SUMMARY
Brown adipose tissue (BAT) protects against obesity by promoting energy expenditure via uncoupled respiration. To uncover BAT-specific long non-coding RNAs (lncRNAs), we used RNA-seq to reconstruct de novo transcriptomes of mouse brown, inguinal white, and epididymal white fat and identified ~1500 lncRNAs, including 127 BAT-restricted loci induced during differentiation and often targeted by key regulators PPARγ, C/EBPα and C/EBPβ. One of them, lnc-BATE1, is required for establishment and maintenance of BAT identity and thermogenic capacity. lnc-BATE1 inhibition impairs concurrent activation of brown fat and repression of white fat genes, and is partially rescued by exogenous lnc-BATE1 with mutated siRNA-targeting sites, demonstrating a function in trans. We show that lnc-BATE1 binds heterogeneous nuclear ribonucleoprotein U and that both are required for brown adipogenesis. Our work provides an annotated catalog for the study of fat depot-selective lncRNAs, available online, and establishes lnc-BATE1 as a novel regulator of BAT development and physiology.
Graphical Abstract
INTRODUCTION
Brown adipose tissue (BAT), which is specialized for energy expenditure and heat generation, is an attractive therapeutic target for obesity. BAT is densely packed with mitochondria expressing high levels of Uncoupling protein 1 (UCP1), which facilitates proton leakage to uncouple respiration from ATP synthesis. In rodents, BAT is activated by overfeeding as a physiological response to limit weight gain (Rothwell and Stock, 1979). Mice deficient in BAT activity are susceptible to obesity and diabetes (Feldmann et al., 2009; Hamann et al., 1996; Lowell et al., 1993), while mice with increased BAT activity or increased numbers of thermogenic adipocytes within their white fat are healthy and lean (Bostrom et al., 2012; Chiang et al., 2009; Seale et al., 2011). In humans, recent studies have demonstrated the presence of active BAT in adults (Cypess et al., 2009; Nedergaard et al., 2007; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). Human BAT activity correlates positively with resting metabolic rate and negatively with body mass index (Cypess et al., 2009; Saito et al., 2009), suggesting that it contributes to body weight variability among individuals. Understanding the mechanisms underlying BAT development is thus an area of immense interest.
Previous studies have revealed many protein regulators of BAT development (Kajimura et al., 2010; Villarroya and Vidal-Puig, 2013). We and others have shown that microRNAs can also regulate BAT lineage determination and browning of white fat (Chen et al., 2013; Mori et al., 2010; Sun and Trajkovski, 2014; Sun et al., 2011; Trajkovski et al., 2012). Identifying novel RNA regulators of BAT development thus represents an attractive opportunity for finding new therapeutic targets against obesity.
Long non-coding RNAs (lncRNAs) are increasingly recognized as an additional layer of regulation during cell development and disease (Alvarez-Dominguez et al., 2014a; Fatica and Bozzoni, 2013; Hu et al., 2012). We previously showed that a set of lncRNAs common to white and brown fat are essential for adipogenesis (Sun et al., 2013). One of them, lnc-RAP1 (Firre), is exclusively nuclear and interacts with the nuclear matrix factor hnRNP U to mediate transchromosomal interactions between loci encoding adipogenic factors (Hacisuleyman et al., 2014). Our knowledge of lncRNAs that selectively modulate BAT development and physiology, however, remains limited (Zhao et al., 2014).
Here, we integrate genome-wide surveys of transcription by RNA-seq and chromatin state by ChIP-seq to comprehensively characterize lncRNAs active in mouse brown, inguinal white, and epididymal white adipose tissues (BAT, iWAT and eWAT, respectively). We uncover >1000 previously unannotated lncRNA genes, including 127 with BAT-restricted expression, many of which are induced during differentiation and are targeted by key adipogenic factors PPARγ, C/EBPα and C/EBPβ. One of them, lnc-BATE1, is a BAT-selective lncRNA required for proper development and maintenance of mature, thermogenic brown adipocytes. lnc-BATE1 acts in trans to selectively sustain the core BAT gene program and repress WAT-selective genes, and binds hnRNP-U, which is also required for brown adipogenesis. Our work thus provides a roadmap for the discovery of fat depot-selective lncRNAs regulating adipocyte lineage-specific development and function, which can be readily implemented through an online resource (https://sites.google.com/site/sunleilab/data/lncrnas).
RESULTS
Global Discovery of Adipose lncRNAs
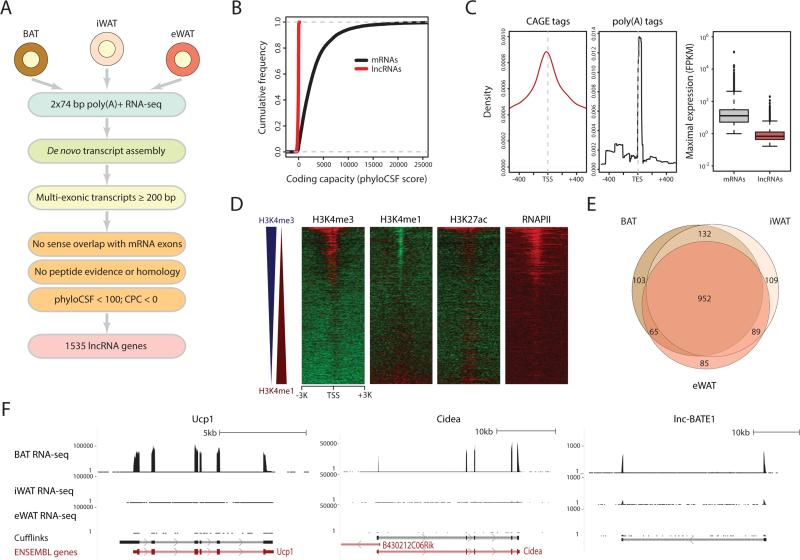
Our previous work on lncRNAs important for white and brown adipogenesis was limited to existing gene annotations (Sun et al., 2013), which suffer from incompleteness and inaccuracy. To better define lncRNAs active in adipose in vivo, including those restricted to different subtypes, we set out to reconstruct de novo the transcriptome of primary mouse BAT, iWAT and eWAT (Figure 1A). We performed paired-end sequencing of long poly (A)-selected RNAs from each tissue and mapped ~half a billion reads to the mouse genome (Table S1). We then used Cufflinks (Trapnell et al., 2010) to assemble gene and transcript models and quantify their expression. As a measure of quality, we examined expression estimates for genes annotated by Ensembl (Flicek et al., 2014) and found high precision and reproducibility in our data (Figures S1A and S1B).
Figure 1. Global Discovery of Adipose Tissue lncRNAs.
(A) lncRNA discovery pipeline. See text and Supplemental Experimental Procedures.
(B) Coding capacity of adipose-expressed mRNAs and lncRNAs as estimated by phyloCSF (Lin et al., 2011).
