Abstract
C-type natriuretic peptide (CNP) stimulates endochondrial ossification by activating the transmembrane guanylyl cyclase, natriuretic peptide receptor-B (NPR-B). Recently, a spontaneous autosomal recessive mutation that causes severe dwarfism in mice was identified. The mutant, called long bone abnormality (lbab), contains a single point mutation that converts an arginine to a glycine in a conserved coding region of the CNP gene, but how this mutation affects CNP activity has not been reported. Here, we determined that thirty to greater than one hundred-fold more CNPlbab was required to activate NPR-B as compared to wild-type CNP in whole cell cGMP elevation and membrane guanylyl cyclase assays. The reduced ability of CNPlbab to activate NPR-B was explained, at least in part, by decreased binding since ten-fold more CNPlbab than wild-type CNP was required to compete with [125I][Tyr0]CNP for receptor binding. Molecular modeling suggested that the conserved arginine is critical for binding to an equally conserved acidic pocket in NPR-B. These results indicate that reduced binding to and activation of NPR-B causes dwarfism in lbab−/− mice.
Keywords: Endochondrial ossification, Cyclic-GMP, Guanylyl cyclase B, Type II cGMP dependant protein kinase, Nppc
1. INTRODUCTION
Natriuretic peptides (NPs) are structurally related pleiotropic factors that regulate the cardiovascular, skeletal, and possibly other systems [25]. Atrial natriuretic peptide (ANP) reduces blood pressure by stimulating renal sodium and water excretion [11], reducing intravascular volume [8], and relaxing vascular smooth muscle [32]. Infusion of synthetic B-type natriuretic peptide (BNP) into animals or humans elicits similar responses as ANP, but gene deletion studies in mice indicate that BNP is not required for the maintenance of normal blood pressure [29]. C-type natriuretic peptide (CNP) is also a vasorelaxant [10], but its most obvious function is to stimulate endochondral ossification, resulting in the lengthening of long bones [19, 28].
Natriuretic peptides exert their functions via interaction with single membrane-spanning guanylyl cyclases called natriuretic peptide receptors. Peptide binding activates their catalytic domains resulting in elevated intracellular cGMP concentrations. Both ANP and BNP bind and activate natriuretic peptide receptor A (NPR-A/GC-A) [6, 27], whereas CNP selectively activates natriuretic peptide receptor B (NPR-B/GC-B) [16]. ANP, BNP, CNP, as well as osteocrin, also bind the natriuretic peptide clearance receptor (NPR-C), which controls local concentrations of these peptides through constitutive receptor-mediated internalization and degradation [18].
CNP is translated as a preprohormone that has multiple biologically active forms. Tissues primarily contain a 53-amino acid form of CNP, but an amino-terminal deleted 22-amino acid form of CNP is the predominant circulating species found in blood and cerebral spinal fluid (Fig. 1). Both the 53- and 22-residue forms have similar NPR-B activation and binding profiles [35].
Fig 1.
Cartoon schematic of the primary amino acid structure of atrial natriuretic peptide (ANP), B-type-natriuretic peptide (BNP), the 53- and 22- amino acid forms of C-type natriuretic peptide (CNP), and the peptide encoded by the lbab mutation in Nppc (CNPlbab). The shading indicates residues conserved among family members, the dark bars indicate disulfide bridges, and the asterisk indicates the arginine-to-glycine substitution present in CNPlbab.
CNP is postulated to regulate long bone growth through its induction of endochondral ossification [23, 34]. CNP-dependent activation of NPR-B produces cGMP, which activates the type II cGMP dependent protein kinase, PKGII. Functional inactivation of the genes encoding CNP [7], NPR-B[28, 30], or PKGII [5, 24] produce dwarfism. The relevant substrates for PKGII in the bone growth pathway have not been determined, but transgenic overexpression of CNP was shown to inhibit MAP kinase activity and partially compensate for fibroblast growth factor receptor-3-dependent dwarfism in a mouse achondroplasia model [17, 33].
