Abstract
Uncovering the complexity of mesenchymal stem cell (MSC) differentiation requires novel methods to capture the dynamics of the process in a quantitative and high-throughput manner. To this end, we developed a lentiviral array (LVA) of reporters to capture the dynamics of gene and pathway activity during MSC differentiation into adipogenic, chondrogenic, and osteogenic lineages. Our results identified signature promoters and pathways with unique activation profile for each MSC lineage. In combination with chemical inhibitors, lineage-specific reporters predicted the effects of signaling pathway perturbations on MSC differentiation. Interestingly, some pathways were critical for differentiation into all lineages, while others had differential effects on each lineage. Our study suggests that when combined with large chemical or siRNA libraries, the reporter LVA can be used to uncover novel genes and signaling pathways affecting complex biological processes such as stem cell differentiation or reprogramming.
Introduction
DNA or oligonucleotide microarrays have long been established as the experimental tool for monitoring genome-wide transcription of cells or tissues (i.e., mRNA abundance measurements) under different conditions or environmental stimuli. However, acquisition of dynamic information by microarrays is limited due to high labor and gene chip costs. In addition, microarray profiling cannot provide dynamic measurements in real time, because it requires sample destruction. Most important, mRNA abundance does not necessarily correlate with activity rendering it difficult to draw firm conclusions implicating a particular gene product (mRNA or protein) in the biological process under study. On the other hand, reporter assays measure the activity of a gene promoter (P) or a transcription factor (TF) binding site (Response Element (RE)) using reporter proteins, such as luciferase or green fluorescence protein (GFP). RE is defined as a consensus binding site in the promoter region of a gene where a specific TF binds and regulates transcription. In this context, measurements of the reporter activity reflect the activity of the upstream signaling pathway that leads to TF activation (e.g., phosphorylation), nuclear translocation and binding to RE initiating transcription of the reporter.
Transfected cell arrays that rely on promoter activation of a reporter gene showed high sensitivity and the ability to acquire kinetic data in real time.1,2,3,4,5,6 However, due to low transfection efficiencies, acquiring stably transfected cells requires long-term drug selection limiting the application of this approach to cell lines as opposed to primary cells. Also, the transient nature of transfection makes it difficult to follow cells for a long time (days to weeks) as required for stem cell differentiation. Therefore, development of novel strategies that enable high throughput, real-time, and quantitative measurements of pathway activation would greatly facilitate understanding of complex biological processes, such as stem cell fate commitment, and may also serve as platforms for screening siRNAs, small molecules, or therapeutics.
To this end, our group developed a lentiviral dual promoter vector (LVDP) that enables real-time monitoring of gene expression using two reporter genes from two independent promoters;7,8,9 (i) the P/RE of interest driving expression of a reporter protein (e.g., ZsGreen) and indicates promoter or pathway activation; and (ii) a constitutive promoter (e.g., CMV or PGK) driving expression of a second reporter (e.g., DsRed) that is used to measure transduction efficiency as well as internal normalization of the data, which thus becomes independent of the transduction efficiency or the number of gene copies per cell.7,8 To achieve this goal, polyadenylation, terminator, and insulator sequences were introduced between the two transcriptional units to minimize promoter interference, and ensure that expression of each reporter gene is independent from the other.9 Therefore, the LVDP enables quantitative measurements of reporter activity.
Here, we developed a set of LVDP vectors carrying P/RE that are activated by stimuli that induce MSC differentiation along the adipogenic, osteogenic, or chondrogenic lineages. An array of lentiviruses (lentivirus array, LVA) each carrying one distinct P/RE driving ZsGreen expression were used to transduce MSC in a 384-well format and monitor the dynamics of gene or pathway activation during differentiation using fluorescence imaging. Our experiments revealed signature reporters with distinct dynamic profiles for each MSC lineage. In addition, using a set of chemical inhibitors, we determined the effects of signaling pathways in this complex process. Interestingly, some pathways affected all lineages in a similar way, while others exhibited differential effects on each lineage. The results of the P/RE reporter activity were verified by functional differentiation assays, indicating that this technology may be applied for screening of large siRNA or chemical inhibitor libraries to identify novel pharmaceutical targets and decipher the role of signaling pathways during differentiation.
Results
Lentiviral vector for tracking MSC tri-lineage differentiation
Previously, our group developed a lentiviral dual promoter vector (LVDP) that enables quantitative measurements of promoter activity independent of virus titer using fluorescence microscopy (Figure 1a).9 In this study, we developed a small library of LVDPs to monitor gene and pathway activation during MSC differentiation. To this end, literature-reported promoters (-P) of adipogenic, chondrogenic, or osteogenic genes were subcloned into LVDP. Additionally, a number of transcription factor consensus-binding sites or RE were also cloned to capture signaling pathways that are activated during MSC differentiation. The entire list of P and RE (P/RE) that were cloned into the LVDP is shown in Table 1.
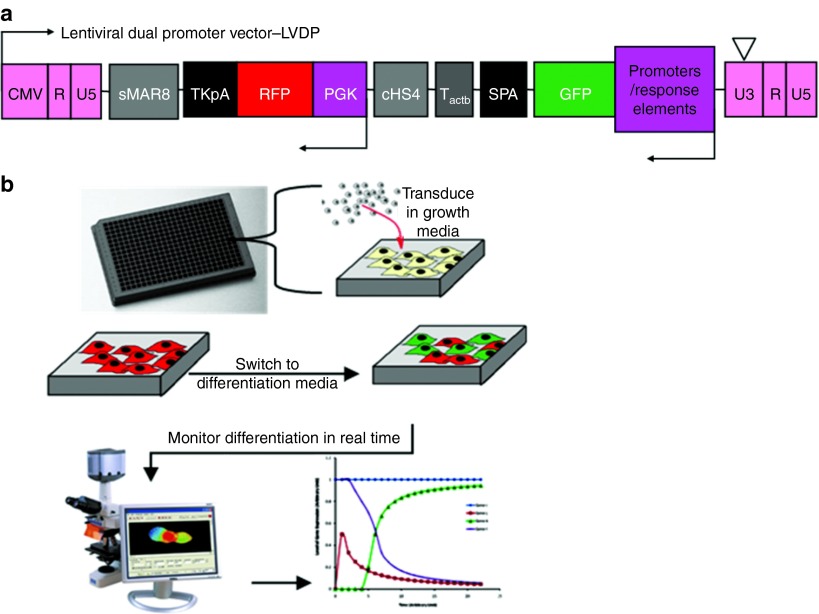
Figure 1.
Experiment setup. (a) Schematic of lentiviral dual promoter vector (LVDP). All P/RE shown in this study were subcloned into LVDP. (b) Schematic of experiment procedure. MSCs were seeded in a 384-well plate, transduced with P/RE carrying lentiviruses and were kept in GM until they reached confluence. Subsequently, they were treated with DM or GM and images of live cells were obtained daily.