(C) Density of CAGE tags (left) and poly(A) tags (center) within 1kb of lncRNA transcription start sites (TSS) or end sites (TES), respectively. (Right) Box plots of maximal gene-level expression distributions for adipose-expressed mRNAs (maximal FPKM >1) and lncRNAs (maximal FPKM >0.1).
(D) Evidence of histone marking, open chromatin, and RNA Pol II binding within TSS ±3kb regions of adipose- expressed lncRNAs. Color intensity represents the log2 signal enrichment over input. Heat maps are sorted by the difference in enrichment for H3K4me3 and H3K4me1, depicted by blue and red triangles to the left, respectively.
(E) Overlap between lncRNAs detected (FPKM >0) in BAT, iWAT and eWAT.
(F) Examples of BAT-restricted mRNAs and lncRNAs. Tracks depict RNA-seq signal for poly(A)+ RNA from BAT, iWAT and eWAT as density of mapped reads. Bottom tracks depict de novo transcript models by Cufflinks and Ensembl gene annotations. Left-to-right arrows indicate transcripts in the plus strand; right-to-left arrows indicate transcripts in the minus strand.
As many as 30% of the transcribed genomic segments in our samples mapped outside of annotated loci (Figure S1C), presenting a large opportunity for gene discovery. To define lncRNAs with high confidence, we focused on transcripts with evidence of splicing that do not intersect known mRNA exons in the same strand, and implemented a stringent pipeline to evaluate their coding capacity (Figure 1A, Supplemental Experimental Procedures). This analysis classified the BAT, iWAT and eWAT transcriptomes into 13342 known mRNA genes, 1535 lncRNA genes, and 566 genes of unclear coding potential based on our criteria. Our lncRNAs do not appear to encode peptides, as evidenced by mass spectrometry, by ribosome profiling, and by computational assessment of coding capacity (Figures 1B, S1D and S1E). We further confirmed our ability to delineate authentic lncRNA units by finding specific enrichment for 5’ CAGE and 3’ poly(A) tags at their transcription start and end sites, respectively (Figure 1C). Importantly, 1237 lncRNA transcripts from 1032 loci do not intersect Ensembl, RefSeq or UCSC annotations, highlighting the necessity of our de novo reconstruction approach. Overall, ~90% of our lncRNAs are supported by at least one other source of unbiased experimental evidence in addition to RNA-seq (Figure S1G; Supplemental Experimental Procedures), globally validating our lncRNA models.
Analysis of the properties of adipose lncRNAs revealed that they are globally lower-expressed than mRNAs, yet share the same promoter marks of active transcription (Figures 1C, 1D and S1H), consistent with being independent Pol II transcripts. About half of the lncRNAs originate from active enhancer elements defined by a high H3K4me1/H3K4me3 ratio, as expected (Natoli and Andrau, 2012). As characteristic of mouse (Guttman et al., 2009) and human lncRNAs (Cabili et al., 2011), adipose lncRNAs have fewer exons and are thus shorter than mRNAs, and they show higher primary sequence conservation at promoters vs. exons (Figures S1I-S1L). Importantly, 297 out of 1535 lncRNA genes are detectable (FPKM >0) in only one of the three adipose subtypes examined (Figure 1E), despite comparable coverage across samples (Figure S1F), indicating substantial depot-restricted expression. About a third of these loci are exclusive to BAT and resemble genes encoding key BAT-intrinsic proteins, as illustrated by lnc-BATE1 (Figure 1F), a lncRNA that we focus on later because of its remarkable BAT specificity and induction during brown adipogenesis (see below). Thus, we provide a comprehensive catalog of bona fide and mostly unannotated adipose lncRNAs (Table S2), many of which may contribute to development or function of distinct adipocyte lineages.
Adipose Tissue-specific lncRNAs and their Regulation
To examine the tissue specificity of adipose-expressed lncRNAs, we quantified their levels across 30 primary tissues from the mouse ENCODE project (Stamatoyannopoulos et al., 2012) (Figure 2A). We scored the specificity of each gene to each tissue by its fractional expression level (Supplemental Experimental Procedures), and found greater tissue specificity of lncRNAs vs. mRNAs (Figure S2A), as expected (Cabili et al., 2011). We then used an empirical threshold to define tissue-restricted genes and selected those with an adipose subtype as the tissue of maximal specificity (Supplemental Experimental Procedures). This yielded 127 BAT-, 81 iWAT-, and 240 eWAT-specific lncRNAs (Figure S2B and Table S2). Thus, we also find greater fat depot specificity among lncRNAs (~30%) than among protein-coding genes (7%), as illustrated by lnc-BATE1, which is highly abundant in BAT but not in any of the other tissues examined (Figure S2C).
Figure 2. Adipose tissue-specific lncRNAs and their regulation.
(A) Abundance of adipose-expressed mRNAs (13342) and lncRNAs (1535) across 30 tissues from ENCODE, based on our de novo gene models. Color intensity represents the fractional expression across all the tissues examined (see Supplemental Experimental Procedures).
(B) Proportion of BAT-specific and eWAT-specific lncRNAs with promoter-proximal (TSS ±3kb) BAT- or eWAT-specific PPARγ binding (Rajakumari et al., 2013), as determined by peaks of ChIP-seq signal enrichment. ***p <0.001 (Kolmogorov-Smirnov test).
(C) Examples of BAT- and eWAT-restricted lncRNAs showing BAT- or eWAT-specific PPARγ promoter-proximal binding, respectively. UCP1, a BAT-restricted mRNA locus targeted by PPARγ specifically in BAT, is shown for comparison. Tracks depict RNA-seq signal for poly(A)+ RNA from BAT and eWAT as density of mapped reads (black) and ChIP-seq signal for PPARγ binding in BAT and eWAT as density of processed signal enrichment (purple). Peaks of signal enrichment are shown in gray under the ChIP-seq tracks. Bottom tracks depict de novo transcript models by Cufflinks and Ensembl gene annotations as in Figure 1F.
(D) Expression dynamics of BAT-specific and iWAT-specific lncRNAs during brown adipogenesis. Shown are abundance estimates (FPKM) from poly(A)+ RNA-seq of brown preadipocytes (D0) and cultured brown adipocytes (D8) (Sun et al., 2013), based on our de novo gene models.
(E) Dynamic changes in promoter-proximal chromatin marking and transcription factor binding among BAT-specific lncRNAs during brown adipogenesis. Shown are changes in ChIP-signal for binding of C/EBPα, C/EBPβ, PPARγ, and RNA Pol II, as well as H3K27ac, H3K4me1, and H3K4me2 marking, between immortalized brown pre-adipocytes before (D0) and after (D2) adipogenic induction (Lee et al., 2013). Changes are log2 ratios of normalized read counts within TSS ±3kb regions.
