Abstract
Transcriptional control of microRNAs (miRNA) by cell signaling pathways, especially in the context of growth factor regulation, is a widely recognized phenomenon with broad-reaching implications. However, several recent studies indicate that not just transcription, but also processing of miRNAs is subject to regulation as part of an integrated physiological response to various stimuli and environmental changes. The canonical miRNA biogenesis pathway; sequential steps of nucleolytic cleavage by the RNase III enzymes Drosha and Dicer, are emerging regulatory hubs for the modulation of miRNA expression as part of both physiological and pathological responses. In this article we use well-characterized growth-factor signaling pathways such as transforming growth factor-β (TGF-β), Protein Kinase B (PKB, also known as Akt) and extracellular-signal-regulated kinase (ERK) to illustrate how basic cell signaling pathways modulate the activities of these components of the miRNA biogenesis pathway to achieve optimal miRNA expression patterns.
Introduction: growth factors and miRNA maturation
One emerging paradigm in the study of cell signaling pathways is the role of post-transcriptional RNA regulation in modulating gene expression. A primary player in this regulation is microRNA (miRNA); small ~22 nucleotide (nt) RNA molecules that bind target mRNAs, usually in the 3′ untranslated regions (UTR), and inhibit their expression. Since their original description as mediators of development in C. elegans [1] miRNAs have proven integral components of nearly every aspect of biology. Perhaps nowhere is this importance illustrated as clearly as in the modulation of growth factor signaling pathways. Driven by the quest to discover novel targets for cancer therapeutics, knowledge of miRNA activity in response to growth factors has grown nearly exponentially.
Detailed molecular analyses have revealed a highly structured process of miRNA biogenesis involving sequential processing of long mRNA-like transcripts down into the ~22 nt single stranded RNA (ssRNA) effector molecule that is the mature miRNA. As the details of this processing pathway have emerged, it has become increasingly apparent that for each step in the miRNA biogenesis pathway, there exist alternative strategies through which they may be regulated or circumvented by cell signaling. Here we describe how growth factor signaling pathways utilize both the canonical and non-canonical miRNA biogenesis pathway to achieve the intricate balance necessary to respond appropriately to growth factor signaling pathways. We further speculate that the emerging interest in miRNA precursor stability (box 1) will eventually also be tied into growth factor signaling biology. In this article we do not discuss how individual miRNAs contribute to their respective pathways, as those subjects are extensively reviewed elsewhere [2, 3].
Box 1. New avenues for control - primary and precursor stability.
In addition to altering the activity of miRNA-processing enzymes, miRNA biogenesis can be modulated by altering the stability of both pri-miRNAs and pre-miRNAs. By limiting the availability of pri-miRNAs or pre-miRNAs for processing, the downstream effect of miRNA-mediated gene regulation is affected. Pri-miRNA levels can be altered by RNA editing. The adenosine deaminase acting on RNA (ADAR) proteins are capable of deaminating adenosines on single-stranded RNA molecules [49]. Upon editing of pri-miR-142 by ADAR, pri-miR-142 is degraded by Tudor-SN, a component of RISC and also a ribonuclease [49].
Alternatively, pre-miRNAs can be destabilized. Expression of the RNA-binding protein is often inversely correlated with expression of members of the let-7 family of miRNAs. This is because Lin28 can bind the stem-loop of pre-let-7 and recruit the uridylatransferase Zcchc11 (TUT4) [50, 51]. Uridylation of pre-let-7 by Zcchc11 destabilizes pre-let-7, which results in degradation of the precursor and thus lower levels of mature let-7. To date neither pri-miRNA nor pre-miRNA destabilizing mechanisms have been linked to growth factor signaling pathways. However, expression of Zcchc11 can promote proliferation in different transformed cell lines [52], while ADAR has been shown to promote proliferation in astrocytomas [53]. Therefore, it is likely that these mechanisms of miRNA regulation will soon be placed in the wider context of growth factor signaling pathways under either physiological or pathological conditions.
