SUMMARY
Mutations affecting spliceosomal proteins are the most common class of mutations in patients with myelodysplastic syndromes (MDS), yet their role in MDS pathogenesis has not been delineated. Here we report that mutations affecting the splicing factor SRSF2 directly impair hematopoietic differentiation in vivo, which is not due to SRSF2 loss of function. By contrast, SRSF2 mutations alter SRSF2’s normal sequence-specific RNA binding activity, thereby altering recognition of specific exonic splicing enhancer motifs to drive recurrent mis-splicing of key hematopoietic regulators. This includes SRSF2 mutation-dependent splicing of EZH2 that triggers nonsense-mediated decay, which, in turn, results in impaired hematopoietic differentiation. These data provide a mechanistic link between a mutant spliceosomal protein, alterations in splicing of key regulators, and impaired hematopoiesis.
INTRODUCTION
Somatic mutations in genes encoding components of the spliceosome have been identified in a spectrum of human malignancies, including ~60% of patients with myelodysplastic syndromes (MDS) (Bejar et al., 2012; Papaemmanuil et al., 2013; Yoshida et al., 2011). These mutations occur most commonly in SF3B1 (Splicing Factor 3b Subunit 1), SRSF2 (Serine/arginine-Rich Splicing Factor 2), and U2AF1 (U2 Small Nuclear RNA Auxiliary Factor 1) and almost always as heterozygous missense mutations that are mutually exclusive (Papaemmanuil et al., 2011; Wang et al., 2011; Yoshida et al., 2011). While the genetic data in MDS suggests that these alterations are critical to disease pathogenesis, it remains unknown how these mutations contribute to MDS and if they are sufficient to induce MDS.
Recent studies have suggested that mutations in the spliceosomal gene U2AF1 alter RNA splicing (Brooks et al., 2014; Graubert et al., 2012; Ilagan et al., 2014; Przychodzen et al., 2013; Quesada et al., 2012) and studies of gene expression in primary patient samples with and without U2AF1 mutations have been performed in an effort to identify downstream mis-spliced genes that might contribute to abnormal hematopoiesis (Brooks et al., 2014; Graubert et al., 2012; Ilagan et al., 2014). However it remains unknown how these mutations contribute to hematopoietic transformation. To date, no studies have investigated the in vivo effects of spliceosomal mutations expressed from the endogenous locus in the correct cellular context, which might allow delineation of how these alleles contribute to MDS pathogenesis.
To test whether spliceosomal gene mutations are sufficient to drive MDS, and determine how altered RNA splicing contributes to transformation in vivo, we studied the biological and transcriptional consequences of mutations in SRSF2. SRSF2 mutations occur in 20–30% of MDS and ~50% of chronic myelomonocytic leukemia (CMML) patients (Papaemmanuil et al., 2013; Yoshida et al., 2011). SRSF2 is a member of the serine/arginine-rich (SR) protein family that contributes to both constitutive and alternative splicing by binding to exonic splicing enhancer (ESE) sequences within pre-mRNA through its RNA recognition motif domain (RRM) (Graveley and Maniatis, 1998; Liu et al., 2000; Schaal and Maniatis, 1999; Zahler et al., 2004). SRSF2 mutations are consistently associated with adverse outcome amongst MDS and AML patients (Papaemmanuil et al., 2013; Vannucchi et al., 2013; Zhang et al., 2012). Despite the clinical importance of SRSF2 mutations, to date there have been no studies of the functional impact of SRSF2 mutations on hematopoiesis or splicing. Here, we studied the biological and transcriptional effects of somatic expression of the common SRSF2 P95H mutation in the hematopoietic compartment.
RESULTS
Srsf2P95H mutant mice develop MDS, a phenotype distinct from mice with heterozygous or homozygous loss of Srsf2
Given the genetic heterogeneity of primary patient samples as well as the fact that stable overexpression of spliceosomal proteins, even in wild-type (WT) form, is poorly tolerated (Lareau et al., 2007), we first generated a murine model for conditional expression of the commonly occurring SRSF2 P95H mutation from the endogenous murine locus of Srsf2 (Figure 1A and Figure S1A–B). Mice heterozygous for the Srsf2P95H allele (Srsf2P95H/WT) were crossed to Mx1-cre transgenic mice (Kuhn et al., 1995) on a C57BL/6 background to allow for inducible expression of Cre recombinase following intraperitoneal injection of polyinosine-polycytosine (pIpC) (12 µg/g every other day for three days by injection as previously described (Moran-Crusio et al., 2011)) (Figure S1C–D and Supplemental Experimental Procedures). mRNA sequencing (RNA-seq) analysis of hematopoietic stem/progenitor cells (HSPCs) two weeks after the last pIpC injection of 6-week-old Mx1-cre Srsf2P95H/WT and Mx1-cre Srsf2 WT control mice confirmed heterozygous expression of the mutant allele in equal proportion to the remaining WT Srsf2 allele in Mx1-cre Srsf2P95H/WT mice (Figure 1B).
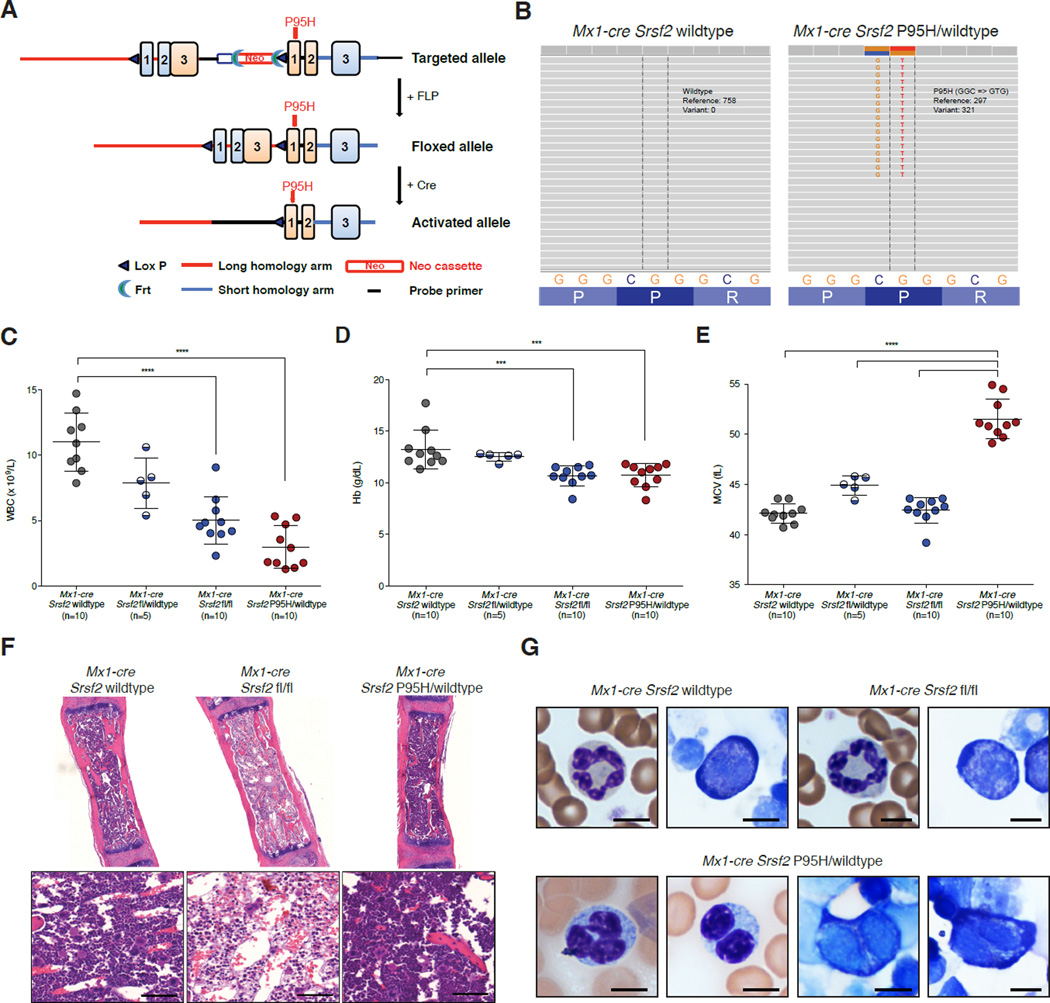
Figure 1. Conditional expression of Srsf2P95H results in myeloid dysplasia, a phenotype distinct from heterozygous or homozygous loss of Srsf2.