To investigate the regulatory basis for adipose subtype-selective lncRNA expression, we first examined global occupancy maps of PPARγ, a master adipogenic TF, assessed by ChIP-seq in primary BAT and eWAT (Rajakumari et al., 2013). We found that PPARγ targets the promoters of 754 (~50%) lncRNAs in BAT or eWAT, as indicated by binding within their TSS ± 3kb regions (Figure S2D). Importantly, BAT-selective lncRNAs are enriched for BAT-specific PPARγ promoter binding, whereas eWAT-selective lncRNAs are enriched for eWAT-specific binding, as seen for key depot-specific proteins (Figures 2B and 2C). Among depot-specific lncRNAs whose promoters are bound by PPARγ in both BAT and eWAT, we still found stronger PPARγ binding in their tissue of selective expression (Figures S2D and S2E).
We then focused on lncRNAs active in BAT, for which profiles of expression, histone marking, and TF binding during brown adipogenesis are available (Lee et al., 2013; Sun et al., 2013) (Figure S2F). As expected from their depot-specific regulation, BAT-selective lncRNAs are specifically enriched for induction during brown adipogenesis, with 49 (38%) upregulated >2-fold (Figure 2D). lncRNA activation is reflected at the chromatin level and correlates with binding of C/EBPα, C/EBPβ and PPARγ early during differentiation (Figures 2E, S2F and S2G). The most predictive activation event is C/EBPα targeting, most of which represents new binding events at differentiation day 2, while co-targeting by C/EBPα, C/EBPβ and PPARγ is associated with the strongest induction levels (Figure S2G). These findings characterize multiple BAT-selective lncRNAs that are targeted by common adipogenic TFs, often in a BAT-specific manner, and show dynamic regulation during differentiation.
Validation of BAT-selective lncRNAs
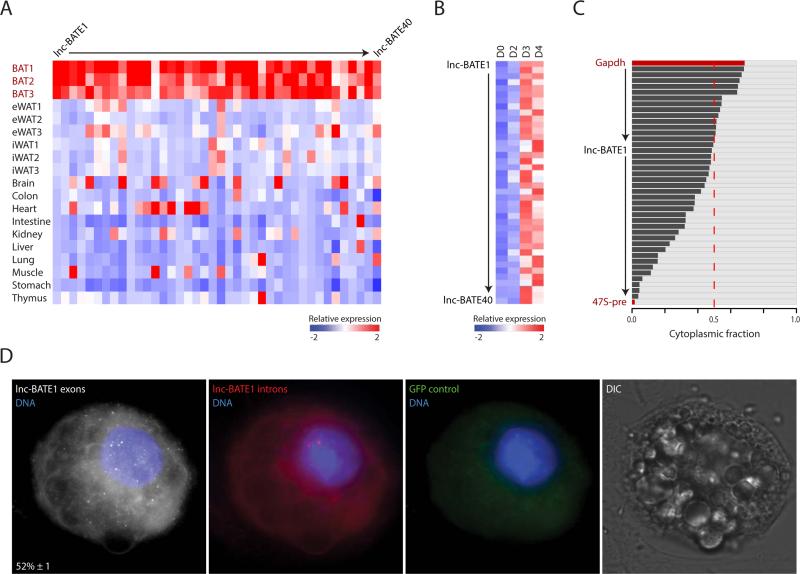
To focus our validation efforts, we ranked candidate lncRNAs by their BAT specificity score, upregulation during brown adipogenesis, and abundance, and chose the top 40 for qPCR-based validation. For 38 out of 40 candidates, we confirmed significantly higher expression in BAT vs. the average expression across 12 major organs, with 26 lncRNAs highest-expressed in BAT (Figure 3A). We also monitored expression during brown adipogenesis in culture, and verified that all 40 candidates were upregulated (Figure 3B). Next, to examine subcellular distribution, we isolated RNA from cytoplasmic and nuclear fractions of mature brown adipocytes and quantified lncRNA abundance by qPCR (Figure 3C). Most candidates (27 out of 40) were enriched in the nucleus, with four of them closely resembling the 47S pre-rRNA at >90% nuclear retention, consistent with previous observations (Alvarez-Dominguez et al., 2014b). Others, including lnc-BATE1, were similarly abundant in the nucleus and in the cytoplasm.
Figure 3. Validation of BAT-selective lncRNAs.
(A) Validation of 40 lncRNAs in BAT, eWAT and iWAT (n =3) and across 10 tissue samples by qPCR. Color intensity represents column mean-centered expression.
(B) Induction of 40 BAT lncRNAs during brown adipogenesis. Expression values during a 4-day differentiation time course of cultured mouse pre-adipocytes were determined by qPCR (n =3). Color intensity represents row mean-centered expression.
(C) Subcellular localization of 40 BAT lncRNAs. The relative proportion of cytoplasmic (black) and nuclear (gray) expression was assessed by qPCR (n =3). GAPDH mRNA and 47S pre-rRNA represent predominantly cytoplasmic and predominantly nuclear controls, respectively. Rows are sorted from highest to lowest cytoplasmic fraction.
(D) Detection of lnc-BATE1 transcripts by single-molecule RNA FISH. Shown are maximum z-stack projections of fluorescence microscopy images. lncRNA molecules and DNA staining are pseudocolored as indicated. Shown at the bottom left panel corner for lnc-BATE1 exons is the mean ±SEM (n =2) percent of nuclear-localized transcripts. GFP control indicates background fluorescence measured in the GFP channel. DIC indicates imagining in the differential interference contrast channel.