Background: the miRNA biogenesis pipeline
A distinct set of rules governed by a specific series of proteins mediates the development of miRNAs from long, mRNA-like, primary (pri-miRNA) transcripts into the short [20–22 base pairs (bp)] single-stranded mature miRNA molecules that are physiologically relevant (Figure 1A, Reviewed in [4]). Briefly, this process involves sequential processing of the transcript by protein complexes primarily identified by the inclusion of a specific type III RNase enzyme. The biogenesis of miRNA begins with a long primary transcript known as pri-miRNA which bears a 7-methylguanosine cap and a poly-(A) tail. Pri-miRNA contains one or more stable stem-loop structures which encode mature miRNA(s). In the first processing step, the RNAse III enzyme Drosha, in complex with DeGiorgio Critical Region 8 (DGCR8 also known as Pasha) together with several other cofactors binds and cleaves the stem-loop region of pri-miRNA to generate a precursor miRNA (pre-miRNA) [4–6]. Pre-miRNA, usually <100 bp long, contains only the stem-loop portion of pri-miRNA and encodes what will become the mature miRNA in either its 5′ or 3′ arm extending out from the stem loop along with a second, nearly complementary, sequence on the opposing arm (Figure 1B). This structure is highly conserved among all miRNAs and is the basis for its recognition and export from the nucleus by Exportin 5 (EXP5) [7].
Figure 1. Basic components of the miRNA biogenesis pathway.
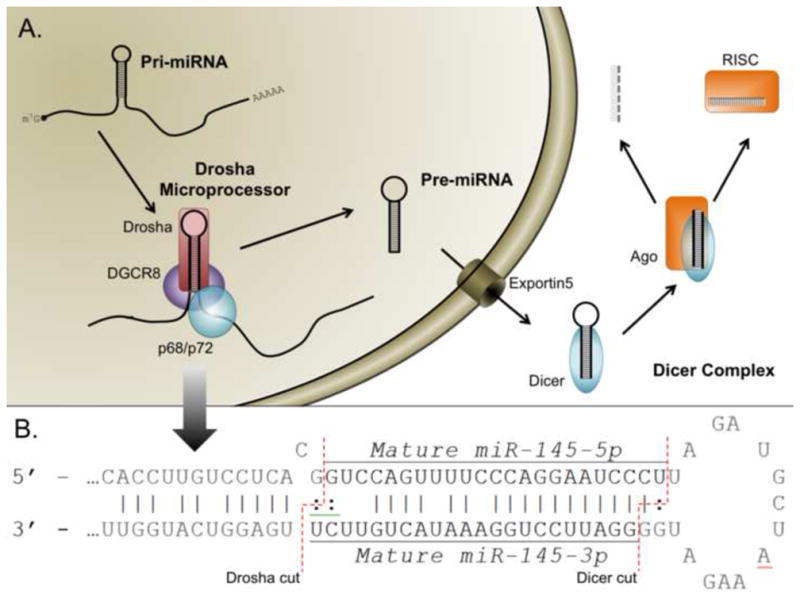
miRNA begins as a long primary transcript (pri-miRNA) with a structure resembling mRNA. Pri-miRNAs contain a 5′ cap (m7-G) and a polyA tail at the 3′ end (AAAA). Pri-miRNA undergoes two sequential processing steps. First, the Drosha microprocessor complex which is composed of Drosha, DGCR8 and DEAD-Box RNA helicases (p68 or p72) cleaves pri-miRNA to generate precursor miRNA (pre-miRNA). Following cleavage by the Drosha microprocessor, the resulting pre-miRNA precursor maintains a stereotypical stem-loop structure. Following the export of pre-miRNA from the nucleus by exportin 5 (EXP5), Dicer processes the pre-miRNA into double stranded mature miRNA. Mature miRNA will be loaded into Argonaute proteins (Ago), which separates mature miRNA into two single stranded miRNAs. (B) Future miRNAs adopt a stereotypical stem-loop structure within the pri-miRNA sequence (shown here for miR-145). The mature sequence is encoded on one side of the loop and the degraded * strand on the other. More recent nomenclature has moved to calling each strand 5p and 3p according to which side of the strand they occupy. Here, the dotted lines indicate the site of future cleavage by Dicer and Drosha.
When in complex, Drosha and DGCR8 comprise the, “microprocessor complex” and can variably interact with two DEAD-box helicase proteins p68 (also called DDX5) and p72 (also called DDX17) whose expression is required for the biogenesis of some, though not all, miRNAs [8]. What role these RNA helicases play in miRNA biogenesis is presently unknown. Interestingly, some subset of miRNAs that require p72 for maturation, do not require p68; suggesting there exists heterogeneity in the composition of this important structure [8].