Table 1. List of P/RE that were subcloned into lentiviral dual promoter vector and their targeted lineages.

To test our system, we employed human MSCs from two different anatomic locations namely bone marrow (BM) or hair follicle (HF). The latter were derived in our laboratory and were shown to be clonally multipotent as individual cells could be coaxed to differentiate into fat, bone, cartilage and SMC.10,11,12,13 BM-MSC and HF-MSC were transduced in 384-well plates using our LVDP recombinant viruses. After transduction, cells were allowed to reach confluency, and were subsequently induced to differentiate into osteo-, adipo- or chondro-genic lineages. Dynamic profiles of each reporter were generated by daily measurements of the green and red fluorescence intensity of live cells using an automated, customized, and high-throughput image analysis protocol. We established an image analysis pipeline for the open source software CellProfiler to automatically identify all fluorescent cells in every image and quantify the total fluorescence intensity. All data were subsequently categorized (according to P/RE, differentiation treatment and time), analyzed (normalization, mean and variance analysis, statistical analysis, etc.) and plotted as graphs or heatmaps using the algorithms we developed in Matlab.
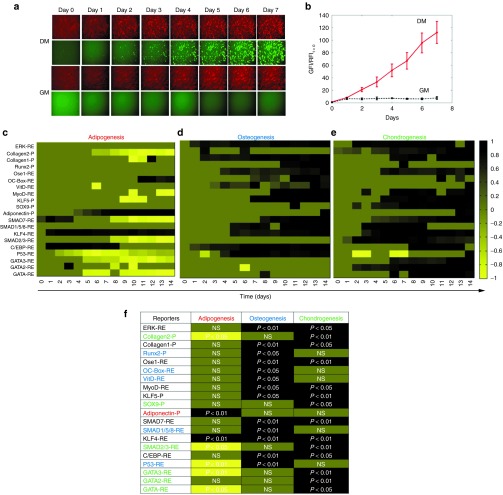
The effect of differentiation on each reporter was compared to that of cells cultured in growth medium (GM) (Figure 1b). As an example, Figure 2a illustrates the activation of Adiponectin-P during adipogenic induction of human BM-MSC. From day 0 to day 7, the red fluorescence intensity (RFI) remained unaffected by differentiation medium (DM) (compared to GM) as expected, since DsRed was driven by the constitutively active PGK promoter. On the other hand, the green fluorescence intensity (GFI) increased under differentiation conditions as ZsGreen was driven by the Adiponectin-P. The normalized data captured the fold increase of the Adiponectin-P activity over time in culture (Figure 2b).
Figure 2.
Dynamic P/RE activity data during stem cell differentiation. (a,b) Example Adiponectin-P activity in BM-MSCs over time after induction of adipogenesis: (a) Fluorescence images; (b) Kinetics of normalized fluorescence intensity dynamics shown as fold increase as compared to t = 0 when differentiation was induced. (c–e) Illustration of reporter activation in the form of heatmaps during adipo-(c), osteo- (d), or chondro-genic (e) differentiation for 14 days. (f) Graphic illustration of the activation of each reporter during differentiation as resulted by statistical analysis with the AUC method. All normalized fluorescence intensities were further normalized to the maximum fold increase (or decrease) of between DM and GM treatment. The colors range from black denoting highly upregulated to yellow denoting highly downregulated P/RE activity. Dark green color represents no significant difference between DM and GM. Red label: reporter for adipogenesis, Blue label: reporter for osteogenesis and Green label: reporter for chondrogenesis. NS, not significant difference
The dynamic data are summarized as heatmaps representing differentiation of human HF-MSCs into all three lineages (Figure 2c–e). The fold changes of reporter activity in DM over GM are indicated with a range of colors from yellow to black, representing significantly decreased or increased activity, respectively. The heatmaps allow for simultaneous comparison of the response of P/RE under the three differentiation conditions. In addition, we performed statistical analysis with the AUC method, which provided collective information for the entire kinetic curve (Figure 2f). Each P/RE is color labeled as red, blue, or green to represent adipo-, osteo- or chondro-genenic lineage, respectively, based on the following criterion: a P/RE is considered a reporter for a particular lineage if it is activated during differentiation into that lineage, but shows no difference or is downregulated during differentiation into the other two lineages. The maximum fold difference between DM and GM and the exact day when it occurred are also depicted in Supplementary Table S1. Finally, the dynamic differentiation profiles of the reporters are shown in Figure 3 and Supplementary Figure S1.
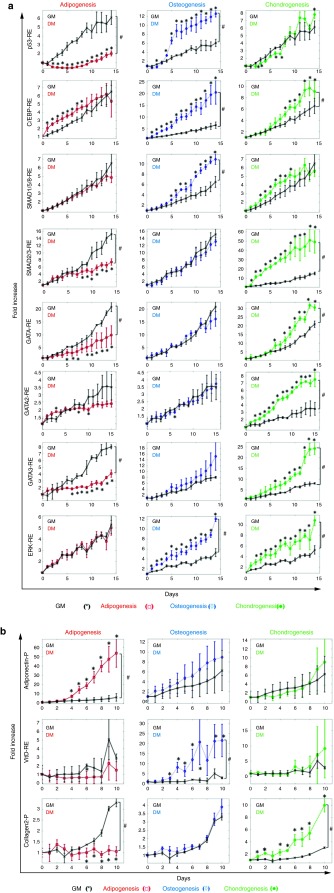
Figure 3.
Dynamics of P/RE activity during adipo, osteo, or condrogenic differentiation. (a) Normalized activity of the following RE: p53-RE, C/EBP-RE, SMAD1/5/8-RE, SMAD2/3-RE, GATA-RE, GATA2-RE, GATA3-RE and ERK-RE; (b) Normalized activity of adiponectin-P, vitaminD-RE and collagen2-P when cells were treated with adipogenic (red lines, □), osteogenic (blue lines, ◊) and chondrogenic (green lines, •) differentiation medium, respectively; control cells were cultured in GM (black lines x). Normalized fluorescence intensity was plotted as fold-increase over day 0, when differentiation was induced. All values represent the mean ± SD of triplicate samples in a representative experiment (n = 3). * denotes P < 0.05 between DM and GM using individual as calculated by t-test; and # denotes P < 0.05 between DM and GM as calculated by the AUC method.
Signature responses mapping differentiation
Our results revealed pathways that were differentially regulated during differentiation across the three lineages (Figure 3a). First, p53-RE was rapidly down-regulated during early stages of adipogenic differentiation reaching a minimum on day 6 (−7.81 ± 1.34-fold compared to GM) and increasing slightly up to day 14. In contrast, p53-RE was significantly upregulated during osteogenic differentiation exhibiting maximal upregulation on day 6 postinduction (2.62 ± 0.41-fold compared to GM). On the other hand, chondrogenic differentiation did not affect the p53-RE activity (AUC, P > 0.05), suggesting that the response of this pathway is unique to each lineage.
