Abstract
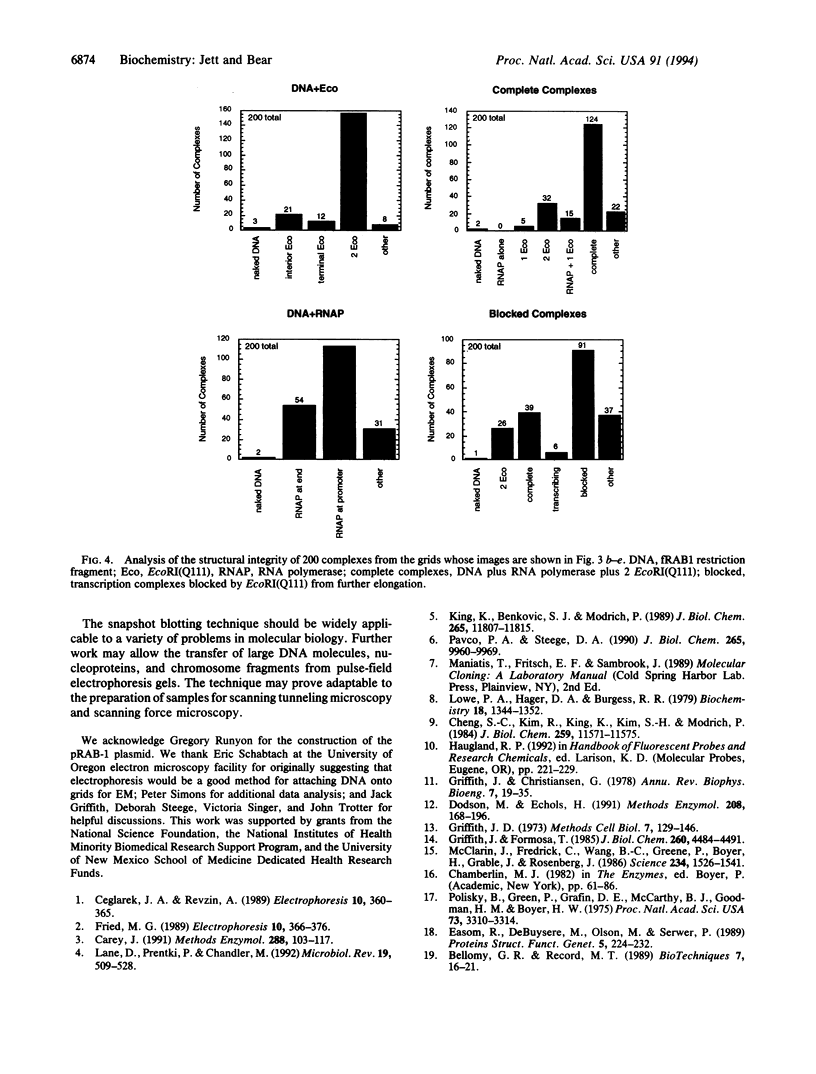
We present a technique, "snapshot blotting," for the electrophoretic transfer of nucleic acids and nucleoprotein complexes in gel electrophoresis bands onto highly stable carbon film-coated grids for imaging by electron microscopy. The method permits structural analysis of macromolecular species that have been resolved by a gel mobility-shift assay. To demonstrate the efficiency and integrity of the transfer process for a multiprotein-DNA assembly, we have imaged various species of a prokaryotic transcription complex, using the cleavage-defective EcoRI(Q111) protein as an orientation marker and as a blockade of transcription elongation. Snapshot blotting should be of great utility in the structural characterization of nucleic acids and protein-nucleic acid interactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellomy G. R., Record M. T., Jr A method for horizontal polyacrylamide slab gel electrophoresis. Biotechniques. 1989 Jan;7(1):16, 19-21. [PubMed] [Google Scholar]
- Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- Ceglarek J. A., Revzin A. Studies of DNA-protein interactions by gel electrophoresis. Electrophoresis. 1989 May-Jun;10(5-6):360–365. doi: 10.1002/elps.1150100514. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Kim R., King K., Kim S. H., Modrich P. Isolation of gram quantities of EcoRI restriction and modification enzymes from an overproducing strain. J Biol Chem. 1984 Sep 25;259(18):11571–11575. [PubMed] [Google Scholar]
- Dodson M., Echols H. Electron microscopy of protein-DNA complexes. Methods Enzymol. 1991;208:168–196. doi: 10.1016/0076-6879(91)08013-8. [DOI] [PubMed] [Google Scholar]
- Easom R. A., DeBuysere M. S., Olson M. S., Serwer P. Size determination of multienzyme complexes using two-dimensional agarose gel electrophoresis. Proteins. 1989;5(3):224–232. doi: 10.1002/prot.340050306. [DOI] [PubMed] [Google Scholar]
- Fried M. G. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis. 1989 May-Jun;10(5-6):366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Electron microscopic visualization of DNA in association with cellular components. Methods Cell Biol. 1973;7:129–146. doi: 10.1016/s0091-679x(08)61774-4. [DOI] [PubMed] [Google Scholar]
- Griffith J., Formosa T. The uvsX protein of bacteriophage T4 arranges single-stranded and double-stranded DNA into similar helical nucleoprotein filaments. J Biol Chem. 1985 Apr 10;260(7):4484–4491. [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Lane D., Prentki P., Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992 Dec;56(4):509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990 Jun 15;265(17):9960–9969. [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]