Once in the cytoplasm, the pre-miRNA stem-loop is processed into a dsRNA duplex containing only the 5′ (5p) and 3′ (3p) miRNA molecules by a second RNAse III enzyme named Dicer [9, 10]. After processing, one strand of the miRNA gets loaded into one of four argonaute (Ago) proteins [11]. These proteins mediate the effector function of the miRNAs either through inhibiting translation of the target mRNA or, in the case of Ago2, by directly degrading the mRNA transcript [11]. The remaining miRNA strand, termed the star or passenger strand and denoted by a *, is degraded.
Despite the highly ordered nature of this maturation process, multiple components of the developmental pathway are open to manipulation or deviation. The dissection of these deviations has proved extremely important in studying both the function and significance of miRNAs under physiological conditions. Growth factor signaling pathways utilize this typified biogenesis pathway to exert various biological effects.
Control of miRNA biogenesis by DNA binding proteins: Smads at the Drosha microprocessor
Perhaps the best understood, if not the most highly regulated, step of miRNA biogenesis is at the point of pri-to-pre-miRNA processing by the Drosha microprocessor complex. Canonically, increases in mature and pre-miRNA levels stem from higher pri-miRNA expression, usually as a result of increased transcription. However, this is not always the case. In 2009, it was reported that when pulmonary artery smooth muscle cells (PASMCs) are stimulated with ligands of the transforming growth factor β (TGF-β) family, such as TGF-β and bone morphogenetic protein 4 (BMP4), pri-miR-21 levels are unchanged while mature miR-21 expression increases significantly [12]. This result suggested a transcription-independent mechanism of miR-21 regulation which was confirmed by the induction of miR-21 even in the presence of the transcriptional inhibitor α-amanatin [12]. Biochemical analysis showed that the increase in mature miR-21 is caused by faster turnover in the biogenesis of miR-21 mediated by the nuclear translocation of Smad proteins [12, 13].
Smads represent the primary signal-transduction molecules of the TGF-β family of growth factors. Receptor activation by TGF-β family ligands induces phosphorylation of several Smad proteins collectively called the Receptor-specific Smads (R-Smads) [14]. Phosphorylated R-Smads form a complex with the common-Smad (co-Smad), Smad4, and translocate to the nucleus where they act as transcriptional regulators to promote or inhibit gene expression [14]. R-Smad/co-Smad hetero-dimerization is required for the transcriptional regulation mediated by TGF-β signaling. Surprisingly, Davis, et al. [12] found that Smad4 is dispensable for TGF-β induced induction of miR-21. Furthermore, stimulation with TGF-β ligands induced an association between Smads and the RNA helicase p68, indicating a direct recruitment of R-Smads to the Drosha microprocessor complex [12].
More recent miRNA expression profiling experiments revealed that stimulation of PASMCs with either TGF-β or the related BMP4 induces the expression of approximately 20 miRNAs [13]. Following stimulation, many of these miRNAs are bound directly by Smad proteins through a 5-base motif that closely resembles the Smad DNA binding element (SBE; 5′-CAGAC-3′) in a region overlapping the encoded mature miRNA [13]. Thus, R-Smads associate with pri-miRNA in a sequence specific manner and facilitate the Drosha microprocessor activity to enhance mature miRNA expression (Figure 2A). Interestingly, in silico analyses have found that pre-miRNA sequences encode a higher number of transcription factor biding sites than would be predicted by chance [15]. These results suggest that the mechanism for Smad-dependent regulation of pri-miRNAs may represent a widespread, though underappreciated, strategy for controlling miRNA expression.
Figure 2. Integration of miRNA into cell growth factor signaling pathways.
