To the editor:
Ex vivo gene therapy is one of the current strategies being tested to treat genodermatoses such as epidermolysis bullosa (EB).1 In fact, Mavilio et al. proved the feasibility of this therapeutic modality in a patient with the junctional form of EB (JEB).2 Efforts are now being directed toward the development of efficient approaches minimizing potential genotoxic effects due to vector-induced insertional mutagenesis. Gene correction by gene editing through nuclease-facilitated homologous recombination (HR) has recently been proven to be achievable on recessive dystrophic EB cells that were subsequently reprogrammed to induced pluripotent stem cells (iPSCs) and differentiated to collagen VII–expressing keratinocytes.3 We have also demonstrated the feasibility of zinc-finger nuclease–facilitated, HR-mediated insertion of a marker gene into the intron 1 of the PPP1R12C gene (AAVS1 locus) in a limited number of human epidermal repopulating cells that, upon grafting, persisted as small foci in skin regenerated in immunodeficient mice.4 In this study we report that engraftment and persistent skin regeneration can be achieved with an expanded stem cell clone isolated from AAVS1 gene–targeted human keratinocytes.
We first determined whether sorted enhanced green fluorescent protein (EGFP)-positive cells, present in low proportion (<0.1%) of the HR-targeted human keratinocytes (ref. 4 and Supplementary Figure S1a online), could be isolated and expanded in culture. Our previous attempts at growing EGFP-positive, FACS-selected cells from a bulk population containing <5% of EGFP-positive human keratinocytes had failed. To overcome this limitation, we supplemented our culture medium with the ROCK inhibitor Y-27632 to enhance the expansion of sorted keratinocytes with stem features.5,6 Only one of the very few GFP-positive keratinocyte colonies derived from cells recovered from the cell-sorting procedure maintained an undifferentiated phenotype and grew larger than two centimeters in diameter, consistent with the characteristics of an epidermal stem cell derivative or holoclone.7,8 The rest of the colonies grew no larger than 2–3 mm in diameter after 15 days in culture (data not shown). Nonviable, differentiating keratinocyte colonies were scraped out from the flask, and the large colony was harvested and cells expanded to perform EGFP fluorescence and DNA analysis, and grafting. Detailed materials and methods are given in the Supplementary Materials and Methods online).
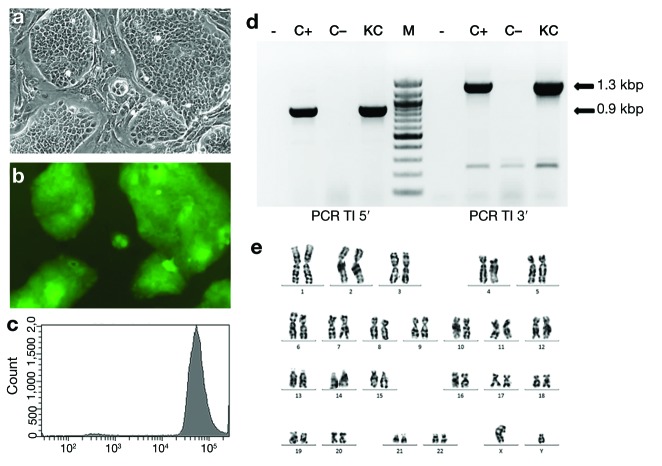
In contrast to a polyclonal EGFP-expressing keratinocyte population after standard retroviral transduction, the expanded clone displayed, under the microscope, a homogeneous morphology and fluorescence (Figure 1a,b and Supplementary Figure S1b,c) and a sharp peak of fluorescence, detected by flow cytometry, indicative of uniform EGFP expression (Figure 1c). PCR analysis of DNA isolated from the clone showed amplified DNA bands consistent with “on target” integration of the PGK-EGFP, HR-cassette in the AAVS1 locus (Figure 1d).4 Southern blot analysis confirmed targeted integration and the absence of additional “off target” HR cassette integrations (Supplementary Figure S2a,b). Cytogenetic analysis revealed a normal (46,XY) karyotype with no detectable structural chromosomal abnormalities (Figure 1e). Bioengineered skin equivalents prepared with the expanded-clone keratinocytes were grafted to immunodeficient mice. Four weeks after grafting, human skin regeneration was confirmed by detection of EGFP epifluorescence in five out of six transplanted mice.
Figure 1.
