Abstract
The lantibiotic nisin is an antimicrobial peptide that is widely used as a food preservative to combat food-borne pathogens1. Nisin contains dehydroalanine and dehydrobutyrine residues that are formed via dehydration of Ser/Thr by the lantibiotic dehydratase NisB2. Recent biochemical studies revealed that NisB glutamylates Ser/Thr side chains as part of the dehydration process3. However, the molecular mechanism by which NisB utilizes glutamate to catalyze dehydration remains unresolved. Here we show that this process involves glutamyl-tRNAGlu to activate Ser/Thr residues. In addition, the 2.9 Å crystal structure of NisB in complex with its substrate peptide NisA reveals the presence of two separate domains that catalyze the Ser/Thr glutamylation and glutamate elimination steps. The co-crystal structure also provides the first insights into substrate recognition by lantibiotic dehydratases. Our findings demonstrate a non-anticipated role for aminoacyl-tRNA in the formation of dehydroamino acids in lantibiotics, and serve as a basis for the functional characterization of the many lantibiotic-like dehydratases involved in the biosynthesis of other classes of natural products.
Bacterial resistance to currently used antibiotics is a growing health threat. A potential solution to this emerging problem is the development of new antibiotics with multiple modes of action that would challenge bacterial resistance mechanisms. For instance, the lantibiotic nisin has been used worldwide in the food industry for over 40 years without substantial development of resistance1,4. This unique property is thought to be a consequence of nisin’s dual mode of action: pore formation in bacterial cell membranes and stalling of peptidoglycan biosynthesis by sequestering the cell wall precursor lipid II5–7.
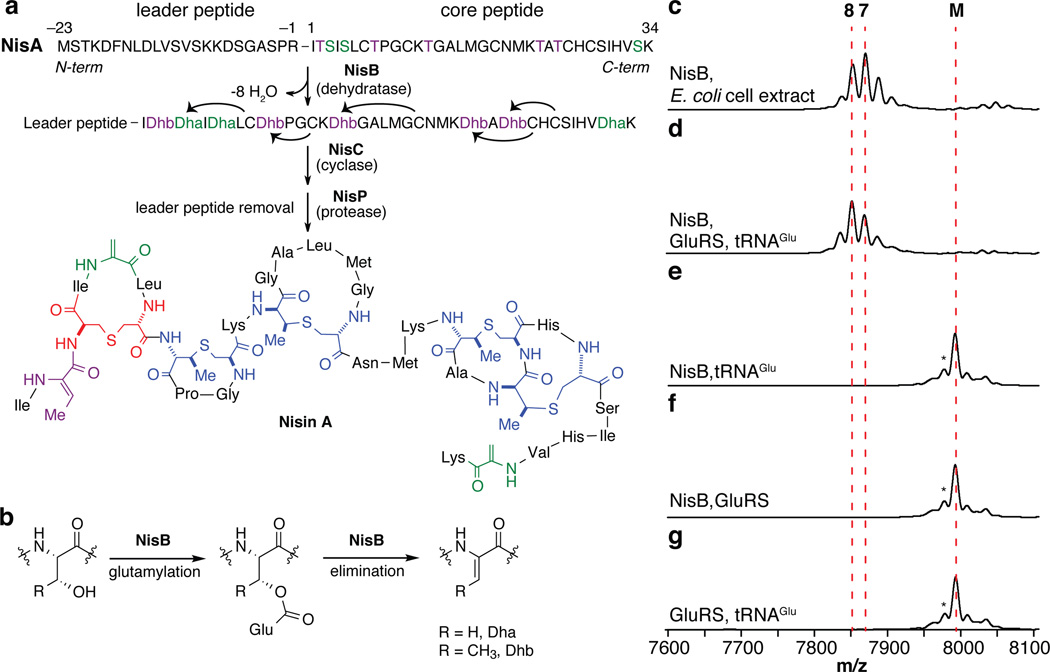
Lantibiotics are lanthionine-containing antimicrobial peptides8. Nisin is generated from a ribosomally synthesized linear precursor peptide termed NisA9. The dehydratase NisB then dehydrates eight serines and threonines in the NisA core region yielding dehydroalanine (Dha) and dehydrobutyrine (Dhb) residues, respectively (Fig. 1a)2. The cyclase NisC subsequently catalyzes the formation of five lanthionine and methyllanthionine cross-links by the nucleophilic addition of cysteinyl thiols to Dha and Dhb, respectively (Fig. 1a)10. In the final maturation step, the lantibiotic protease NisP removes a leader peptide, which is important for recognition by NisB and NisC, to yield the mature lantibiotic11.
