Abstract
The current study aimed to examine the gene specific mechanisms by which the actions of the vitamin D receptor (VDR) are distorted in prostate cancer. Transcriptional responses toward the VDR ligand, 1α,25(OH)2D3, were examined in non-malignant prostate epithelial cells (RWPE-1) and compared to the 1α,25(OH)2D3-recalcitrant prostate cancer cells (PC-3). Time resolved transcriptional studies for two VDR target genes revealed selective attenuation and repression of VDR transcriptional responses in PC-3 cells. For example, responses in PC-3 cells revealed suppressed responsiveness of IGFBP3 and G0S2. Furthermore, Chromatin Immunoprecipitation (ChIP) assays revealed that suppressed transcriptional responses in PC-3 cells of IGFBP3 and G0S2 were associated with selective VDR-induced NCOR1 enrichment at VDR-binding regions on target-gene promoter regions. We propose that VDR inappropriately recruits co-repressors in prostate cancer cells. Subsequent direct and indirect mechanisms may induce local DNA methylation and stable transcriptional silencing. Thus a transient epigenetic process mediated by co-repressor binding, namely, the control of H3K9 acetylation, is distorted to favor a more stable epigenetic event, namely DNA methylation.
Keywords: NCOR1, Prostate cancer, Epigenetics, VDR
1. Introduction
Epigenetic mechanisms are central to the evolution of malignant phenotypes. The androgen receptor (AR) [1] exerts a profound control on the growth and differentiation of normal prostate. Its cellular actions have been studied extensively; for example the process of androgen withdrawal was exploited to define apoptosis [2]. The AR co-operates with WNT and mTOR pathways [3,4] to induce prostate epithelial cell proliferation. Equally, other nuclear receptors, such VDR, PPARs, and RARs, exert mitotic restraint, at least in part by antagonizing WNT signaling and activation of cell cycle arrest through regulation of gene targets such as CDKN1A (encodes p21(waf1/cip1)), IGFBP3 [5–11] and G0S2 [12,13]. In prostate cancer (CaP) the central actions of the AR are exploited in androgen deprivation therapy (ADT) to derive significant clinical benefit. Ultimately this is not sustained and treatment failure following ADT is characterized by ADT-recurrent CaP (ADT-RCaP), which is invariably lethal.
The impact of ADT on the malignant cell presents a formidable environment that cancer cells must adapt to. This process is multifaceted and includes loss of mitogenic signals downstream of the AR, triggering apoptosis, hypoxia (due to endothelial cell collapse) and inflammation that has an associated mileu of cytokine and other signals. Central aspects of the escape mechanisms to this restraint include increasing intrinsic AR ligand production and AR signaling capacity. However the transcriptional actions of the AR in ADT-RCaP are not merely a re-iteration of the normal AR transcriptome, but rather represent a fundamentally different transcriptome. Epigenetic events are central to the evolution of the altered AR signaling capacity.
The AR transcriptional program evolves toward increased targeting of proliferative gene promoters and decreased targeting of pro-differentiation genes [14,15]. For example, the oncogenic actions of the TMPRSS2/ETS fusion, a common event in CaP [16], are critical precisely because the TMPRSS2 promoter is sustained in an AR responsive state. More recently genome-wide ChIP-chip and ChIP-Seq approaches have revealed considerable variability in the targeted transcriptional networks [17–19]. For example in CaP, as the disease progresses, there are altered levels of H3K4me1 and 2 on gene enhancer regions in the ADT-RCaP state, where cells have evolved resistance to anti-androgen therapies. In this new state the targeted increase of H3K4me1 and 2 at different enhancer regions allows the cells to initiate a different AR transcriptional program [20].
These events are not unique to CaP. In a range of solid tumors and myeloid leukemia, nuclear receptors that normally exert mitotic restraint, such as the VDR, RARs and PPARs, become skewed, with selective suppression of gene targets associated with antiproliferative actions [21–26]. Thus RARs, PPARs and the VDR display altered transcriptomes in CaP as a result of distorted epigenetic events (reviewed in Ref. [27]). Dissecting and exploiting the epigenetic mechanisms contributing to altered nuclear receptor function offer significant therapeutic promise. Therefore the development of CaP provides a key system to study the evolution of the malignant epi-genome, and defining these mechanisms is of clinical significance.
Loss and gain of function of transcriptional co-activators and co-repressors associates with transcriptional rigidity. Co-activators and co-repressors each display both loss and gain of function, and can result in similar phenotypes. Thus the loss of a co-activator can lead to suppressed ability of a transcription factor to trans-activate a given target. Similarly the gain of function of co-repressors can limit transactivation ability and enhance trans-repression. The opposite patterns will in turn enhance the trans-activation function. Compared to their co-activator cousins, the co-repressors are somewhat under-explored. Ambiguity remains over how and to what extent these actions are distorted in cancer (reviewed in Ref. [27]). The sheer diversity of transcription factors and co-repressors interactions contributes significantly to this uncertainty. This in turn is compounded by the fact that there are functionally different co-repressor isoforms [28–30] and that co-repressor actions appear specific to each phase of the cell cycle [31–33].