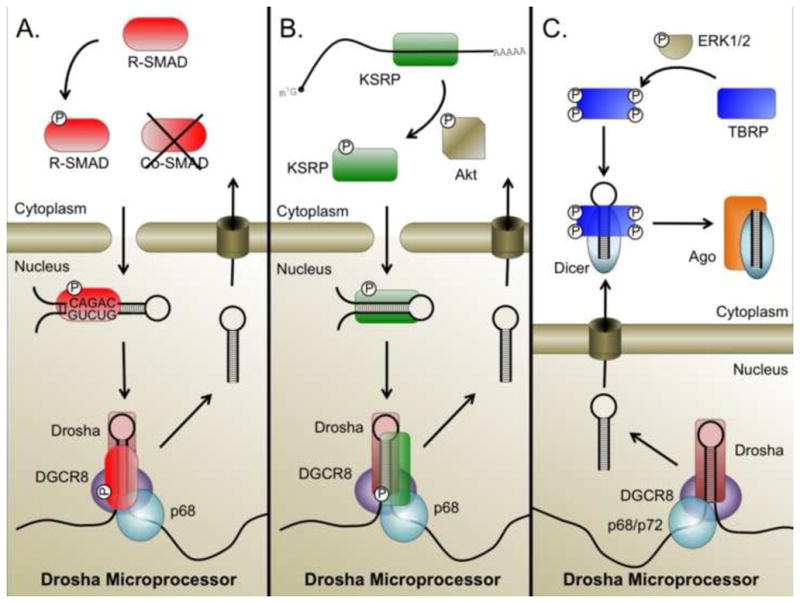
(A) Activation of receptor Smads by TGF-β family ligands leads to their translocation into the nucleus. In the nucleus, R-Smads bind, independent of the transcriptionally necessary co-Smad, to a conserved sequence in pri-miRNAs which they recruit to the Drosha microprocessor complex and facilitate pri- to pre-miRNA maturation. (B) PI3K-mediated activation of Akt induces the phosphorylation of KSRP. Phosphorylated KSRP is dissociated from mRNA and associates with targeted pri-miRNA sequence to recruit the Drosha microprocessor and facilitate pri- to pre-miRNA processing. (C) Activation of MAPK/ERK induces phophorylation of TRBP, which is subsequently recruited to Dicer and promotes the processing of miRNAs associated with cell proliferation.
A similar mechanism has been described for miRNA processing regulation by the tumor suppressor p53 [16]. Following DNA damage, p53 translocates to the nucleus where it binds p68 along with various pri-miRNAs to facilitate their biogenesis. For both the Smad and p53 pathways, knockdown of p68 by siRNA is sufficient to inhibit miRNA maturation [12, 16], suggesting that p68 is an essential factor for pri-miRNA processing. Whereas Smad proteins bind pri-miRNAs by a distinct sequence-specific mechanism [13], it is unclear how p53 specifically recognizes a set of pri-miRNAs.
Protein recruitment to the Drosha microprocessor complex does not always facilitate miRNA processing. Following stimulation with estradiol (E2), estrogen receptor α (ERα) also associates with p68 and the Drosha microprocessor [17]. However, unlike the previous examples, ERα inhibits pri-miRNA processing by Drosha [17]. Following E2 stimulation, nearly all miRNAs are affected, suggesting that ER-α act as a general regulator of miRNA expression unlike the more specific induction achieved by p53 and Smads. It is also unclear whether p72 (DDX17) plays a similar role to p68 in the induction of pri-miRNA processing by Smads, p53, or ERα.
Control of the Drosha microprocessor by RNA binding proteins: KSRP activation by Akt
Like the DNA binding proteins described above, RNA binding proteins are also known to facilitate pri-to-pre-miRNA processing. The singe-stranded RNA binding KH-type splicing regulatory protein (KSRP) performs various cellular activities, such as facilitating alternative splicing and binding to the 3′ UTR of mRNAs to recruit the exosome and expedite their degradation [18]. In addition, KSRP binds the terminal loop of a subset of pri-miRNAs and facilitates their processing. Though the microprocessor activity of KSRP was originally attributed to direct interaction with a GGG motif in the terminal loop of let-7a [19], KSRP activity has since been found to modulate a broader repertoire of miRNAs include miR-155 [20], miR-16 [21] and miR-206 [22], suggesting that KSRP acts to enhance the catalytic activity of the Drosha microprocessor through a sequence-independent mechanism.
