyurea
- NHEJ
nonhomologous end joining
- PBS
phosphate-buffered saline
- RNR
ribonucleotide reductase
- siRNA
small interfering RNA
Authorship Contributions
Participated in research design: M.-C. Chen, Zhou, Zhang, Yuan, Un, Wu, Malkas, M.C.-T. Hu, Yen.
Conducted experiments: M.-C. Chen, Zhang, Yuan, S. Hu, Chou, C.-H. Chen, Wu, Wang, Li, Su, Chung.
Performed data analysis: M.-C. Chen, Zhou, Zhang, Yuan, Chou, C.-H. Chen, Wu, X. Liu, Smith, Li, Warden, Z. Liu, Su, Chung.
Wrote or contributed to the writing of the manuscript: M.-C. Chen, Un, Chou, C.-H. Chen, Wu, Smith, Warden, M.C.-T. Hu, Yen.
Footnotes
This research was supported by the National Cancer Institute of the National Institutes of Health [Grants R01-CA127541, R01-CA113859, P30-CA033572]; Taiwan Medical University grant [TMU102-AE1-B43]; and by the Avon Foundation for Women grant [02-2013-051]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Abbas T, Keaton MA, Dutta A. (2013) Genomic instability in cancer. Cold Spring Harb Perspect Biol 5:a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli E, Sharma VB, Hartman AR, Lin PS, McPherson L, Ford JM. (2011) Enhanced sensitivity to cisplatin and gemcitabine in Brca1-deficient murine mammary epithelial cells (Abstract). BMC Pharmacol 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87:757–766. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 57:289–300. [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YM, Park SH, Tsai WB, Wang SY, Ikeda MA, Berek JS, Chen DJ, Hu MC. (2012) FOXO3 signalling links ATM to the p53 apoptotic pathway following DNA damage (Abstract). Nat Commun 3:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin NJ. (2012) DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 12:801–817. [DOI] [PubMed] [Google Scholar]
- DelloRusso C, Welcsh PL, Wang W, Garcia RL, King MC, Swisher EM. (2007) Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res 5:35–45. [DOI] [PubMed] [Google Scholar]
- Deng CX, Brodie SG. (2000) Roles of BRCA1 and its interacting proteins. BioEssays 22:728–737. [DOI] [PubMed] [Google Scholar]
- Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, Issaeva N, Sleeth K, Sharma RA, Helleday T. (2010) Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res 70:5389–5398. [DOI] [PubMed] [Google Scholar]
- Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA 92:2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehlmann R. (2003) Current CML therapy: progress and dilemma. Leukemia 17:1010–1012. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. (2008) DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8:193–204. [DOI] [PubMed] [Google Scholar]
- Hu T, Chung YM, Guan M, Ma M, Ma J, Berek JS, Hu MC. (2014) Reprogramming ovarian and breast cancer cells into non-cancerous cells by low-dose metformin or SN-38 through FOXO3 activation (Abstract). Sci Rep 4:5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. [DOI] [PubMed] [Google Scholar]
- Keshelava N, Frgala T, Krejsa J, Kalous O, Reynolds CP. (2005) DIMSCAN: a microcomputer fluorescence-based cytotoxicity assay for preclinical testing of combination chemotherapy. Methods Mol Med 110:139–153. [DOI] [PubMed] [Google Scholar]
- Lin ZP, Lee Y, Lin F, Belcourt MF, Li P, Cory JG, Glazer PM, Sartorelli AC. (2011) Reduced level of ribonucleotide reductase R2 subunits increases dependence on homologous recombination repair of cisplatin-induced DNA damage. Mol Pharmacol 80:1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZP, Ratner ES, Whicker ME, Lee Y, Sartorelli AC. (2014) Triapine disrupts CtIP-mediated homologous recombination repair and sensitizes ovarian cancer cells to PARP and topoisomerase inhibitors. Mol Cancer Res 12:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugeri-Saccà M, Bartucci M, De Maria R. (2012) DNA damage repair pathways in cancer stem cells. Mol Cancer Ther 11:1627–1636. [DOI] [PubMed] [Google Scholar]
- Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, et al. (2003) Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol Cancer Ther 2:371–382. [PubMed] [Google Scholar]
- Moeller BJ, Pasqualini R, Arap W. (2009) Targeting cancer-specific synthetic lethality in double-strand DNA break repair. Cell Cycle 8:1872–1876. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. (2001) Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15:3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt OS. (2008) Hydroxyurea for the treatment of sickle cell anemia. N Engl J Med 358:1362–1369. [DOI] [PubMed] [Google Scholar]
- Reichard P, Ehrenberg A. (1983) Ribonucleotide reductase: a radical enzyme. Science 221:514–519. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kühne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16:715–724. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Zhou B, Chu B, Yen Y. (2006) Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets 6:409–431. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Zhu L, Qiu W, Yuan YC, Xi B, Yen Y. (2004) In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res 64:1–6. [DOI] [PubMed] [Google Scholar]
- Shewach DS, Lawrence TS. (2007) Antimetabolite radiosensitizers. J Clin Oncol 25:4043–4050. [DOI] [PubMed] [Google Scholar]
- Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, Blum JL, Schneider NR, Wistuba II, Shay JW, et al. (1998) Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res 58:3237–3242. [PubMed] [Google Scholar]
- Un F, Qi C, Prosser M, Wang N, Zhou B, Bronner C, Yen Y. (2006) Modulating ICBP90 to suppress human ribonucleotide reductase M2 induction restores sensitivity to hydroxyurea cytotoxicity. Anticancer Res 26 (4B):2761–2767. [PubMed] [Google Scholar]
- Westerfield M. (1993) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio), M. Westerfield, Eugene, OR. [Google Scholar]
- Wooster R, Weber BL. (2003) Breast and ovarian cancer. N Engl J Med 348:2339–2347. [DOI] [PubMed] [Google Scholar]
- Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y. (2003) Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res 63:980–986. [PubMed] [Google Scholar]
- Young CW, Hodas S. (1964) Hydroxyurea: Inhibitory Effect on DNA Metabolism. Science 146:1172–1174. [DOI] [PubMed] [Google Scholar]
- Zhou B, Su L, Hu S, Hu W, Yip ML, Wu J, Gaur S, Smith DL, Yuan YC, Synold TW, et al. (2013) A small-molecule blocking ribonucleotide reductase holoenzyme formation inhibits cancer cell growth and overcomes drug resistance. Cancer Res 73:6484–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.