The proto-typical co-repressors NCOR1 and NCOR2/SMRT were cloned in 1995 using nuclear receptor as bait [34,35], and both proteins exist in large multimeric complexes (~2.0 MDa) [36] with histone deacetylases and other histone modifying enzymes (reviewed in Ref. [37]). These complexes are recruited to many different transcription factors, to repress gene activity. The importance of targeted basal repression by co-repressors is evident in the lethality of the Ncor1−/− and Ncor2/Smrt−/− mice [38].
Evidence has also emerged that NCOR1 and NCOR2/SMRT complexes are dynamically recruited to activated transcription factors leading to active transrepression [39], for example associated with suppression of inflammation [40]. Similarly, co-repressor induced transrepression of the glucocorticoid receptor has been established on a genome-wide scale [41]. Finally, de-repression occurs whereby loss of co-repressor association, following activated transcription factor, leads to up-regulation of target genes independently of the sustained presence of the transcription factor [42]. The first direct measurement of the genome-wide distribution of NCOR2/SMRT has established basal and activated distribution during adipogenesis and identified repression of key differentiation programs and hinted at more dynamic interactions with euchromatic regions than hitherto suspected [43,44].
Well-established oncogenic roles for NCOR1 and NCOR2/SMRT have been elucidated in acute promyelocytic leukemia that results from a fusion between RARα, and either the promyelocytic leukemia (PML) or promyelocytic leukemia zinc finger (PLZF) genes [23]. Both chimeric proteins sustain NCOR1 interactions and consequently RARα-mediated cell differentiation is blocked, in part, as a result of maintaining a condensed chromatin structure around the promoters of RARα target genes that govern normal hematopoietic differentiation [45,46]. The importance of inappropriate NCOR1 binding in these disease states has been exploited to stratify patients to tailored therapies. Furthermore the ability of steroidal nuclear receptor such as the AR and ERα to bind NCOR1 and NCOR2/SMRT is important to therapeutic exploitation with receptor antagonists in prostate and breast cancer. Therefore co-repressors appear to play roles in firstly driving critical oncogenic events, but secondly providing a rational targeted strategy toward the key histone modifying enzymes contained within the complex.
Expression profiling in solid tumors has revealed altered NCOR1 and NCOR2/SMRT expression and localization, for example in breast, bladder, and prostate cancers [21,24,26,47,48]. However, to date, uncertainty remains over their precise role in solid tumors, especially in the case of breast and prostate cancers where the etiology of disease is intimately driven by the actions of steroid nuclear receptors. In CaP cells, elevated levels of NCOR2/SMRT have been detected and suppress VDR responsiveness [21]. Similarly, PPAR actions are disrupted and can be targeted selectively by using HDAC inhibitor co-treatments [32,49]. More specifically, elevated NCOR1, and to a lesser extent NCOR2/SMRT correlated with, and functionally drove, the selective insensitivity of PPARα,γ receptors toward dietary derived and therapeutic ligands most clearly in ADT-RCaP cells [32]. Elevated levels of NCOR1 occur in ERα negative breast cancer cells and in turn attenuate anti-mitotic actions of VDR. Again, this molecular lesion can be targeted in ERα negative breast cancer cell lines with co-treatments of VDR ligand (e.g. 1α,25(OH)2D3) plus HDAC inhibitors resulting in selective re-expression of VDR target genes, notably VDUP1 and GADD45A [24]. Together, the studies in breast and prostate cancer suggest that NR shows specificity in their interactions with co-repressors. NCOR1 appears to be involved in the regulation of receptors such as the VDR and PPARs and NCOR2/SMRT with steroid nuclear receptors; this may reflect the emergent specificities of receptor interactions observed in the murine co-repressor knockout models [50,38,51,52].
In contrast, a parallel literature has revealed loss of NCOR1 and NCOR2/SMRT is associated with the ADT-RCaP phenotype and enhances AR transcriptional programs [53,54]. Similar roles for NCOR1 and NCOR2/SMRT appear in the development of breast cancer and Tamoxifen resistance [47]. In contrast, increased NCOR1 and NCOR2/SMRT expression in CaP suppresses the responsiveness of other nuclear receptors that usually exert mitotic restraint, such as VDR and PPARα,γ [32,47–49]. Thus, in CaP progression, there are conflicting selection pressures on co-repressor expression and recruitment. To address this conflict we examined the kinetics of NCOR1 recruitment to genes that display differing transcriptional responsiveness toward 1α,25(OH)2D3 in non-malignant and malignant prostate epithelial cells.