The activity of KSRP in regulating the Drosha microprocessor is closely tied to growth factor signaling. Mutational analyses of KSRP have revealed that phosphorylation of KSRP at various Serine residues including Ser193 [22] as well as Ser274 and Ser670 can enhance its pri-miRNA binding affinity and promote pri-miRNA processing [21]. During differentiation of myoblasts, Briata, et al. [22] found that phosphorylation of Ser193 mediates a switch of KSRP affinity from mRNA to pri-miRNA in vitro. They further found that this phosphorylation is Akt-dependent; constitutive activation of Akt2 was sufficient to enhance the production of myogenic miRNAs in a KSRP-dependent manner [22]. Conversely, Akt2-deficient cells grown in differentiation media displayed increased expression of pri-miRNAs, but not of the mature miRNAs, suggesting a block of pri-to pre-miRNA processing as a result of blocked KSRP phosphorylation by Akt2 [22]. Consistent with this idea, miRNA expression was significantly altered in the muscles of KSRP null mice following cardiac injury by administration of cardiotoxin [22]. These studies identify KSRP phosphorylation as an integral part of the Akt pathway (Figure 2B) and suggest that Akt signaling may activate a miRNA-processing cascade and mediate cell type-specific outcomes. KSRP-mediated miRNA biogenesis can also be regulated by other, non-growth factor, pathways as indicated by the finding that DNA-damage induced phosphorylation at Ser274 and Ser670 by ATM can also promote maturation of specific miRNAs [21].
The positive effect of KSRP on the biogenesis of let-7a-1 can be counteracted by the splicing factor heteronuclear ribonucleoprotein A1 (hnRNPA1) which binds the same region of pre-let-7a stem-loop and displaces KSRP [23]. Interestingly, hnRNPA1 can also promote the expression of a number of miRNAs [24] raising the intriguing possibility that the Drosha microprocessor components can alternate between agonistic and antagonistic roles in the context of specific miRNA biogenesis.
mirtrons
As described above, pri-to-pre-miRNA processing by the Drosha microprocessor complex is a critical regulatory step in the life of miRNAs. However, some miRNAs avoid this step by utilizing alternative processing pathways. In both flies [25] and humans [26] mirtrons, pre-miRNAs generated directly from the spliced introns of mRNA transcripts, have been identified by deep-sequencing and validated as functional. Because they are derived directly by splicing as a fully formed pre-miRNA-like stem-loop structure these sequences are able to bypass the microprocessor [25, 26]. To our knowledge no studies have yet placed mirtron development and/or activity into a physiological context. Therefore, how these miRNAs contribute to cellular functions remains unknown. However, this may be changing, as the development of Drosha/DGCR8-deficient embryonic stem (ES) cell lines will make the analysis of such non-canonical miRNAs more straightforward [27, 28]. Furthermore, one recent study identified 237 mouse and 240 human splicing-derived mirtrons indicating the potential importance of this understudied class of miRNAs [29].
Control at the Dicer complex: regulation of TRBP by MAPK/ERK signaling
The pre-to-mature miRNA processing complex, of which the catalytic core is Dicer, can also serve as an important site for regulation of miRNA biogenesis. In fact, KSRP was originally described as an interacting protein of the Dicer complex where it acts through a sequence-specific association with the pre-miRNA stem-loop [20] TO promote maturation [19, 20]. It is unclear, however, whether Akt-mediated phosphorylation of KSRP plays a role in controlling Dicer processing.
Similarly, The human immunodeficiency virus (HIV)-1 transactivating response RNA binding protein (TRBP) is an important component of the Dicer complex [30, 31]. However, the activity of TRBP in this process is not constitutive. Rather, a recent publication identified four Serine residues, of which phosphorylation was required to promote this interaction [32]. miRNA microarray analysis comparing HEK293 cells expressing either a non-phosphorylatable TRBP with a phospho-mimic TRBP identified a variety of growth-associated miRNAs, including miR-17, miR-20a and miR-92a, that were induced upon phosphorylation. Conversely, the constitutively active protein uniformly decreased expression of various let-7 species, a family of miRNA with well-documented growth-suppressive functions [33]. The kinases responsible for phosphorylating TRBP were identified as the extracellular signal-regulated kinase, ERK1/2 [32]. Similar to the Akt pathway, the mitogen-activated kinase (MAPK)/ERK pathway is a central modulator of numerous cellular pathways and is commonly mutated in cancers [34]. Thus, TRBP recruitment to the Dicer complex may act as a regulatory switch activated by MAPK/ERK signaling to promote expression of growth-promoting miRNAs (Figure 2C) [32].
