Abstract
The contribution of disulfide bridges to the thermostability of a type A feruloyl esterase (AuFaeA) from Aspergillus usamii E001 was studied by introducing an extra disulfide bridge or eliminating a native one from the enzyme. MODIP and DbD, two computational tools that can predict the possible disulfide bridges in proteins for thermostability improvement, and molecular dynamics (MD) simulations were used to design the extra disulfide bridge. One residue pair A126-N152 was chosen, and the respective amino acid residues were mutated to cysteine. The wild-type AuFaeA and its variants were expressed in Pichia pastoris GS115. The temperature optimum of the recombinant (re-) AuFaeAA126C-N152C was increased by 6°C compared to that of re-AuFaeA. The thermal inactivation half-lives of re-AuFaeAA126C-N152C at 55 and 60°C were 188 and 40 min, which were 12.5- and 10-folds longer than those of re-AuFaeA. The catalytic efficiency (k cat/K m) of re-AuFaeAA126C-N152C was similar to that of re-AuFaeA. Additionally, after elimination of each native disulfide bridge in AuFaeA, a great decrease in expression level and at least 10°C decrease in thermal stability of recombinant AuEaeA variants were also observed.
Introduction
A disulfide bridge is formed by the oxidation of two thiols each from two cysteines, thus linking the two cysteines and their respective main peptide chains, which can restrict the motion of the unfolded, random coil of protein or stabilize the folded state of protein [1,2]. One disulfide bridge can contribute 2.3–5.2 kcal/mol to the thermodynamic stability of proteins [1,3]. The contribution of disulfide bridges to the stability of proteins can be measured, to a certain extent, by the change in protein thermostability upon introduction or elimination of one or more disulfide bridges. Considerable evidence has demonstrated the thermostability effects of engineered disulfide bridges in protein. For example, upon introduction of a disulfide bridge A162C-K308C in the lipase B (CalB) from Candida antarctica, the half-life of the enzyme was increased by 4.5-fold at 50°C [4]. The optimum temperature of xylanase (TLX) from Thermomyces lanuginosus increased approximately 10°C upon introduction of a disulfide bridge Q1C-Q24C [5]. Conversely, the absence of a disulfide bridge contributed to the increased conformational flexibility and thermolability of the lipase (PFL) from Pseudomonas fragi [6,7].
Feruloyl esterases (FAEs, EC 3.1.1.73) cleave the ester bond between polysaccharides and hydroxycinnamic acid in hemicellulose networks for the subsequent hydrolysis of hemicellulose by hemicellulose-digesting enzymes. Their potential for degradation and reutilization of the natural biomass is significant [8]. Based on substrate preference and primary structure homology, FAEs have been classified into four types: type A, B, C and D [9]. Hitherto, crystallographic structure of a type A FAE from Aspergillus niger has been analyzed. The structure of this enzyme is based on an α/β hydrolase fold and consists of a major nine-stranded mixed β-sheet, two minor two-stranded β-sheet arrangements and seven helixes [10,11]. Moreover, there are three disulfide bridges located in the FAE simulating three legs of a tripod [10]. FAEs that are similar to other biomass-degradation enzymes can be applied in several industries, such as animal feed preparation, papermaking, baking, biofuel, and production of bioactive phenolic components [12,13]. Unfortunately, most wild-type FAEs have poor thermostability, which lowers their tolerance to the high temperatures encountered in bioprocesses, as for example in pulp bleaching and feedstuff preparation. Only a limited number of thermostable FAEs have been reported thus far, and these FAEs are mostly from bacteria, such as type A TtFAE from Thermoanaerobacter tengcongensis, and type B Tx-Est1 from Thermobacillus xylanilyticus [14,15]. Generally, the FAEs from fungi are not thermostable, such as the AfFaeA from Aspergillus flavus and AnFaeA from A. niger [16,17]. Accordingly, it is very important to discover more thermophilic FAEs or to improve the thermostability of mesophilic enzymes and proteins by employing the promising strategy of protein engineering [18,19].
The thermostability of AnFaeA was improved through multiple amino acid substitutions by error prone PCR technique [20]. Some rational design methods have been developed and applied to increase the protein thermostability. The lipase CalB was thermally improved by rational design based on the flexibility of the amino acid residues (B-factor values) and RosettaDesign [21]. The rational engineering of disulfide bridges in protein is another promising strategy that has been used to improve the thermostability of T4 lysozyme [22], and Trichoderma reesei endo-1,4-β-xylanase II [23]. These rational protein engineering methods could greatly decrease the heavy workloads of the researcher. In our previous work, a gene AufaeA encoding a mesophilic type A FAE (AuFaeA) from A. usamii was cloned and expressed in Pichia pastoris [24]. In the present study, the contribution of disulfide bridges to AuFaeA thermostability was studied by introducing an extra disulfide bridge designed by computational prediction or eliminating a native one from the enzyme. This work made a first step for further studies on higher thermostability modification of type A FAEs, especially those from fungi, by other methods, such as N- or C-terminus substitution [25,26] and directed evolution [27].
Materials and Methods
Strains, Plasmids and Culture Media
Escherichia coli JM109 and plasmid pUCm-T (Sangon, Shanghai, China) were used for gene cloning and DNA sequencing. A recombinant T-plasmid, pUCm-T-AufaeA, was constructed and preserved in our laboratory [24]. E. coli DH5α and plasmid pPIC9K (Invitrogen, San Diego, CA, USA) were used for constructing the recombinant expression plasmids. E. coli JM109 and DH5α were cultured in the LB medium (10 g/L tryptone, 5 g/L yeast extract and 10 g/L NaCl, pH 7.2). P. pastoris GS115 and its transformants were cultured and methanol-induced in the YPD, MD, YPD with geneticin G418, BMGY and BMMY media, which were prepared as described in the manual of Multi-Copy Pichia Expression Kit (Invitrogen).
Analysis of Primary and Three Dimensional Structures
Homology sequence search at the NCBI website (http://www.ncbi.nlm.nih.gov/) was performed using the BLAST server, while homology alignment of type A FAE primary structures was analyzed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Using the crystal structure of type A FAE from A. niger (AnFaeA, PDB code: 1USW) as a template, three dimensional (3D) structures of A. usamii AuFaeA and its variants, sharing high primary-structure identities with A. niger AnFaeA, were homologically modeled and optimized using the MODELLER 9.9 program (http://salilab.org/modeller/). The B-factor values of AnFaeA (1USW) were analyzed by B-FITTER software [28]. The 3D structure was visualized using PyMOL software (http://pymol.org).