(A) Depiction of the Srsf2P95H allele. (B) RNA-seq of LSK cells in Mx1-cre Srsf2WT and Mx1-cre Srsf2 P95H/WT mice. (C) White blood cell (WBC) count, (D) hemoglobin (Hb), and (E) mean corpuscular volume (MCV) of red blood cells of CD45.1 recipient mice 18 weeks following noncompetitive transplantation of bone marrow from CD45.2+ Mx1-cre Srsf2WT, Mx1-cre Srsf2fl/WT, Mx1-cre Srsf2fl/fl, and Mx1- cre Srsf2 P95H/WT mice (n=10 mice/genotype for all genotypes except Mx1-cre Srsf2fl/WT where n=5; pIpC was administered to recipient mice 4 weeks following transplantation). (F) H&E staining of femurs (Bars: 50 µm) and (G) peripheral blood smears from Mx1-cre Srsf2WT, Mx1-cre Srsf2fl/fl or Mx1-cre Srsf2 P95H/WT mice (Bars: 10 µm). A representative neutrophil (left) and erythroid precursor (right) is shown for Srsf2 WT and KO mice. Mx1-cre Srsf2P95H cells were marked by hypolobated and hypogranulated neutrophils (left 2 photos) and nuclear irregularities as well as cytoplasmic vacuolization and blebbing of erythroid precursors (2 rightmost photos). Error bars represent mean ± SD; ***p < 0.001; ****p < 0.0001. See also Figure S1.
It is currently unknown if the heterozygous SRSF2 P95H mutation confers a gain-offunction, haploinsufficient loss-of-function, or dominant-negative loss-of-function. We therefore compared expression of the Srsf2P95H mutation with conditional loss of Srsf2 in vivo (Wang et al., 2001). Bone marrow (BM) mononuclear cells (MNCs) from 6-week-old CD45.2 Mx1-cre Srsf2 WT, Mx1-cre Srsf2fl/WT (heterozygous floxed mice for inducible deletion of 1 copy of Srsf2), Mx1-cre Srsf2fl/fl (homozygous floxed mice for inducible deletion of both copies of Srsf2), and Mx1-cre Srsf2 P95H/WT were transplanted into lethally irradiated congenic CD45.1 recipient mice followed by pIpC injection 4 weeks later (note that all mice were treated with pIpC to control for any potential phenotypic effects of pIpC administration on biological or splicing phenotypes). This was done to assess for the phenotypic effects of Srsf2 deletion or mutation in a hematopoietic cell-autonomous manner. Western blot (WB) analysis revealed deletion of Srsf2 in BM MNCs from Mx1-cre Srsf2fl/fl mice and normal total Srsf2 levels in Mx1-cre Srsf2 P95H/WT BM MNCs (Figure S1E). Significant leukopenia and anemia were seen in mice with homozygous Srsf2 deletion or heterozygous expression of the P95H mutation 18 weeks post-transplant (Figure 1C–D) that was also seen at earlier time points (Figure S1F–G). The presence of similar cytopenias in mice bearing homozygous Srsf2 deletion and heterozygous Srsf2P95H point mutation suggested a possible dominant-negative function imposed by the P95H mutation. However, the anemia in Srsf2P95H mice was characterized by increased mean corpuscular volume (MCV) of red blood cells relative to WT mice or mice with loss of 1–2 copies of Srsf2 (Figure 1E). Moreover, histological assessment of mice 14 weeks post-pIpC revealed prominent BM aplasia in Srsf2 homozygous knockout (KO) mice, whereas mice expressing the heterozygous P95H mutation had normal BM cellularity (Figure 1F). Platelet counts were normal in Srsf2P95H mutant mice at all time points examined (Figure S1H).
Given that macrocytic anemia, a hallmark of anemia in MDS, was present in Srsf2P95H mutant mice we next performed cytological examination of peripheral blood and bone marrow smears from Mx1-cre Srsf2 WT, Mx1-cre Srsf2fl/fl, and Mx1-cre Srsf2 P95H/WT mice to assess for morphologic dysplasia. This revealed prominent myeloid and erythroid dysplasia in Srsf2P95H mice but not in Mx1-cre Srsf2 WT or Mx1-cre Srsf2fl/fl mice (Figure 1G and Figure S1I). Myeloid dysplasia was apparent based on detection of hypolobated and hypogranulated neutrophils while erythroid dysplasia was evident based on nuclear irregularities and cytoplasmic vacuolization and blebbing in erythroid precursors. Overall, these results indicate that mutations in Srsf2P95H result in morphologic dysplasia and cytopenias with preserved marrow cellularity, features which are characteristic of human MDS, whereas complete loss of Srsf2 is incompatible with hematopoiesis.
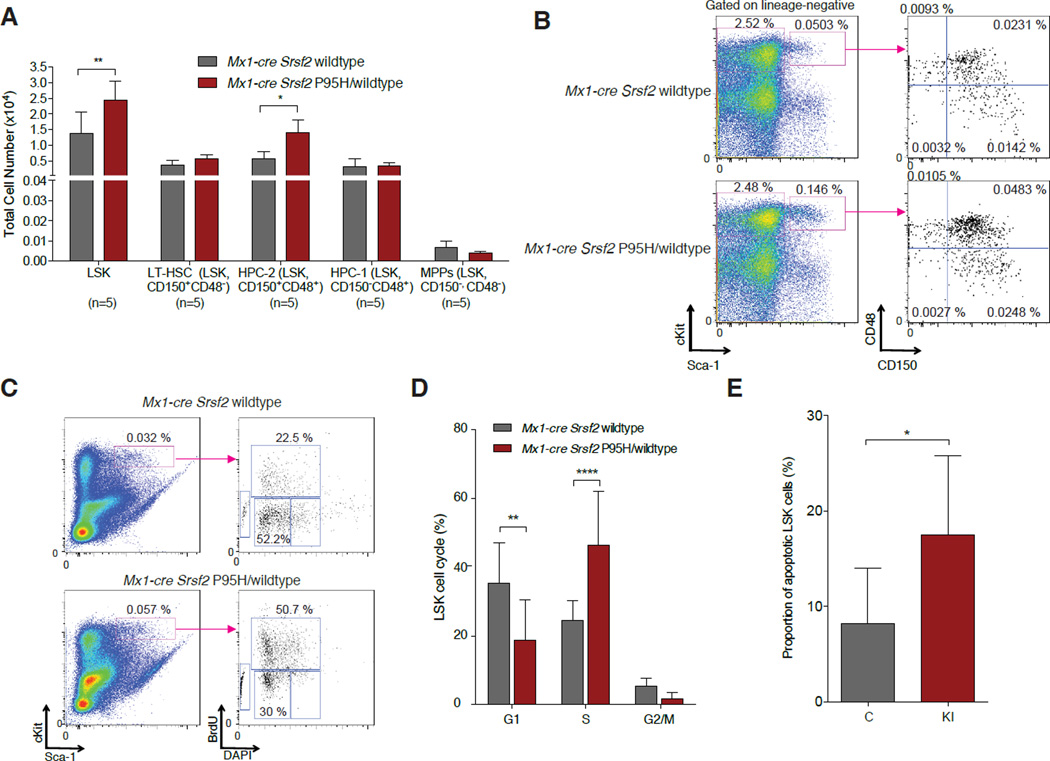
Given that mutations in SRSF2 occur as early genetic events in MDS pathogenesis (Papaemmanuil et al., 2013) and that MDS is characterized by expansion of HSPCs, we next examined HSPC numbers and function in Srsf2P95H mice. Analysis of CD45.2+ HSPC subsets from Mx1-cre Srsf2 P95H/WT mice and littermate controls 14 weeks after pIpC injection revealed expansion of lineage-negative Sca1+ c-Kit+ (LSK) and restricted hematopoietic progenitor cells (LSK CD48+ CD150+; hematopoietic progenitor cell fraction 2 (HPC-2) (Oguro et al., 2013)) in mutant mice relative to controls (Figure 2A–B). Similar LSK expansion was seen in spleens of Srsf2P95H mutant mice (although splenomegaly was not observed up to 20 weeks post-pIpC) (Figure S2A–B). Because the detection of increased HSPCs in Srsf2P95H mutant mice appeared paradoxical given the decreased peripheral blood counts in these same mice, we next examined cell cycle kinetics and apoptosis of Srsf2 mutant HSPCs. Indeed, Srsf2P95H LSK cells were characterized by an increase in the proportion of cells in S-phase as well as in early apoptosis (Figure 2C–E). Despite HSPC expansion in Srsf2P95H mutant mice, purified LSK cells from mice with homozygous Srsf2 deletion or heterozygous Srsf2P95H mutation had similarly impaired colony formation and serial re-plating capacity in vitro (Figure S2C).