Fig. 1. Biosynthesis of the lantibiotic nisin.
(a) Post-translational modifications involved in nisin biosynthesis. NisB dehydrates Ser/Thr residues in NisA via (b) glutamylation forming Dha and Dhb, respectively. NisC catalyzes the formation of lanthionines (red) and methyllanthionines (blue). NisP removes the leader peptide. Negative numbers represent the position of an amino acid in the leader peptide with respect to the core region. MALDI-TOF MS analysis of His6-NisA incubated with Glu, ATP and His6-NisB in the presence of E. coli (c) cell extract, (d) His6-GluRS and tRNAGlu, (e) tRNAGlu, (f) His6-GluRS, or (g) GluRS and tRNAGlu in the absence of His6-NisB. (M) Unmodified (7993 m/z, calc. 7996 m/z), (7) seven-fold dehydrated (7870 m/z, calc. 7870 m/z), and (8) eight-fold dehydrated (7853 m/z, calc. 7852 m/z) His6-NisA. (*) Peak resulting from laser-induced deamination (Extended Data Fig. 6).
Twenty-six years since the characterization of the first lantibiotic gene cluster12, the mechanism by which lantibiotic dehydratases (LanB) introduce dehydroamino acids in class I lantibiotics like nisin has remained enigmatic. Recently, NisB was shown to dehydrate NisA via an unprecedented glutamylation mechanism (Fig. 1b)3. However, NisB was only active in the presence of an unknown component within Escherichia coli cell extract3. Herein, we identify glutamyl-tRNAGlu as the key component needed to catalyze the formation of dehydroamino acids in class I lantibiotics. In addition, we report the co-crystal structure of NisA bound to NisB, which provides key information on the glutamyl-tRNAGlu dependent esterification of Ser/Thr residues in NisA and offers the first insights into leader peptide binding and substrate recognition by lantibiotic dehydratases.
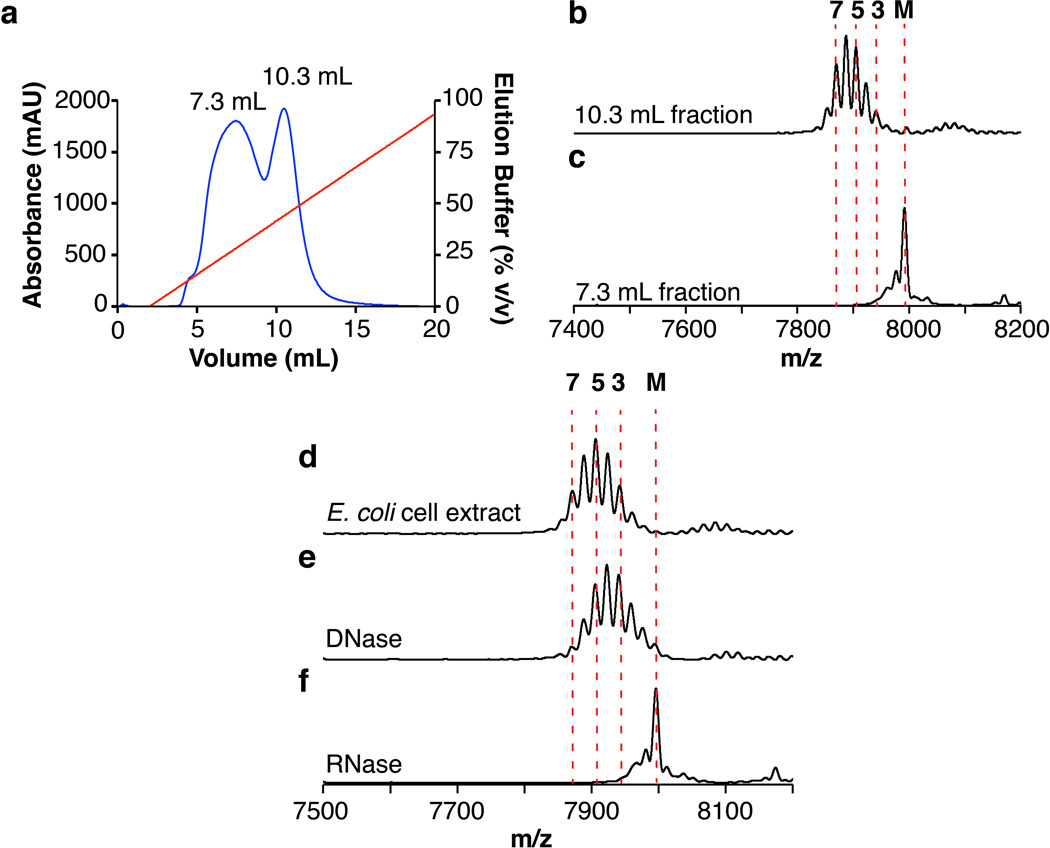
In the previously proposed dehydration mechanism3, glutamate needs to be activated prior to the formation of an ester linkage with the side chain of Ser/Thr residues in NisA. To identify the required component for activation, anion exchange chromatographic fractions of E. coli cell extracts were analyzed for supporting NisB-catalyzed dehydration of NisA by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Extended Data Fig. 1). Surprisingly, the A260/A280 ratio for the fraction supporting NisB activity was 1.7, suggesting the presence of nucleic acids. Treatment of E. coli cell extract with DNase did not prevent NisB-catalyzed dehydration of NisA but treatment with RNase abolished activity (Extended Data Fig. 1).