Location of the Disulfide Bridge Sites
Modeling of Disulfide Bonds in Proteins (MODIP, http://caps.ncbs.res.in/dsdbase/modip.html) and Disulfide by Design (DbD, http://cptweb.cpt.wayne.edu/DbD2/) were used to detect amino acid pairs, in the 3D structure of AuFaeA, where the disulfide bridges were likely to form. Based on the physicochemical properties of the located amino acids and their positions in the 3D structure of AuFaeA, several pairs of amino acids were selected for substitution with cysteine residues, creating a series of candidate variants of AuFaeA.
Introduction of an Extra Disulfide Bridge
The root mean square deviation (RMSD) value is an important index for estimating the thermostability of a protein conformation. RMSD was defined as the Cα-atomic fluctuation parameter of a protein from its original conformation to the changed one at a high temperature and at a certain time. In addition, RMSD had a negative correlation with the thermostability of a protein [29]. To predict the thermostability of AuFaeA and its candidate variants with extra disulfide bridges, their 3D structures were modeled and subjected to molecular dynamics (MD) simulation processes, respectively, at 500 K for 10 ns using GROMACS 4.5 package (http://www.gromacs.org/). The RMSD value was calculated using g_rms software from GROMACS 4.5 package, and statistical analysis was performed using Origin 9 software (http://www.originlab.com/).
Based on the computational prediction, AuFaeAA126C-N152C, a variant of AuFaeA, with the smallest RMSD value was selected. The variant-encoding gene, AufaeA A126C-N152C, was constructed by synchronously mutating Ala126- and Asn152-encoding codons of AufaeA into Cys126- and Cys152-encoding ones. Using pUCm-T-AufaeA as a template, AufaeA A126C-N152C was amplified using a QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with two pairs of forward-reverse PCR primers, A126C and N152C (Table 1), and inserted into plasmid pUCm-T. The resultant recombinant T-plasmid, named pUCm-T-AufaeA A126C-N152C, was transformed into E. coli JM109 and confirmed by DNA sequencing.
Table 1. Primers used for site-directed mutagenesis.
| Primer | Primer sequence (5’-3’) a | Size (bp) |
|---|---|---|
| A126C | ATCCGGACTATTGCCTTACCGTGACA | 26 |
| TGTCACGGTAAGGCAATAGTCCGGAT | 26 | |
| N152C | GCGACATATGACTGCGTCCGTCTGTAC | 27 |
| GTACAGACGGACGCAGTCATATGTCGC | 27 | |
| C29T | ACGCCGACCTAACTAATATTCCATCGACT | 29 |
| AGTCGATGGAATATTAGTTAGGTCGGCGT | 29 | |
| C91T | ACTCTACCTCAAACTAACGATTGCG | 25 |
| CGCAATCGTTAGTTTGAGGTAGAGT | 25 | |
| C234T | ACTGGGGATGAAGTACAGACTTGTGAGGCA | 30 |
| TGCCTCACAAGTCTGTACTTCATCCCCAGT | 30 |
a The mutant codons are boxed.
Elimination of the Native Disulfide Bridges
The contribution of native disulfide bridges to the thermostability of AuFaeA was investigated by constructing three single variants: AuFaeAC29T, AuFaeAC91T and AuFaeAC234T. These mutations were designed by substituting three cysteine residues, Cys29, Cys91 and Cys234, with the corresponding threonine ones, respectively. Their encoding genes, AufaeA C29T, AufaeA C91T and AufaeA C234T, were constructed by site-directed mutagenesis with three pairs of forward-reverse primers C29T, C91T and C234T (Table 1), respectively, using a QuikChange Mutagenesis Kit. The target PCR products were separately purified, and then inserted into pUCm-T. The resultant recombinant T-plasmids, named pUCm-T-AufaeA C29T,-AufaeA C91T and-AufaeA C234T, were transformed into E. coli JM109 and confirmed by DNA sequencing.
Enzyme Activity and Protein Assays
The substrate, p-nitrophenyl ferulate (pNPF), was synthesized with a single-step method as reported previously [30]. FAE activity was determined by measuring the amount of p-nitrophenol (pNP) released from pNPF as described [31], with minor modification. Briefly, the reaction mixture (8 volumes of 100 mM Na2HPO4–citric acid buffer (pH 5.5) containing 2.5% (v/v) Triton X-100, 1 volume of 10 mM pNPF in dimethyl sulfoxide and 1 volume of suitably diluted enzyme) was incubated at 40°C for 10 min. The released pNP was measured at 410 nm using a spectrophotometer. One unit (U) of FAE activity was defined as the amount of enzyme that released 1 μmol pNP per minute under the standard assay conditions as stated above.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the method by Laemmli [32]. The separated peptide bands were visualized by staining with Coomassie Brilliant Blue R-250 (Sigma, St. Louis, MO, USA), and molecular weights were estimated in comparison to the standard protein markers using Quantity One software. The protein content was measured with a BCA-200 Protein Assay Kit (Pierce, Rockford, IL, USA), using bovine serum albumin as the standard.
Expression and Purification of the re-FAEs
AufaeA and its mutant genes were excised from their recombinant T-plasmids by digestion with EcoRI and NotI, respectively. The isolated genes were inserted into pPIC9K, and then transformed into E. coli DH5α. The resultant recombinant expression plasmids, designated as pPIC9K-AufaeA,-AufaeA A126C-N152C,-AufaeA C29T,-AufaeA C91T and-AufaeA C234T, were separately linearized with SalI and electroporated into P. pastoris GS115. All P. pastoris transformants were respectively inoculated on YPD plates with increasing concentrations of G418 for the screening of multiple copies of integrated FAE genes. P. pastoris transformed with pPIC9K without AufaeA or any of the mutants was used as a control (P. pastoris GSC). Expression of respective FAE genes in P. pastoris was performed according to the instructions of Multi-Copy Pichia Expression Kit (Invitrogen) with minor modification [33].
The transformed P. pastoris was induced by 1% methanol for 72 h, and the expressed re-FAE in the supernatant was salted out by adding ammonium sulfate (NH4)2SO4 to 75% saturation. The collected precipitate was dissolved and dialyzed in 20 mM Na2HPO4–citric acid buffer (pH 5.5). The dialyzed solution was concentrated by ultrafiltration using a 10-kDa cut-off membrane (Millipore, Billerica, MA, USA) and was loaded on a Sephadex G-50 column (Amersham Pharmacia Biotech, Uppsala, Sweden; 1.6 × 80 cm), followed by elution with the same buffer at a flow rate of 0.4 mL/min. Aliquots of 2 mL eluent containing re-FAE were pooled and concentrated for further studies.