Figure 2. Conditional expression of Srsf2P95H results in expansion of hematopoietic stem and progenitor cells with increased cell proliferation and apoptosis.
(A) Enumeration and (B) FACS analysis of BM LSK cells, long-term hematopoietic stem cells (LT-HSC), restricted hematopoietic progenitor cell fractions 1 (HPC-1) and 2 (HPC-2) and multipotent progenitor (MPP) cells (Oguro et al., 2013) in 12-week old Mx1-cre Srsf2 WT and Mx1-cre Srsf2 P95H/WT mice (n=5 mice/genotype). (C) Cell cycle analysis of LSK cells from Mx1-cre Srsf2WT or Mx1-cre Srsf2 P95H/WT mice with in vivo BrdU administration. Representative FACS plot analysis showing gating on LSK cells followed by BrdU versus DAPI stain is shown on the left. (D) Relative quantification of the percentage of LSK cells in S, G2M, and G1 phase is shown on the right (n=8 mice per group). (E) Relative quantification of the percentage of Annexin V+/DAPI- LSK cells (n=8 mice/genotype). Error bars represent mean ± SD; *p < 0.05; **p < 0.01; ****p < 0.0001. See also Figure S2.
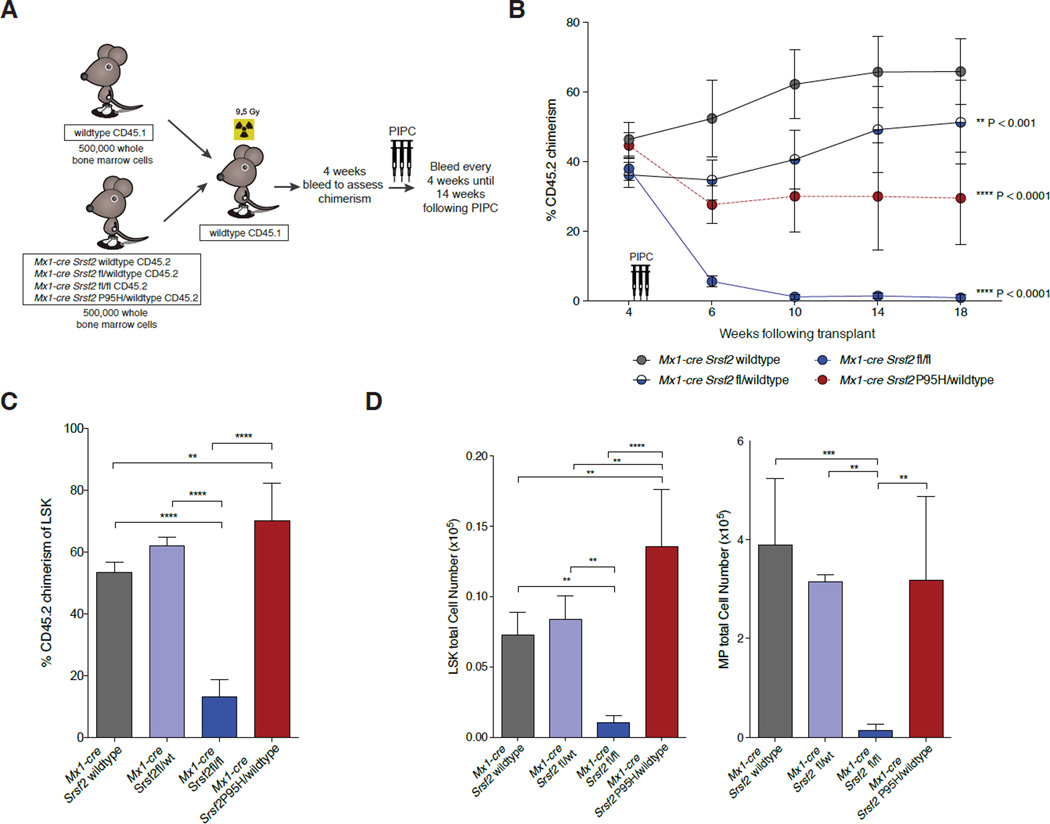
To assess the functional effects of Srsf2 alterations on HSC self-renewal in vivo, we next compared Srsf2 heterozygous KO, homozygous KO, and heterozygous P95H mutant mice in competitive transplantation assays (Figure 3A). Equal numbers of BM MNCs from CD45.1 WT mice and CD45.2 Mx1-cre Srsf2 WT, Mx1-cre Srsf2fl/WT, Mx1-cre Srsf2fl/fl, or Mx1-cre Srsf2 P95H/WT mice were transplanted into lethally irradiated CD45.1 mice followed by pIpC injection 4 weeks later. Assessment of peripheral blood chimerism monthly thereafter revealed a complete loss of CD45.2 chimerism in mice transplanted with Mx1-cre Srsf2fl/fl cells and significant decrease in chimerism in mice transplanted with Mx1-cre Srsf2 P95H/WT cells (Figure 3B, Figure S3A–B). However, analysis of BM LSK chimerism 18 weeks post-transplant revealed an increase in CD45.2+ HSPCs derived from Srsf2P95H mice relative to other groups and near complete absence of CD45.2+ HSPCs from Srsf2fl/fl mice (Figure 3C–D, Figure S3C). Serial competitive transplantation of whole bone marrow from Srsf2P95H, Srsf2 heterozyous KO, and Srsf2 WT primary recipient transplanted mice continued to reveal impaired reconstitution capacity of Srsf2P95H mutant mice relative to Srsf2 heterozygous KO or control mice (Figure S3D). Of note, colony assays and competitive transplantation experiments were performed using multiple genotypes of control mice (Cre-negative Srsf2P95H mice as well as Mx1-cre Srsf2 WT mice; Figure S2C and S3E) to control for any possible confounding effect of Cre expression or the presence of the unexcised P95H knockin allele.
Figure 3. Srsf2P95H mutation impairs hematopoietic stem cell self-renewal in a manner distinct from Srsf2 loss.
(A) Depiction of competitive bone marrow (BM) transplantation assay. (B) Percentage of CD45.2+ chimerism in the peripheral blood of recipient mice (n=10 mice/genotype). (C) Chimerism and (D) flow cytometric enumeration of CD45.2+ LSK (left) and myeloid progenitor (MP; lineage-negative Sca1-c-Kit+) (right) cells in BM of Mx1-cre Srsf2WT, Mx1-cre Srsf2fl/WT, Mx1-cre Srsf2fl/fl and Mx1-cre Srsf2 P95H/WT mice 14 weeks after pIpC injection. Error bars represent mean ± SD; **p< 0.001, ***p < 0.0002; ****p< 0.0001. See also Figure S3.
The fact that Mx1-cre Srsf2 P95H/WT mice had an increase in HSPCs despite impaired formation of mature peripheral blood cells suggested that mutant Srsf2 was associated with impaired HSPC differentiation. Flow cytometric analysis of mature and intermediate precursor cell subsets in Srsf2P95H mice was therefore performed to identify the stage of impaired hematopoiesis. This revealed that peripheral leukopenia was predominantly due to decreased peripheral blood B-cells, evident at all stages of B lymphopoiesis following the transition of preproB to pro-B cells, in Srsf2P95H mice relative to controls (Figure S3F–G). Moreover, immunophenotypic analysis of intermediate hematopoietic progenitors (Pronk et al., 2007) revealed deficits in early erythroid progenitors in Srsf2P95H mice relative to controls, initiating at the pre-MegE and pre-CFUE stages (Figure S3H–I). Given prior data that homozygous deletion of Srsf2 resulted in defective T cell maturation and CD45 splicing (Wang et al., 2001), we also examined thymic T cell differentiation and CD45 isoform expression in Srsf2P95H mice relative to controls (Figure S3J–K). This revealed no effect of Srsf2P95H mutation on thymic T cell maturation or protein expression of the specific CD45 isoforms previously identified to be downregulated with homozygous deletion of Srsf2 (Wang et al., 2001).