lnc-BATE1 is Required for Brown Adipocyte Development, Function, and Maintenance
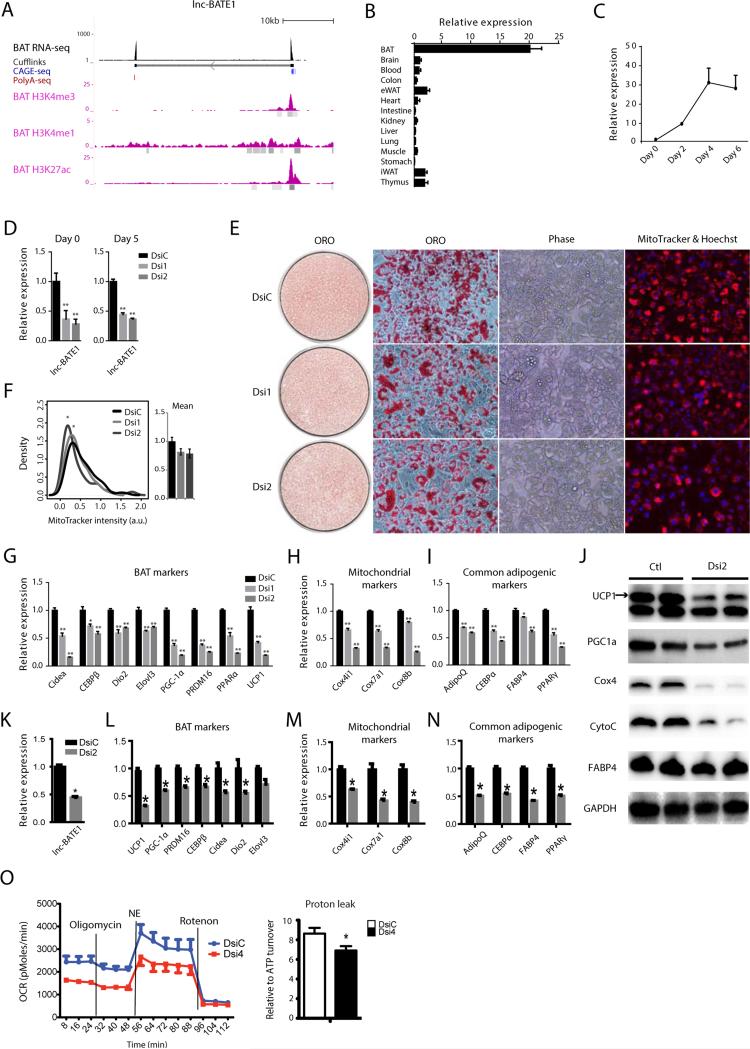
Our ranking of adipose lncRNAs by their abundance, regulation and depot selectivity identified lnc-BATE1 as a top candidate modulator of brown adipogenesis. lnc-BATE1 is an independent intergenic locus targeted by C/EBPα, C/EBPβ, and PPARγ giving rise to polyadenylated transcripts spliced from two exons (Figures 4A and S2H), coincident with RefSeq gene NR_077224. 5’ and 3’ RACE revealed transcript variants with slightly different transcription start sites and a common termination site (Figures S3A and S3B). lnc-BATE1 is equally distributed between cytosol and nucleus, as evidenced by cell fractionation and by single-molecule RNA FISH, which additionally indicated mean levels of 18 ± 2 transcripts per cell (Figure 3C, 3D and S3C). Importantly, lnc-BATE1 is enriched 10-20-fold in brown vs. white adipocytes and is upregulated 30-fold during brown adipogenesis (Figures 4B and 4C).
Figure 4. lnc-BATE1 is required for brown adipocyte development, function, and maintenance.
(A) lnc-BATE1 locus map. Track 1 depicts BAT poly(A)+ RNA-seq signal as density of mapped reads. Track 2 depicts de novo transcript models by Cufflinks. Tracks 3-4 display RNA 5’capping and 3’polyadenylation sites as evidenced by CAGE tags (blue) and poly(A) tags (red), respectively; only tags from the strand of transcription are shown. Tracks 5-7 display ENCODE BAT ChIP-seq signal from H3K4me3, H3K4me1 and H3K27ac marks, respectively, as density of processed signal enrichment; peaks of signal enrichment are shown in gray under each track.
(B) Expression of lnc-BATE1 across 14 mouse tissues assessed by qPCR.
(C) Expression of lnc-BATE1 during the course of brown adipogenesis in culture assessed by qPCR.
(D) Expression of lnc-BATE1 in cultured brown adipocytes transfected with DsiRNA control (DsiC) or DsiRNAs targeting lnc-BATE1 (Dsi1 and Dsi2) and collected for qPCR at differentiation days 0 and 5.
(E) Representative images of DsiRNA-treated cultured brown adipocytes at differentiation day 5 labelled with Oil red O (ORO, red) or MitoTracker® Deep Red FM (red) plus Hoechst (blue), respectively.
(F) Quantification of integrated density signal of MitoTracker® fluorescence in individual cells from (E). Signal distributions are shown to the left and their mean values to the right.
(G-I) Expression of BAT (G), mitochondrial (H), and common adipogenic markers (I) in DsiRNA-treated cultured day 5 brown adipocytes.
(J) Protein levels of BAT, mitochondrial and common adipogenic markers assessed by western blot on cell lysates from DsiRNA-treated cultured day 5 brown adipocytes.
(K-N) Knockdown of lnc-BATE1 in mature brown adipocytes 72h post-transfection with DsiRNAs (K) impairs expression of BAT (L), mitochondrial (M), and common adipogenic markers (N).
(O) Representative metabolic flux curves from cultured DsiRNA-treated day 5 brown adipocytes treated with the adrenergic agent norepinephrine (NE) and 2% BSA. Oxygen consumption rates (OCR) are mean ±SEM. and are normalized by protein concentration.
Error bars are mean ± SEM., n=3. *P ≤0.05, **P ≤0.01.
To investigate lnc-BATE1 function, we designed Dicer-substrate siRNAs (DsiRNAs) and transfected them into primary brown pre-adipocytes, followed by induction of differentiation. Over 70% knockdown was achieved at differentiation day 0, and ~60% remained at day 5 (Figure 4D). lnc-BATE1 KD resulted in limited changes in lipid accumulation and cell morphology during differentiation (Figure 4E), but significantly downregulated mRNA levels of all brown fat markers examined, including Cidea, C/EBPβ, PGC1α, PRDM16, PPARα, and UCP1 (Figure 4G), as well as mitochondrial markers (Cox4i, Cox7a and Cox8b) (Figure 4H). General adipogenic markers (AdipoQ, C/EBPα, Fabp4, and PPPARγ) were also downregulated but to a lesser extent (Figure 4I), consistent with the limited effects on general adipocyte differentiation. Immunoblotting further confirmed reduced protein levels of BAT-selective genes (UCP1, PGC1α) and mitochondrial markers (Cox4, CytoC) (Figure 4J). lnc-BATE1 KD by traditional siRNAs or by shRNAs targeting different sites gave very similar results (Figures S3D-J), indicating that lnc-BATE1 KD phenotypes are unlikely due to RNAi off-target effects. In contrast to the dramatic downregulation of BAT markers, lnc-BATE1 KD led to upregulation of WAT-selective genes (see below; Figures 5E-G).
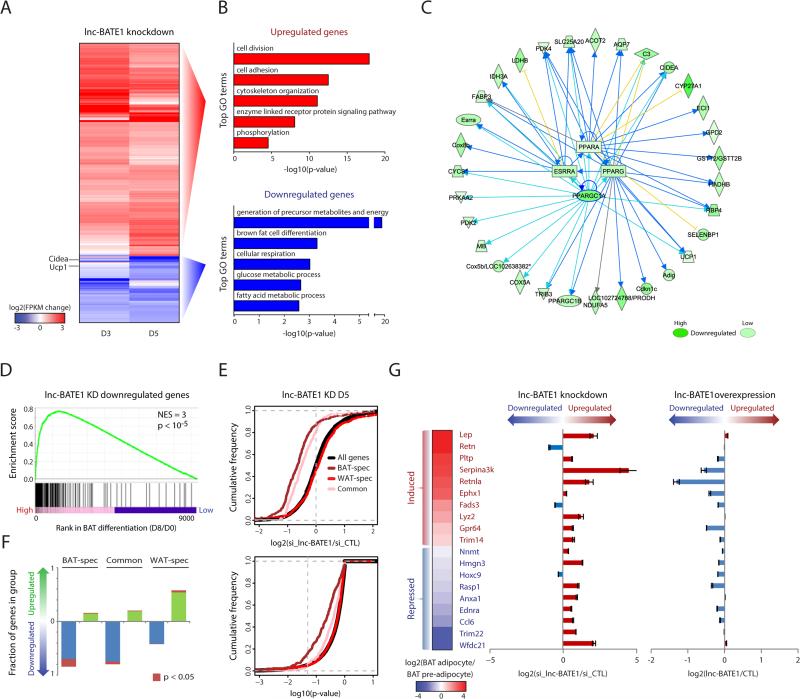
Figure 5. lnc-BATE1 mediates concurrent activation of the brown fat and suppression of the white fat gene programs.