C/EBP-RE was significantly activated during osteogenesis C/EBP-RE (3.5 ± 0.44 fold compared to GM; AUC, P < 0.05). Interestingly, the same RE was activated—albeit to a lesser extent—during the early stages (0–8 days) of adipogenic differentiation (1.72 ± 0.35-fold compared to GM, P < 0.05) and the late stages of chondrogenic differentiation (after day 8) reaching a maximum on day 12 (1.71 ± 0.21-fold, P < 0.05). As expected, the SMAD1/5/8-RE exhibited high activity over the entire span of osteogenic differentiation (AUC, P < 0.05) but was only marginally upregulated under chondrogenic conditions (AUC, P > 0.05) and showed no response to adipogenic induction (AUC, P > 0.05).
The TGF-β pathway is critical for chondrogenic differentiation and as a result the SMAD2/3-RE was significantly upregulated during chondrogenesis, reaching 5.89 ± 0.50-fold on day 7. On the other hand, the SMAD2/3-RE activity decreased in adipogenic media (−2.16 ± 0.27-fold on day 13 as compared to GM), but showed no change during osteogenic induction (AUC, P > 0.05). Surprisingly, chondrogenic induction significantly up-regulated the GATA-REs resulting in 1.88 ± 0.24-fold increase on day 10 for GATA-RE, 2.32 ± 0.18-fold increase on day 12 for GATA2-RE and 3.07 ± 0.37-fold increase on day 13 for GATA3-RE. On the other hand, two out of three GATA-REs decreased during adipogenesis (GATA-RE: −2.10 ± 0.86-fold on day 14, and GATA3-RE: −2.88 ± 0.28-fold on day 12) but showed no response to osteogenic induction (AUC, P > 0.05). Finally, ERK-RE was continuously upregulated during both osteogenic and chondrogenic differentiation (AUC, P < 0.05) but showed no response to adipogenic induction (AUC, P > 0.05).
Unique identifiers for lineage specificity
In addition to signature pathways describing MSC differentiation, our screening also uncovered lineage-identifying P/RE reporters (Figure 3b). During the 10-day experiment, Adiponectin-P activity was highly upregulated during adipogenesis, (9.62 ± 2.81-fold with respect to GM on day 9; AUC, P < 0.05) but showed no significant change during osteogenic or chondrogenic induction (AUC, P > 0.05). Similarly VitaminD-RE exhibited significant up-regulation during osteogenesis (11.84 ± 6.46-fold with respect to GM on day 7; AUC, P < 0.05) but no significant response to adipogenesis or chondrogensis (AUC, P > 0.05). Lastly, the collagen2-P was significantly up-regulated during chondrogenesis (3.31 ± 0.37-fold with respect to GM on day 9; AUC, P < 0.05), did not respond to osteogenesis (AUC, P > 0.05) and decreased in adipogenesis (AUC, P < 0.05).
LVDP reporters as screening tool to identify pathways involved in MSC differentiation
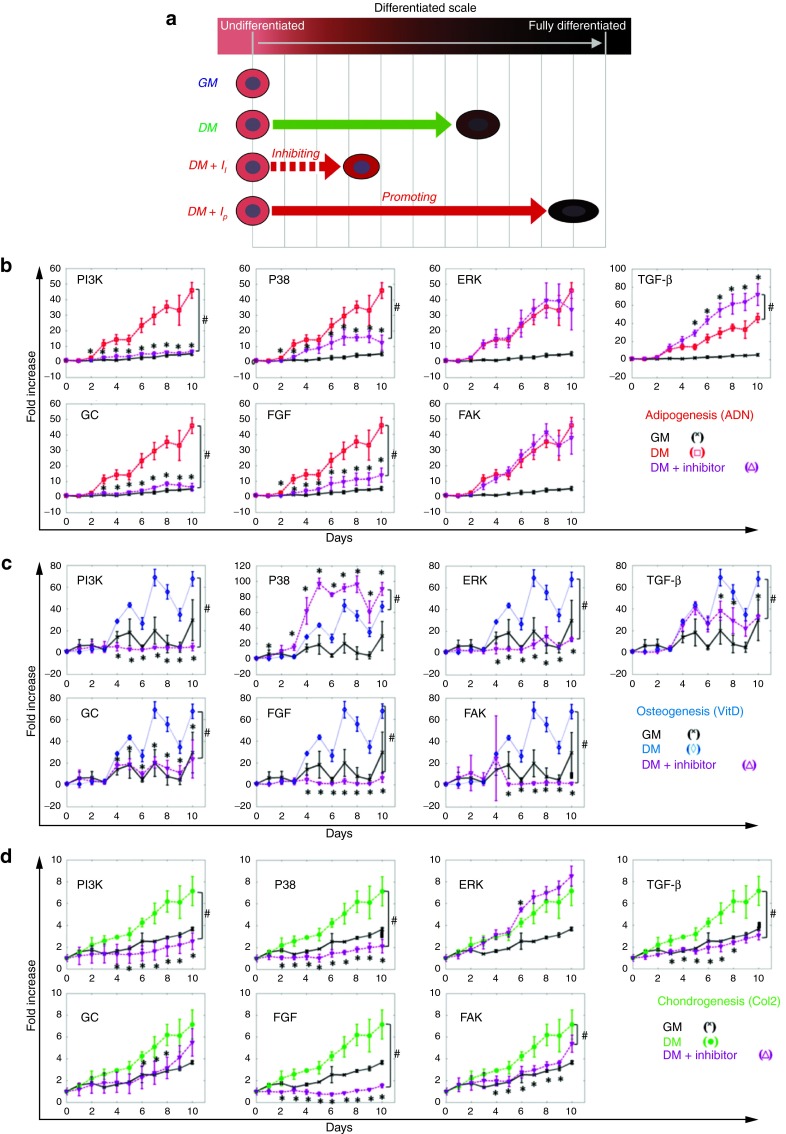
Next, we examined whether our reporters can effectively predict the differentiation outcome of MSC when the signaling network is perturbed. As proof of concept, we applied well-studied chemical inhibitors that target specific cellular pathways and subsequently, we monitored the reporter activation kinetics over time. We expected that inhibition of a pathway that is necessary for differentiation would decrease reporter activation, whereas inhibition of a pathway that blocks differentiation would further increase it (Figure 4a).
Figure 4.