Collectively, biological analysis of Srsf2P95H mutant mice identified phenotypes distinct from mice with partial or complete loss of Srsf2, suggesting that SRSF2 mutations alter SRSF2’s normal function rather than resulting in haploinsufficiency or a dominant-negative function. Of note, despite the impaired hematopoietic differentiation, increase in HSPC subsets, and morphologic dysplasia in Srsf2P95H/WT mice, no Srsf2P95H mutant mice developed acute myeloid leukemia in up to 70 weeks of observation.
SRSF2 Mutations Are Associated with Global Alterations of Gene Expression and Splicing
We next sought to identify the transcriptional and post-transcriptional alterations caused by SRSF2 mutations through RNA-seq of purified LSK and myeloid progenitor (MP; lineage-negative Sca1- c-Kit+) populations. This was performed 4 weeks after pIpC administration. In unsupervised cluster analysis based on coding gene expression, samples clustered first by cell type, then genotype (Figure S4A). Expression of several hematopoietic regulators was altered in Srsf2P95H mutant cells, including up-regulation of Gfi1, Cebpe, and Hoxb2 in LSK cells, down-regulation of Gata1 and Gata2 in MP cells, and down-regulation of Cdkn1a in both populations. In addition, we observed preferential down- versus up-regulation of the expression of coding genes in Srsf2 mutant cells relative to WT (Figure S4B–C). Gene Ontology (GO) analysis revealed an enrichment for down-regulation of genes in both LSK and MP cells involved in the regulation of cell cycle, proliferation, differentiation, and apoptosis (up-regulated genes were not enriched for these processes; Figure S4D)
To identify changes in splicing driven by SRSF2 mutations that might contribute to disease, we augmented our mouse data with RNA-seq data from primary CMML (n=13; 3 with SRSF2 mutation) and acute myeloid leukemia (AML; n=9; 5 with SRSF2 mutation) patient samples (Table S1), as well as K562 cells ectopically expressing an empty vector or a single allele of SRSF2 (WT, P95H, P95L, P95R). In all sequenced patients with SRSF2 mutations, the WT and mutant alleles were expressed at similar levels (Table S1), as was the case for the Srsf2P95H mouse cells (Figure 1). Similarly, isogenic K562 cells with lentiviral expression of WT or mutant SRSF2 cells expressed WT and mutant SRSF2 at roughly equal levels (Figure S4E–G). We quantified global changes in splicing of ~125,000 alternative splicing events and ~160,000 constitutive splice junctions associated with SRSF2 mutations in these five datasets (LSK, MP, CMML, AML, K562). We required a minimum change in isoform ratio of 10% to call an event differentially spliced (where a change in isoform ratio is defined as an absolute, rather than relative, quantity as the increase or decrease in the percentage of all mRNAs transcribed from the parent gene that follow a given splicing pattern). In all datasets, SRSF2 mutations were associated with differential splicing of all classes of splicing events, as well as novel alternative splicing and intron retention of splice junctions annotated as constitutively spliced. However, only a relatively small fraction of alternatively spliced events of any class were affected by SRSF2 mutations (Figure S4H). SRSF2 mutations were associated with a mild bias towards exon skipping, but did not lead to globally increased levels of predicted substrates for degradation by nonsense-mediated decay.
SRSF2 Mutations Alter Exonic Splicing Enhancer Preference While SRSF2 Loss Does Not
Since SRSF2 normally recognizes ESE elements within the pre-mRNA to promote exon recognition (Graveley and Maniatis, 1998; Liu et al., 2000; Schaal and Maniatis, 1999; Zahler et al., 2004), we hypothesized that SRSF2 mutations might alter its normal sequence-specific activity. To test this, we performed an ab initio motif identification screen. We quantified the occurrence of each possible k-mer (k = 4, 5, 6) within cassette exons that were differentially spliced in Srsf2P95H MP cells, and identified k-mers that were enriched or depleted in cassette exons promoted versus repressed in Srsf2P95H cells. We identified enriched and depleted motifs using a non-parametric (Kolmogorov-Smirnov) statistical test with a p-value threshold of 0.05. Significantly enriched k-mers were C-rich, while depleted k-mers were G-rich (Figure S4I–J). We then performed an identical analysis using our K562 data, which likewise identified CCAG and GGTG as the most enriched and depleted consensus motifs, respectively (Figure 4A–B). A recent solution structure of SRSF2 in complex with RNA revealed that SRSF2 has a consensus motif of SSNG (where “S” represents C or G) and efficiently recognizes both CCNG and GGNG (Daubner et al., 2012). Therefore, our ab initio analysis suggested that mutations affecting the P95 residue may alter SRSF2's ability to recognize variants of its normal SSNG motif.
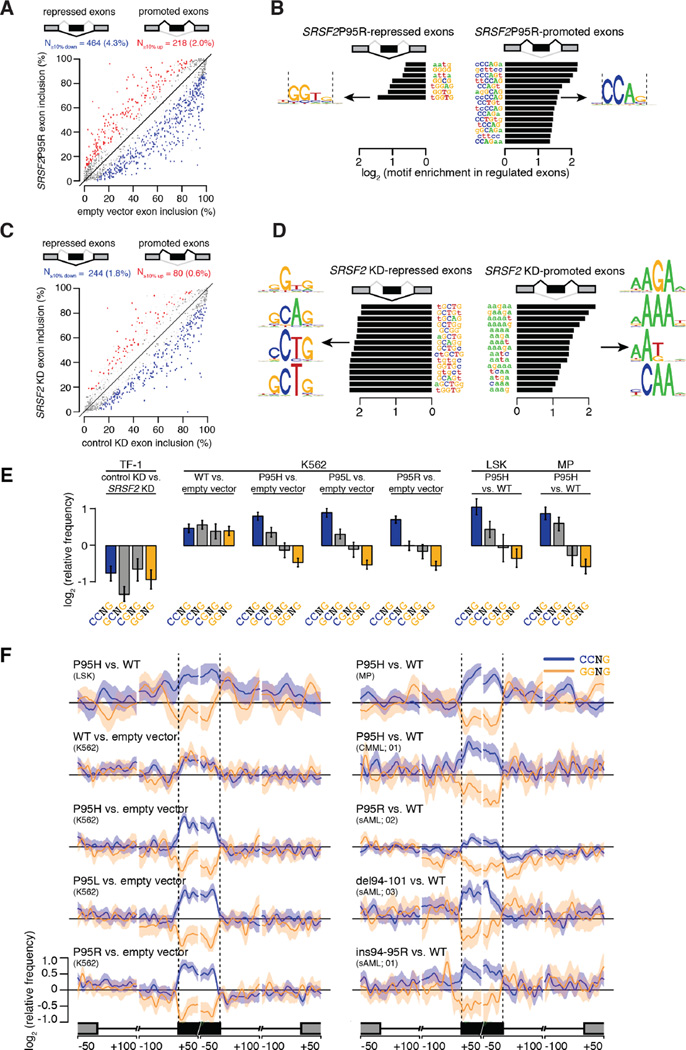
Figure 4. SRSF2 mutations alter exonic splicing enhancer preference.
(A) Scatter plot of cassette exon inclusion in K562 cells expressing empty vector or SRSF2 P95R. Percentages, percent of alternatively spliced cassette exons with increased or decreased inclusion. Red and blue dots represent individual cassette exons that are promoted or repressed in SRSF2 P95R versus empty vector cells, respectively. Promoted and repressed cassette exons are defined as those whose inclusion levels are increased or decreased by ≥10% with a Bayes factor ≥5, as estimated by Wagenmakers’s framework (Wagenmakers et al., 2010). (B) Enriched (right) and depleted (left) k-mers in cassette exons promoted versus repressed in SRSF2 P95R versus WT cells. (C) Scatter plot of cassette exon inclusion in TF-1 cells following transfection with a siRNA against SRSF2 or a control non-targeting siRNA (“KD”, knockdown). Percentages, percent of alternatively spliced cassette exons with increased or decreased inclusion. (D) Enriched (right) and depleted (left) k-mers in cassette exons promoted versus repressed in SRSF2 KD versus control cells. (E) Mean enrichment of all variants of the SSNG motif in cassette exons promoted versus repressed in TF-1 cells following SRSF2 knockdown and K562, LSK, and MP cells expressing WT or mutant SRSF2. Error bars, 95% confidence intervals estimated by bootstrapping. (F) Relative frequency of CCNG and GGNG motifs in cassette exons promoted versus repressed by SRSF2 mutations in LSK and MP cells (top), K562 cells (left), and primary AML and CMML samples with or without SRSF2 mutations (right; sample numbers correspond to patient identifiers in Table S1). Shading, 95% confidence interval by bootstrapping. The cartoon illustrates a portion of a meta-gene containing the differentially spliced cassette exon; from left to right, the features are the upstream exon (gray box) and intron (black line), the cassette exon (black box, vertical dashed lines), and the downstream intron (black line) and exon (gray box). Horizontal axis, genomic coordinates defined with respect to the 5' and 3' splice sites, where 0 is the splice site itself. Vertical axis, relative frequency of the indicated motifs over genomic loci containing cassette exons promoted versus repressed by SRSF2 mutations (log scale). See also Figure S4.