In humans, familial homozygous loss-of-function mutations in the gene encoding NPR-B, Npr2, lead to a form of dwarfism known as acromesomelic dysplasia type Maroteaux (AMDM), which is characterized by disproportionately short arms and legs after birth [2]. Individuals that are heterozygous for mutations in NPR-B have normal limb proportions but are significantly shorter than comparable individuals without the mutation [22]. Recently, an AMDM patient was shown to have a novel mutation in NPR-B which allowed the receptor to bind CNP, but abolished cGMP signaling by the receptor in a dominant negative fashion [13].
In 1996, researchers at The Jackson Laboratory identified a spontaneous autosomal recessive mouse mutation characterized by overall smaller body size and proportional dwarfing of all organs and long bones [31]. They called this mutation, lbab, for “long bone abnormality” and mapped the mutation to chromosome 1 of the mouse genome. Recently Jiao and colleagues further characterized this mutation and found that the lbab phenotype was associated with a single point mutation in the Nppc gene [15]. A C to G transversion was found in exon 2 of Nppc that results in the substitution of a glycine for an arginine at position 117 in proCNP. An absolutely conserved D-R-I sequence is present within the ring structure of all natriuretic peptides. The lbab mutation changes this sequence to D-G-I. After processing, the 22-amino acid peptide encoded by the lbab mutation differs from by a single amino acid in position 13 (R13G) and is referred to henceforth as CNPlbab (Fig. 1). Jiao and colleagues speculated that this point mutation results in loss of function but provided no experimental data to support this hypothesis.
The purpose of this study was to determine if the peptide encoded by the lbab mutation is less biologically active than the wild-type peptide. This was evaluated by whole cell ligand binding as well as whole cell cGMP elevation and membrane guanylyl cyclase assays. We found that the single amino acid difference between the mutant and wild-type forms of CNP dramatically reduce its ability to bind and activate NPR-B.
2. MATERIALS AND METHODS
2.1. Reagents
CNPlbab (GLSKGCFGLKLDGIGSMSGLGC, disulfide bridge: 6–22) was synthesized by AnaSpec, Inc (San Jose, CA). [125I][Tyr0]CNP (1–22) was purchased from Phoenix Pharmaceuticals (Phoenix, AZ). [α-32P]GTP was purchased from Perkin Elmer (Waltham, MA).
2.2. Cell Lines
Human embryonic 293 cells lacking any known natriuretic peptide receptor were transfected with 10 µg of pRK5-NPR-B [26] and 1 µg of pWL-neo to confer neomycin resistance. An individual clone stably expressing NPR-B was selected with plastic cloning cylinders after 10–14 d of growth in medium containing 200 µg/ml neomycin. NIH3T3 cells were maintained as previously described [1].
2.3. Preparation of Crude Membranes
293-NPR-B cells were scraped off 10 cm plates with 0.75 ml of phosphatase inhibitor buffer (25 mM Hepes pH 7.4, 20% glycerol, 50mM NaCl, 50 mM NaF, 2 mM EDTA, 0.5 µM microcystin, EDTA-free protease inhibitors (Roche)), sonicated for 1 s with a Misonix Ultrasonic Processor at 4°C, and centrifuged at 20,000 × g for 15 min at 4°C. Membrane pellets were resuspended in phosphatase inhibitor buffer at 5–10 mg protein/ml.
2.4. Guanylyl Cyclase Assays
Guanylyl cyclase assays were performed at 37°C for 3 min in a buffer containing 25 mM Hepes pH 7.4, 50 mM NaCl, 0.1% BSA, 0.5 mM 1-methyl-3-isobutylxanthine, 1 mM GTP, 0.5 µM microcystin, 1 mM EDTA, 1–2 µCi of [α-32P]GTP, and 5 mM MgCl2 with or without CNP. Reactions were started by the addition of 80 µl of the above reagents to 50–200 µg of crude membrane protein suspended in 20 µl of phosphatase inhibitor buffer. Reactions were stopped by the addition of 0.5 ml 110 mM ZnOAc and 0.5 ml 110 mM NaCO3 on ice. Cyclic-cGMP accumulation was determined as described previously [4].