Thiol Titration
Guanidine hydrochloride (5.5 M, Sigma) was added for 15 min to denature the purified re-AuFaeA or re-AuFaeAA126C-N152C (final concentration of 2 mg/mL) [34]. Thiol titration was performed by incubating thirty parts of denatured re-FAE with one part of 4 mg/mL dithionitrobenzoic acid in 0.25 M Tris-HCl buffer (pH 8.0) for 15 min at 25°C, and the absorbance was measured at 410 nm in a spectrophotometer [35]. The wild-type re-AuFaeA containing one free cysteine at position 235 was used as a control. The free cysteine content of each re-FAE was calculated from a cysteine concentration-OD410 standard formula, which was established by testing the OD410 of cysteine concentration at a range from 0 to 0.4 mM. The number of disulfide bridges in the re-FAEs was deduced from the different value of the number of total cysteines in protein primary structure and free ones in 3D structure at the tested concentration.
Temperature Optimum and Thermal Inactivation Half-life
Purified re-AuFaeA and its variants were functioned in the standard enzyme activity assay conditions except the changed temperatures to measure the temperature optima, and were incubated in the absence of substrate at different temperatures for 60 min or longer time to estimate their thermostability. The thermal inactivation half-life (t 1/2) was defined as the time when the residual activity of the re-FAE, determined under the standard conditions, was 50% of its original activity.
Enzyme Kinetic Parameters
The hydrolytic reaction rates (U/mg) of re-AuFaeA and re-AuFaeAA126C-N152C were separately determined under the standard assay conditions, with a wide-range of pNPF concentration (0.5 to 20.0 mM). Data were fitted to the Michaelis-Menten equation to generate K m and k cat values, using Graph-Pad Prism 5.0 software.
Results and Discussion
Location of the Disulfide Bridge Sites
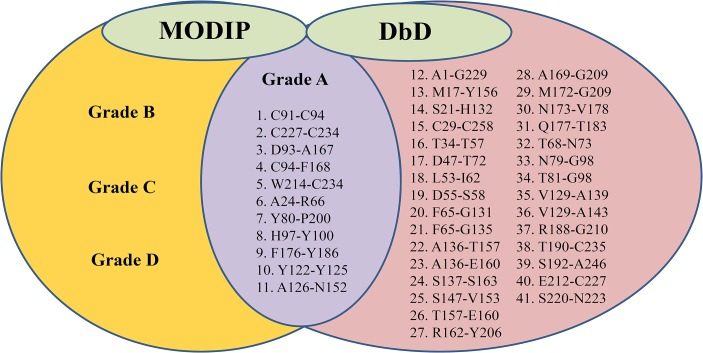
The primary structures of the AuFaeA and AnFaeA (PDB code: 1USW) were highly homologous (98.5%) in identity. Identification of three native disulfide bridges, C29-C258, C91-C94 and C227-C234, in AuFaeA (Fig 1A), was an essential step for engineering the disulfide bridges to improve AuFaeA thermostability. DbD and MODIP have been previously applied to successfully improve the stability of xylanase from Bacillus stearothermophilus, lipase from Rhizomucor miehei and acetylcholinesterase from Drosophila melanogaster [35–37]. In this study, DbD suggested 41 pairs of amino acid sites for possible disulfide bridges (Fig 2), while 69 amino acid pairs were ranked from grades A to D by MODIP (data not shown) based on their likelihood to form disulfide bridges. Eleven pairs of amino acids were identified by both DbD and MODIP for their highest possibilities to form disulfide bridges (Fig 2). Upon further analysis, 6 predicted pairs, C91-C94, C227-C234, D93-A167, C94-F168, W214-C234 and H97-Y100, were rejected as follows: the first two pairs were native disulfide bridges; the following three pairs were located near the native ones and could affect the protein conformation; and the last pair was located near a catalytic triad with a distance of 6 Å, which could influence the enzymatic activity. Ultimately, the remaining 5 pairs, A24-R66, Y80-P200, F176-Y186, Y122-Y125 and A126-N152, were evaluated by MD simulation.
Fig 1. The modeled 3D structures of AuFaeA and AuFaeAA126C-N152C.
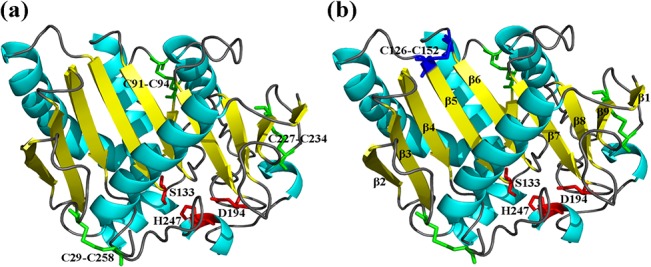
The 3D structures of AuFaeA and AuFaeAA126C-N152C were modeled using the crystal structure of AnFaeA (PDB code: 1USW) as template. (a) The catalytic triad S133-H247-D194 and three native disulfide bridges C29-C258, C91-C94, C227-C234 are separately marked in red and green. (b) The extra disulfide bridge C126-C152 located in AuFaeAA126C-N152C is marked in blue.
Fig 2. Alignment of amino acid sites for disulfide bridges predicted by MODIP and DbD.
The amino acid sites predicted by DbD are shown in the right (pink) oval. The amino acid sites predicted by MODIP are shown in the left (yellow) oval. The middle (purple) oval shows 11 amino acid sites predicted by both DbD and MODIP that have the highest possibilities of disulfide bridge formation.
Introduction of the Extra Disulfide Bridge
The RMSD value is an important index for evaluating the conformational flexibility of protein at high temperature [29]. A hybrid xylanase AEXynM, with an increased melting temperature (T m) of 34°C, was more thermostable than its wild type, which was consistent with the evaluation of RMSD values [26]. In this study, the RMSD values of AuFaeAA24C-R66C, AuFaeAY80C-P200C, AuFaeAF176C-Y186C and AuFaeAY122C-Y125C (selected as an example shown in Fig 3) were higher, while those of AuFaeAA126C-N152C were lower than those of AuFaeA, after equilibration (Fig 3A). Simultaneously, the distributions of RMSD values of AuFaeAA126C-N152C, AuFaeAY122C-Y125C and AuFaeA were statistically analyzed and were mainly concentrated on 0.475 Å, 0.825 Å and 0.525 Å, respectively (Fig 3B). The data indicated that AuFaeAA126C-N152C was less flexible and more rigid than AuFaeA and AuFaeAY122C-Y125C. Because the rigidity of a protein was positively related to its thermostability [38], the variant AuFaeAA126C-N152C was predicted to be more thermostable than the wild-type AuFaeA. Therefore, the amino acid pair A126-N152 was selected for substitution with cysteine residues to construct the extra disulfide bridge in AuFaeA. After site-directed mutagenesis, the mutant gene was obtained and confirmed by DNA sequencing.