Another component of the Dicer complex, the protein kinase R activating protein (PACT) also acts as an important activator of pre-miRNA processing. Although PACT is not essential for miRNA maturation, inhibition of PACT decreases expression levels of multiple mature miRNAs [35]. It remains unclear whether this interaction can be controlled by growth factor signaling pathways. Interestingly, knocking down any components of the Dicer complex, (i.e. Dicer, PACT, or TRBP) inhibits the levels of both pre-miRNAs and mature miRNAs [36]. This suggests the existence of some positive feedback loop by which mature miRNAs modulate the levels of their pri- and/or pre-miRNAs. One such mechanism was recently described for let-7 expression in C. elegans where the Ago2 homolog, ALG-1 can bind a conserved region in the 3′ end of pri-let-7 to promote its processing [37]. This process is dependent on expression of mature let-7, and thus the mature miRNA acts through a positive feedback loop to promote its own biogenesis.
Dicer-independent maturation of pre-miRNA
As with the Drosha microprocessor complex, the Dicer complex may also be bypassed in miRNA biogenesis. Deep sequencing of both zebra fish [38] and mice [39] deficient in Dicer showed an enrichment of one specific mature miRNA: miR-451. Pre-miR-451 has a unique, highly conserved structure that allows it to associate with Ago2, which contains RNase activity. Loading of pre-miR-451 into Ago2 results in slicing of pre-miR-451 and generates a 30 nt intermediate miR-451, which is then further trimmed to become a mature miRNA [38, 39].
Pathological significance of growth factor regulated miRNAs
Aberrant expression of some miRNAs controlled by growth factor signaling pathways have been reported and implicated in the pathogenesis of disorders such as cancer and cardiovascular disease,. For example, miR-21 is known as an “onco-miR” because it is robustly upregulated in nearly all tumor samples [40] and it can propagate oncogenic activities by targeting tumor suppressors such as programmed cell death protein 4 (PDCD4) [41], tropomycin1 (TPM1) [42] and phosphatase and tensin homolog (PTEN) [43]. As activation of TGF-β signaling is often associated with various tumor types, it is intriguing to speculate that increased expression of miR-21 is, in part, due to aberrant activation of TGF-β signal and warrants anti-TGF-β therapies [44]. Similar to tumor samples, upregulatoin of miR-21 can be initiated by cardiac stress and may lead to cardiac hypertrophy or cardiac fibrosis ina MAPK/ERK pathway-dependent manner [40].
Other miRNAs may act as tumor suppressors. The let-7 family of miRNAs targets several oncogenes including c-Myc [45], numerous members of the Ras (N-Ras, K-Ras, and H-Ras) family of oncogenes [46] and high mobility group A2 (HMGA2) [47]. Biogenesis of let-7 is inhibited by ERK1/2-mediated phosphorylation of TRBP and unlike miR-21, let-7 expression is often lower in tumor samples than in normal tissues [33]. In some tumors, the Let-7 target c-Myc promotes the expression of Lin28b, a negative regulator of let-7, therefore, the c-Myc-let-7 axis forms a positive feedback loop to activate multiple oncogenic pathways. We speculate that future studies will uncover many more examples of human diseases that are initiated, and/or driven by aberrant miRNA expression as a result of misregulation of growth factor controlled miRNA biogenesis pathways.
Conclusion
As summarized in this article, control of miRNA biogenesis is an integral component of cell biological activities (Figure 3). Aberrations in miRNA biogenesis result in abnormal expression of multiple miRNAs, which can contribute to developmental defects and human diseases. Recent findings illustrate the importance of understanding not only transcriptional regulation but also post-transcriptional regulation of miRNA biogenesis. The existence of potential regulatory mechanisms was originally suggested in a 2006 study by Thompson, et al., who identified a substantial correlation between the primary and mature miRNA species expressed in normal cells but not in tumor samples [48]. It is clear that the ability to balance miRNA expression is an important part of normal growth factor signaling and that maintaining this balance can be achieved at every step of the miRNA biogenesis pathway.
Figure 3. Integration of biological signaling pathways at the sites of miRNA biogenesis.