2.5. Whole Cell Stimulations
Cells plated in poly-D-lysine coated 48-well plates were incubated overnight in serum-free media. Medium was aspirated and 0.2 ml DMEM containing 1 mM 1-methyl-3-isobutylxanthine (IBMX) was added and incubated for 10 min. Medium was aspirated and cells were treated with DMEM containing 1 mM IBMX with or without natriuretic peptide for 1 to 5 min. Treatment medium was aspirated and the reaction was stopped with 0.2 ml ice-cold ethanol. An aliquot of the supernatant was dried in a centrifugal vacuum concentrator and analyzed for cGMP content using a [125I]-radioimmunoassay kit from Perkin Elmer as per manufacturer’s instructions.
2.6. Radioligand Binding Assays
Cells in 24-well plates were washed with DMEM and then incubated with 0.2% BSA in DMEM at 37°C for 1 h. Medium was aspirated and 150 µl of binding medium containing 75 pM [125I][Try0]CNP and 1% BSA, alone or with unlabeled ligand, was added to the well. The plate was incubated for 1 h at 4°C and then medium was aspirated and wells were washed with 0.5 ml ice-cold PBS. The PBS was aspirated and 0.5 ml 1 N NaOH was added to the well to remove cells. The supernatant was transferred to glass tubes and the amount of [125I][Tyr0]CNP present was assessed in a gamma counter. Nonspecific binding was determined by repeating the above assay in the presence of 1 µM unlabeled CNP. Specific binding was calculated by subtracting the nonspecific binding from the total binding.
2.7. Modeling of NPR-B With CNP
Modeller-9.3 was used to model the NPR-B/CNP structure based upon the crystal structure of NPR-A bound to ANP solved by Ogawa and colleagues (RCSB Protein Data Bank access code 1T34) [21]. Numbering of human NPR-B starts with initiation methionine as position 1. This model includes the entire extracellular domain, residues Arg23-Gly464. Spatial constraints were created from the sequence alignment between NPR-B and NPR-A. The structure was then randomized and minimized upon the constraints. Figures were drawn with the PyMol program [9].
3. RESULTS