Fig 3. Calculation and distribution of the RMSD values.
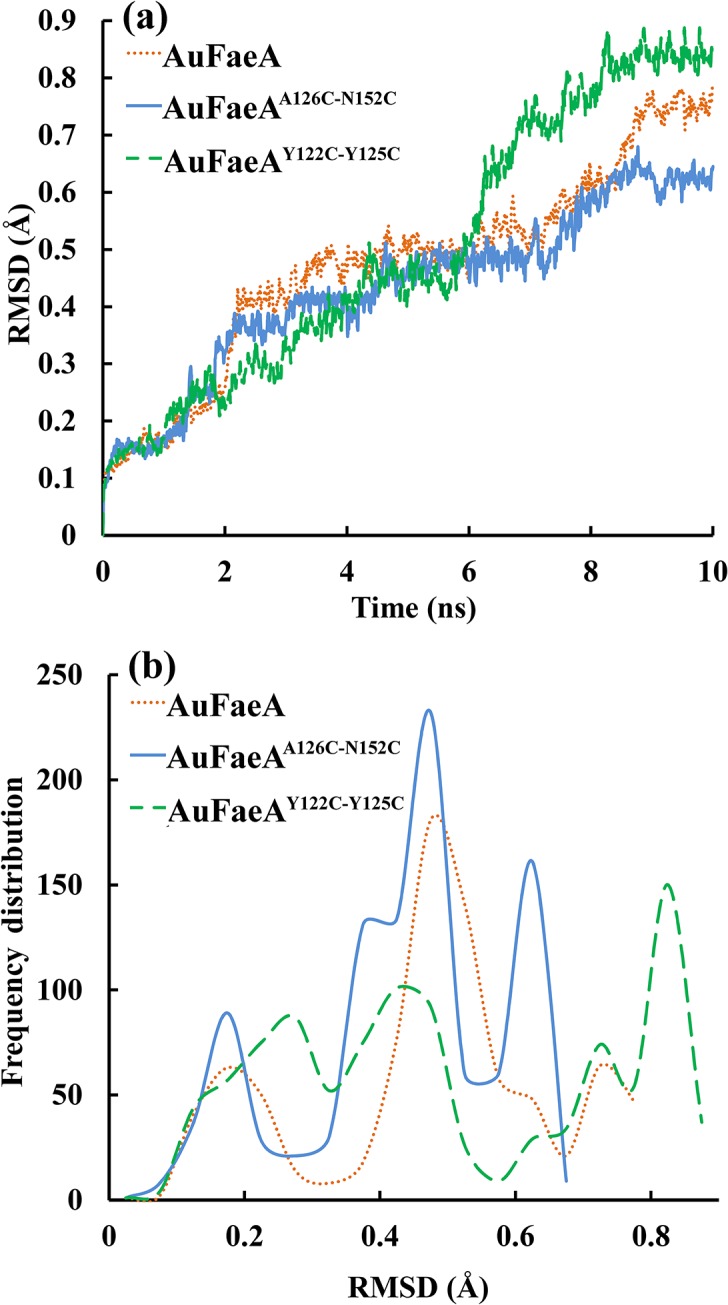
Graphical representation of: (a) RMSD values of AuFaeA (dotted line), AuFaeAA126C-N152C (solid line), and AuFaeAY122C-Y125C (dashed line), respectively, after MD simulation processes at 500 K for 10 ns. (b) Distribution of RMSD values of AuFaeA (dotted line), AuFaeAA126C-N152C (solid line), and AuFaeAY122C-Y125C (dashed line). MD, molecular dynamics; RMSD, root mean square deviation.
Expression and Purification of the re-AuFaeA and re-AuFaeAA126C-N152C
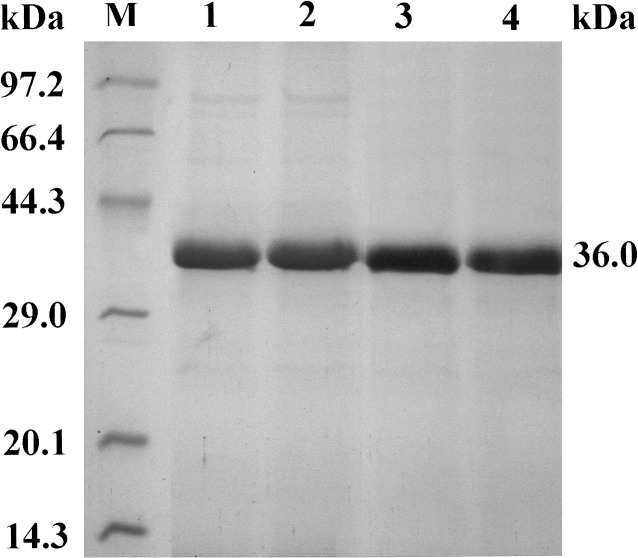
An advantage of the P. pastoris expression system is the high purity of the expressed recombinant protein, as described in the Multi-Copy Pichia Expression Kit (Invitrogen, USA). Purities of recombinant A. usamii xylanase and A. sulphureus β-mannanase expressed in P. pastoris GS115 and X-33 have been reported to be as high as 90 and 97%, respectively [39,40]. In this work, after the transformed P. pastoris was induced by 1.0% methanol for 72 h, the content of expressed re-AuFaeA or re-AuFaeAA126C-N152C was more than 88% in the cultured supernatant (Fig 4, lane 1 and 2). The activities of the expressed re-AuFaeA and re-AuFaeAA126C-N152C were 11.03 and 13.73 U/mL, respectively, which were slightly lower than that (16.6 U/mL) of re-AnFaeA [41]. The specific activities of purified re-AuFaeA and re-AuFaeAA126C-N152C, towards pNPF, were 50.2 and 59.7 U/mg, respectively. SDS-PAGE assay displayed single protein bands of purified re-AuFaeA and re-AuFaeAA126C-N152C, with apparent molecular weights of approximately 36.0 kDa (Fig 4, lane 3 and 4).
Fig 4. SDS-PAGE analysis of the re-AuFaeA and re-AuFaeAA126C-N152C.
Lane M, standard protein marker; lane 1 and 2, the cultured supernatants of re-AuFaeA and re-AuFaeAA126C-N152C; lane 3 and 4, the purified re-AuFaeA and re-AuFaeAA126C-N152C with same molecular weights of 36.0 kDa.