2. Results
2.1. Altered regulation of IGFBP3 and G0S2 in prostate epithelial cells
Non-malignant and malignant prostate cell lines display a range of anti-proliferative responses toward 1α,25(OH)2D3. Non-malignant prostate epithelial RWPE-1 cell is highly responsive toward 1α,25(OH)2D3 [31] whereas the PC-3 CaP cell line, derived from a metastasis [55], is recalcitrant to the anti-proliferative actions of 1α,25(OH)2D3 [21,32,56].
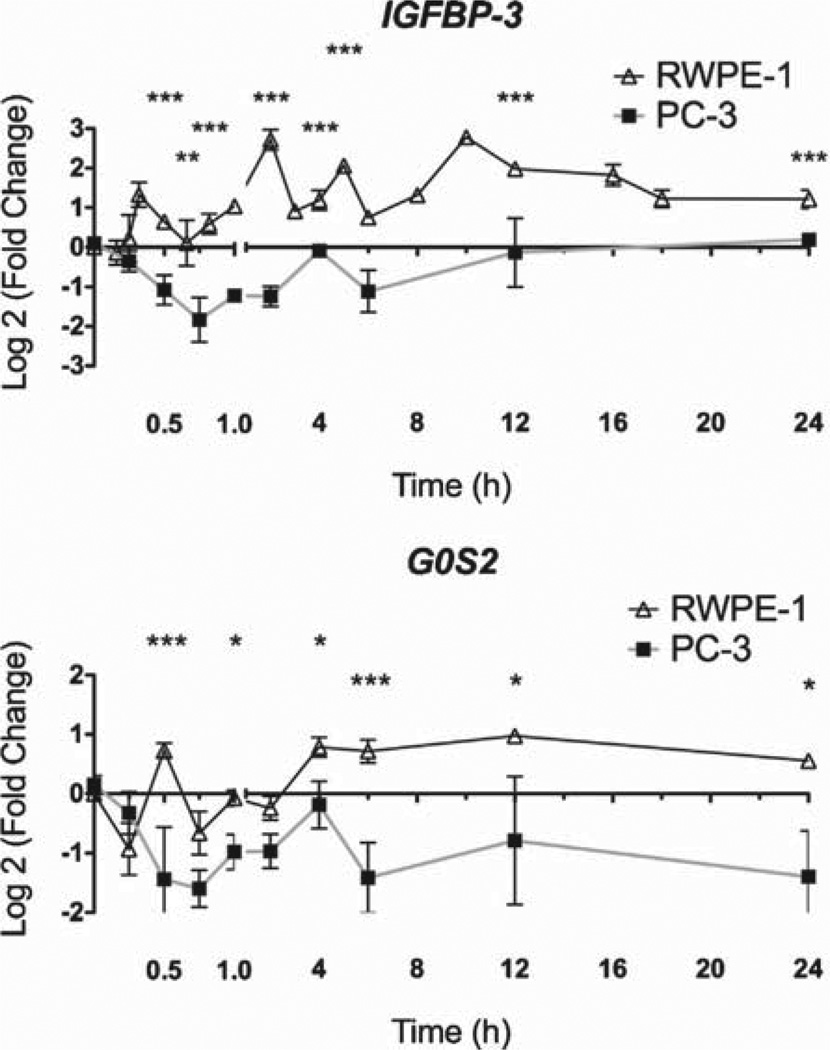
As a functional indicator of 1α,25(OH)2D3 actions, VDR-mediated gene regulatory actions were examined in RWPE-1 and PC-3 cells. Time-resolved regulation studies were undertaken with IGFBP3 and G0S2. The time-resolved kinetics in RWPE-1 and PC-3 cells for the genes are shown in Fig. 1. The kinetics of IGFBP3 and G0S2 mRNA regulation were highly pronounced in RWPE-1 cells. In contrast, the mRNA was significantly reduced in PC-3 at multiple time points. Together these data indicate that gene regulation by 1α,25(OH)2D3 was most dynamic in cells that were most responsive to the anti-proliferative effects (RWPE-1 cells). By comparison, in PC-3 cells, the mRNA regulation profiles were selectively attenuated.
Fig. 1.
Dynamic regulation of VDR target genes. Panel A. RWPE-1 and PC-3 cells were treated with 1α,25(OH)2D3 (100 nM) or EtOH and mRNA was extracted at the indicated time points, and accumulation of IGFBP3 and G0S2 was measured using TaqMan Q-RT-PCR. Accumulation of each target is given as log2 (fold change). Each data point represented the mean of triplicate experiments in triplicate wells ±S.E.M. (*p < 0.05, **p < 0.01, ***p < 0.001).