(A) Expression change of 1,014 mRNAs that are differentially expressed (P <0.05, DESeq) in cultured brown adipocytes upon lnc-BATE1 KD, collected at differentiation days 3 (D3) and 5 (D5). Changes are log2 expression (FPKM) ratios over control siRNA.
(B) Top 5 non-redundant gene ontology (GO) biological process terms enriched (P <0.05, Fisher's test) among mRNA genes that show significantly higher (top) or lower (bottom) expression (P <0.05, DESeq) upon lnc-BATE1 KD relative to control.
(C) Network diagram of top upstream transcription regulators whose inhibition best explains genes downregulated (P <0.05, DESeq) upon lnc-BATE1 KD, along with their known direct targets. Arrows and blocked lines indicate transcriptional activation and repression, respectively. Blue and yellow lines indicate whether the predicted inhibition of the upstream regulator is consistent or inconsistent with the state of the downstream molecule, respectively; gray lines generated no prediction. Highlighted blue lines emphasize PGC-1α relationships.
(D) Gene set enrichment analysis for overlap between genes depleted upon lnc-BATE1 KD and the BAT differentiation gene signature published previously (Sun et al., 2013). NES, normalized enrichment score; p, empirical p-value.
(E) Cumulative density distributions of expression changes (top) and p-values for these changes (bottom) for all expressed protein-coding genes and for BAT-specific, WAT-specific and common adipogenic genes in lnc-BATE1 siRNA-treated cultured day 5 brown adipocytes. Changes are log2 expression (FPKM) ratios relative to control siRNA. Vertical gray line denotes the P <0.05 significance threshold (bottom).
(F) Proportion of BAT-specific, WAT-specific and common adipogenic genes that are upregulated (log2 expr. change vs. control >0) or downregulated (log2 expr. change vs. control <0) in lnc-BATE1 siRNA-treated cultured day 5 brown adipocytes.
(G) lnc-BATE1 mediates repression of WAT marker genes. Expression change of select WAT markers during brown adipogenesis in culture, shown as the log2 expression ratio between brown adipocytes (Day 6) and pre-adipocytes (Day 0) (left). Expression change in cultured day 3 brown adipocytes transfected with siRNA targeting lnc-BATE1, relative to control siRNA (middle). Expression change in cultured day 5 white adipocytes expressing ectopic lnc-BATE1, relative to GFP control (right).
Error bars are mean ±SEM., n ≥3.
Impaired BAT marker expression upon lnc-BATE1 loss could be due to preferential disruption of the BAT gene program or be an indirect effect of poor cell differentiation. To distinguish between these possibilities, we depleted lnc-BATE1 in mature brown adipocytes, using an electroporation method that yielded ~60% knockdown (Figure 4K). We observed no evident changes in cell morphology 3 days post-transfection (not shown), but found a significant reduction in BAT, mitochondrial, and common adipogenic markers (Figures 4L-N). Thus, lnc-BATE1 is essential for establishing the gene program of developing brown adipocytes and for its maintenance in mature ones.
lnc-BATE1 loss also affected mitochondrial biogenesis, as indicated by decreased mitochondrial staining (Figures 4E and 4F), suppression of mitochondrial genes (Figures 4H, 4M and S3J), and loss of Ucp1 protein (Figure 4J). To examine whether lnc-BATE1 KD alters thermogenesis, we measured oxygen consumption in the presence of the adrenergic agent norepinephrine (NE) and 2% BSA to specifically measure Ucp1-dependent uncoupled respiration (Li et al., 2014) (Figure 4O). lnc-BATE1 KD cells exhibited generally lower oxygen consumption, consistent with lower mitochondrial content, but also showed specific impairment of their relative NE-stimulated respiration, consistent with reduced Ucp1 accumulation and activity. These data demonstrate that lnc-BATE1 is essential during brown adipogenesis for induction of multiple mitochondrial proteins, including Ucp1, and for thermogenesis in brown adipocytes.
To assess if lnc-BATE1 is induced during browning of white fat, we examined its expression in inguinal WAT of mice exposed to 4oC for 1 week. We found that lnc-BATE1 is upregulated 3- to 4-fold (Figure S4A), suggesting a role in WAT browning. To test this, we used retroviral shRNAs to infect primary inguinal white pre-adipocytes, followed by induction of differentiation in the absence (Figure S4B-D) or presence of NE (Figure S4E). Similar to the effects on brown adipogenesis, lnc-BATE1 KD resulted in limited effects on lipid accumulation and cell morphology (not shown), but impaired expression of the examined BAT, mitochondrial and, to a lesser extent, common adipogenic markers (Figures S4C). In contrast, 4 out of 7 WAT-selective genes were significantly upregulated (Figure S4D). In the presence of NE, we further found that induction of thermogenic genes UCP1 and PGC1α is blunted by lnc-BATE1 KD (Figure S4E). Similar effects were observed in cultured epididymal adipocytes (Figure S4F and S4G).
To study the impact of lnc-BATE1 gain of function on brown adipogenesis, we cloned lnc-BATE1 and introduced it into brown pre-adipocytes via retroviral transduction, followed by induction of differentiation. We could not find any significant changes in lipid accumulation, cell morphology (not shown), or enhancement of BAT marker gene expression, however, under standard or limited differentiation conditions (Figures S5A and S5B), suggesting that normal lnc-BATE1 levels suffice to maximally stimulate brown adipogenesis and excess ectopic expression has no further effect. Similarly, overexpressing lnc-BATE1 in primary inguinal or epididymal white pre-adipocytes followed by induction of differentiation did not significantly impact BAT-selective genes (Figures S5C-E), indicating that lnc-BATE1 gain of function is insufficient to promote browning. Finally, we tested if lnc-BATE1 functions in brown adipocyte lineage determination from myoblast progenitors by ectopically expressing it in C2C12 myoblasts followed by induction of differentiation, but did not find evident effects in cell morphology (not shown) or in expression of the myogenic markers examined (Figure S5F). Thus, lnc-BATE1 is a BAT-selective factor necessary but not sufficient for brown adipocyte development, function, and maintenance.