Formation of the Extra Disulfide Bridge
Introduction of the engineered disulfide bridge was verified by thiol titration of the purified re-AuFaeA and re-AuFaeAA126C-N152C at concentrations of 2 mg/mL (0.056 mM) under the denaturing conditions. Based on the cysteine standard formula y = 0.8727x + 1.3972 (x: cysteine concentration, mM; y: OD410), the OD410 at the cysteine concentration of 0.056 mM was 1.446. The detected OD410 (1.452 and 1.437) of purified re-AuFaeA and re-AuFaeAA126C-N152C were almost unanimous with the standard value (Table 2). Since re-AuFaeA containing one free cysteine at position 235 was used as the control, its absorption of 1.437 indicated that AuFaeAA126C-N152C also had one free cysteine at the same position. In other words, with the exception of the native disulfide bridges, a new variant with an extra disulfide bridge C126-C152 was successfully created.
Table 2. Deduced number of free cysteines and disulfide bridges in re-AuFaeA and re-AuFaeAA126C-N152C.
| Enzyme (2 mg/mL) | OD410 after DTNB treatment | Deduced number of free cysteines* | Deduced total number of disulfide bridges |
|---|---|---|---|
| re-AuFaeA | 1.452 | 1 | 3 |
| re-AuFaeAA126C-N152C | 1.437 | 1 | 4 |
* Based on the cysteine standard formula y = 0.8727x + 1.3972 (x: cysteine concentration, mM; y: OD410), the OD410 at the cysteine concentration of 0.056 mM was 1.446.
Thermal Properties of the re-AuFaeA and re-AuFaeAA126C-N152C
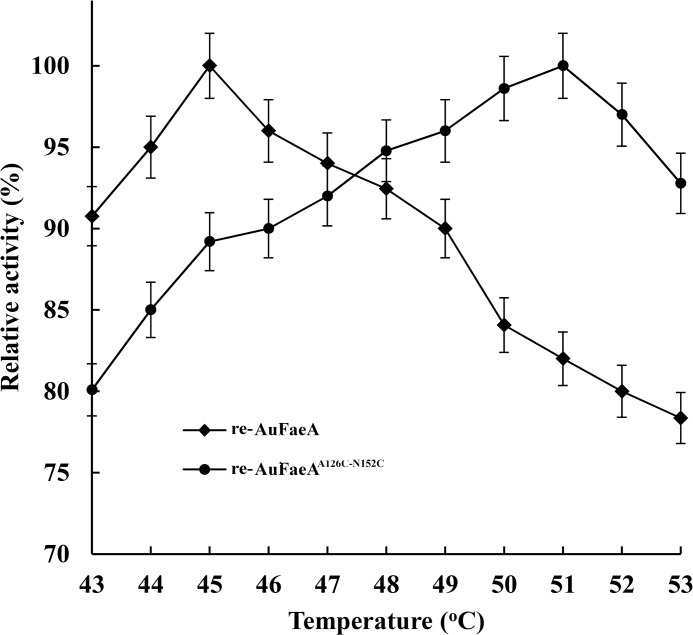
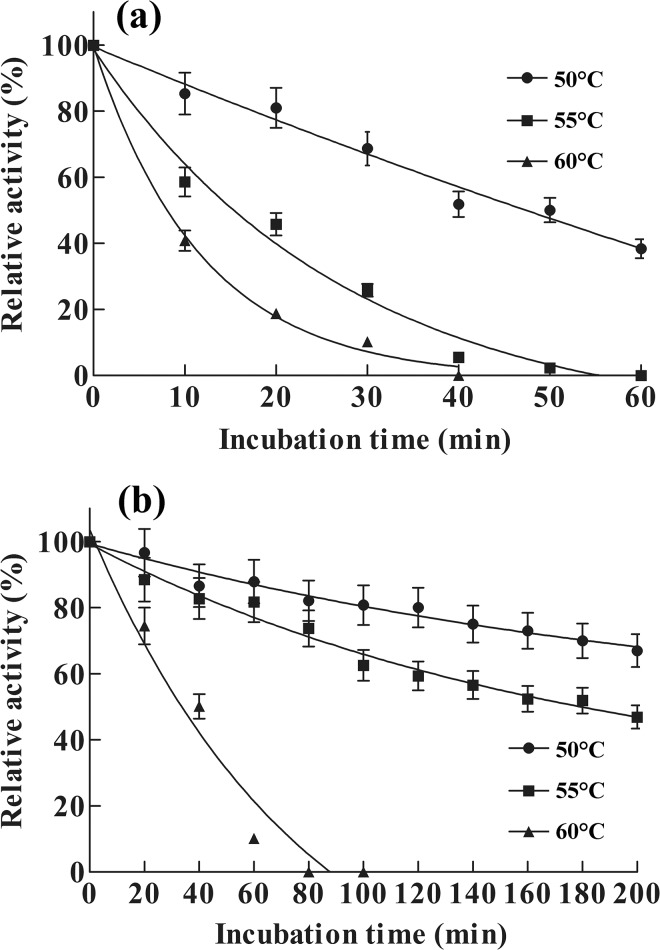
The temperature optimum of re-AuFaeAA126C-N152C was 51°C (Fig 5), which was 6°C higher than that of re-AuFaeA and even higher than those (43–47°C) of recombinant AnFaeA variants [17]. The thermal inactivation half-lives (t 1/2) of re-AuFaeA at 50, 55 and 60°C were 50, 15 and 4 min (Fig 6A), respectively. It entirely lost its activity at 60°C for 40 min. The half-lives (t 1/2) of re-AuFaeAA126C-N152C at 55 and 60°C were 188 and 40 min, which were 12.5- and 10-folds longer than those of re-AuFaeA (Fig 6B), respectively. These results suggested that a properly engineered disulfide bridge is capable of improving the type A feruloyl esterase thermostability.
Fig 5. Temperature optima of the re-AuFaeA and re-AuFaeAA126C-N152C.
The temperature optima were measured under the standard assay conditions, with variable temperatures ranging from 43 to 53°C.
Fig 6. Thermostability of the re-AuFaeA and re-AuFaeAA126C-N152C.
Thermostability of (a) re-AuFaeA and (b) re-AuFaeAA126C-N152C were evaluated by incubating in the absence of substrate at temperatures ranging from 50 to 60°C for different time, respectively, and the residual enzyme activities were measured under the standard assay conditions.