2.2. Temporal distribution of NCOR1 to target genes is altered in 1α,25(OH)2D3-recalcitrant cells
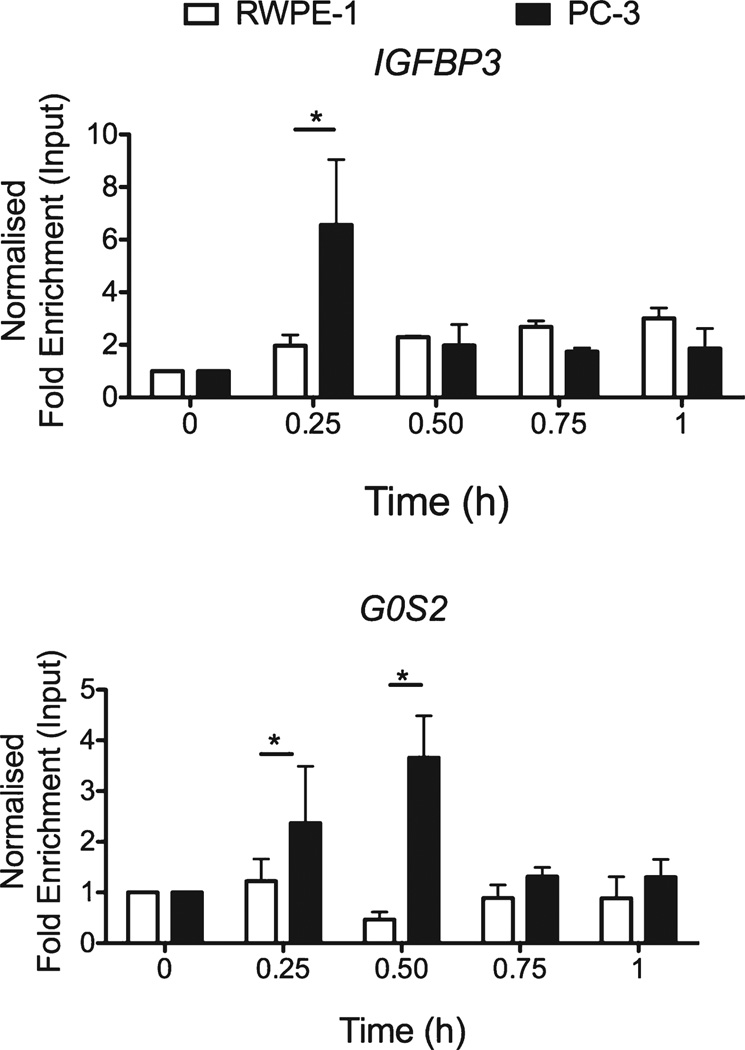
Q-ChIP was undertaken to examine recruitment of NCOR1 to the TSS of G0S2 and a well-characterized VDRE on IGFBP3 [57] in PC-3 cells compared to RWPE-1 cells (Fig. 2). Enhanced 1α,25(OH)2D3-regulated NCOR1 recruitment was evident in 1α,25(OH)2D3-recalcitrant PC-3 cells compared to 1α,25(OH)2D3-sensitive RWPE-1 at these regions examined. This occurred rapidly on both genes in PC-3 cells within the first hour of exposure to 1α,25(OH)2D3, compared to RWPE-1 cells, NCOR1 appeared to cycle off and be subsequently recruited back at later time points; at 24 h on the IGFBP3 response element and at 2 and 12 h on the TSS of G0S2 (data not shown). These findings suggest the underlying mechanisms of recruitment of NCOR1 in response to VDR activation differ significantly between the two cell types.
Fig. 2.
NCOR1 differentially associates with target genes. RWPE-1 and PC-3 cells were treated with 1α,25(OH)2D3 (100 nM) or EtOH for indicated time points. Association of NCOR1 was measured at each region using X-ChIP with ChIP grade antibodies and normalized, and given as fold enrichment over input [31]. Enrichment was measured using Q-PCR with primers specific to these regions that amplified products <150 bp. All measurements were performed in technical duplicate and biological triplicate (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Discussion
The VDR governs and influences anti-mitotic and prodifferentiation transcriptional programs and these actions are distorted in CaP cells [10]. Therefore dissecting the 1α,25(OH)2D3-recalcitrant phenotype is of potential clinical significance. To address this aim, we examined whether the co-repressor protein NCOR1 was differentially recruited to target genes that are known to regulate these anti-mitotic transcriptional programs.