To further explore this hypothesis, we compared the relative enrichment of all four SSNG variants in cassette exons that were differentially spliced upon depletion of SRSF2, overexpression of WT SRSF2, or expression of mutant SRSF2. SRSF2 depletion—achieved by knock down of endogenous SRSF2 in the absence of mutant protein expression (Figure S4K)—caused preferential skipping of cassette exons, consistent with SRSF2’s canonical role in promoting exon recognition (Figure 4C). Ab initio motif analyses identified both C- and G-rich variants of the SSNG motif as the most enriched motifs in cassette exons that were repressed following SRSF2 depletion (Figure 4D). Quantitation of the enrichment of each SSNG variant revealed that all were associated with exon repression following knock down; in contrast, overexpression of WT SRSF2 was associated with enrichment of each SSNG variant (Figure 4E). This data suggests that different SSNG variants function as equally efficacious SRSF2-dependent ESEs, consistent with SRSF2’s in vitro binding specificity (Daubner et al., 2012). In contrast, K562 cells as well as LSK and MP cells expressing mutant Srsf2 exhibited enrichment for CCNG and depletion for GGNG in exons that were promoted versus repressed (Figure 4E).
To test whether this motif enrichment and depletion was due to ESE activity, we computed the spatial distribution of CCNG and GGNG motifs across genomic loci containing cassette exons that were promoted or repressed in association with SRSF2 mutations. CCNG and GGNG were respectively enriched and depleted specifically over cassette exons, and not over the flanking introns or exons. We observed similar motif preferences and distributions in patient transcriptomes (Figure 4F). As CCNG/GGNG motifs were not consistently enriched/depleted in introns flanking differentially spliced cassette exons, and we were unable to identify enriched motifs with ab initio searches in introns, we conclude that differential cassette exon splicing is likely due primarily to altered recognition of exonic motifs. Together, these data reveal spatially restricted enrichment of specific ESEs in association with SRSF2 mutations, and suggest that SRSF2 mutations cause alteration rather than loss of normal ESE recognition activity.
SRSF2 Proline 95 Mutations Alter RNA Binding Specificity By Changing the Conformation of Both RRM Termini
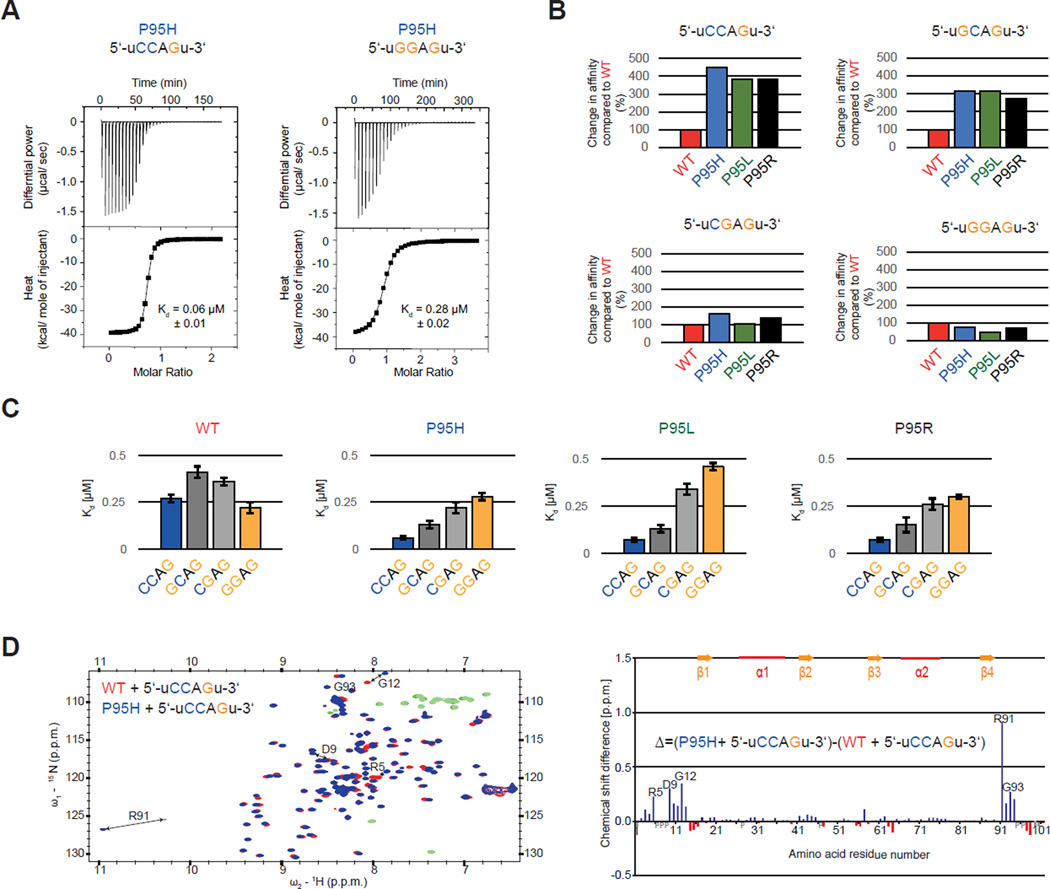
We next tested whether this association between SRSF2 mutations and enrichment/depletion of specific ESEs was due to altered SRSF2:RNA interactions. We purified SRSF2’s RNA recognition motif domain (RRM) as previously described and performed isothermal titration calorimetry (ITC) with the RNA ligand 5'-uCCAGu-3', an optimal SRSF2 target according to the SSNG consensus sequence (Daubner et al., 2012). All three P95 mutations resulted in an increase in binding affinity of 3.9–4.5-fold relative to WT SRSF2 (Figure 5A–B and Figure S5A), consistent with the enrichment for CCNG motifs that we observed in exons promoted by SRSF2 mutations (Figure 4B). We next tested whether P95 mutations resulted in altered RNA binding specificity. In contrast to 5'-uCCAGu-3' RNA, ITC measurements revealed that all three P95 mutants exhibited a 1.2–2.1-fold decrease in binding affinity to the 5'-uGGAGu-3' RNA relative to WT SRSF2 (Figure 5A–B and Figure S5B). ITC measurements using the RNA sequences 5'-uGCAGu-3' and 5'-uCGAGu-3' revealed that G>C substitutions at the second motif position resulted in larger increases in binding affinity than at the first motif position (2.6–3.4-fold versus 1.1–1.8-fold; Figure 5B and Figure S5C–D). The RNA binding preferences measured by ITC were remarkably consistent with ESE enrichment identified by RNA-seq. For each mutant, the level of motif enrichment (Figure 4E) was roughly proportional to the affinity increase (Figure 5C), and the enrichment and affinity measurement supported the same relative preference for each specific motif (CC > GC > CG > GG). This strongly supports the notion that splicing changes caused by P95 mutations are the result of an altered sequence-specificity of the SRSF2 RRM.
Figure 5. Proline 95 mutations change RNA-binding specificity of the SRSF2 RNA recognition motif domain (RRM) in vitro and lead to relocation of the N- and C-termini.
(A) ITC raw data and binding curve for SRSF2 RRM P95H mutant with 5’-uCCAGu-3’ and 5’-uGGAGu-3’ RNA. (B) Change in RNA-binding affinity (%) for SRSF2 RRM P95H (blue), P95L (green) and P95R (black) mutants compared to WT (red) (Daubner et al., 2012), using RNA targets 5’-uCCAGu-3’, 5’-uGCAGu-3’, 5’-uCGAGu-3’ and 5’-uGGAGu-3’. (C) Change in RNA-binding specificity of SRSF2 RRM WT, P95H, P95L and P95R with 5’-UCCAGU-3’ (blue), 5’- UGCAGU-3’ (dark grey), 5’-UCGAGU-3’ (light grey) and 5’-UGGAGU-3’ RNA (orange). (D) (left) Overlay of 2D [15N-1H] HSQCs of wild type (red) and P95H mutant (blue) bound to 5’-UCCAGU-3’ RNA, with negative peaks in green (WT) and light-green (mutant). (right) Difference of the chemical shift perturbations of P95H mutant and wild type. Positive values (blue) with a higher perturbation with the P95H mutant and negative values (red) with a higher perturbation with the WT are shown. Missing assignments are marked with grey bars and proline with a grey P. Residues with the highest difference are depicted in both graph and spectra. See also Figure S5.