Morphological, molecular, and cytogenetic characteristics of AAVS1 gene–targeted keratinocyte clone. (a) Microscopic appearance (phase contrast) of the clone grown in the presence of lethally irradiated feeder layer cells. (b) Green fluorescent protein (GFP) fluorescence corresponding to the cells shown in a. (c) Flow cytometry fluorescence analysis of clone cells showing a sharp peak of fluorescence indicative of uniform GFP expression. (d) PCR analysis of transgene cassette integration (TI) within the AAVS1 intron 1 locus. The 1.3-kbp and 0.9-kbp bands (arrows) demonstrate targeted integration of the cassette at 3′ and 5′ genome junctions, respectively. Lane –, mock PCR amplification; lane C+, PCR amplification of DNA from a positive control; lane C–, PCR amplification from normal human DNA; lane KC, PCR amplification of DNA from AAVS1-targeted keratinocyte clone. (e) Cytogenetic analysis of the clonal, gene-targeted keratinocytes. The GTG banding shows a normal 46,XY karyotype.
Twelve weeks after grafting, when approximately three epidermal turnover cycles should have occurred, macroscopic inspection was performed (Figure 2a,b) and skin biopsy samples taken from engrafted skin for histological examination and for PCR analysis to confirm AAVS1 locus–targeted transgene integration. The analysis revealed regeneration of a normal, EGFP-fluorescent human skin (Figure 2c) with typical histological architecture and proper expression of differentiation markers (Figure 2d and Supplementary Figure S3). The molecular analysis showed that the grafts rendered the correct transgene integration PCR amplicons, exactly as the cells in culture, ruling out selection of putative off-target or gene rearrangement events in vivo (Supplementary Figure S2c). Overall, we could demonstrate the feasibility of selecting gene-targeted epidermal cells with long-term repopulating ability consistent with stem cell features.
Figure 2.
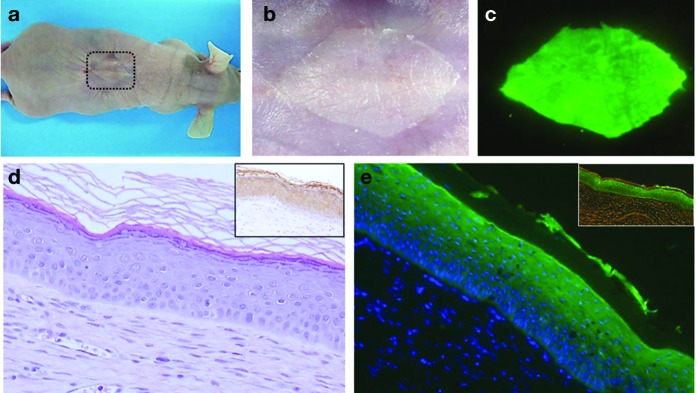
Macroscopic and microscopic appearance of the human skin regenerated from clonal gene-targeted keratinocytes. (a) A representative engrafted mouse showing human regenerated skin (dotted square) 12 weeks after grafting of bioengineered skin carrying AAVS1 locus–targeted clonal keratinocytes. (b) Macroscopic appearance (with illumination) of the regenerated skin shown depicted by the dotted square in the engrafted mouse of panel a. (c) Enhanced green fluorescent protein (EGFP) epifluorescence (blue-light illumination) of the engrafted skin shown in panel b. (d) Microphotograph (H&E-stained) of a skin section from a 12-week-old graft showing normal human skin architecture. The inset (right upper) shows the normal expression of involucrin using a human-specific monoclonal antibody to involucrin. (e) EGFP fluorescence detection in a frozen section (paraformaldehyde-fixed) from the graft analyzed in panel d. DAPI staining shows nuclei. The inset shows the direct EGFP fluorescence observed via fluorescence phase-contrast microscopy. DAPI, 4',6-diamidino-2-phenylindole.
Therapeutic approaches such as HR using primary human keratinocytes require individual clonal analysis not only to characterize the gene-targeting event but also to assess the regenerative capacity of individual clone(s) using stringent in vivo assays. Recently Melo et al. reported LAMA3 correction in JEB human keratinocytes through AAV-mediated HR.9 However, in their short-term (5 weeks after grafting) in vivo assessment of skin regeneration conducted with pools of keratinocytes, targeting of bona fide epidermal stem cells required for permanent gene correction could not be confirmed. Our previous long-term in vivo study showing the persistence of small (AAVS1-targeted), EGFP-positive skin foci suggested the epidermal stem cell nature of the cells originating them.4 However, because these foci comprised <1% of the total graft surface, the definitive competence for whole epidermal regeneration, needing robust proliferative capacity of targeted cell clones, had not been strictly proven. We used an optimized in vivo approach, previously employed to establish humanized skin models and therapeutic options for several genodermatoses10–12 and to assess the safety and regenerative capacity of isolated holoclones genetically modified with retroviral vectors.13 Here this system allowed us to demonstrate that epidermal stem cells subjected to transient expression of DNA nucleases preserve their epidermal repopulating competence in vivo.