lnc-BATE1 Mediates Concurrent Activation of the Brown Fat and Suppression of the White Fat Gene Expression Programs
To gain further insights into lnc-BATE1 function from global gene expression analysis, we performed RNA-seq in differentiating DsiRNA-treated brown adipocytes, and identified 1,014 differentially expressed genes (P <0.05, DESeq), comprising 781 enriched and 233 depleted in lnc-BATE1 KD vs. control cells (Figure 5A). Higher-expressed genes were enriched for general functions in cell division, adhesion and signaling processes that are normally suppressed during adipogenesis (Figures 5B top and S6A), whereas lower-expressed ones comprised genes specifically linked to brown adipogenesis and mitochondrial biogenesis and function, which fail to be activated upon lnc-BATE1 loss (Figures 5B bottom and S6B). Gene set enrichment analysis (Subramanian et al., 2005) of depleted genes further indicated significant overlap with the gene signature activated during brown adipogenesis published previously (Sun et al., 2013) (Figure 5D). We thus sought to computationally identify upstream regulators that may be responsible for suppression of these genes upon lnc-BATE1 KD (Supplemental Experimental Procedures and Table S3). Pathway analysis identified PGC1α, ESRRα, PPARα, and PPARγ as the top TFs whose inhibition would explain downregulation of 64 genes (P <10−14 – P <10−5, Fisher's test) (Figure 5C). Independent gene set enrichment analysis further showed that genes activated by these factors are significantly depleted upon lnc-BATE1 KD (P <10−5 for all; Figure S6C). These results indicate that lnc-BATE1 is required for a genetic program associated with brown adipogenesis.
Suppression of the brown adipogenesis program upon lnc-BATE1 KD could be due to suppression of genes important for adipogenesis in general. To test this possibility, we defined groups of BAT-specific, WAT-specific, and common adipogenic protein-coding genes based on their tissue specificity scores (Table S2; Supplemental Experimental Procedures), and studied the impact of lnc-BATE1 KD on their expression (Figures 5E, 5F, S6D and S6E). We found that, in general, inhibiting lnc-BATE1 leads to repression of BAT-selective or common adipogenic genes that are normally activated during brown adipogenesis, but there is a more profound influence on BAT-selective genes (~15% downregulated at P<0.05, DESeq) than on common adipogenic ones (~6% downregulated at P<0.05, DESeq), indicating that lnc-BATE1 indeed acts as a BAT-selective regulator. In contrast, WAT-selective genes were mostly upregulated upon lnc-BATE1 KD, whether they are normally repressed or activated during brown adipocyte differentiation (Figures 5F and S6E), indicating that their higher expression is not merely due to impaired adipogenesis. To validate this finding, we focused on 20 of the most widely-used WAT markers (Kajimura et al., 2008; Seale et al., 2007; Villanueva et al., 2013; Walden et al., 2012) (Figures 5G and S6F), and confirmed by qPCR their upregulation upon lnc-BATE1 KD by either siRNAs (16 out of 19 genes upregulated, mean 3.4-fold) or shRNAs (16 out of 19 genes upregulated, mean 2-fold). Accordingly, lnc-BATE1 gain of function in iWAT adipocyte culture led to their general downregulation, which was specifically anticorrelated with their upregulation upon lnc-BATE1 inhibition in BAT adipocyte culture (Pearson's r= -28 to r= -46, P <0.05, t-test). Thus, lnc-BATE1 specifically mediates concurrent activation of the core BAT gene program and repression of WAT genes.
lnc-BATE1 Functions in trans
lncRNAs can function in cis or in trans during cell differentiation (Fatica and Bozzoni, 2013; Hu et al., 2012). To distinguish between these possibilities, we first examined the expression of genes neighboring lnc-BATE1 within a ~1.75Mb window (Figure S7A). We found no correlation in the tissue expression patterns of these genes vs. lncBATE-1 (Figure S7B), and showed that their levels are unaffected by its depletion (Figures S7C and S7D), indicating that lnc-BATE1 does not regulate its neighbors in cis.
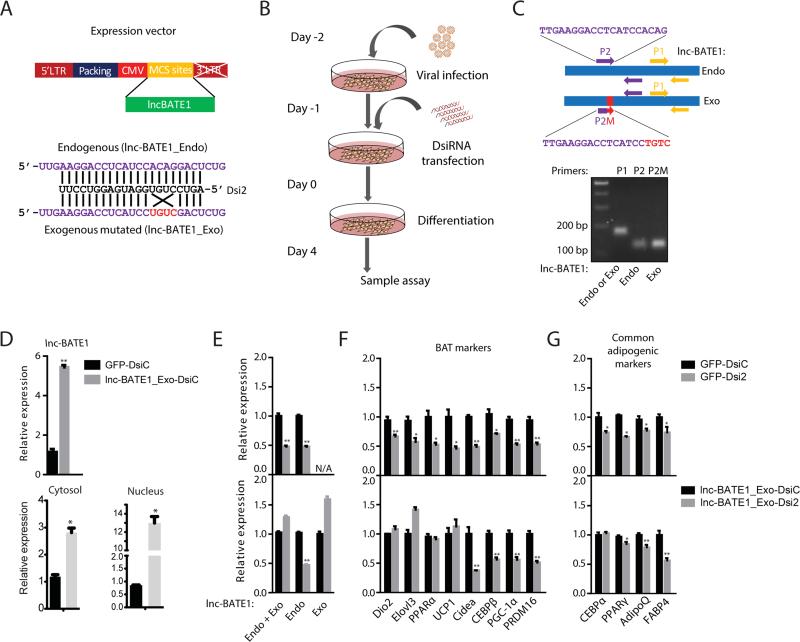
To investigate if lnc-BATE1 functions in trans, we tested whether the defects of lnc-BATE1 KD cells could be rescued by ectopically-expressed lnc-BATE1 that escapes DsiRNA targeting. We thus constructed an exogenous mutated lnc-BATE1 (lnc-BATE1_Exo) with a 4nt mutation at the DsiRNA2 targeting site designed to abolish KD (Figure 6A), and transduced it or GFP control into brown pre-adipocytes prior to DsiRNA transfection and subsequent induction of differentiation (Figure 6B). lnc-BATE1 expression was assessed with primers specific to endogenous or exogenous variants or common to both (Figure 6C). Introducing lnc-BATE1_Exo, which localized to both nucleus and cytoplasm, increased total lnc-BATE1 levels by >5-fold (Figure 6D). As expected, Dsi2 inhibited endogenous but not exogenous lnc-BATE1 (Figure 6E), and in GFP control cells impaired expression of 8 BAT markers and, to a lesser extent, of 4 common adipogenic genes (Figures 6F and 6G top panels). In cells expressing lnc-BATE1_Exo, however, we found rescued expression for half of the examined BAT markers, including Dio2, Elovl3, PPARα and UCP1, and for the common adipogenic factors C/EBPα and PPARγ (Figures 6F and 6G bottom panels). These results demonstrate RNA-based function and indicate that lnc-BATE1 can act in trans to modulate brown adipogenesis.
Figure 6. Exogenous siRNA-resistant lnc-BATE1 partially rescues gene suppression in brown adipocytes depleted of endogenous lnc-BATE1.