As a starting point to these questions the current study examined differential mRNA regulation of two VDR target genes in different prostate cell models. These approaches revealed that 1α,25(OH)2D3 regulated expression was attenuated and even repressed compared to vehicle controls in PC-3 cells that are recalcitrant to the anti-mitotic actions of 1α,25(OH)2D3, compared to RWPE-1 cells. Building on these studies, we examined the binding of NCOR1 following VDR activation and revealed that 1α,25(OH)2D3 induced greater NCOR1 association on the IGFBP3 and G0S2 promoters in PC-3 cells, compared to RWPE-1 cells. Thus NCOR1 was sustained and enriched at VDR binding sites to different extents and at different time points. Probably reflecting looping events, the TSS showed sustained NCOR1 enrichment throughout the time course [58].
It is now over 30 years since the initial reports demonstrated the anticancer actions of 1α,25(OH)2D3 [59–61]. Following these studies, anti-proliferative effects were demonstrated in a wide variety of cancer cell lines, including those from prostate [5,62–64], as well as xenograft and transgenic CaP models [65,66]. As the anti-cancer effects of the ligand emerged, large-scale epidemiological studies found inverse associations between circulating 25(OH)D3 and cancer risk and advanced disease [67–75]. However, while in vitro, in vivo and epidemiological data support links between replete VDR signaling, growth restraint and broad anticancer activities, clinical exploitation of this receptor has been limited. A significant impediment to translation remains the inability to predict accurately which patients will respond to either chemoprevention or chemotherapy strategies centered on vitamin D compounds. The mechanisms that drive this resistant phenotype are often illusive and probably involve multiple aspects of disruption. Key mechanisms include gene amplification of the 1α,25(OH)2D3 metabolizing enzyme CYP24A1 [76] and repression of the VDR by more general repressors such as SNAIL [77]. The process of inappropriate co-repressor recruitment leading to stable gene silencing also contributes to this phenotype and in particular may shed light on why the VDR and other nuclear receptors are often expressed in non-malignant and retained in malignant prostate epithelial cells [32].
The process of inappropriate co-repressor recruitment may lead to stable gene silencing and in turn shed light on why the VDR and other nuclear receptors are expressed in non-malignant and retained in malignant prostate epithelial cells seemingly independent of the antiproliferative response [32].
The differential recruitment of co-repressors also addresses another ambiguity in their cancer biology, namely the impact of altered expression of co-repressors. Increased NCOR1 and NCOR2/SMRT expression occurs also in breast and bladder cancer and suppressed the responsiveness of nuclear receptors that exert mitotic restraint, such as VDR and PPARα,γ [21,24,26,32,47–49,78]. In contrast, other studies have shown that down-regulated NCOR1 and NCOR2/SMRT enhanced AR transcriptional programs in CaP [53,54,79]. The current study suggests that distorted co-repressor recruitment may provide a route for the selective silencing of critical transcriptional programs and thereby allow CaP cells to escape VDR-regulated mitotic restraint.
There is compelling evidence that histone and DNA methylation processes disrupt transcriptional actions, both alone and together. For example, one consequence of NCOR1 and NCOR2/SMRT association at target genes is the loss of H3K9ac and accumulation of H3K9me2, allowing the potential for hypermethylation at adjacent CpG regions. Further links exist between NCOR1 and DNA methylation through its interaction with KAISO [80]. Similarly regions of H3K9 and −K27 methylation, have the potential to recruit heterochromatin binding protein 1 (HP1) [81]. The recruitment of HP1 in turn re-enforces the recruitment of H3K9 methylase (KMT1A/SUV39H1) [82] and DNA methyltransferases (DNMTs) [83]; enzymes that add and sustain repressive methylation marks to histones and CpG (reviewed in Ref. [84]). Thus the processes of repressive histone modifications and DNA methylation become self-reinforcing. To complement stable DNA methylation of genomic regions, transcription by nuclear receptors appears to be associated with dynamic changes in DNA methylation [85–87]. It therefore seems possible that one consequence of increased co-repressor recruitment to target genes is the loss of H3K9ac and the accumulation of H3K9me2 that may facilitate hypermethylation at adjacent CpG regions. Thus co-repressor actions can direct DNA methylation either through the histone modifications that they drive or through the physical association of DNA methyltransferases. In this manner the inappropriate recruitment of co-repressors may attract the DNA methylation machinery and drive selective and stable gene silencing. It is interesting to note also that the VDR has also been shown to regulate specific histone demethylases [88] adding a further layer of complexity to these relationships and the specificity of gene locus targeting.
4. Materials and methods
4.1. Agents
1α,25(OH)2D3 (gift of Dr. Milan Uskokovic, BioXell S.p.A., Italy) stored as 1 mM stocks in ethanol.
4.2. Q-RT-PCR
RNA was isolated using TRizol (Invitrogen). Target gene expression was quantitated on an ABI 7900 (Applied Biosystems®) machine using TaqMan assays. Measurements were performed in technical and biological triplicate. The statistical significance was calculated using Student’s t-test.