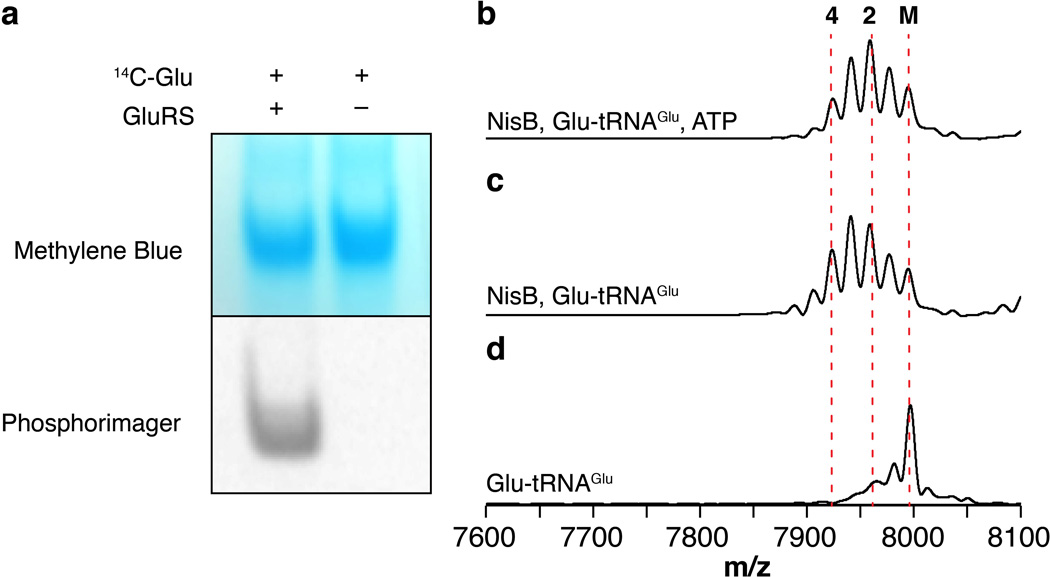
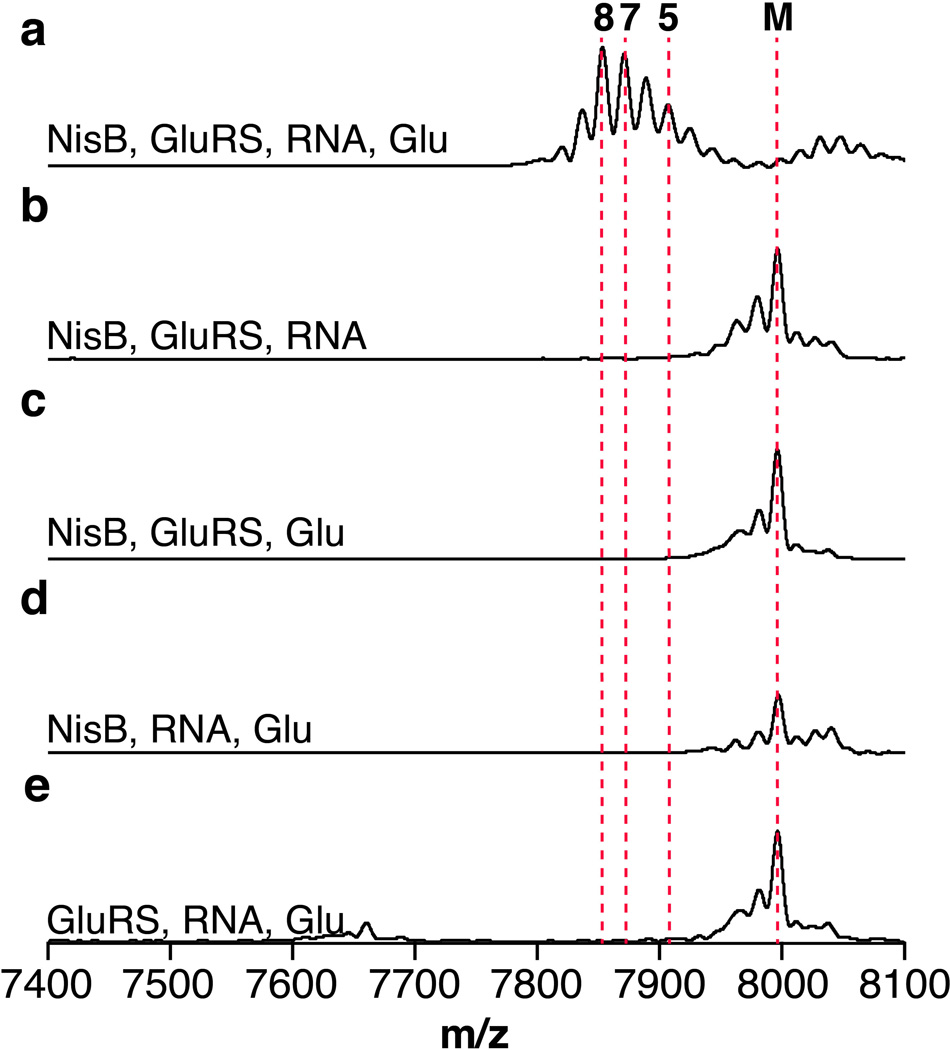
The requirements for glutamate and RNA suggested the possibility of a glutamyl-tRNAGlu dependent dehydration process. We therefore cloned, expressed, and purified glutamyl-tRNA synthetase (GluRS) as well as tRNAGlu from E. coli. MALDI-TOF MS analysis of NisA after incubation with NisB, Glu, ATP, GluRS and tRNAGlu revealed up to eight dehydrations in NisA (Fig. 1c–d). No activity was observed when GluRS, tRNAGlu, or NisB were omitted from the reaction assay (Fig. 1e–g). The inability of NisB to dehydrate NisA in the absence of GluRS (Fig. 1e) demonstrates that NisB does not aminoacylate tRNAGlu but instead utilizes the glutamyl-tRNAGlu synthesized in situ by GluRS. Dehydration assays with purified glutamyl-tRNAGlu and subsequent MALDI-TOF MS analysis confirmed this conclusion and also showed that ATP was not required for activity (Extended Data Fig. 2). This observation suggests that dehydration of Ser/Thr residues located at different positions within NisA is not driven by the consumption of energy.
The observed requirement for E. coli glutamyl-tRNAGlu raised the question whether the nisin producing organism, Lactococcus lactis, employs the same tRNA-dependent biosynthetic strategy. MALDI-TOF MS analysis of NisA treated with NisB, Glu, ATP, and both GluRS and total RNA from L. lactis revealed up to eight dehydrations in NisA (Extended Data Fig. 3), confirming that NisB can utilize glutamyl-tRNAGlu from L. lactis.