(A) Construction of exogenous siRNA-resistant lnc-BATE1 mutant (lnc-BATE1_Exo) from the endogenous transcript (lnc-BATE1_Endo).
(B) Schematic illustration of procedure used for rescue experiments.
(C) Design of qPCR primer pairs and agarose gel image of the resulting PCR products. Lane 2: lnc-BATE1_Endo or _Exo amplified by P1 primer pair; lane 3: lnc-BATE1_Endo amplified by P2 primer pair; lane 4: lnc-BATE1_Exo amplified by P2M primer pair.
(D) Expression (top) and localization (bottom) of total lnc-BATE1 in brown adipocytes infected with GFP control viruses or with lnc-BATE1_Exo viruses prior to transfection with control DsiRNA (DsiC).
(E-G) Expression of endogenous or exogenous lnc-BATE1 (E), brown adipocyte markers (F) and general adipogenic markers (G) in brown adipocytes infected with GFP control virus or with lnc-BATE1_Exo virus prior to transfection with control DsiRNA (DsiC) or lnc-BATE1 DsiRNA (Dsi2).
Error bars are SEM., n =3. *P ≤0.05, **P ≤0.01.
lnc-BATE1 Interacts with hnRNPU, which is Required for Brown Adipocyte Development
lncRNAs can function by binding proteins to form functional complexes (Rinn and Chang, 2012). We thus sought to identify protein partners of lnc-BATE1 via RNA pull-down in nuclear and cytosolic lysates (Experimental Procedures). However, we did not find any interactions specific to lnc-BATE1 (not shown), suggesting that its protein partners are either of low abundance or are masked by non-specific interactions with proteins of similar molecular weight.
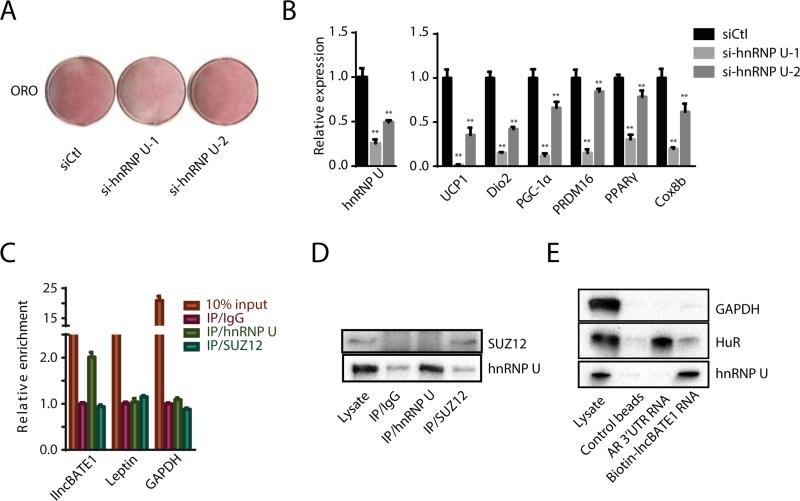
We next sought to examine by RNA immunoprecipitation (RIP) proteins known to bind lncRNAs in adipocytes. We previously showed that the nuclear matrix factor hnRNP U is required for the proper localization of Firre, a lncRNA essential for white adipogenesis, to its targets (Hacisuleyman et al., 2014; Sun et al., 2013). Interestingly, we found a putative hnRNP U binding motif within lnc-BATE1 (Figure S7E), suggesting that the two interact. To explore this possibility, we first asked whether hnRNP U contributes to brown adipogenesis. hnRNP U KD with siRNAs in differentiating brown adipocytes significantly impaired lipid droplet accumulation (Figure 7A) and BAT marker gene expression (Figure 7B), indicating that it is needed for brown adipogenesis. We then performed RIP against hnRNP U, and detected a specific interaction with lnc-BATE1 (Figures 7C and 7D). In contrast, RIP against SUZ12, a PRC2 subunit that binds a wide range of lncRNAs non-specifically (Davidovich et al., 2013; Kaneko et al., 2013), did not enrich for lnc-BATE1. These results were confirmed by RNA pull-down and immunoblotting (Figure 7E). Binding of androgen receptor (AR) 3’UTR RNA to HuR protein served as positive control for these studies and as negative control for hnRNP U binding. As expected, hnRNP U and HuR were enriched by lnc-BATE1 and AR 3’UTR RNA, respectively, whereas Gapdh housekeeping protein was not (Figure 7E). These findings demonstrate a specific and direct interaction between lnc-BATE1 and hnRNP U, suggesting that they form a functional ribonucleoprotein complex to regulate brown adipogenesis.
Figure 7. lnc-BATE1 interacts with hnRNP U, which is required for brown adipocyte differentiation.
(A) Oil red O staining of brown adipocytes differentiated in culture upon siRNA-mediated hnRNP U KD.
(B) Expression of hnRNP U and marker genes in cultured brown adipocytes following hnRNP U KD, quantified by qPCR.
(C-D) Association between endogenous lnc-BATE1 and hnRNP U in the nucleus of cultured brown adipocytes. RNA immunoprecipitation (RIP) enrichment was assessed as RNA associated to hnRNP U or Suz12 relative to IgG control by qPCR (C) or Western blot (D).
(E) lnc-BATE1 and hnRNP U specifically interact in vitro. Western blots for biotin-RNA pull-down show specific interaction between lnc-BATE1 and hnRNP U but not GAPDH or HuR protein, which specifically interacts with androgen receptor (AR) 3’UTR RNA.
Error bars are SEM., n =3. *P ≤0.05, **P ≤0.01.
DISCUSSION
Elucidating factors controlling development of distinct types of fat is crucial for finding new targets to treat metabolic disorders. In particular, factors that selectively promote brown adipogenesis are of key interest as potential targets for obesity. Here, we present the first comprehensive catalog of lncRNAs active across different adipose tissues, including ~450 that are highly subtype-selective, providing a valuable resource for the discovery of lncRNAs with adipocyte lineage-specific functions. This resource is available online (https://sites.google.com/site/sunleilab/data/lncrnas) and can be used to efficiently identify functional lncRNAs based on their tissue expression, specificity, and regulation features, as illustrated by our work showing that lnc-BATE1, a lncRNA chosen based on these features, is required for the brown adipocyte phenotype.