4.3. ChIP protocols
X-ChIP was used to measure the association of NCOR1 binding as described previously [31]. Briefly, chromatin from 1.5 × 106 mid-exponential cells was cross-linked. Pre-cleared inputs were immunoprecipited with NCOR1 (Abcam ab24552). Complexes were recovered using magnetic beads, washed, cross-linking was reversed and further cleared DNA was recovered by standard precipitation approaches. 25 ng DNA was used per Q-PCR reaction using SYBRgreen with pre-optimized primers as described previously [31]. Student’s t-test was used to calculate the significant enrichment.
Acknowledgements
1α25(OH)2D3 was a gift from Dr. Milan Uskokovic (BioXell S.p.A., Italy). M.J.C. acknowledges the support of NucSys, a European Community FP6 – Marie Curie Research Training Network, the Biotechnology and Biological Sciences Research Council and support in part from National Institute of Health grants R01 CA095367-06 and 2R01-CA-095045-06. M.J.C. also acknowledges support, in part, of the NCI Cancer Center Support grant to the Roswell Park Cancer Institute [CA016056] and the Alliance Foundation of Roswell Park Cancer Institute. B.M.T. acknowledges the support of a program grant C1015/A9077 from Cancer Research UK.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Lubahn DB, et al. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240(4850):327–330. doi: 10.1126/science.3353727. [DOI] [PubMed] [Google Scholar]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. British Journal of Cancer. 1972;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, et al. Synergistic effects of coactivators GRIP1 and beta-catenin on gene activation: cross-talk between androgen receptor and Wnt signaling pathways. Journal of Biological Chemistry. 2004;279(6):4212–4220. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- 4.Yang X, et al. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene. 2006;25(24):3436–3444. doi: 10.1038/sj.onc.1209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell MJ, et al. Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. Journal of Molecular Endocrinology. 1997;19(1):15–27. doi: 10.1677/jme.0.0190015. [DOI] [PubMed] [Google Scholar]
- 6.Khanim FL, et al. Altered SMRT levels disrupt vitamin D(3) receptor signalling in prostate cancer cells. Oncogene. 2004;23(40):6712–6725. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 7.Kubota T, et al. 19-Nor-26,27-bishomo-vitamin D3 analogs: a unique class of potent inhibitors of proliferation of prostate, breast, and hematopoietic cancer cells. Cancer Research. 1998;58(15):3370–3375. [PubMed] [Google Scholar]
- 8.Liu M, et al. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes and Development. 1996;10(2):142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 9.Saramaki A, et al. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Research. 2006;34(2):543–554. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne J, Campbell MJ. The vitamin D receptor in cancer. Proceedings of the Nutrition Society. 2008;67(2):115–127. doi: 10.1017/S0029665108006964. [DOI] [PubMed] [Google Scholar]
- 11.Palmer HG, et al. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. Journal of Cell Biology. 2001;154(2):369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitareewan S, et al. G0S2 is an all-trans-retinoic acid target gene. International Journal of Oncology. 2008;33(2):397–404. [PMC free article] [PubMed] [Google Scholar]
- 13.Zandbergen F, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochemical Journal. 2005;392(Pt. 2):313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendriksen PJ, et al. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Research. 2006;66(10):5012–5020. doi: 10.1158/0008-5472.CAN-05-3082. [DOI] [PubMed] [Google Scholar]
- 15.Taneja SS, et al. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. Journal of Biological Chemistry. 2004;279(14):13944–13952. doi: 10.1074/jbc.M306576200. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 17.Anderson SP, et al. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor {alpha}, retinoid × receptor and liver × receptor in mouse liver. Molecular Pharmacology. 2004;66(6):1440–1452. doi: 10.1124/mol.104.005496. [DOI] [PubMed] [Google Scholar]
- 18.Bookout AL, et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handschin C, Meyer UA. Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Archives of Biochemistry and Biophysics. 2005;433(2):387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanim FL, et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23(40):6712–6725. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 22.Rashid SF, et al. Synergistic growth inhibition of prostate cancer cells by 1 alpha, 25 dihydroxyvitamin D(3) and its 19-nor-hexafluoride analogs in combination with either sodium butyrate or trichostatin A. Oncogene. 2001;20(15):1860–1872. doi: 10.1038/sj.onc.1204269. [DOI] [PubMed] [Google Scholar]
- 23.Lin RJ, et al. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391(6669):811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 24.Banwell CM, et al. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clinical Cancer Research. 2006;12(7 Pt. 1):2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 25.Ting HJ, et al. Increased expression of corepressors in aggressive androgen-independent prostate cancer cells results in loss of 1alpha, 25-dihydroxyvitamin D3 responsiveness. Molecular Cancer Research. 2007;5(9):967–980. doi: 10.1158/1541-7786.MCR-06-0318. [DOI] [PubMed] [Google Scholar]