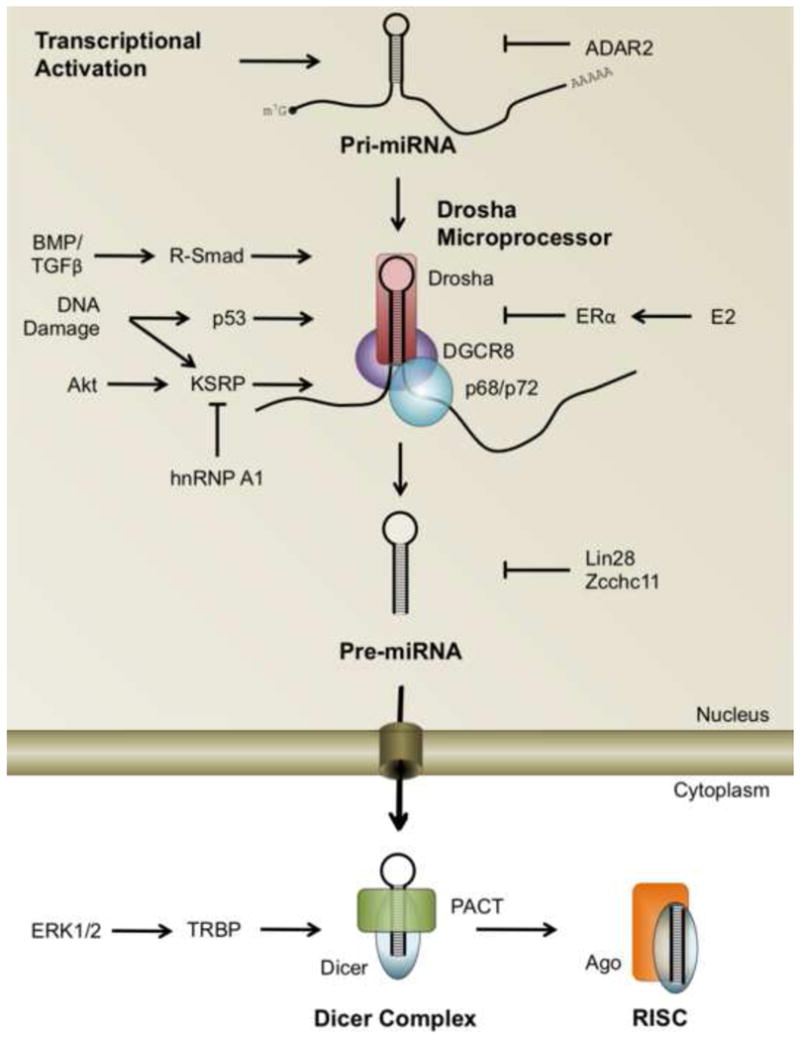
miRNA biogenesis pathways are emerging as critical components of gene regulatory mechanism controlled by growth factor signaling (and other) pathways. Activation or inactivation of the Drosha microprocessor by DNA binding proteins (Smads, p53, and ERα), RNA binding proteins (KSRP) or TBRP has been well documented. However, mechanisms of regulation of many other proteins in the miRNA biogenesis pathway by different signaling pathways remain to be uncovered.
The examples discussed here represent the first insights into how miRNA biogenesis is integrated into growth factor signaling pathways. Future studies will likely identify additional pathways affecting these important signal transduction networks. Furthermore, the mechanisms underlying the specificity of regulated miRNAs by different signaling pathways remain to be elucidated. We anticipate that in the future these directions will provide insights into the pathogenesis of various human disorders and present novel therapeutic targets.