We characterize lnc-BATE1 as a novel BAT-selective factor that has limited roles on general adipocyte differentiation but serves critical brown adipocyte-selective functions. Indeed, lnc-BATE1 is essential for the formation and maintenance of mature brown adipocytes capable of thermogenesis. A different type of thermogenic adipocytes, termed “beige” or “brite”, have been shown to form within white fat depots, in response to cold stress or other stimuli, but share many components of the BAT gene program (Petrovic et al., 2010; Schulz et al., 2011; Wu et al., 2012). We find that lnc-BATE1 is upregulated during cold-induced beige adipocyte expansion and its loss impairs induction of BAT-selective genes, suggesting a broader role for lnc-BATE1 in general thermogenic programming. In support of this notion, our loss-of-function and gain-of-function studies indicate that lnc-BATE1 can act in trans not only to sustain a thermogenic phenotype, but also to suppress WAT-selective gene programming.
lncRNAs often function by partnering with proteins such as chromatin modifiers and RNA binding factors (Wang and Chang, 2011). For instance, hnRNP U is responsible for localization of lncRNAs Xist and Firre to the subnuclear domains where they function(Hacisuleyman et al., 2014; Hasegawa et al., 2010). We find that lnc-BATE1 directly interacts with hnRNP U, which is also required for brown adipogenesis, suggesting that it may form a functional ribonucleoprotein complex with lnc-BATE1 to exert its function in a cell type-specific manner. However, hnRNP U can recognize a wide array of substrates and participates in many aspects of RNA metabolism, and lnc-BATE1's presence in both the nucleus and cytosol suggests additional cytosolic protein or RNA partners. The functional impact of its specific interaction with hnRNP U on BAT development and function thus warrants further investigation. Collectively, our work provides a basis for the study of adipose tissue-selective lncRNAs and demonstrates their importance as BAT-specific regulators, which may be exploited for selective stimulation of BAT development for therapeutic use.
EXPERIMENTAL PROCEDURES
Tissue isolation and Cell Culture
Primary fat tissues were isolated from 8-week-old B/C mice, and primary brown and white preadipocytes were isolated from 3~4-week-old mice and differentiated in culture as described (Sun et al., 2011). 293T cells and C2C12 myoblasts were maintained in DMEM plus 10% or 20% FBS, respectively. C2C12 cells were differentiated in DMEM with 2% horse serum.
RNA-seq
Total RNA from BAT, iWAT and eWAT samples was isolated using a Qiagen kit. Sequencing libraries were prepared as described (Sun et al., 2011) and sequenced on the Illumina HiSeq2000 platform (see Supplemental Experimental Procedures for analysis details). RNA-seq data from this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE66686).
Single-molecule RNA FISH
Single-molecule RNA FISH, fluorescence microscopy, image acquisition and analysis were conducted as described (Alvarez-Dominguez et al., 2014b) (see Supplemental Experimental Procedures for details).
lncRNA Knockdown
Pre-adipocytes at ~80% confluence were transfected with 100nM siRNAs or DsiRNAs. 6~8 hours later, cells were recovered in full culture medium, grown to confluence, and induced to differentiate as described (Sun et al., 2013). For shRNA-mediated knockdown, cells at ~60% confluence were infected with shRNA retroviruses and induced to differentiate 48h post-infection. siRNA knockdown in mature brown adipocytes was performed as described (Rajakumari et al., 2013) (see Supplemental Experimental Procedures for details).
Plasmid and Retroviral Transduction
lncRNA expression plasmids or shRNA viral plasmids were co-transfected with retroviral packaging vector pCL-Eco into 293T cells using FuGENE6 (Promega), and viruses were collected at 48h and 72h post-transfection. Cells were induced to differentiate 48h post-infection and collected for downstream analysis at indicated times.
Oil-Red-O, Hoechst and Mitotracker Staining
5-day differentiated brown adipocytes were stained with 100mM Mitotracker Red FM and 1:5000 dilution of Hoechst at 37°C for 40min. ORO staining was performed as described (Sun et al., 2011) (see Supplemental Experimental Procedures for details).
Extracellular Flux Analysis
5-day differentiated RNAi-treated brown adipocytes were applied to an Extracellular Flux Analyzer (Seahorse bioscience) and analyzed for oxygen consumption rates according to the manufacturer's instructions (see Supplemental Experimental Procedures for details).
lncRNA Cloning
To ectopically express lnc-BATE1, 3 different variants were cloned into a modified pSIRENRetroQ-ZsGreen Vector (Clontech). To make exogenous mutated lnc-BATE1, mutated nucleotides were introduced by PCR amplification of overlapping products harboring mutated nucleotides. lnc-BATE1 shRNA plasmids were made by inserting annealed oligos into the pMKO vector.
RNA Immunoprecipitation
Nuclei from 4-day differentiated brown adipocytes were isolated and nuclear or cytosolic lysates were prepared, treated with 300 U/ml RNase inhibitor, and incubated with 5ug of indicated antibody. RNA-protein complexes were immunoprecipitated with protein A/G beads and 20% were kept for western blot and the rest used for RNA extraction (see Supplemental Experimental Procedures for details).
RNA Pull-down
Biotin-labeled lnc-BATE1 and androgen receptor 3’UTR RNA were transcribed using a MEGAscript kit (Life Technologies). Biotinylated RNAs were purified with a NucAway spin column as described (Tsai et al., 2010) and incubated with brown adipocyte nuclear lysate for 3h at 4°C. Beads were then washed and boiled in SDS buffer for 5min at 95°C, and the retrieved protein visualized by immunoblotting (see Supplemental Experimental Procedures for details).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Singapore NRF fellowship (NRF-2011NRF-NRFF 001-025) to L.S, National Institute of Health (NIH) grants DK047618, DK068348 and 5P01 HL066105 to HFL. This research is also supported by the Singapore National Research Foundation under its CBRG grant (CBRG14may001) and administrated by the Singapore Ministry of Health's National Medical Research Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.A., Z.B., D.X., B.Y., K.L., N.S., L.S., M.Y., Y. L. and S.C. performed research. J.A., Z.B, H.L, and L.S., designed the project, interpreted the results, and wrote the manuscript. D.X., B.Y., K.L., N.S., L.S., and S.C. contributed discussions.
REFERENCES
- Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. International journal of hematology. 2014a;99:531–541. doi: 10.1007/s12185-014-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014b;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SH, Bazuine M, Lumeng CN, Geletka LM, Mowers J, White NM, Ma JT, Zhou J, Qi N, Westcott D, et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell. 2009;138:961–975. doi: 10.1016/j.cell.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2013;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann A, Flier JS, Lowell BB. Decreased brown fat markedly enhances susceptibility to diet-induced obesity, diabetes, and hyperlipidemia. Endocrinology. 1996;137:21–29. doi: 10.1210/endo.137.1.8536614. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Gene Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fromme T, Schweizer S, Schottl T, Klingenspor M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite/beige adipocytes. EMBO Rep. 2014;15:1069–1076. doi: 10.15252/embr.201438775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2010;10:e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. Noncoding Transcription at Enhancers: General Principles and Functional Models. Annual Review of Genetics. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American journal of physiology Endocrinology and metabolism. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Trajkovski M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism: clinical and experimental. 2014;63:272–282. doi: 10.1016/j.metabol.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, Hu Y, Peng X, Feng X, Saez E, et al. Adipose Subtype-Selective Recruitment of TLE3 or Prdm16 by PPAR gamma Specifies Lipid Storage versus Thermogenic Gene Programs. Cell Metabolism. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya F, Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metab. 2013;17:638–643. doi: 10.1016/j.cmet.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. American journal of physiology Endocrinology and metabolism. 2012;302:E19–31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.