P95 is located at the C-terminal end of the SRSF2 RRM and the published solution structure of SRSF2 in complex with 5'-uCCAGu-3' revealed extensive contacts of P95 with the second cytosine (Figure S5E), emphasized by several intermolecular Nuclear Overhauser Effects (NOEs) (Daubner et al., 2012). To test whether SRSF2’s RNA binding surface was altered by P95 mutations, we conducted nuclear magnetic resonance (NMR) titration with the SRSF2 P95H RRM and the 5'-uCCAGu-3' RNA, and assigned the backbone of this complex using standard heteronuclear NMR experiments. Mapping of the chemical shift perturbations revealed that the RNA-binding surface of the RRM is not disturbed by the P95H mutation. However, both termini experienced large changes in their environment (Figure 5D), an observation that held true for all three P95 mutations (Figure S5F). Consistent with our ESE and ITC analyses, this relocation of termini primarily affected the second cytosine, which exhibited the largest chemical shift perturbations of its proton resonances (Figure S5G–H). Smaller changes of chemical shifts were observed when P95 mutants were bound to 5’-uGGAGu-3’ (Figure S5H). Together, our experiments indicate that SRSF2 mutations change SRSF2’s normal RNA-binding affinity and specificity in vitro, likely explaining the widespread alterations in ESE preference that we observed in vivo.
Mutant SRSF2 Promotes Mis-splicing and Degradation of EZH2
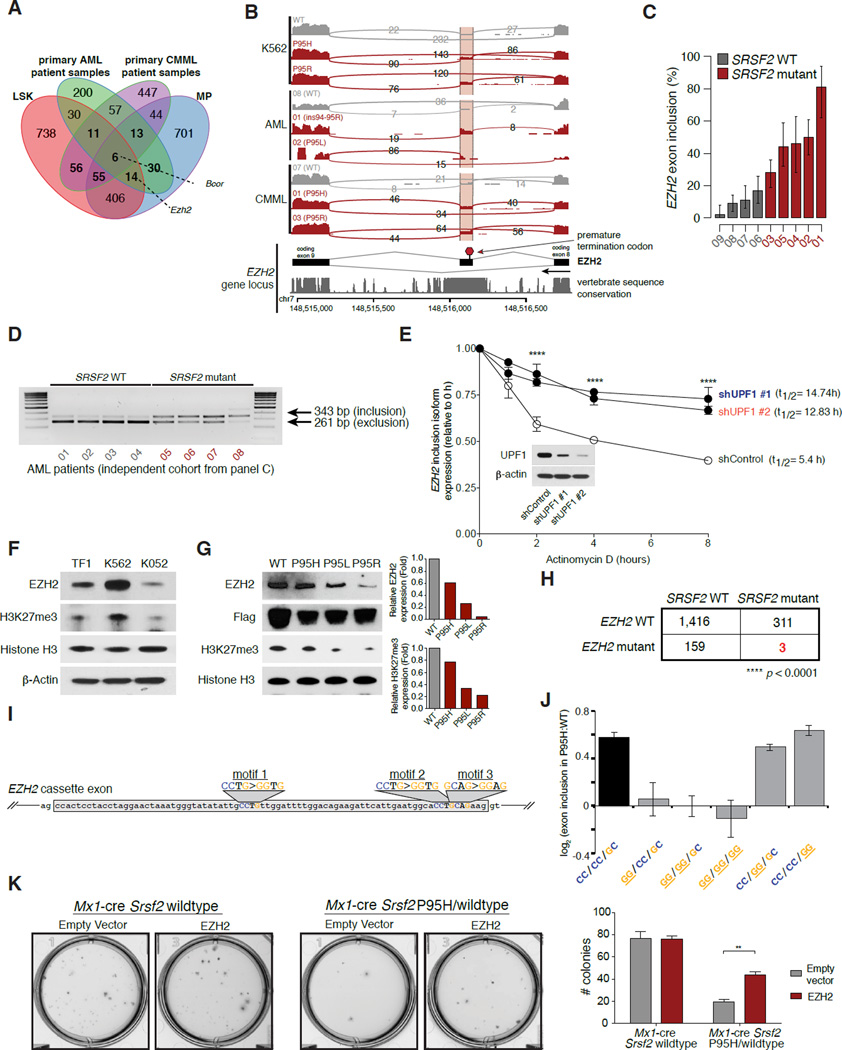
We next used our transcriptome data to identify common changes in splicing driven by SRSF2 mutations that might contribute to disease. Intersection of differentially spliced genes in LSK, MP, CMML, and AML samples identified 75 genes differentially spliced in association with SRSF2 mutations in both LSK and MP cells and at least one primary patient cohort, as well as an additional 97 (LSK) and 87 (MP) genes differentially spliced in one mouse cell population, but not the other, as well as a patient cohort (Figure 6A, Table S2–5). Many of these genes have known importance in myeloid malignancies. For example, SRSF2 mutations promoted inclusion of a highly conserved “poison” cassette exon of EZH2 (Enhancer of zeste homolog 2), and repressed a frame-preserving cassette exon of BCOR (BCL6 corepressor) (Figure 6A and Figure S6A–B). Of note, we did not identify altered splicing of CD45 in SRSF2 mutant cells (Supplemental Tables 2–5) which was previously noted as altered in murine Srsf2 KO hematopoietic cells (Wang et al., 2001).
Figure 6. SRSF2-mutant primary murine and patient samples exhibit convergent splicing alterations.
(A) Intersection of genes exhibiting differential splicing in SRSF2 mutant versus WT mouse LSK and MP cells and primary AML and CMML samples (restricted to orthologous genes). (B) IGV/sashimi plot illustrating the EZH2 cassette exon promoted by SRSF2 mutations in multiple datasets analyzed here (top; patient numbers listed in the Sashimi plot correspond to numbers in Table S1 detailing patient characteristics). The DNA sequence conservation of the locus, as estimated by phastCons (Siepel et al., 2005), across 30 vertebrate species is shown in the track below the Sashimi plot. (C) Bar plot describing the percentage of EZH2 transcripts harboring a specific cassette exon in SRSF2 mutant relative to WT primary AML samples from RNA-seq data. Error bars, 95% confidence intervals. (D) RT-PCR of EZH2 exon inclusion event in an independent set of SRSF2 WT and mutant AML samples. (E) qRT-PCR of EZH2 inclusion isoform in SRSF2 P95H mutant cell line K052 cells with or without UPF1 knockdown and actinomycin D treatment. (F) Western blot (WB) analysis for EZH2 and histone H3 lysine 27 trimethylation (H3K27me3) in SRSF2/EZH2 WT (TF-1, K562) and SRSF2 P95H mutant/EZH2 WT (K052) AML cell lines. (G) WB analysis for EZH2, H3K27me3, and FLAG epitope in K562 cells with lentiviral overexpression of N-terminal FLAG tagged SRSF2 WT, SRSF2P95H, SRSF2 P95L, or SRSF2 P95R (left). Relative quantification of EZH2 protein expression by WB to total histone H3 expression in K562 cells expressing SRSF2 mutants relative to WT is shown on right. (H) EZH2 and SRSF2 mutations are mutually exclusive in sequencing of >1,000 MDS patients (Bejar et al., 2012; Ernst et al., 2010; Haferlach et al., 2014; Muto et al., 2013; Papaemmanuil et al., 2013). (I) Cartoon of EZH2 cassette exon, with SSNG motifs highlighted and mutations to GG equivalents shown. (J) EZH2 cassette exon inclusion for minigenes containing the endogenous cassette exon or a cassette exon with mutation of motifs 1, 2, and/or 3 to the GG equivalent. (K) Photographs (left) and enumeration (right) of c-Kit+/ZsGreen1+ cells from Srsf2 WT or Srsf2P95H mice 14 days after overexpression of empty vector or EZH2 cDNA and plating in methylcellulose media. See also Figure S6 and Table S1–S5.