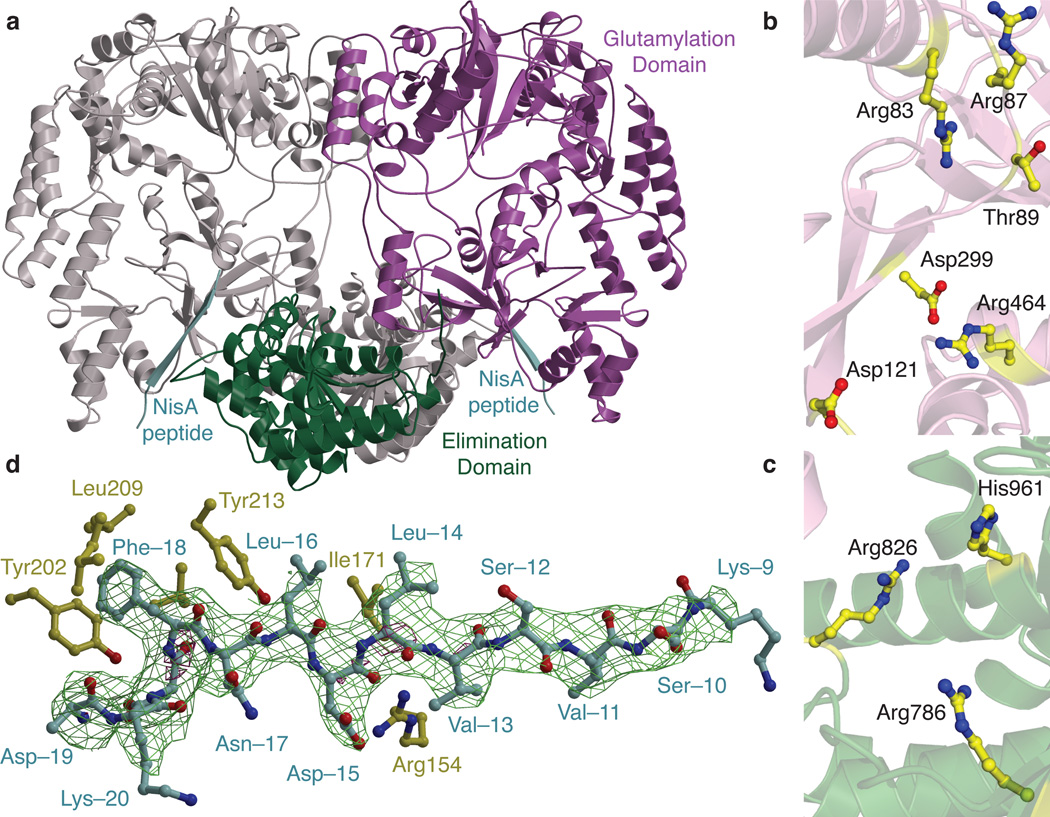
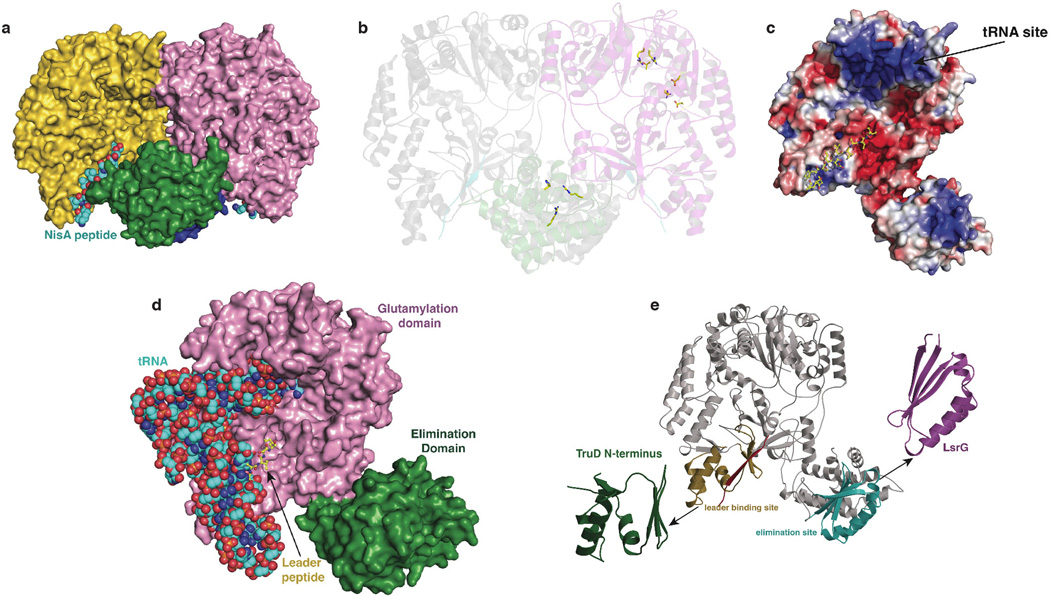
To obtain structural insights into the dehydration process, we determined the co-crystal structure of NisB in complex with its substrate peptide NisA to 2.9 Å resolution (Extended Data Table 1). The overall structure of the 117 kDa dehydratase reveals a dimer with a bifurcate cleft-like structure in the center (Extended Data Fig. 4). Residues spanning Ser5–Ile706 comprise a multi-domain amino-terminal region and Arg716–Glu990 form a single carboxy-terminal domain (Fig. 2a). Residues covering Ille706–Arg716 and Val893–Gly919 were not modeled in the final structure due to lack of electron-density.
Fig. 2. Crystal structure of the lantibiotic dehydratase NisB.