Response of P/RE reporters during pathway inhibition. (a) Schematic of experimental design. (b–d) Dynamics of P/RE activity during adipogenic (red lines, □), osteogenic (blue lines, ◊), or chondrogenic (green lines, •) differentiation in the presence or absence of the indicated chemical inhibitor (I). Purple (Δ) and black lines (x) represent the response of cells in DM+I and GM, respectively. Normalized fluorescence intensity was plotted as fold-increase over day 0, when differentiation was induced. All values represent the mean ± SD of triplicate samples in a representative experiment (n = 3). * denotes P < 0.05 between DM+I and DM as calculated by t-test; and # denotes P < 0.05 between DM+I and DM as calculated by the AUC method.
The inhibitors included targets of cellular signaling pathways such as MAPKs, PI3Kinase, FAK as well as TGF-β, FGF and Glucocorticoid receptor mediated pathways. First, we employed western blot analysis and reporter assay, to verify that at the indicated concentration each inhibitor blocked the corresponding signaling pathway effectively (Supplementary Figure S3). The entire list of inhibitors along with their corresponding concentrations is listed in Supplementary Table S2. As lineage identifying reporters we applied Adiponectin-P for adipogenesis, VitaminD-RE for osteogenesis and Collagen2-P for chondrogenesis since these reporters showed high-fold activity and specificity for their respective lineage. In addition, for adipogenesis we used BM-MSCs whereas for osteogenesis and chondrogenesis HF-MSCs, as we observed that BM-MSCs were more effective in differentiating along the adipogenic lineage based on the Adiponectin-P activity and Oil Red O staining (Supplementary Figure S2).
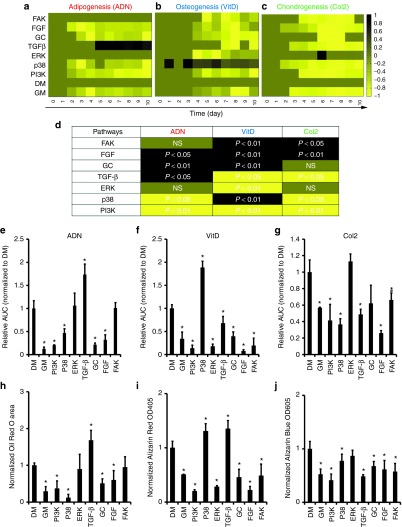
For each lineage, MSCs were transduced with the corresponding reporters, and then they were coaxed to differentiate in the presence of the indicated inhibitor. Cells were imaged in both green and red fluorescence channels daily for a period of 10 consecutive days. Cells growing in GM or inhibitor-free DM served as controls. For each inhibitor, the normalized fold increase of each promoter was plotted as a function of time and the representative kinetic plots were listed in (Figure 4b–d). The data were summarized in the form of heatmaps (Figure 5a–c). In addition, Supplementary Table S3 illustrates the maximum fold difference with respect to DM and the day that this occurs. The pairwise comparisons of the kinetic profiles under differentiation or growth conditions were performed using the AUC method. The effect of each inhibitor/pathway on each lineage was depicted by color and the P values were included (Figure 5d). The integrated response of each of the three reporters namely Adiponectin-P, VitaminD-RE, and Collagen2-P were calculated in the presence of the indicated inhibitors (Figure 5e–f) and compared with functional differentiation assays, for adipogenic (Oil Red O), osteogenic (Alizarin Red), or chondrogenic (Alcian Blue) lineage respectively (Figure 5h–j). The corresponding images of differentiated cells are shown in Supplementary Figure S4.
Figure 5.
P/RE responses and functional differentiation assays in the presence of pathway inhibitors. (a-c) Dynamic data of P/RE activity in the presence of inhibitors of the indicated pathways summarized as heatmaps for (a) adipogenic; (b) osteogenic; or (c) chondrogenic lineage. All normalized fluorescence intensities were further normalized to the maximum fold increase (or decrease) between DM and DM+I treatments. (d) Graphic illustration of the activation of each lineage specific reporter during pathway inhibition as resulted by statistical analysis with the AUC method. The colors range from black denoting highly upregulated to yellow denoting highly downregulated P/RE activity. Dark green color represents no significant difference between DM and DM+I. (e-g) Integrated response of (e) Adiponectin-P (ADN), (f) VitaminD-RE (VitD), or (g) Collagen2-P (Col2). (h-j) Measurement of the extent of differentiation into (h) adipogenic; (i) osteogenic; or (j) chondrogenic lineage. For Oil Red O staining, the area occupied by red oil droplets was measured in at least 10 images from 3 independent wells in each condition. For Alcian blue and Alizarin red staining, the dyes were extracted from the cells and absorbance was measured at OD605 and OD405, respectively. All values were normalized to those of cells in DM and plotted as mean ± SD of triplicate samples in a representative experiment (n = 3). The symbol (*) denotes P < 0.05 between DM and DM+I as calculated by the t-test; and # denotes P < 0.05 between DM and DM+I as calculated by the AUC method. NS, not significant difference.
FGF-R, PI3K, and glucocorticoid-R are essential pathways for MSC differentiation
Our results show that blocking the FGF-R resulted in complete inhibition of osteogenesis, chondrogenesis and adipogenesis as evidenced by the loss of activity of VitaminD-RE, Collagen 2-P and adiponectin-P, respectively. Similarly, PI3K and Glucocorticoid-R inhibition decreased the responses of all three reporters as well. The reporter activity data were verified by functional assays measuring the extent of differentiation at the end of the experiment.
TGF-βRI, p38, ERK, and FAK inhibition differentially regulate MSC differentiation
As shown in (Figure 3a), the SMAD2/3-RE was strongly upregulated during chondrogenesis but was down-regulated during adipogenesis, suggesting that the TGF-β pathway might promote chondrogenesis but inhibit adipogenesis. Indeed, blocking the activity of TGF-βRI and consequently the phosphorylation of SMAD2/3 inhibited chondrogenesis but promoted adipogenesis as seen by the collagen2-P and adiponectin-P activity, respectively. Functional assays confirmed the reporter results showing significant increase in oil-droplet formation and corresponding decrease in glycosaminoglycan accumulation.
We also perturbed the MAPK pathway using specific inhibitors for ERK and p38. Interestingly, ERK inhibition led to inhibition of VitaminD-RE, which was accompanied by decreased Alizarin Red staining. On the other hand, ERK inhibition had no significant effect on adipogenesis or chondrogenesis. Interestingly, p38 inhibition decreased the activation of adiponectin-P and collagen2-P but increased vitaminD-RE activity. In agreement, we observed significantly decreased oil droplet formation and glycosaminoglycan accumulation but increased calcium deposition. Finally, FAK inhibition abrogated the activity of osteogenic and chondrogenic reporters, in agreement with the functional assays. These results indicate that our reporters monitor MSC differentiation accurately and can be used to identify genes and pathways that regulate the process, as well as to discover pharmacological regulators of differentiation.