3.1. Reduced potency of CNPlbab in elevating whole cell cGMP concentrations
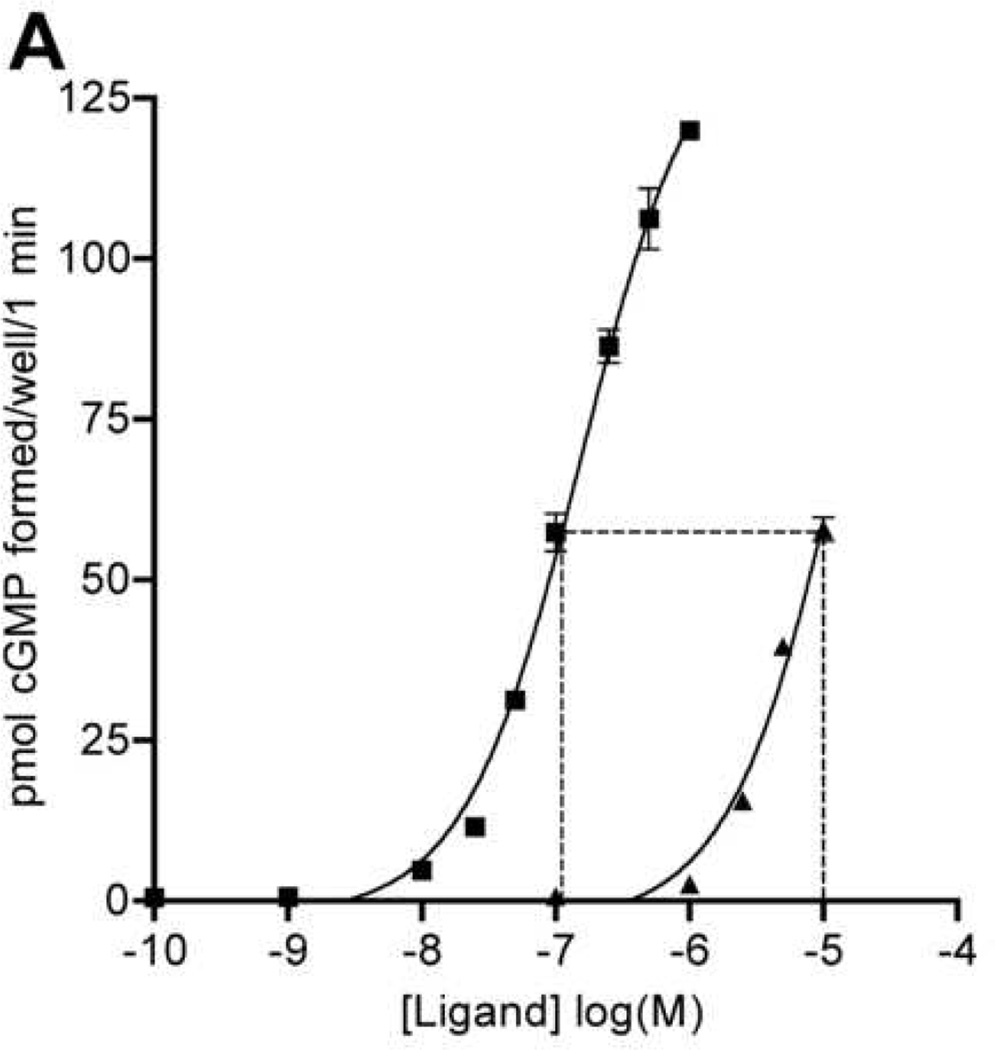
Cells overexpressing the rat NPR-B receptor (293-NPR-B, Fig. 2A) or cells endogenously expressing the mouse NPR-B receptor (NIH3T3, Fig. 2B) were stimulated with increasing concentrations of CNP or CNPlbab and intracellular cGMP concentrations were estimated by radioimmunoassay. Although cells were incubated with as much as 1 µM and 10 µM concentrations of the wild-type and mutant forms of CNP, saturation was not achieved for either peptide, consistent with a previous report for wild-type CNP and human NPR-B [16]. Importantly, the concentration-response curve for CNPlbab was significantly shifted to the right compared to wild-type CNP in 293 cells expressing rat NPR-B. The stimulation observed with 10 µM CNPlbab was equivalent to the level of activation observed with 115 nM CNP, an approximately 85-fold difference. Whole cell stimulations of NIH3T3 cells demonstrated an exaggerated trend with 10 µM CNPlbab, yielding the same level of activation as about 30 nM concentrations of wild-type CNP, about a 300-fold difference. Thus, with respect to both the rat and the mouse NPR-B receptors, CNPlbab has a markedly diminished ability to elevate intracellular cGMP concentrations compared to the wild-type molecule.
Fig 2.
Effect of increasing concentrations of CNP (■) or CNPlbab (▲) on cGMP generation in intact cells and membrane preparations. (A) One-minute stimulations of 293-NPR-B cells stably overexpressing rat NPR-B. Results are expressed as the mean of triplicate determinations ± SEM. This data is representative of three separate trials. (B) Five-minute stimulations of mouse NIH3T3 cells endogenously expressing NPR-B. Results are expressed as the mean of triplicate determinations ± SEM. This data is representative of two separate trials. (C) Three-minute stimulation of membrane preparations from 293-NPR-B cells. Results are expressed as the mean of three separate experiments ± SEM. Each individual experiment contained duplicate data points.