Moreover, we can confirm that a strategy based on skin regeneration from single or a limited number of gene-corrected clones is realistic. However, given the targeting efficiencies achieved with current gene-editing tools, robust methods of cell selection are still required. This could be achieved by using either a Cre/loxP removable selection cassette or a constitutive marker compatible with clinical applications.
With the advent of novel gene-editing techniques and other strategies, such as the derivation of true corrected human epidermal stem cells from iPSCs capable of permanent skin regeneration, clonal analyses assessing their efficacy are likely to be required. Our study paves the way for the development of such future clonal correction-based therapeutic approaches.
SUPPLEMENTARY MATERIAL Supplementary Materials and Methods Supplementary Figure S1. Morphological and fluorescence features of AAVS1-targeted holoclone cells. Supplementary Figure S2. Molecular analyses of AAVS1-targeted holoclone cells and derived grafts. Supplementary Figure S3. Expression of epidermal differentiation markers in AAVS1-targeted holoclone grafts.
Acknowledgments
We are indebted to Jesus Martinez and Edilia de Almeida for animal care and to Nuria Illera, Almudena Holguin, and María Luisa Retamosa for mouse xenotransplantation. F.L. was supported by grants PI11/01225 from Instituto de Salud Carlos III and S2010/BMD-2359 from Comunidad de Madrid. M.D.R. was supported by grant S2010/BMD-2420 from Comunidad de Madrid. A.R. is supported by grants from the Italian Ministry of University and Research (FIRB 2008).
Supplementary Material
References
- Uitto J. Rare heritable skin diseases: targets for regenerative medicine. J Invest Dermatol. 2012;132:2485–2488. doi: 10.1038/jid.2012.334. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A.et al. (2006Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells Nat Med 121397–1402. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L.et al. (2013TALEN-based gene correction for epidermolysis bullosa Mol Ther 211151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio A, Miselli F, Lombardo A, Marconi A, Malagoli Tagliazucchi G, Gonçalves MA.et al. (2013Targeted gene addition in human epithelial stem cells by zinc-finger nuclease–mediated homologous recombination Mol Ther 211695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D, Matsushita N, Toki F, Higashiyama S. Efficient expansion of human keratinocyte stem/progenitor cells carrying a transgene with lentiviral vector. Stem Cell Res Ther. 2013;4:127. doi: 10.1186/scrt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Duan H, Wang Y, Qu M, Yang L., and, Xie L. ROCK inhibitor Y-27632 increases the cloning efficiency of limbal stem/progenitor cells by improving their adherence and ROS-scavenging capacity. Tissue Eng Part C Methods. 2013;19:531–537. doi: 10.1089/ten.tec.2012.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathor MB, Ferrari G, Dellambra E, Cilli M, Mavilio F, Cancedda R.et al. (1996Clonal analysis of stably transduced human epidermal stem cells in culture Proc Natl Acad Sci USA 9310371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y., and, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SP, Lisowski L, Bashkirova E, Zhen HH, Chu K, Keene DR.et al. (2014Somatic correction of junctional epidermolysis bullosa by a highly recombinogenic AAV variant Mol Ther 22725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M, Llames S, García E, Meana A, Cuadrado N, Recasens M.et al. (2010In vivo assessment of acute UVB responses in normal and xeroderma pigmentosum (XP-C) skin-humanized mouse models Am J Pathol 177865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di WL, Larcher F, Semenova E, Talbot GE, Harper JI, Del Rio M.et al. (2011Ex-vivo gene therapy restores LEKTI activity and corrects the architecture of Netherton syndrome-derived skin grafts Mol Ther 19408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufenvenne K, Rice RH, Hausser I, Oji V, Hennies HC, Rio MD.et al. (2012Long-term faithful recapitulation of transglutaminase 1–deficient lamellar ichthyosis in a skin-humanized mouse model, and insights from proteomic studies J Invest Dermatol 1321918–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher F, Dellambra E, Rico L, Bondanza S, Murillas R, Cattoglio C.et al. (2007Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy Mol Ther 151670–1676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.