Kinetic Parameters
The kinetic parameters of re-AuFaeA and re-AuFaeAA126C-N152C were characterized with purified enzymes, using pNPF as the substrate (Table 3). The re-AuFaeAA126C-N152C increased Michaelis constant (K m) by approximately 1.6-fold compared to that of the re-AuFaeA, indicating a possible decrease in substrate affinity. However, the re-AuFaeAA126C-N152C also increased catalytic turnover frequency (k cat) by approximately 1.6-fold compared with that of re-AuFaeA. Consequently, the catalytic efficiency (k cat/K m) of re-AuFaeAA126C-N152C was similar to that of re-AuFaeA. Since the kinetics of both enzymes were done at 40°C showing equal activity, the mutant enzyme was expected to show higher activity at higher temperature.
Table 3. Kinetic parameters based on Michaelis-Menten equation.
| Enzyme | K m (mM) | V max (U mg-1) | k cat (min-1) | k cat/K m (mM-1 min-1) |
|---|---|---|---|---|
| re-AuFaeA | 3.62±0.08 | 225±2.0 | 8093±72 | 2235±72 |
| re-AuFaeAA126C-N152C | 5.98±0.12 | 356±2.0 | 12805±72 | 2141±56 |
Elimination of the Native Disulfide Bridge
It has been reported that, after removal of the disulfide bridge constraints on the Ribonuclease T1 flexibility, the enzyme’s folding and functional abilities were maintained, but its stability was decreased [42]. Similarly, it was deduced that the three native disulfide bridges, C29-C258, C91-94 and C227-C234 located in AuFaeA were most likely contributing to its thermostability. Using site-directed mutagenesis, three different mutants were generated each having one natural cysteine mutated to threonine.
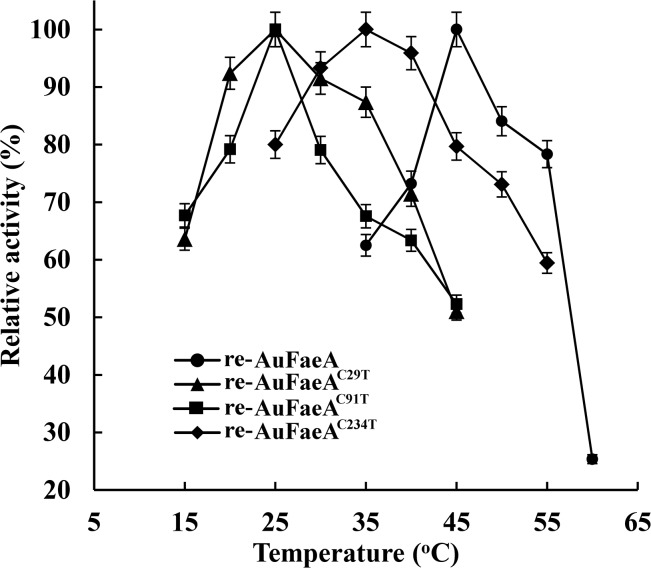
The activities of the expressed re-AuFaeAC29T, re-AuFaeAC91T and re-AuFaeAC234T in the cultured supernatants were up to 0.84, 0.42 and 0.45 U/mL, respectively, which were much lower than that (11.03 U/mL) of re-AuFaeA. The temperature optima of re-AuFaeAC29T, re-AuFaeAC91T and re-AuFaeAC234T were 25, 25 and 35°C, respectively (Fig 7), which were at least 10°C lower than that of re-AuFaeA. Their thermostabilities were also significantly decreased (below 25°C). These results demonstrated the importance of each native disulfide bridge to the thermostability of re-AuFaeA.
Fig 7. Temperature optima of the re-AuFaeA, re-AuFaeAC29T, re-AuFaeAC91T and re-AuFaeAC234T.
The temperature optima were measured under the standard assay conditions, with varying temperatures ranging from 15 to 60°C.
Analysis of the 3D Structures of AuFaeA and Its Variants
The predicted 3D structures of AuFaeA and AuFaeAA126C-N152C consist of a major nine-stranded mixed β-sheet, one minor two-stranded β-sheet arrangement and seven helixes. Three conserved residues in AuFaeA, viz. S133, D194 and H247, constitute a characteristic catalytic triad as that in AnFaeA [11], in which S133 acts as the nucleophile, H247 as the proton acceptor/donor and D194 as the residue stabilizing the histidine. Three native disulfide bridges located in AuFaeA were expected to stabilize its 3D structure from three directions (Fig 1A). Therefore, after the elimination of each native disulfide bridge, the structural conformation of AuFaeA was changed. The region was no longer rigid relatively, which resulted in the significantly decreased thermostability of AuFaeA. Inevitably, the introduced threonine could affect protein stability, because it introduces an additional methyl group, which could cause steric clashes that could further destabilize the enzymes. However, compared to the effect from the elimination of the native disulfide bridges on the stability of AuFaeA, the effect from the introduced methyl group could be weak. The results indicated that native disulfide bridges in type A FAEs from fungi could play an irreplaceable role in the protein stability.
Compared the native disulfide bridges to the engineered one in the AuFaeA, the native bridges mainly stabilize regions with long loops, while the engineered C126-C152 binds the end of the β5 and β6 strands and could thus stabilize the edge of the large β-sheet (Fig 1B). This indicates that at high temperature the protein may unfold from the edge of the β-sheet but that this could be prevented by the disulfide bridge. Additionally, the B-factor values, namely the atomic displacement parameters, of AnFaeA (1USW) amino acid residues were analyzed by B-FITTER software (Fig 8). Compared the B-factors of engineered and native disulfide bridge positions in 1USW, the regions around amino acid sits 126 and 152 have high B-factor values, which indicates that these regions are more mobile. This fits into the strategy of stabilizing proteins by making mutations in regions with high B-factors [28]. It could be deduced that the engineered disulfide bridges in weak spots of the FAEs or other proteins, to some extent, could make them much more thermostable.
Fig 8. The B-factor values of amino acid residues of AnFaeA (PDB code: 1USW).
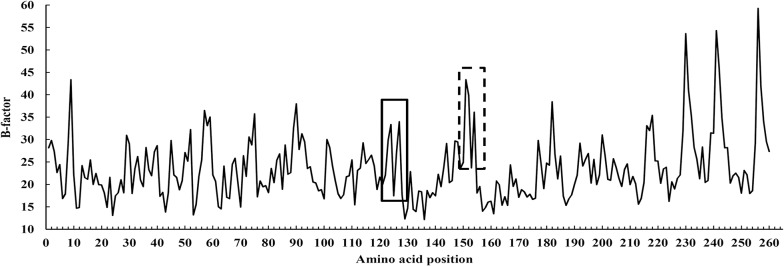
The regions around amino acid residues 126 and 152 with high B-factor values are marked in solid and dashed boxes.