3.2. Reduced potency of CNPlbab in stimulating NPR-B guanylyl cyclase activity
Crude membrane preparations from 293-NPR-B cells were used to access the ability of CNP and CNPlbab to directly activate NPR-B in broken cell preparations (Fig. 2C). Concentration-response curves generated by CNPlbab were shifted to the right of those generated with wild-type CNP. The activity seen with 10 µM CNPlbab was equivalent to the activation seen with 275 nM CNP, amounting to a 36-fold difference. Hence, the decreased cGMP production observed in whole cells (Fig. 2A and 2B) is a direct result of the diminished ability of CNPlbab to activate the NPR-B receptor.
3.3. CNPlbab has reduced affinity to NPR-B
Using a competitive binding assay, the affinity of CNP and CNPlbab for rat NPR-B was examined using 293-NPR-B cells. Specific binding of 75 pM [125I][Tyr0]CNP was progressively inhibited when increasing amounts of CNP or CNPlbab were included in the binding assay (Fig. 3). The concentrations needed for 50% inhibition of specific [125I][Tyr0]CNP binding to NPR-B (IC50) were 370 pM and 3900 pM for CNP and CNPlbab, respectively. Thus, CNPlbab binds NPR-B at least ten-fold less avidly than wild-type CNP binds NPR-B.
Fig 3.
Reduced binding of CNPlbab to NPR-B. Specific binding of [125I][Tyr0]CNP to rat NPR-B stably expressed in 293 cells was competed with wild-type CNP (■) or CNPlbab (▲). Cells were incubated with DMEM containing 1% BSA and 75 pM [125I][Tyr0]CNP, alone or in the presence of the indicated concentrations of nonradiolabeled CNP or nonradiolabeled CNPlbab at 4°C for 1 h. Results are expressed as the counts per minute specifically bound at each peptide concentration (B) divided by counts per minute specifically bound in the absence of displacing peptide (B0). Results are the mean of four trials that were assayed in triplicate. The vertical bar within each symbol represents the standard error of the mean.
4. DISCUSSION
In this study, the ability of CNPlbab to activate and bind to NPR-B was examined. In our experiments, the capacity of CNPlbab to activate both mouse and rat NPR-B in whole cells and in crude membrane guanylyl cyclase assays was markedly reduced. Similarly, whole cell binding assays indicated that ten-fold more CNPlbab compared to the wild-type peptide was required to compete with [125I][Tyr0]-CNP for binding to NPR-B in whole cells. Hence, we conclude that the lbab mutation in Nppc causes dwarfism because it encodes an amino acid substitution that decreases the ability of CNP to bind and activate its cognate receptor, NPR-B.
One interesting aspect of our data is that we saw varying degrees of diminished responses to CNPlbab in our assays. We saw a 10-fold decrease in binding affinity, but an 85-fold decrease in whole cell cGMP accumulation with CNPlbab. One might expect to see similar fold shifts across all assays. However, that was not the case. Although there may be two separate effects of the single amino acid change seen in CNPlbab—decreased binding affinity as well as decreased ability to activate NPR-B once bound—we cannot separate these effects due to the differences in assay conditions as previously noted by Garbers and colleagues [12].