In order to identify potential functional consequences of recurrent mis-splicing, we focused on the splicing event in EZH2. SRSF2 mutant cells exhibited preferential inclusion of a “poison” cassette exon, which introduces a premature termination codon predicted to result in nonsense-mediated decay (NMD) of EZH2 (Figure 6B–C). Both the poison exon itself and its flanking intronic sequences exhibited high sequence conservation across vertebrates-exceeding the sequence conservation exhibited by the upstream and downstream constitutive coding exons themselves- which is a common feature of physiologically important splicing events (Lareau et al., 2007; Ni et al., 2007) (Figure 6B).
We validated this EZH2 splicing change using both qualitative and quantitative isoform-specific RT-PCR in leukemia cell lines that were WT or mutant for SRSF2 (Figure S6C–D), as well as in an independent panel of primary AML patient samples with or without SRSF2 mutations (n = 8; 4 with SRSF2 mutations; Figure 6D; Figure S6E).
Next, to confirm whether the cassette exon promoted by SRSF2 mutations triggers degradation by NMD, we measured the half-life of the inclusion isoform of EZH2 in SRSF2 P95H cells transfected with a control or anti-UPF1 (a required NMD factor) shRNA following transcriptional shutoff with actinomycin D (t Hoen et al., 2011) (Figure 6E and Figure S6F–G). The fact that the mRNA half-life of the inclusion isoform of EZH2 was lengthened by UPF1 knockdown in these experiments suggests that this particular isoform of EZH2, which is promoted by mutant SRSF2, undergoes NMD. The half-life of a well-characterized NMD substrate of SRSF3 (Lareau et al., 2007; Ni et al., 2007) similarly increased following UPF1 knockdown, confirming that UPF1 knockdown effectively inhibited NMD (Figure S6H).
Next, to identify whether the protein product of EZH2 is altered in SRSF2 mutant cells, we performed WB analysis of a panel of human AML cell lines WT (TF-1, K562) or mutant for SRSF2 (K052) (all WT for EZH2). This revealed lower EZH2 protein levels as well as lower global levels of histone H3 lysine 27 trimethylation (H3K27me3; a methylation mark placed by EZH2) in SRSF2 mutant K052 cells (Figure 6F). To further validate this finding in an isogenic context, we performed WB analysis in K562 cells ectopically expressing WT SRSF2, or SRSF2 P95H/L/R mutant cDNA. This analysis revealed consistent down-regulation of EZH2 protein expression as well as global H3K27me3 in all three SRSF2-mutant samples compared with SRSF2 WT K562 cells (Figure 6G).
Consistent with SRSF2 mutations promoting a disabling splicing change in EZH2, EZH2 loss-of-function mutations are common in MDS. In an analysis of >1,800 MDS patients where EZH2 and SRSF2 were both sequenced, EZH2 loss-of-function mutations were mutually exclusive with SRSF2 mutations (p<0.0001; (Bejar et al., 2012; Ernst et al., 2010; Haferlach et al., 2014; Muto et al., 2013; Papaemmanuil et al., 2013) (Figure 6H).
The above data strongly link SRSF2 mutations to disabling splicing of EZH2. We next sought to examine whether the change in RNA ESE preference induced by SRSF2 mutations caused EZH2 mis-splicing. We therefore cloned the genomic locus containing the EZH2 poison exon and flanking introns and constitutive exons to create a minigene that recapitulates this splicing event. We identified three potential SRSF2-dependent SSNG motifs in the poison exon (CCTG, CCTG, GCAG), one or more of which we expected to be better recognized by mutant SRSF2 than WT SRSF2. We then mutated each motif to the corresponding GG equivalent, both separately and in combination (Figure 6I). Measuring cassette exon recognition in K562 cells expressing WT or mutant SRSF2, we found that the first motif was required for robust splicing change in SRSF2-mutant cells, such that the mutation CCTG>GGTG prevented an increase in poison exon recognition (Figure 6J). We conclude that SRSF2 mutations induce a disabling splicing change in EZH2 in an ESE-dependent manner consistent with altered RNA recognition activity.
We next sought to test whether restoring normally spliced EZH2 mRNA could rescue hematopoiesis in SRSF2-mutant cells. EZH2 full-length cDNA or an empty vector (both in a retroviral ZsGreen1 vector) were overexpressed in c-Kit+ Srsf2P95H or WT cells followed by assessment of methylcellulose colony formation of c-Kit+/ZsGreen1+ cells. EZH2 cDNA was equally overexpressed in Srsf2 mutant and WT cells (Figure S6I) and Srsf2P95H mutant cells overexpressing full-length EZH2 experienced a ~50% increase in colony formation relative to Srsf2P95H mutant cells expressing an empty vector (Figure 6K and Figure S6J). In contrast, EZH2 overexpression had no substantial effect on initial colony formation in Srsf2 WT cells (Figure 6K and Figure S6I). These data identify that restoration of normally spliced EZH2 mRNA in SRSF2 mutant cells at least partially rescues the hematopoietic defects induced by mutant SRSF2.
DISCUSSION
The consistent occurrence of heterozygous point mutations affecting highly restricted residues of spliceosomal proteins strongly suggests a gain-of-function or dominant-negative activity for these mutations in malignant transformation. Here we identify an effect of the SRSF2 P95H mutation distinct from loss of SRSF2, and reveal that mutations in SRSF2 confer an alteration in function that results in key aspects of MDS. This includes an increase in HSPCs in Srsf2P95H mutant mice with impaired differentiation, altered cell cycle kinetics, and increased apoptosis resulting in peripheral cytopenias and morphologic dysplasia. By contrast, WT Srsf2 appears to be constitutively required for hematopoiesis.
Transcriptional analysis of SRSF2 mutant cells revealed that SRSF2 mutations result in genome-wide alterations in ESE preference in both human and murine cells. Biochemical analysis of the interaction of SRSF2 with RNA in cell-free in vitro assays identified an analogous change in specificity of interactions between SRSF2 and pre-mRNA induced by SRSF2 mutations. This altered interaction of mutant SRSF2 with RNA appears to be due to an effect of SRSF2 P95H/L/R mutations on the conformations of the termini of SRSF2’s RRM domain as revealed by NMR spectroscopy. Our genomic and biochemical assays indicate that SRSF2 mutations cause alteration, rather than loss-of-function, driving preferential recognition of cassette exons containing C- versus G-rich ESEs.
The altered pre-mRNA recognition activity of mutant SRSF2 likely underlies the mis-splicing of key transcriptional regulators—several of which have previously been implicated in MDS pathogenesis. This includes promotion of a “poison” exon of EZH2 that undergoes NMD and results in reduced EZH2 protein expression in SRSF2 mutant cells. Loss-of-function mutations in EZH2 occur in the same exact spectrum of myeloid malignancies as SRSF2 mutations (Ernst et al., 2010; Nikoloski et al., 2010) and loss of Ezh2 has been functionally linked to MDS development in vivo (Muto et al., 2013). Moreover, SRSF2 and EZH2 mutations are mutually exclusive in MDS patients (Haferlach et al., 2014; Papaemmanuil et al., 2013), but the basis for this observation was previously unknown. The data here provide a mechanistic basis for this mutual exclusivity as SRSF2 mutations functionally reduce EZH2 protein expression.
In addition to the effects of mutant SRSF2 on EZH2 splicing and protein expression, a number of other genes of known importance in hematopoiesis and malignancy were also consistently differentially spliced in isogenic human cells, primary patient samples, and murine cells bearing mutant SRSF2. These include additional genes mutated in MDS (such as BCOR), genes with importance in hematopoietic stem cell self-renewal (such as IKAROS), and genes critical for cell survival (such as CASPASE 8). Future efforts to understand the functional effects of each of these specific splicing events will be important in further delineating the effects of mutant SRSF2 on MDS pathogenesis as well as possibly providing novel means for therapeutic targeting of SRSF2-mutant cells.
Our studies, which reveal both mechanistic splicing alterations and specific mis-spliced isoforms in SRSF2-mutant cells, may inform therapeutic opportunities for targeting SRSF2-mutant cells. For example, the observations that mutant SRSF2 promotes inclusion of a poison exon in an ESE-dependent manner, and that restoration of normally spliced EZH2 mRNA partially rescues defective hematopoiesis in SRSF2-mutant cells, suggest that normal cellular function may be at least partially restored by manipulating specific pathologic splicing events.