Conclusions
Effects of disulfide bridges on the thermostability of AuFaeA were investigated by either introducing an extra disulfide bridge to, or by eliminating each native disulfide bridge from AuFaeA. Firstly, five pairs of amino acids (A24-R66, Y80-P200, F176-Y186, Y122-Y125 and A126-N152) that can form disulfide bridges in AuFaeA with the highest possibility were located by both MODIP and DbD. Then, the variant AuFaeAA126C-N152C obtained by mutating A126 and N152 to C126 and C152 was predicted to be more thermostable than the wild-type AuFaeA by MD simulation. Finally, AuFaeA, AuFaeAA126C-N152C and three variants (AuFaeAC29T, AuFaeAC91T and AuFaeAC234T) each eliminating a native disulfide bridge were expressed in P. pastoris GS115. Experimental results with the disulfide bridge mutants confirmed that the disulfide bridges contribute significantly to the thermostability of AuFaeA. The computational prediction combined with site-directed mutagenesis to enhance the thermostability and to explore the thermostable mechanism of AuFaeA also can be applied in other proteins or enzymes.
Acknowledgments
The authors are grateful to Prof. Xianzhang Wu (School of Biotechnology, Jiangnan University, Jiangsu, China) for providing technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was financially supported by the Fundamental Research Fund for the Central Universities of China (No. JUDCF13011, JUSRP51412B) (http://yjsb.jiangnan.edu.cn/, http://www.moe.gov.cn/) and the Postgraduate Innovation Training Project of Jiangsu (No. CXZZ13_0757) (http://www.ec.js.edu.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dombkowski AA, Sultana KZ, Craig DB. Protein disulfide engineering. FEBS Lett. 2014; 588: 206–212. 10.1016/j.febslet.2013.11.024 [DOI] [PubMed] [Google Scholar]
- 2. van Beek HL, Wijma Hj, Fromont L, Janssen DB, Fraaije MW. Stabilization of cyclohexanone monooxygenase by a computationally designed disulfide bond spanning only one residue. FEBS Open Bio. 2014; 4: 168–174. 10.1016/j.fob.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tidor B, Karplus M. The contribution of cross-links to protein stability: a normal mode analysis of the configurational entropy of the native state. Proteins 1993; 15: 71–79. [DOI] [PubMed] [Google Scholar]
- 4. Le QAT, Joo JC, Yoo YJ, Kim YH. Development of thermostable Candida Antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol Bioeng. 2012; 109: 867–876. 10.1002/bit.24371 [DOI] [PubMed] [Google Scholar]
- 5. Wang YW, Fu Z, Huang HQ, Zhang HS, Yao B, Xiong HR, et al. Improved thermal performance of Thermomyces lanuginosus GH11 xylanase by engineering of an N-terminal disulfide bridge. Bioresour Technol. 2012; 112: 275–279. 10.1016/j.biortech.2012.02.092 [DOI] [PubMed] [Google Scholar]
- 6. Alquati C, Gioia LD, Santarossa G, Alberghina L, Fantucci P, Lotti M. The cold-active lipase of Pseudomonas fragi: heterologous expression, biochemical characterization and molecular modeling. Eur J Biochem. 2002; 269: 3321–3328. [DOI] [PubMed] [Google Scholar]
- 7. Mansfeld J, Vriend G, Dijkstra BW, Veltman OR, Van den Burg B, Venema G, et al. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J Biol Chem 1997; 272: 11152–11156. [DOI] [PubMed] [Google Scholar]
- 8. Faulds CB. What can feruloyl esterases do for us? Phytochem Rev. 2010; 9: 121–132. [Google Scholar]
- 9. Crepin VF, Faulds CB, Connerton IF. Functional classification of the microbial esterases. Appl Microbiol Biotechnol. 2004; 63: 647–652. [DOI] [PubMed] [Google Scholar]
- 10. Hermoso JA, Sanz-Aparicio J, Molina R, Juge N, González R, Faulds CB. The crystal structure of feruloyl esterase A from Aspergillus niger suggests evolutive functional convergence in feruloyl esterase family. J Mol Biol. 2004; 338: 495–506. [DOI] [PubMed] [Google Scholar]
- 11. McAuley KE, Svendsen A, Patkar SA, Wilson KS. Structure of a feruloyl esterase from Aspergillus niger . Acta Crystallogr D Biol Crystallogy. 2004; 60: 878–887. [DOI] [PubMed] [Google Scholar]
- 12. Wu M, Abokitse K, Grosse S, Leisch H, Lau PCK. New feruloyl esterases to access phenolic acids from grass biomass. Appl Biochem Biotechnol. 2012; 168: 129–143. [DOI] [PubMed] [Google Scholar]
- 13. Koseki T, Fushinobu S, Ardiansyah, Shirakawa H, Komai M. Occurrence, properties, and applications of feruloyl esterases. Appl Microbiol Biotechnol. 2009; 84: 803–810. 10.1007/s00253-009-2148-8 [DOI] [PubMed] [Google Scholar]
- 14. Abokitse K, Wu M, Bergeron H, Grosse S, Lau PCK. Thermostable feruloyl esterase for the bioproduction of ferulic acid from triticale bran. Appl Microbiol Biotechnol. 2010; 87: 195–203. 10.1007/s00253-010-2441-6 [DOI] [PubMed] [Google Scholar]
- 15. Rakotoarivonina H, Hermant B, Chabbert B, Touzel JP, Remond C. A thermostable feruloyl-esterase from the hemicellulolytic bacterium Thermobacillus xylanilyticus releases phenolic acids from non-pretreated plant cell walls. Appl Microbiol Biotechnol. 2011; 90: 541–552. 10.1007/s00253-011-3103-z [DOI] [PubMed] [Google Scholar]
- 16. Zhang SB, Zhai HC, Wang L, Yu GH. Expression, purification and characterization of a feruloyl esterase A from Aspergillus flavus . Protein Expr Purif. 2013; 92: 36–40. 10.1016/j.pep.2013.08.009 [DOI] [PubMed] [Google Scholar]
- 17. Zhang SB, Wu ZL. Identification of amino acid residues responsible for increased thermostability of feruloyl eaterase A from Aspergillus niger using the PoPMuSiC algorithm. Bioresour Technol. 2011; 102: 2093–2096. 10.1016/j.biortech.2010.08.019 [DOI] [PubMed] [Google Scholar]
- 18. Joo JC, Pohkrel S, Pack SP, Yoo YJ. Thermostabilization of Bacillus circulans xylanase via computational design of a flexible surface cavity. J Biotechnol. 2010; 146: 31–39. 10.1016/j.jbiotec.2009.12.021 [DOI] [PubMed] [Google Scholar]