In order to elucidate the basis for the decreased ability of CNPlbab to bind and activate NPR-B, we sought to identify critical molecular interactions between CNP and NPR-B via molecular modeling. A previous molecular model of human NPR-B complexed to CNP was created by He and colleagues [14], which focused on interactions that provided ligand specificity. The mutation in CNPlbab occurs in a highly conserved region, which was not discussed in the previous model. Unfortunately, the coordinates for this model were not deposited into the RSCB database. Hence, we created a new structural model of human NPR-B bound to either CNP or CNPlbab based on the crystal structure of the rat ANP/NPR-A complex. Like NPR-A [21], NPR-B is thought to function as a head-to-head dimer, with two-fold symmetry in the ligand binding pocket. By examining the interactions of the R13G mutation of CNPlbab compared to wild-type CNP, we have identified key differences in the molecular interaction of the ligands with the receptor. For wild-type CNP, Arg13 fits tightly into an acidic pocket, with its positively charged guanidinium group juxtaposed to two acidic residues, Asp176 and Glu77, of the receptor. An additional interaction between the planar ring of Tyr103 of the receptor and CNP further stabilizes the interaction (Fig. 4A). When CNPlbab is bound to NPR-B, the hydrogen side chain of the glycine residue is neither long enough to fill the receptor pocket nor is it appropriately charged to ion pair with the acidic residues lining the base of the pocket (Fig. 4B). In solution, CNP does not have a well-defined structure but is forced into a particular conformation when it binds the receptor. Although Gly13 in CNPlbab affords the peptide greater conformational flexibility, it does not allow for the favorable electrostatic interactions with Asp176 or Glu77 of NPR-B envisioned for wild-type CNP. Much like Arg13 is absolutely conserved in CNP across all species, the importance of Asp176, Glu 77, and Tyr103 in the receptor-ligand interaction is underscored by the fact that all three residues are absolutely conserved in all mammalian homologs of NPR-B whose primary amino acid sequence is deposited in the NCBI database (data not shown). The disruption of these critical points-of-contact are likely to reduce binding of CNP to NPR-B, but this remains to be quantified.
Fig 4.
Theoretical modeling of CNP and CNPlbab binding to human NPR-B. Van der Waals surfaces are portrayed around all residues, with CNP depicted as thick yellow sticks and NPR-B colored according to electrostatic potential, with red indicating acidic regions and blue indicating basic regions. A bound chloride ion is shown as a light blue sphere. (A) Wild-type CNP (with arginine at position 13) is shown modeled with human NPR-B. Human NPR-B residues that have the potential to interact with this arginine are displayed as thick sticks and labeled in white. (B) CNPlbab (with glycine at position 13) is shown modeled with human NPR-B.
In conclusion, we have demonstrated a blunted cellular and in vitro response to the version of CNP encoded by the murine lbab allele. To our knowledge, this is the first report to characterize a functional consequence of a single point mutation in a ligand for any guanylyl cyclase. At physiologic concentrations, the mutated natriuretic peptide CNPlbab is unlikely to cause significant elevations in cGMP, which would fail to trigger the down stream signal transduction pathways that normally ensue after CNP release in bone tissue. Recently, human chromosomal 2:7 translocations located close to the NPPC gene were identified that result in elevated CNP mRNA expression, increased CNP protein levels, and skeletal overgrowth [3, 20]. Whether mutations exist in the coding region of CNP that modify human skeletal growth has yet to be reported, but based on the lbab−/− mouse, we believe this scenario is likely.
ACKNOWLEGDEMENTS
This work was funded by grants from the University of Minnesota Medical School, NIH grant (AI057585) to CAE, and a grant-in-aid from the Graduate School of the University of Minnesota. ARY was supported in part by a fellowship from the 3M Corporation.
GLOSSARY
- CNP
C-type natriuretic peptide
- NPR-B/GC-B
Natriuretic peptide receptor B
- NP
natriuretic peptide
- ANP
atrial natriuretic peptide
- BNP
B-type natriuretic peptide
- NPR-A/GC-A
natriuretic peptide receptor A
- NPR-C
natriuretic peptide clearance receptor
- PKGII
type II cGMP dependant protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrea R. Yoder, Email: yoder013@umn.edu.
Andrew C. Kruse, Email: krus0114@umn.edu.
Cathleen A. Earhart, Email: earhart@umn.edu.
Douglas H. Ohlendorf, Email: ohlen@umn.edu.
Lincoln R. Potter, Email: potter@umn.edu.