- 26.Abedin SA, et al. Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis. 2009;30(3):449–456. doi: 10.1093/carcin/bgp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battaglia S, Maguire O, Campbell MJ. Transcription factor co-repressors in cancer biology: roles and targeting. International Journal of Cancer. 2010;126(11):2511–2519. doi: 10.1002/ijc.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mengeling BJ, et al. SMRTepsilon, a corepressor variant, interacts with a restricted subset of nuclear receptors, including the retinoic acid receptors alpha and beta. Molecular and Cellular Endocrinology. 2012;351(2):306–316. doi: 10.1016/j.mce.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodson ML, Jonas BA, Privalsky ML. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. Journal of Biological Chemistry. 2005;280(9):7493–7503. doi: 10.1074/jbc.M411514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorne JL, et al. Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Research. 2011;39(6):2045–2056. doi: 10.1093/nar/gkq875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battaglia S, et al. Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010;31(9):1650–1660. doi: 10.1093/carcin/bgq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altintas DM, et al. Cell cycle regulated expression of NCoR might control cyclic expression of androgen responsive genes in an immortalized prostate cell line. Molecular and Cellular Endocrinology. 2011;332(1–2):149–162. doi: 10.1016/j.mce.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Horlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377(6548):397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO Journal. 2000;19(16):4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perissi V, et al. Deconstructing repression: evolving models of co-repressor action. Nature Reviews Genetics. 2010;11(2):109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 38.Jepsen K, et al. Cooperative regulation in development by SMRT and FOXP1. Genes and Development. 2008;22(6):740–745. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu C, et al. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. Journal of Biological Chemistry. 2005;280(14):13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 40.Tiefenbach J, et al. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Molecular Biology of the Cell. 2006;17(4):1643–1651. doi: 10.1091/mbc.E05-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surjit M, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145(2):224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 42.Heikkinen S, et al. Nuclear hormone 1alpha, 25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Research. 2011;39(21):9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raghav SK, et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Molecular Cell. 2012;46(3):335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Barish GD, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metabolism. 2012;15(4):554–562. doi: 10.1016/j.cmet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller-Tidow C, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116(18):3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoemme C, et al. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood. 2008;111(5):2887–2895. doi: 10.1182/blood-2007-03-079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Girault I, et al. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clinical Cancer Research. 2003;9(4):1259–1266. [PubMed] [Google Scholar]
- 48.Kim JY, Son YL, Lee YC. Involvement of SMRT corepressor in transcriptional repression by the vitamin D receptor. Molecular Endocrinology. 2009;23(2):251–264. doi: 10.1210/me.2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang TH, Szabo E. Enhanced growth inhibition by combination differentiation therapy with ligands of peroxisome proliferator-activated receptor-gamma and inhibitors of histone deacetylase in adenocarcinoma of the lung. Clinical Cancer Research. 2002;8(4):1206–1212. [PubMed] [Google Scholar]
- 50.Ghisletti S, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes and Development. 2009;23(6):681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa S, et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(40):14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102(6):753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 53.Liao G, et al. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. Journal of Biological Chemistry. 2003;278(7):5052–5061. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 54.Hodgson MC, et al. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. Journal of Biological Chemistry. 2005;280(8):6511–6519. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 55.Kaighn ME, et al. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Investigative Urology. 1979;17(1):16–23. [PubMed] [Google Scholar]
- 56.Campbell MJ, et al. The anti-proliferative effects of 1alpha, 25(OH)2D3 on breast and prostate cancer cells are associated with induction of BRCA1 gene expression. Oncogene. 2000;19(44):5091–5097. doi: 10.1038/sj.onc.1203888. [DOI] [PubMed] [Google Scholar]