Discussion
The process of stem cell differentiation is complex and involves multiple signaling pathways that are sometimes activated in more than one lineage. In addition, the effect of soluble factors, extracellular matrix molecules and in general the MSC microenvironment play important and sometime conflicting roles in driving differentiation along one or more lineages. Understanding this process would benefit from a system that enables quantitative measurements of the dynamics of gene and pathway activation in noninvasive and high-throughput manner as stem cells differentiate in response to various stimuli. To this end, we developed a set of lentivirus dual promoter vectors (LVDP) each carrying two independent gene cassettes. The P/RE of interest drives expression of one reporter protein (ZsGreen), and a constitutive promoter drives expression of a second reporter (DsRed), which is used to measure transduction efficiency and for data normalization. The P/RE were chosen to represent each of the three MSC lineages. This novel lentiviral array (LVA) empowered monitoring of MSC differentiation by following the dynamic activity of multiple P/RE using quantitative fluorescence microscopy.
It is well known that chondrocytes and osteocytes are derived from a common progenitor (osteo-chondro progenitor) suggesting that they share common pathways during the early stages of differentiation.14 Other studies have shown a reciprocal relationship between adipogenesis and osteo/chondrogenesis since many factors that promote fat inhibit bone or cartilage formation and vice versa.15,16 Indeed, our results show differential regulation of several transcription factor consensus binding sites, e.g., p53-RE, SMAD-RE, GATA-RE and C/EBP-RE during adipogenesis, versus chondrogenesis and osteogenesis. Several RE exhibited differential activity in adipogenesis versus osteogenesis or chondrogenesis, suggesting that they might be good reporters that discriminate between lineages. Specifically, the p53-RE was significantly down-regulated during adipogenesis, but it was highly activated during osteogenesis. In agreement with our results, the p53 pathway was found to be downregulated during adipogenesis17 and p53 knockout MSCs showed enhanced adipogenic potential.18 In contrast to adipogenesis, the role of p53 in osteogenesis remains controversial, as p53 null osteoblasts or MSCs exhibited accelerated osteogenic differentiation,19,20,21 while p53 knockdown was shown to inhibit osteogenesis of muscle cells.18 Our results demonstrate that MSCs activated the p53-RE suggesting a positive role of p53 in osteogenesis.
The SMAD2/3-RE was also downregulated during adipogenesis, while it was activated during chondrogenesis. This result is in agreement with previous studies that reported inhibition of adipogenesis22 and enhancement of chondrogenesis23 by the SMAD2/3 pathway. Indeed, we showed that blocking the phosphorylation of SMAD2/3 by a chemical inhibitor (SB431549) enhanced adipogenesis whereas completely abrogated chondrogenesis. On the other hand, the SMAD1/5/8-RE was activated during osteogenic and chondrogenic differentiation, in agreement with studies showing enhanced osteogenesis24 and chondrogenesis through this pathway.25 We also showed that GATA-REs were also highly activated during chondrogenesis and downregulated during adipogenesis. In agreement, previous studies have shown that both GATA2 and 3 were downregulated during adipogenesis, while overexpression of GATAs abrogated oil droplet formation and terminal differentiation.26 In addition, GATA consensus binding sequences have been identified in chondrogenic promoters, such as SOX-927 and cartilage-derived retinoic acid-sensitive protein,28 but little is known about the functional role of GATAs in chondrogenesis.
Interestingly, C/EBP-RE was activated during MSC differentiation irrespective of the lineage commitment, albeit the kinetics of activation were different for each lineage. C/EBPs are important transcription factors that are composed of six members (α, β, γ, δ, ɛ, ζ). Whereas C/EBPα, β, and δ were shown to be critical for adipogenic differentiation, C/EBP β and δ were also reported to play a role in chondrogenesis and osteogenesis. In fact, previous studies have shown that C/EBPβ and δ were activated first during adipogenesis and regulated the expression of C/EBPα. Once expressed, C/EBPα activated the expression of multiple adipose-specific genes.15 During osteogenesis C/EBPβ and δ synergized with RUNX-2 and induced bone formation.29 However in chondrogenesis, C/EBPβ was important for the late stages of differentiation and in particular for hypertrophic differentiation.30 Notably, our measurements of C/EBP-RE activity captured both the early activation of the C/EBP pathway during adipogenesis as well as the late activation (after day 5) during chondrogenesis.
In addition to pathway activation, our lentiviral P/RE arrays revealed reporters that are activated only in one lineage. Specifically, we found that adiponectin-P, VitD-RE and collagen 2-P were activated only during adipogenic, osteogenic, and chondrogenic differentiation, respectively. This prompted us to hypothesize that these reporters could be used to predict the outcome of differentiation into each lineage and therefore, they could be used to determine the role of biochemical pathways in this process. To obtain proof of concept, we screened a set of chemical inhibitors and evaluated the effect of pathway inhibition on reporter activity as well as lineage commitment as evidenced by functional differentiation assays.
We found that the glucocorticoid receptor antagonist, the FGF receptor inhibitor and the PI3K inhibitor significantly decreased differentiation toward all three lineages. In line with these results, others have shown that supplementation with FGF and glucocorticoids in the differentiation medium enhanced adipogenic,31,32 osteogenic,33,34 and chondrogenic35,36 differentiation. Our results are also in agreement with studies highlighting the importance of the PI3 kinase pathway in MSC differentiation.37,38,39 Interestingly, members of the mitogen-activated protein kinase (MAPK) pathway had significant but differential effects on MSC differentiation. Whereas, inhibition of ERK significantly reduced osteogenic differentiation, it had no effect on adipogenesis in agreement with other studies.40,41 On the other hand, p38 inhibition decreased chondrogenic and adipogenic differentiation, but enhanced osteogenesis significantly, identifying the p38 pathway as inhibitory of bone formation.42,43,44 Finally, inhibition of FAK diminished both chondrogenesis and osteogenesis, but had no effect on adipogenesis.45,46 The reporter activity data were supported by functional differentiation assays suggesting that our reporters faithfully represented terminal differentiation along their respective lineages. Of note, multiple carefully selected P/RE reporters may be used to dissect the role of various pathways at different temporal stages (early, intermediate, and late) of stem cell differentiation.
Collectively, our results demonstrate that LVA can be applied to live cells to monitor stem cell differentiation in real time as well as to uncover pathways that regulate this complex process. Although the LVA system was employed to study MSC differentiation, it can also be adopted to monitor differentiation or reprogramming of other adult stem cells, cancer stem cells or induced pluripotent stem cells (iPSC). In addition to soluble factors, the LVA is ideal for monitoring stem cell differentiation in response to extracellular matrix signals or biomaterials of varying chemical composition or mechanical stiffness.47,48,49,50 Applications in cancer biology or other areas of biology and bioengineering should also be feasible. Notably, the combination of the lentiviral reporters with chemical libraries has the potential to discover novel chemical inducers of differentiation or reprogramming; while the combination with the siRNA libraries can lead to the discovery of novel genes that participate in lineage specification or reprogramming. The lentiviral reporters may be particularly useful for real-time measurements of P/RE activity, especially in experiments involving a small number of cells or even individual cells where use of conventional methods, e.g., RT-PCR, western blots (WB) or differentiation assays may be difficult. Finally, the dynamic information gained from this approach may facilitate generation of mathematical models to predict the outcome of the differentiation process from the dynamics of the underlying signaling pathways.