REFERENCES
- 1.Abbey SE, Potter LR. Lysophosphatidic acid inhibits C-type natriuretic peptide activation of guanylyl cyclase-B. Endocrinology. 2003;144:240–246. doi: 10.1210/en.2002-220702. [DOI] [PubMed] [Google Scholar]
- 2.Bartels CF, Bukulmez H, Padayatti P, Rhee DK, van Ravenswaaij-Arts C, Pauli RM, et al. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bocciardi R, Giorda R, Buttgereit J, Gimelli S, Divizia MT, Beri S, et al. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum Mutat. 2007;28:724–731. doi: 10.1002/humu.20511. [DOI] [PubMed] [Google Scholar]
- 4.Bryan PM, Potter LR. The atrial natriuretic peptide receptor (NPR-A/GC-A) is dephosphorylated by distinct microcystin-sensitive and magnesium-dependent protein phosphatases. J Biol Chem. 2002;277:16041–16047. doi: 10.1074/jbc.M110626200. [DOI] [PubMed] [Google Scholar]
- 5.Chikuda H, Kugimiya F, Hoshi K, Ikeda T, Ogasawara T, Shimoaka T, et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418–2429. doi: 10.1101/gad.1224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinkers M, Garbers DL, Chang MS, Lowe DG, Chin HM, Goeddel DV, et al. A membrane form of guanylate cyclase is an atrial natriuretic peptide receptor. Nature. 1989;338:78–83. doi: 10.1038/338078a0. [DOI] [PubMed] [Google Scholar]
- 7.Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, et al. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci U S A. 2001;98:4016–4021. doi: 10.1073/pnas.071389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bold AJ, de Bold ML, Sarda IR. Functional-morphological studies on in vitro cardionatrin release. J Hypertens Suppl. 1986;4:S3–S7. [PubMed] [Google Scholar]
- 9.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 10.Drewett JG, Fendly BM, Garbers DL, Lowe DG. Natriuretic peptide receptor-B (guanylyl cyclase-B) mediates C-type natriuretic peptide relaxation of precontracted rat aorta. J Biol Chem. 1995;270:4668–4674. doi: 10.1074/jbc.270.9.4668. [DOI] [PubMed] [Google Scholar]
- 11.Forssmann WG, Richter R, Meyer M. The endocrine heart and natriuretic peptides: histochemistry, cell biology, and functional aspects of the renal urodilatin system. Histochem Cell Biol. 1998;110:335–357. doi: 10.1007/s004180050295. [DOI] [PubMed] [Google Scholar]
- 12.Garbers DL, Koesling D, Schultz G. Guanylyl cyclase receptors. Mol Biol Cell. 1994;5:1–5. doi: 10.1091/mbc.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachiya R, Ohashi Y, Kamei Y, Suganami T, Mochizuki H, Mitsui N, et al. Intact kinase homology domain of natriuretic peptide receptor-B is essential for skeletal development. J Clin Endocrinol Metab. 2007;92:4009–4014. doi: 10.1210/jc.2007-1101. [DOI] [PubMed] [Google Scholar]
- 14.He XL, Dukkipati A, Garcia KC. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J Mol Biol. 2006;361:698–714. doi: 10.1016/j.jmb.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 15.Jiao Y, Yan J, Jiao F, Yang H, Donahue LR, Li X, et al. A single nucleotide mutation in Nppc is associated with a long bone abnormality in lbab mice. BMC Genet. 2007;8:16. doi: 10.1186/1471-2156-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koller KJ, Lowe DG, Bennett GL, Minamino N, Kangawa K, Matsuo H, et al. Selective activation of the B natriuretic peptide receptor by C-type natriuretic peptide (CNP) Science. 1991;252:120–123. doi: 10.1126/science.1672777. [DOI] [PubMed] [Google Scholar]
- 17.Krejci P, Masri B, Fontaine V, Mekikian PB, Weis M, Prats H, et al. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J Cell Sci. 2005;118:5089–5100. doi: 10.1242/jcs.02618. [DOI] [PubMed] [Google Scholar]
