Abstract
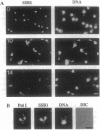
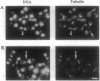
SRP1, a suppressor of certain temperature-sensitive mutations in RNA polymerase I in Saccharomyces cerevisiae, encodes a protein that is associated with nuclear pores. By using a system of conditional SRP1 expression and by isolating temperature-sensitive srp1 mutants, we have demonstrated that Srp1p is essential for maintenance of the crescent-shaped nucleolar structure, RNA transcription, and the proper functions of microtubules as inferred from analysis of nuclear division/segregation and immunofluorescence microscopy of microtubules. Different mutant alleles showed significantly different phenotypes in relation to these apparently multiple functional roles of the protein. We have also found that eight imperfect 42-amino-acid tandem repeats present in Srp1p are similar to the 42-amino-acid repeats in armadillo/plakoglobin/beta-catenin proteins present in adhesive junction complexes of higher eukaryotes. We discuss this similarity in connection with the observed pleiotropic effects of srp1 mutations.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berlin V., Styles C. A., Fink G. R. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol. 1990 Dec;111(6 Pt 1):2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P., Cook P. R., Jackson D. A. Active RNA polymerase I is fixed within the nucleus of HeLa cells. EMBO J. 1990 Jul;9(7):2207–2214. doi: 10.1002/j.1460-2075.1990.tb07390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke W. W., Goldschmidt M. D., Zimbelmann R., Mueller H. M., Schiller D. L., Cowin P. Molecular cloning and amino acid sequence of human plakoglobin, the common junctional plaque protein. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4027–4031. doi: 10.1073/pnas.86.11.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993 Sep;9(9):317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Yamagishi M., Kolb J. M., Nomura M. Suppressor analysis of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae: suppression of mutations in a zinc-binding motif by transposed mutant genes. Mol Cell Biol. 1991 Feb;11(2):746–753. doi: 10.1128/mcb.11.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M. W., Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol Cell Biol. 1993 Apr;13(4):2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Sullivan D. S., Huffaker T., Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992 Nov;119(3):583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Berg S., Reynolds A. B. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994 Mar 11;76(5):789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Peifer M., McCrea P. D., Green K. J., Wieschaus E., Gumbiner B. M. The vertebrate adhesive junction proteins beta-catenin and plakoglobin and the Drosophila segment polarity gene armadillo form a multigene family with similar properties. J Cell Biol. 1992 Aug;118(3):681–691. doi: 10.1083/jcb.118.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Wieschaus E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell. 1990 Dec 21;63(6):1167–1176. doi: 10.1016/0092-8674(90)90413-9. [DOI] [PubMed] [Google Scholar]
- Reynolds A. B., Herbert L., Cleveland J. L., Berg S. T., Gaut J. R. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992 Dec;7(12):2439–2445. [PubMed] [Google Scholar]
- Riggleman B., Wieschaus E., Schedl P. Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 1989 Jan;3(1):96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Boeke J. D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Dodd J., Vu L., Kolb J. M., Buhler J. M., Sentenac A., Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., McGrew J. T., Schroeder A. J., Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Aug;13(8):4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Oakes M., Yamaghishi M., Dodd J. A., Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Dec;12(12):5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumori T., Murayama N., Yamazoe Y., Abe A., Nogi Y., Fukasawa T., Kato R. Expression of a human P-450IIC gene in yeast cells using galactose-inducible expression system. Mol Pharmacol. 1989 Apr;35(4):443–449. [PubMed] [Google Scholar]