Materials and Methods
Cloning and lentivirus preparation. The specific design and optimization of LVDP are described previously8,9 (Figure 1a). Promoter regions of genes were extracted from human genomic DNA (Promega, Madison, WI) or from promoter constructs generously provided by other laboratories (Supplementary Tables S4 and S5) using PCR. PCR fragments were amplified by Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) following manufacturer's protocols. The amplified PCR product was inserted into LVDP upstream of a Green Fluorescent reporter (stable/destabilized ZsGreen) using ClaI and AgeI restriction sites. Similarly, three to six repeats of transcription factor response element (RE) motifs were cloned into LVDP, upstream of a minimal CMV and the reporter ZsGreen region using BsiWI and BstBI restriction sites. All restriction enzymes were ordered from Fermentas/Thermo Fisher Scientific (Pittsburgh, PA). All cloning products were confirmed by sequencing with ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA). For detailed information concerning promoter locations, transcription factor binding motifs and the primer designs (see Supplementary Tables S4 and S5). The resulting lentiviral vectors were pseudotyped with VSV-G and virus production was carried out using the calcium phosphate precipitation method in 293T/17 cell line as detailed in ref. 8. Titers of lentiviral preparations were determined using 293T/17 cells and ranged between 107 and 108 IFU/ml.
Cell culture, differentiation, and experimental setup. Human bone marrow MSCs (29-year-old male) were obtained from Stem Cell Technologies (Vancouver, Canada) whereas human hair follicle MSCs were isolated from a 73-year-old male donor as described previously.10 MSCs were seeded in a 384-well plate at a density of 6,000 cells/cm2 and transduced the next day with the different lentiviral reporter constructs. MSCs were cultured with growth medium (DMEM supplemented with 10% MSC qualified FBS and 1 ng/ml bFGF, GIBCO, Grand Island, NY) until confluence. Next day, the growth medium was replaced with the appropriate differentiation medium, which was replenished every 3 days thereafter. For adipogenesis, MSCs were cultured with DMEM supplemented with 10%MSC FBS, 0.5 mmol/l of isobutyl-methylxanthine (Sigma, St Louis, MO), 1 μmol/l of dexamethasone (Sigma), 10 μmol/l of insulin (Sigma) and 200 μmol/l of indomethacin (MP Biomedicals, Solon, OH). For osteogenesis, DMEM was supplemented with 10% MSC FBS, 10 mmol/l of β-glycerolphosphate (Alfa Aesar, Ward Hill, MA), 50 μg/ml of ascorbate-2-phosphate (Sigma), 100 nmol/l of dexamethasone, 20 ng/ml of BMP-2 (Genescript) and 2 ng/ml of bFGF. For chondrogenesis, MSCs were cultured with DMEM supplemented with 10% MSC FBS, 6.25 μg/ml of insulin, 10 ng/ml TGF-β1 (US Biological, Swampscott, MA) and 50 nmol/l of ascorbate-2-phosphate. As negative control, MSCs were cultured in DMEM supplemented with 10% MSC qualified FBS. The schematic representation of experimental setup is illustrated in (Figure 1b).
For the inhibitor screening, chemical inhibitors were added to each of the three differentiation media and replenished every three days. The chemical compounds and respective concentrations are listed in Supplementary Table S2 whereas the experimental setup is illustrated in (Figure 4a).
Image acquisition and data analysis. The kinetics of the reporter construct activation during differentiation was captured using the Zeiss Observer microscope equipped with an automated stage (Carl Zeiss, Thornwood, NY) at ×5 magnification. Images of every well were captured using GFP and DsRED2 filter sets at distinct time intervals (1 day). The captured images were exported in the JPEG grayscale format and processed using Cell Profiler (BROAD Institute, http://www.cellprofiler.org). The positions of florescent cells were first identified with the MoG Adaptive threshold method, and the total fluorescence intensity of each image was calculated as the sum of intensities of the identified regions. The total green (GFI) and red fluorescent intensity (RFI) were quantified by the software Cell Profiler using object identification modules to recognize transduced cells. The RFI at day 0, which corresponds to transduction efficiency in different wells was used for normalization such that the ratio GFI/RFI can represents reporter activity independent of virus titer as we showed previously.7,8 The results were further normalized with that at day 0 (day of induction of differentiation) and were reported as fold increase.
Functional differentiation assays. HF-MSCs and BM-MSCs were seeded at a density of 6,000 cells/cm2 in growth medium. Upon reaching confluence, cells were induced to differentiate toward the adipogenic, osteogenic and chondrogenic lineage in the presence or absence of the chemical inhibitors. To match the lentiviral experiment, the same final concentrations of the inhibitors were used (Table 4). The degree of adipogenic, chondrogenic, or osteogenic differentiation was assessed by Oil Red O, Alcian Blue or Alizarin Red staining, respectively as described previously.10 Color images were obtained using the Zeiss AxioObserver and presented as mosaic images. For Oil Red O staining quantification, the area of the oil droplets was quantified by analyzing at least 10 images from three independent wells in each experiment (n = 3).
For Alcian Blue quantification, alcian blue was extracted by overnight treatment with 6 mol/l guanidine-HCl (125 μl/(48)-well, room temperature) and the following day, OD595 was measured using a microplate reader (Synergy4, Biotek, Winooski, VT). The OD595 of pure guanidine-HCl was subtracted as background signal from the OD595 of each sample.
For Alizarin Red staining, the Osteogenesis assay kit (Millipore, Billerica, MA) was used according to the manufacturer's instructions. For Alizarin Red quantification, 10% acetic acid (200 μl/(48)-well) was added to the cell monolayer and incubated for 30 minutes at room temperature under gentle shaking. Subsequently, the cells were scraped from the wells, transferred to 1.5 ml tubes and incubated at 85 °C for 10 minutes to dissolve the dye thoroughly. After centrifugation to remove debris, the pH of each sample was adjusted with 75 μl of 10% ammonium hydroxide to a final pH 4.1–4.5 and the OD405 was measured using a microplate reader (Synergy4, Biotek, Winooski, VT). The OD405 of pure acetic acid/ammonium hydroxide solution (8/3 ratio) was subtracted as background signal from the OD405 of each sample.