- 57.Malinen M, et al. Cyclical regulation of the insulin-like growth factor binding protein 3 gene in response to 1alpha, 25-dihydroxyvitamin D3. Nucleic Acids Research. 2011;39(2):502–512. doi: 10.1093/nar/gkq820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saramaki A, et al. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha, 25-dihydroxyvitamin D3. Journal of Biological Chemistry. 2009;284(12):8073–8082. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colston K, Colston MJ, Feldman D. 1,25-Dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology. 1981;108(3):1083–1086. doi: 10.1210/endo-108-3-1083. [DOI] [PubMed] [Google Scholar]
- 60.Miyaura C, et al. 1 Alpha, 25-dihydroxyvitamin D3 induces differentiation of human myeloid leukemia cells. Biochemical and Biophysical Research Communications. 1981;102(3):937–943. doi: 10.1016/0006-291x(81)91628-4. [DOI] [PubMed] [Google Scholar]
- 61.Abe E, et al. Differentiation of mouse myeloid leukemia cells induced by 1 alpha, 25-dihydroxyvitamin D3. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(8):4990–4994. doi: 10.1073/pnas.78.8.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elstner E, et al. Novel 20-epi-vitamin D3 analog combined with 9-cis-retinoic acid markedly inhibits colony growth of prostate cancer cells. Prostate. 1999;40(3):141–149. doi: 10.1002/(sici)1097-0045(19990801)40:3<141::aid-pros1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 63.Peehl DM, et al. Antiproliferative effects of 1,25-dihydroxyvitamin D3 on primary cultures of human prostatic cells. Cancer Research. 1994;54(3):805–810. [PubMed] [Google Scholar]
- 64.Colston K, et al. 1,25-Dihydroxyvitamin D3 receptors in human epithelial cancer cell lines. Cancer Research. 1982;42(3):856–859. [PubMed] [Google Scholar]
- 65.Blutt SE, et al. A calcitriol analogue EB1089, inhibits the growth of LNCaP tumors in nude mice. Cancer Research. 2000;60(4):779–782. [PubMed] [Google Scholar]
- 66.Banach-Petrosky W, et al. Vitamin D inhibits the formation of prostatic intraepithelial neoplasia in Nkx3.1; Pten mutant mice. Clinical Cancer Research. 2006;12(19):5895–5901. doi: 10.1158/1078-0432.CCR-06-1039. [DOI] [PubMed] [Google Scholar]
- 67.Garland CF, Garland FC. Do sunlight and vitamin D reduce the likelihood of colon cancer? International Journal of Epidemiology. 1980;9(3):227–231. doi: 10.1093/ije/9.3.227. [DOI] [PubMed] [Google Scholar]
- 68.Garland FC, et al. Geographic variation in breast cancer mortality in the United States: a hypothesis involving exposure to solar radiation. Preventive Medicine. 1990;19(6):614–622. doi: 10.1016/0091-7435(90)90058-r. [DOI] [PubMed] [Google Scholar]
- 69.Luscombe CJ, et al. Prostate cancer risk: associations with ultraviolet radiation, tyrosinase and melanocortin-1 receptor genotypes. British Journal of Cancer. 2001;85(10):1504–1509. doi: 10.1054/bjoc.2001.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States) Cancer Causes and Control. 2005;16(2):83–95. doi: 10.1007/s10552-004-1661-4. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz GG, Hulka BS. Is vitamin D deficiency a risk factor for prostate cancer? (Hypothesis) Anticancer Research. 1990;10(5A):1307–1311. [PubMed] [Google Scholar]
- 72.John EM, et al. Sun exposure, vitamin D receptor gene polymorphisms, and risk of advanced prostate cancer. Cancer Research. 2005;65(12):5470–5479. doi: 10.1158/0008-5472.CAN-04-3134. [DOI] [PubMed] [Google Scholar]
- 73.Chen TC, et al. Prostatic 25-hydroxyvitamin D-1alpha-hydroxylase and its implication in prostate cancer. Journal of Cellular Biochemistry. 2003;88(2):315–322. doi: 10.1002/jcb.10342. [DOI] [PubMed] [Google Scholar]
- 74.Hsu JY, et al. Reduced 1alpha-hydroxylase activity in human prostate cancer cells correlates with decreased susceptibility to 25-hydroxyvitamin D3-induced growth inhibition. Cancer Research. 2001;61(7):2852–2856. [PubMed] [Google Scholar]
- 75.Ahonen MH, et al. Prostate cancer risk and prediagnostic serum 25-hydroxyvitamin D levels (Finland) Cancer Causes and Control. 2000;11(9):847–852. doi: 10.1023/a:1008923802001. [DOI] [PubMed] [Google Scholar]
- 76.Albertson DG, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nature Genetics. 2000;25(2):144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 77.Palmer HG, et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nature Medicine. 2004;10(9):917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Z, et al. NCOR1 mRNA is an independent prognostic factor for breast cancer. Cancer Letters. 2006;237(1):123–129. doi: 10.1016/j.canlet.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 79.Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Molecular Endocrinology. 2006;20(5):1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- 80.Yoon HG, et al. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Molecular Cell. 2003;12(3):723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 81.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends in Genetics. 2009;25(3):129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 82.Fujita N, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. Journal of Biological Chemistry. 2003;278(26):24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 83.Esteve PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes and Development. 2006;20(22):3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49(14):2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Molecular Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 86.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 87.Kangaspeska S, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 88.Pereira F, et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Human Molecular Genetics. 2011;20(23):4655–4665. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]