EXPERIMENTAL PROCEDURES
Generation of the Srsf2P95H conditional knock-in mice is described in Supplemental Experimental Procedures. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at Memorial Sloan Kettering Cancer Center.
Patient samples
Studies were approved by the Institutional Review Boards of Memorial Sloan Kettering Cancer Center and Fred Hutchinson Cancer Research Center and conducted in accordance to the Declaration of Helsinki protocol. Informed consents were obtained from all human subjects.
mRNA sequencing
For sorted mouse cell populations, K562 cells, and primary AML and CMML samples, RNA was extracted using Qiagen RNeasy columns. poly(A)-selected, unstranded Illumina libraries were prepared with a modified TruSeq protocol. 0.5× AMPure XP beads were added to the sample library to select for fragments <400 bp, followed by 1× beads to select for fragments >100 bp. These fragments were then amplified with PCR (15 cycles) and separated by gel electrophoresis (2% agarose). 300 bp DNA fragments were isolated and sequenced on the Illumina HiSeq 2000 (~100M 2×49 bp reads per sample).
RNA-seq read mapping
Reads were mapped to the UCSC hg19 (NCBI GRCh37) human genome or UCSC mm10 (NCBI GRCm38) genome assemblies. First, a modified version of RSEM that called Bowtie v1.0.0 with the -v 2 argument was created. This modified RSEM was then called with the arguments --bowtie-m 100 --bowtie-chunkmbs 500 --calc-ci --output-genome-bam on the gene annotation file. Read alignments with mapq scores of 0 and or a splice junction overhang of less than 6 bp were then filtered out. Remaining unaligned reads were then aligned TopHat v2.0.8b with the arguments --bowtie1 --read-mismatches 2 --read-edit-dist 2 --no-mixed --no-discordant --min-anchor-length 6 --splice-mismatches 0 --min- intron-length 10 --max-intron-length 1000000 --min-isoform-fraction 0.0 --no-novel-juncs --no-novel-indels --raw-juncs on the splice junction file (--mate-inner-dist and --mate-std-dev were calculated by mapping to constitutive coding exons with MISO’s exon_utils.py utility). The resulting TopHat alignments were then filtered as for the RSEM-generated alignments. Finally, the RSEM and TopHat aligned were merged to create final BAM files.
Isoform expression measurements
Two different methods were used to quantify isoform ratios. For alternative splicing events from MISO's v2.0 annotation, MISO was used to estimate isoform ratios. For alternative splicing or intron retention of annotated constitutive junctions, junction reads alone were used as previously described (Hubert et al., 2013). To identify differentially expressed events, we required a minimum of 20 identifying reads (supporting either, but not both, isoforms) per event, as well as a change in isoform ratio >= 10%. For the LSK, MP, and K562 data, we used two-sample statistical comparisons (Wagenmakers's framework; Bayes factor >= 5); for the AML and CMML data, we used group statistical comparisons (Mann-Whitney U test; p-value <= 0.05). Real-time PCR used to measure EZH2 cassette exon inclusion described in Supplementary Methods.
RNA-seq data deposition
RNA-seq data from this study are deposited in the Gene Expression Omnibus (GEO) under accession number: GSE65349.
Supplementary Material
HIGHLIGHTS.
Srsf2P95H/wild-type mice develop myelodysplasia while Srsf2-deficient mice do not.
Proline 95 mutations change the RNA binding specificity of SRSF2.
Mutant SRSF2 promotes an isoform of EZH2 that undergoes nonsense-mediated decay.
Restoring EZH2 expression partially rescues hematopoiesis in Srsf2-mutant cells.
SIGNIFICANCE.
Frequent somatic mutations affecting components of the spliceosome have been identified in hematologic malignancies; however, the functional role of these mutations is not known. Here, we identify that commonly occurring mutations in the spliceosomal gene SRSF2 impair hematopoietic differentiation and promote myelodysplasia by altering SRSF2’s preference for specific exonic splicing enhancer motifs. This results in consistent mis-splicing in a manner that promotes the expression of abnormal isoforms of a number of key hematopoietic regulators, some of which have previously been linked to leukemogenesis (including BCOR and EZH2). These data provide a mechanistic basis for the enrichment of spliceosomal mutations in myelodysplasia, and identify altered RNA recognition as an important driver of leukemogenesis.
ACKNOWLEDGMENTS
E.K. is supported by the Worldwide Cancer Research Fund. AR was supported by the NIH/NHLBI (U01 HL099993), NIH/NIDDK (K08 DK082783), the J.P. McCarthy Foundation, and the Storb Foundation. S.H. and O.A.-W. are supported by grants from the Edward P. Evans Foundation. SH was supported by Yale Comprehensive Cancer Center institutional funds. RKB was supported by the Hartwell Innovation Fund, Damon Runyon Cancer Research Foundation (DFS 04-12), Ellison Medical Foundation (AG-NS-1030-13), NIH/NIDDK (R56 DK103854), NIH/NCI recruitment support (P30 CA015704), and Fred Hutchinson Cancer Research Center institutional funds. J.O.I. was supported by a NIH/NCI training grant (T32 CA009657) and NIH/NIDDK pilot study (P30 DK056465). C.L. is supported by a Career Development Award Grant from the Leukemia and Lymphoma Society and an ATIP-Avenir grant from the French Government. O.A.-W. is supported by an NIH K08 Clinical Investigator Award (1K08CA160647-01), a US Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (W81XWH-12-1-0041), the Josie Robertson Investigator Program, and a Damon Runyon Clinical Investigator Award with support from the Evans Foundation. F.H.-T.A. acknowledges support from the NCCR RNA & Disease, funded by the Swiss National Science Foundation and the SNF Sinergia CRSII3_127454. Y.L. and Y.M. were supported by NIH/NIGMS grant R01 GM102869, and by a Senior Research Fellowship grant number 101908/Z/13/Z to Y.M. from the Wellcome Trust. J.D. acknowledges assistance from Dr. Nezih Cereb, HistoGenetics (Ossining, NY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
E.K., J.I., Y.L., G.M.D., F.H.-T.A., S.H., R.K.B, and O.A.-W. designed the study. E.K., S.L., Y.R.C., J.B.M., H.C., M.-K.K., and O.A.-W. performed animal experiments and generated mice. J.I., A.R., M.M. and A.S.Z. generated SRSF2 constructs, K562 cell lines, and CMML RNA-seq data. S.B. and P.S. provided additional SRSF2 constructs. R.K.B. performed RNA-seq analysis. A.R., J.D., and O.A.-W. provided primary patient leukemia samples. C.L. and I.A. provided advice on animal experiments and helped generate RNA-seq data. S.A. and C.Y.P. performed cytopathologic and histopathologic analyses. Y.L., G.M.D., Y.L., Y.M., F.H.-T.A., and S.H. prepared protein and RNA samples for NMR and ITC studies and performed analysis; J.I. created minigenes and conducted splicing assays. E.K., G.M.D., F.H.-T.A., S.H., R.K.B., and O.A.-W. prepared the manuscript with help from all co-authors.
REFERENCES
- Bejar R, Stevenson K, Caughey B, Abdel-Wahab O, Steensma D, Galili N, Raza A, Kantarjian H, Levine R, Neuberg D, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PloS one. 2014;9:e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Molecular cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome research. 2014 doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Molecular and cellular biology. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. The Journal of experimental medicine. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell stem cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RD, Theimer CA, Wu H, Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. Journal of biomolecular NMR. 2004;28:59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell stem cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, Makishima H. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez G, Jares P, Bassaganyas L, Ramsay A, Beà S, Pinyol M, Martínez-Trillos A, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Molecular and cellular biology. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- t Hoen PA, Hirsch M, de Meijer EJ, de Menezes RX, van Ommen GJ, den Dunnen JT. mRNA degradation controls differentiation state-dependent differences in transcript and splice variant abundance. Nucleic acids research. 2011;39:556–566. doi: 10.1093/nar/gkq790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, Finke C, Score J, Gangat N, Mannarelli C, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–1869. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- Wagenmakers EJ, Lodewyckx T, Kuriyal H, Grasman R. Bayesian hypothesis testing for psychologists: a tutorial on the Savage-Dickey method. Cognitive psychology. 2010;60:158–189. doi: 10.1016/j.cogpsych.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Molecular cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Lawrence M, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca D, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato- Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. The Journal of biological chemistry. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, Hricik T, Heguy A, Hedvat C, Gonen M, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.