- 19. Joo JC, Pack SP, Kim YH, Yoo YJ. Thermostabilization of Bacillus circulans xylanase: Computational optimization of unstable residues based on thermal fluctuation analysis. J Biotechnol. 2011; 151: 56–65. 10.1016/j.jbiotec.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 20. Zhang SB, Pei XQ, Wu ZL. Multiple amino acid substitutions significantly improve the thermostability of feruloyl esterase A from Aspergillus niger . Bioresour Technol. 2012; 117: 140–147. 10.1016/j.biortech.2012.04.042 [DOI] [PubMed] [Google Scholar]
- 21. Kim HS, Le QAT, Kim YH. Development of thermostable lipase B from Candida antarctica (CalB) through in silico design employing B-factor and RosettaDesign. Enzyme Microb Technol. 2010; 47: 1–5. [Google Scholar]
- 22. Matsumara M, Mathews BW. Control of enzyme activity by an engineering disulfide bond. Science 1989; 243: 792–794. [DOI] [PubMed] [Google Scholar]
- 23. Turunen O, Ethuaho K, Fenel F, Vehmaanperä J, Wu X, Rouvinen J, et al. A combination of weakly stabilizing mutations with a disulfide bridge in the α-helix region of Trichoderma reesei endo-1,4-bxylanase II increase the thermal stability through synergism. J Biotechnol. 2001; 88: 37–46. [DOI] [PubMed] [Google Scholar]
- 24. Gong YY, Yin X, Zhang HM, Wu MC, Tang CD, Wang JQ, et al. Cloning, expression of a feruloyl esterase from Aspergillus usamii E001 and its applicability in generating ferulic acid from wheat bran. J Ind Microbiol Biotechnol. 2013; 40: 1433–1441. 10.1007/s10295-013-1339-6 [DOI] [PubMed] [Google Scholar]
- 25. Gao SJ, Wang JQ, Wu MC, Zhang HM, Yin X, Li JF. Engineering hyperthermostability into a mesophilic family 11 xylanase from Aspergillus oryzae by in silico design of N-terminus substitution. Biotechnol Bioeng. 2013; 110: 1028–1038. 10.1002/bit.24768 [DOI] [PubMed] [Google Scholar]
- 26. Zhang HM, Li JF, Wang JQ, Yang YJ, Wu MC. Determinants for the improved thermostability of a mesophilic family 11 xylanase predicted by computational methods. Biotechnol Biofuels 2014; 7: 3 10.1186/1754-6834-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eijsink VG, Gåseidnes S, Borchert TV, van den Burg B. Directed evolution of enzyme stability. Biomol Eng. 2005; 22: 21–30. [DOI] [PubMed] [Google Scholar]
- 28. Reetz MT, Carballeira JD, Vogel A. Iterative saturation mutagenesis on the basis of B-Factors as a strategy for increasing protein thermostability. Angew Chem Int Ed. 2006; 45: 7745–7751. [DOI] [PubMed] [Google Scholar]
- 29. Badieyan S, Bevan DR, Zhang C. Study and design of stability in GH5 cellusases. Biotechnol Bioeng. 2012; 109: 31–44. 10.1002/bit.23280 [DOI] [PubMed] [Google Scholar]
- 30. Hegde S, Srinivas P, Muralikrishna G. Singe-step synthesis of 4-nitrophenyl ferulate for spectrophotometric assay of feruloyl esterases. Anal Biochem. 2009; 387: 128–129. 10.1016/j.ab.2009.01.001 [DOI] [PubMed] [Google Scholar]
- 31. Mastihuba V, Kremnický L, Mastihubová M, Willett JL, Côté GL. A spectrophotometric assay for feruloyl esterases. Anal Biochem. 2002; 309: 96–101. [DOI] [PubMed] [Google Scholar]
- 32. Laemmli UK. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature 1970; 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 33. Li JF, Zhao SG, Tang CD, Wang JQ, Wu MC. Cloning and functional expression of an acidophilic β-mannanase gene (Anman5a) from Aspergillus niger LW-1 in Pichia pastoris . J Agric Food Chem. 2012; 60: 765–773. 10.1021/jf2041565 [DOI] [PubMed] [Google Scholar]
- 34. Surzhik MA, Schmidt AE, Glazunov EA, Firsov DL, Petukhov MG. Introduction of additional thiol groups into glucoamylase in Aspergillus awamori and their effect on the thermal stability and catalytic activity of the enzyme. Appl Biochem Microbiol. 2014; 50: 118–124. [DOI] [PubMed] [Google Scholar]
- 35. Jeong MY, Kim S, Yun CW, Choi YJ, Cho SG. Engineering a de novo internal disulfide bridge to improve the thermal stability of xylanase from Bacillus stearothermophilus No. 236. J Biotechnol. 2007; 127: 300–309. [DOI] [PubMed] [Google Scholar]
- 36. Han Z, Han S, Zheng S, Lin Y. Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl Microbiol Biotechnol. 2009; 85: 117–247. 10.1007/s00253-009-2067-8 [DOI] [PubMed] [Google Scholar]
- 37. Siadat OR, Lougarre A, Lamouroux L, Ladurantie C, Fournier D. The effect of engineered disulfide bonds on the stability of Drosophila melanogaster acetylcholinesterase. BMC Biochem. 2006; 7: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radestock S, Gohlke H. Exploiting the link between protein rigidity and thermostability for data-driven protein engineering. Eng Life Sci. 2008; 8: 507–522. 10.1093/protein/gzn026 [DOI] [PubMed] [Google Scholar]
- 39. Zhang HM, Wu MC, Li JF, Gao SJ, Yang YJ. Cloning and expression of a novel xylanase gene (Auxyn11D) from Aspergillus usamii E001 in Pichia pastoris . Appl Biochem Biotechnol. 2012; 167: 2198–2211. 10.1007/s12010-012-9757-x [DOI] [PubMed] [Google Scholar]
- 40. Chen XL, Gao YH, Ding YH, Lu WQ, Li DF. Cloning, functional expression and characterization of Aspergillus sulphureus β-mannanase in Pichia pastoris . J Biotechnol. 2007; 128: 452–461. [DOI] [PubMed] [Google Scholar]
- 41. Zhang SB, Pei XQ, Wu ZL. Cloning and expression of feruloyl esterase A from Aspergillus niger, and establishment of fast activity detection methods. Chin J Appl Environ Biol. 2009; 15: 276–279. [Google Scholar]
- 42. Pace CN, Grimsley GR, Thomson JA, Barnett BJ. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J Biol Chem. 1988; 263: 11820–11825. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.