- 18.Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, et al. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazawa T, Ogawa Y, Chusho H, Yasoda A, Tamura N, Komatsu Y, et al. Cyclic GMP-dependent protein kinase II plays a critical role in C-type natriuretic peptide-mediated endochondral ossification. Endocrinology. 2002;143:3604–3610. doi: 10.1210/en.2002-220307. [DOI] [PubMed] [Google Scholar]
- 20.Moncla A, Missirian C, Cacciagli P, Balzamo E, Legeai-Mallet L, Jouve JL, et al. A cluster of translocation breakpoints in 2q37 is associated with overexpression of NPPC in patients with a similar overgrowth phenotype. Hum Mutat. 2007;28:1183–1188. doi: 10.1002/humu.20611. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa H, Qiu Y, Ogata CM, Misono KS. Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction. J Biol Chem. 2004;279:28625–28631. doi: 10.1074/jbc.M313222200. [DOI] [PubMed] [Google Scholar]
- 22.Olney RC, Bukulmez H, Bartels CF, Prickett TC, Espiner EA, Potter LR, et al. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) are associated with short stature. J Clin Endocrinol Metab. 2006;91:1229–1232. doi: 10.1210/jc.2005-1949. [DOI] [PubMed] [Google Scholar]
- 23.Pejchalova K, Krejci P, Wilcox WR. C-natriuretic peptide: An important regulator of cartilage. Mol Genet Metab. 2007;92:210–215. doi: 10.1016/j.ymgme.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Pfeifer A, Aszodi A, Seidler U, Ruth P, Hofmann F, Fassler R. Intestinal secretory defects and dwarfism in mice lacking cGMP- dependent protein kinase II. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 25.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 26.Potter LR, Hunter T. Identification and characterization of the major phosphorylation sites of the B-type natriuretic peptide receptor. J Biol Chem. 1998;273:15533–15539. doi: 10.1074/jbc.273.25.15533. [DOI] [PubMed] [Google Scholar]
- 27.Suga S, Nakao K, Hosoda K, Mukoyama M, Ogawa Y, Shirakami G, et al. Receptor selectivity of natriuretic peptide family, atrial natriuretic peptide, brain natriuretic peptide, and C-type natriuretic peptide. Endocrinology. 1992;130:229–239. doi: 10.1210/endo.130.1.1309330. [DOI] [PubMed] [Google Scholar]
- 28.Tamura N, Doolittle LK, Hammer RE, Shelton JM, Richardson JA, Garbers DL. Critical roles of the guanylyl cyclase B receptor in endochondral ossification and development of female reproductive organs. Proc Natl Acad Sci U S A. 2004;101:17300–17305. doi: 10.1073/pnas.0407894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, et al. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc Natl Acad Sci U S A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuji T, Kunieda T. A loss-of-function mutation in natriuretic peptide receptor 2 (Npr2) gene is responsible for disproportionate dwarfism in cn/cn mouse. J Biol Chem. 2005;280:14288–14292. doi: 10.1074/jbc.C500024200. [DOI] [PubMed] [Google Scholar]
- 31.Ward-Bailey PF, Harris BS, Donahue LR, Bronson R. Long bone abnormality (lbab): a new mouse mutation on Chromosome 1 causing small size and skeletal abnormalities. [cited 2007 May 11];Mouse Mutant Resource. 2004 Available from: http://www.jax.org/mmr/lbab.html. [Google Scholar]
- 32.Winquist RJ, Faison EP, Waldman SA, Schwartz K, Murad F, Rapoport RM. Atrial natriuretic factor elicits an endothelium-independent relaxation and activates particulate guanylate cyclase in vascular smooth muscle. Proc Natl Acad Sci U S A. 1984;81:7661–7664. doi: 10.1073/pnas.81.23.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, et al. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- 34.Yasoda A, Ogawa Y, Suda M, Tamura N, Mori K, Sakuma Y, et al. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J Biol Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- 35.Yeung VT, Ho SK, Nicholls MG, Cockram CS. Binding of CNP-22 and CNP-53 to cultured mouse astrocytes and effects on cyclic GMP. Peptides. 1996;17:101–106. doi: 10.1016/0196-9781(95)02099-3. [DOI] [PubMed] [Google Scholar]