Western blot. To evaluate the effectiveness of each inhibitor, we performed western blots for the corresponding phosphor-proteins in each pathway. Human MSCs were serum starved overnight and preincubated with the indicated inhibitors. The next day the cells were treated (30 minutes) to activate the pathway of interest in the presence or absence of the corresponding inhibitor. Specifically, cells were treated with PDGF-BB (50 ng/ml), Sorbitol (400 mmol/l), bFGF (20 ng/ml), TGF-β1 (10 ng/ml) or 10% FBS to stimulate the PI3K, p38, ERK, SMAD2, or FAK pathway, respectively. Afterwards, cells were lysed, and the lysates were subjected to western blot using antibodies for each of the following: pAKT, AKT, pMAPKAPK-2, MAPKAPK-2, pERK, ERK, pSMAD2, SMAD2, pFAK, and FAK (1:1,000 diluted in 5% BSA containing TBST, Cell Signaling Technology, Danvers, MA).
Statistical analysis. All experiments were performed at least three times with triplicate samples for each condition. Pairwise comparison was analyzed by two-tailed Student t-test, and the data were considered statistically different when P < 0.05. In kinetic experiments, pairwise comparisons were performed between both individual time points and for the entire kinetic profiles using of the area under each curve (AUC). The AUC of each sample was approximated with trapezoid area estimation:
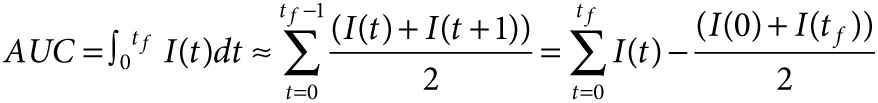 |
where I(t) is the normalized intensity at time t and tf is the final time point.
SUPPLEMENTARY MATERIAL Figure S1. Dynamics of P/RE activity during adipogentic (red lines), osteogenic condition (blue lines), or chondrogenic (green lines) differentiation. Figure S2. P/RE reporters capture the differentiation potentials of HF-MSCs and BM-MSC as shown by functional differentiation staining. Figure S3. Confirmation of pathway inhibition. Figure S4. Representative staining images of MSC that were induced to differentiate in the absence (DM) or presence of the indicated pathway inhibitor. Table S1. Summary of the time, fold increase/decrease and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and GM was maximum (n = 3). Table S2. List of chemical inhibitors, the corresponding pathways, the concentrations used and suppliers. Table S3. Summary of the time, fold increase/decrease, and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and DM+I was maximum (n = 3). Table S4. Cloning information for gene promoters. Table S5. Cloning information for response elements.
Acknowledgments
This work was supported by a grant from the National Science Foundation (CBET-0853993) and the New York State Stem Cell Science (NYSTEM Contract #C024315) to S.T. Andreadis.
Supplementary Material
Dynamics of P/RE activity during adipogentic (red lines), osteogenic condition (blue lines), or chondrogenic (green lines) differentiation.
P/RE reporters capture the differentiation potentials of HF-MSCs and BM-MSC as shown by functional differentiation staining.
Confirmation of pathway inhibition.
Representative staining images of MSC that were induced to differentiate in the absence (DM) or presence of the indicated pathway inhibitor.
ummary of the time, fold increase/decrease and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and GM was maximum (n = 3).
List of chemical inhibitors, the corresponding pathways, the concentrations used and suppliers.
Summary of the time, fold increase/decrease, and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and DM+I was maximum (n = 3).
loning information for gene promoters.
Cloning information for response elements.
References
- Thompson DM, King KR, Wieder KJ, Toner M, Yarmush ML, Jayaraman A. Dynamic gene expression profiling using a microfabricated living cell array. Anal Chem. 2004;76:4098–4103. doi: 10.1021/ac0354241. [DOI] [PubMed] [Google Scholar]
- Wieder KJ, King KR, Thompson DM, Zia C, Yarmush ML, Jayaraman A. Optimization of reporter cells for expression profiling in a microfluidic device. Biomed Microdevices. 2005;7:213–222. doi: 10.1007/s10544-005-3028-3. [DOI] [PubMed] [Google Scholar]
- King KR, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier AK, Ariazi EA, Bellis AD, Bengali Z, Jordan VC, Shea LD. Bioluminescence imaging for assessment and normalization in transfected cell arrays. Biotechnol Bioeng. 2007;98:486–497. doi: 10.1002/bit.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis AD, Peň alver-Bernabé B, Weiss MS, Yarrington ME, Barbolina MV, Pannier AK, et al. Cellular arrays for large-scale analysis of transcription factor activity. Biotechnol Bioeng. 2011;108:395–403. doi: 10.1002/bit.22916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MS, Peñalver Bernabé B, Bellis AD, Broadbelt LJ, Jeruss JS, Shea LD. Dynamic, large-scale profiling of transcription factor activity from live cells in 3D culture. PLoS One. 2010;5:e14026. doi: 10.1371/journal.pone.0014026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimperti S, Lei P, Tian J, Andreadis ST. A novel lentivirus for quantitative assessment of gene knockdown in stem cell differentiation. Gene Ther. 2012;19:1123–1132. doi: 10.1038/gt.2011.208. [DOI] [PubMed] [Google Scholar]
- Tian J, Alimperti S, Lei P, Andreadis ST. Lentiviral microarrays for real-time monitoring of gene expression dynamics. Lab Chip. 2010;10:1967–1975. doi: 10.1039/c003153d. [DOI] [PubMed] [Google Scholar]
- Tian J, Andreadis ST. Independent and high-level dual-gene expression in adult stem-progenitor cells from a single lentiviral vector. Gene Ther. 2009;16:874–884. doi: 10.1038/gt.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai VK, Mistriotis P, Andreadis ST. Clonal multipotency and effect of long-term in vitro expansion on differentiation potential of human hair follicle derived mesenchymal stem cells. Stem Cell Res. 2012;8:74–84. doi: 10.1016/j.scr.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai VK, Mistriotis P, Loh YH, Daley GQ, Andreadis ST. Functional vascular smooth muscle cells derived from human induced pluripotent stem cells via mesenchymal stem cell intermediates. Cardiovasc Res. 2012;96:391–400. doi: 10.1093/cvr/cvs253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Peng HF, Gopinath S, Tian J, Andreadis ST. Derivation of functional smooth muscle cells from multipotent human hair follicle mesenchymal stem cells. Tissue Eng Part A. 2010;16:2553–2564. doi: 10.1089/ten.tea.2009.0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Peng HF, Andreadis ST. Contractile smooth muscle cells derived from hair-follicle stem cells. Cardiovasc Res. 2008;79:24–33. doi: 10.1093/cvr/cvn059. [DOI] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cell Mol Life Sci. 2009;66:236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich SJ, Litteral V, Mayo LD, Tabesh D, Morris D. mdm-2 gene amplification in 3T3-L1 preadipocytes. Differentiation. 1999;64:205–212. doi: 10.1046/j.1432-0436.1999.6440205.x. [DOI] [PubMed] [Google Scholar]