Acknowledgments
We apologize to those colleagues whose references we have omitted from this discussion; due to space restrictions and our focus we were unable to include all articles on this interesting and diverse subject matter. We wish to express our thanks to the members of the Hata/Lagna lab for their suggestions and animated discussion. This work was supported by grants from the National Institute of Health: HL093154 and HL108317, the American Heart Association: 0940095N and the LeDucq foundation Transatlantic network grant to A.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Lee RC, Feinbaum RL, Ambros V. The C, elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nature reviews Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 3.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Yan K-p, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear Export of MicroRNA Precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature cell biology. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 9.Ketting RF, Fischer SEJ, Bernstein E, Sijen T, Hannon GJ, Plasterk RHA. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Development. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint É, Tuschl T, Zamore PD. A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the let-7 Small Temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 12••.Davis BN, Hilyard AC, Lagna G, Hata A. Smad proteins control Drosha-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. The first description of a mechanism for post-transcriptional prom0tion of miRNA maturation by a growth factor signaling pathway. This paper focuses on regulation of miR-21 expression by Smad proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. A thorough analysis of the requirements for Smad-pri-miRNA interaction, proving that this interaction is sequenced-dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Massagué J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 15.Piriyapongsa J, Jordan IK, Conley AB, Ronan T, Smalheiser NR. Transcription factor binding sites are highly enriched within microRNA precursor sequences. Biology Direct. 2011;6:61–72. doi: 10.1186/1745-6150-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 17•.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K-i, Minami Y, O’Malley BW, Kato S. Maturation of MicroRNA Is Hormonally Regulated by a Nuclear Receptor. Molecular Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. These two papers, along with [12], were the first descriptions of post-transcriptional regulation of miRNA biogenesis. [DOI] [PubMed] [Google Scholar]
- 18.Apponi LH, Corbett AH, Pavlath GK. RNA-binding proteins and gene regulation in myogenesis. Trends in Pharmacological Sciences. 2011;32:652–658. doi: 10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. The FASEB Journal. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 21•.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM Kinase Induces MicroRNA Biogenesis in the DNA Damage Response. Molecular Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. Analysis of the different phosphorylated Serine residues on KSRP and how they contribute to miRNA biogeneis in response to DNA damage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Briata P, Lin WJ, Giovarelli M, Pasero M, Chou CF, Trabucchi M, Rosenfeld MG, Chen CY, Gherzi R. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 2012;19:478–487. doi: 10.1038/cdd.2011.117. A paper full of beautiful biochecmical assays showing the switch from mRNA-binding to pri-miRNA-binding induced by Akt-dependent phosphorylation of KSRP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 25.Flynt AS, Greimann JC, Chung W-J, Lima CD, Lai EC. MicroRNA Biogenesis via Splicing and Exosome-Mediated Trimming in Drosophila. Molecular Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJA. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Research. 2012;40:438–448. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim B-M, Choi MY. Non-canonical microRNAs miR-320 and miR-702 promote proliferation in Dgcr8-deficient embryonic stem cells. Biochemical and Biophysical Research Communications. 2012 doi: 10.1016/j.bbrc.2012.1008.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Research. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. Initial paper reporting TRBP acting at the Dicer complex to promote miRNA biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the Human MicroRNA-Generating Complex Mediates MAPK/Erk Signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. This paper disects the role of MAPK/Erk signaling in modulating TRBP activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roush S, Slack FJ. The let-7 family of microRNAs. Trends in Cell Biology. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y, Hur I, Park S-Y, Kim Y-K, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The Role of Dicer Protein Partners in the Processing of MicroRNA Precursors. PLoS ONE. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012 doi: 10.1038/nature11134. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jazbutyte V, Thum T. MicroRNA-21: From Cancer to Cardiovascular Disease. Current Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- 41.Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2007;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 42.Zhu S, Si M-L, Wu H, Mo Y-Y. MicroRNA-21 Targets the Tumor Suppressor Gene Tropomyosin 1 (TPM1) Journal of Biological Chemistry. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 43.Meng F, Henson R, Wehbe–Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X, Wang Z-X, Wang R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biology & Therapy. 2010;10:1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 45.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA Let-7a Down-regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells. Cancer Research. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 46.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS Is Regulated by the let-7 MicroRNA Family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Mayr C, Hemann MT, Bartel DP. Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. This insightful paper used deep-sequencing to show there is not always a relationship between expression of pri-miRNA and their mature sequences and to anticipate the importance of controlling miRNA maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2005;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Blahna MT, Jones MR, Quinton LJ, Matsuura KY, Mizgerd JP. The terminal uridyltransferase enzyme zinc finger CCHC domain containing 11 (ZCCHC11) promotes cellular proliferation independent of its uridyltransferase activity. The Journal of biological chemistry. 2011;286:42381–42389. doi: 10.1074/jbc.M111.259689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O’Connell MA, Gallo A. Down-regulation of RNA Editing in Pediatric Astrocytomas. Journal of Biological Chemistry. 2008;283:7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]