- Molchadsky A, Shats I, Goldfinger N, Pevsner-Fischer M, Olson M, Rinon A, et al. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One. 2008;3:e3707. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesilla-Diaz A, Elvira G, Silva A. p53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp Cell Res. 2009;315:3598–3610. doi: 10.1016/j.yexcr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Choy L, Derynck R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J Biol Chem. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- Mackay AM, Beck SC, Murphy JM, Barry FP, Chichester CO, Pittenger MF. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng. 1998;4:415–428. doi: 10.1089/ten.1998.4.415. [DOI] [PubMed] [Google Scholar]
- Ryoo HM, Lee MH, Kim YJ. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene. 2006;366:51–57. doi: 10.1016/j.gene.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Hellingman CA, Davidson EN, Koevoet W, Vitters EL, van den Berg WB, van Osch GJ, et al. Smad signaling determines chondrogenic differentiation of bone-marrow-derived mesenchymal stem cells: inhibition of Smad1/5/8P prevents terminal differentiation and calcification. Tissue Eng Part A. 2011;17:1157–1167. doi: 10.1089/ten.TEA.2010.0043. [DOI] [PubMed] [Google Scholar]
- Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol Cell Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Koopman P. Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum Mol Genet. 1999;8:691–696. doi: 10.1093/hmg/8.4.691. [DOI] [PubMed] [Google Scholar]
- Bosserhoff AK, Kondo S, Moser M, Dietz UH, Copeland NG, Gilbert DJ, et al. Mouse CD-RAP/MIA gene: structure, chromosomal localization, and expression in cartilage and chondrosarcoma. Dev Dyn. 1997;208:516–525. doi: 10.1002/(SICI)1097-0177(199704)208:4<516::AID-AJA7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, et al. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- Hirata M, Kugimiya F, Fukai A, Ohba S, Kawamura N, Ogasawara T, et al. C/EBPbeta Promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS One. 2009;4:e4543. doi: 10.1371/journal.pone.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauner H, Entenmann G, Wabitsch M, Gaillard D, Ailhaud G, Negrel R, et al. Promoting effect of glucocorticoids on the differentiation of human adipocyte precursor cells cultured in a chemically defined medium. J Clin Invest. 1989;84:1663–1670. doi: 10.1172/JCI114345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer M, Fischbach C, Bauer-Kreisel P, Lieb E, Hacker M, Tessmar J, et al. Basic fibroblast growth factor enhances PPARgamma ligand-induced adipogenesis of mesenchymal stem cells. FEBS Lett. 2004;577:277–283. doi: 10.1016/j.febslet.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Rickard DJ, Sullivan TA, Shenker BJ, Leboy PS, Kazhdan I. Induction of rapid osteoblast differentiation in rat bone marrow stromal cell cultures by dexamethasone and BMP-2. Dev Biol. 1994;161:218–228. doi: 10.1006/dbio.1994.1022. [DOI] [PubMed] [Google Scholar]
- Marie PJ. Fibroblast growth factor signaling controlling bone formation: an update. Gene. 2012;498:1–4. doi: 10.1016/j.gene.2012.01.086. [DOI] [PubMed] [Google Scholar]
- Derfoul A, Perkins GL, Hall DJ, Tuan RS. Glucocorticoids promote chondrogenic differentiation of adult human mesenchymal stem cells by enhancing expression of cartilage extracellular matrix genes. Stem Cells. 2006;24:1487–1495. doi: 10.1634/stemcells.2005-0415. [DOI] [PubMed] [Google Scholar]
- Solchaga LA, Penick K, Porter JD, Goldberg VM, Caplan AI, Welter JF. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203:398–409. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- Sakaue H, Ogawa W, Matsumoto M, Kuroda S, Takata M, Sugimoto T, et al. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J Biol Chem. 1998;273:28945–28952. doi: 10.1074/jbc.273.44.28945. [DOI] [PubMed] [Google Scholar]
- Guntur AR, Rosen CJ. The skeleton: a multi-functional complex organ: new insights into osteoblasts and their role in bone formation: the central role of PI3Kinase. J Endocrinol. 2011;211:123–130. doi: 10.1530/JOE-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka K, Kanematsu T, Takeuchi H, Nakata M, Kikkawa U, Hirata M. Involvement of the phosphoinositide 3-kinase/protein kinase B signaling pathway in insulin/IGF-I-induced chondrogenesis of the mouse embryonal carcinoma-derived cell line ATDC5. Int J Biochem Cell Biol. 2001;33:1094–1103. doi: 10.1016/s1357-2725(01)00067-x. [DOI] [PubMed] [Google Scholar]
- Font de Mora J, Porras A, Ahn N, Santos E. Mitogen-activated protein kinase activation is not necessary for, but antagonizes, 3T3-L1 adipocytic differentiation. Mol Cell Biol. 1997;17:6068–6075. doi: 10.1128/mcb.17.10.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci USA. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal RK, Jaiswal N, Bruder SP, Mbalaviele G, Marshak DR, Pittenger MF. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J Biol Chem. 2000;275:9645–9652. doi: 10.1074/jbc.275.13.9645. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275:4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- Viñals F, López-Rovira T, Rosa JL, Ventura F. Inhibition of PI3K/p70 S6K and p38 MAPK cascades increases osteoblastic differentiation induced by BMP-2. FEBS Lett. 2002;510:99–104. doi: 10.1016/s0014-5793(01)03236-7. [DOI] [PubMed] [Google Scholar]
- Bang OS, Kim EJ, Chung JG, Lee SR, Park TK, Kang SS. Association of focal adhesion kinase with fibronectin and paxillin is required for precartilage condensation of chick mesenchymal cells. Biochem Biophys Res Commun. 2000;278:522–529. doi: 10.1006/bbrc.2000.3831. [DOI] [PubMed] [Google Scholar]
- Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp Cell Res. 2007;313:22–37. doi: 10.1016/j.yexcr.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Suri S, Lee T, Chilton JM, Cooke MT, Chen W, et al. Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat Methods. 2013;10:438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dynamics of P/RE activity during adipogentic (red lines), osteogenic condition (blue lines), or chondrogenic (green lines) differentiation.
P/RE reporters capture the differentiation potentials of HF-MSCs and BM-MSC as shown by functional differentiation staining.
Confirmation of pathway inhibition.
Representative staining images of MSC that were induced to differentiate in the absence (DM) or presence of the indicated pathway inhibitor.
ummary of the time, fold increase/decrease and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and GM was maximum (n = 3).
List of chemical inhibitors, the corresponding pathways, the concentrations used and suppliers.
Summary of the time, fold increase/decrease, and standard deviation (SD) when the normalized fluorescence intensity between the indicated DM and DM+I was maximum (n = 3).
loning information for gene promoters.
Cloning information for response elements.