(a) Overall structure of the NisB homodimer in complex with its substrate peptide NisA showing the disposition of the glutamylation (purple) and glutamate elimination (green) domains while the other monomer is shown in gray. Residues important for glutamylation and glutamate elimination are shown as spheres and the NisA peptide is shown in cyan. (b, c) Residues in NisB essential for either the glutamylation (b) or glutamate elimination activities (c) are clustered in the crystal structure. (d) Simulated annealing omit difference Fourier map (Fo-Fc) contoured to 2.5σ of residues Lys–20 through Lys–9 within the leader peptide of NisA. NisB residues involved in binding of the NisA leader peptide (green) are shown in yellow.
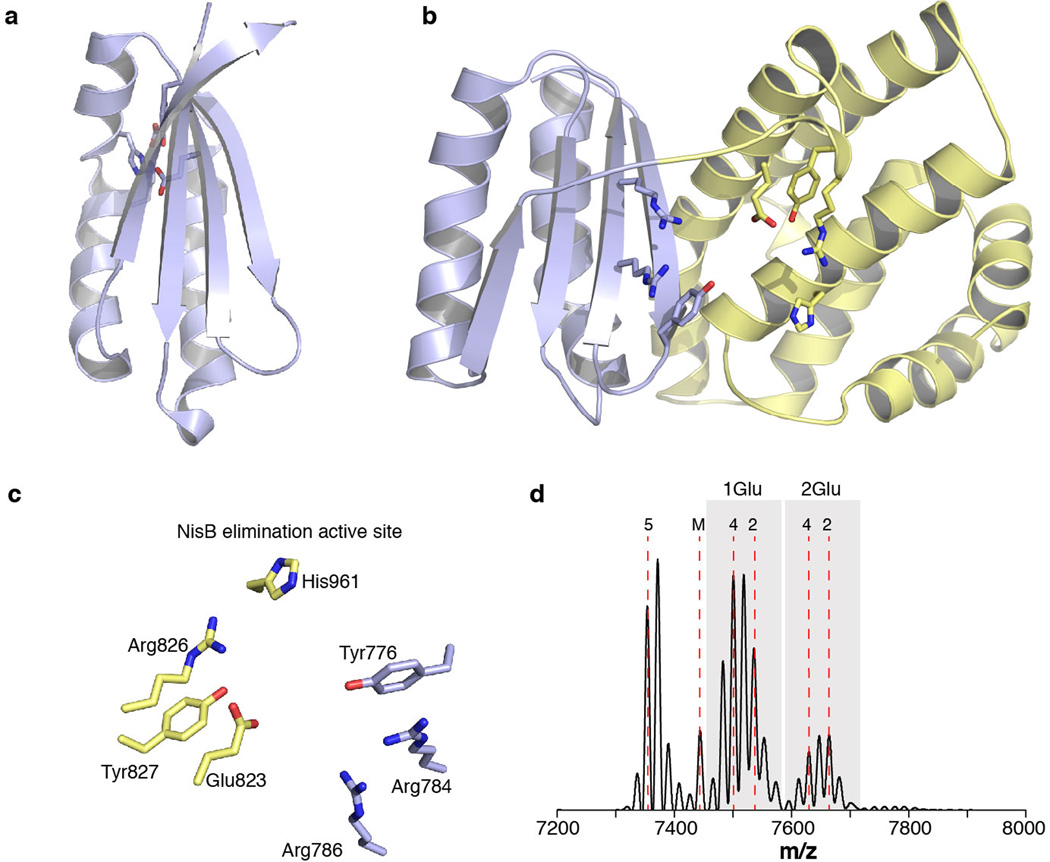
NisB mutations previously shown to interfere with either glutamylation or elimination activity map to distinct clusters in the amino- and carboxy-terminal regions in the structure, respectively (Extended Data Figure 4). Alanine substitutions of NisB residues that abolished glutamylation of NisA but retained in vitro glutamate elimination activity3 are all within a 10 Å radius in the amino-terminal region (Fig. 2b). This 700-residue N-terminal region of NisB can thus be demarcated as the glutamylation domain. Similarly, alanine-scanning studies in NisB demonstrated that Arg786, Arg826, and His961 are important for glutamate elimination, but not for glutamylation activity3. These residues, as well as others important for dehydration based on mutagenesis data3 (Extended Data Fig. 5), cluster to a small site in the carboxy-terminal domain (Fig. 2c), and consequently, the C-terminal 300-residue domain of NisB catalyzes the glutamate elimination step. Interestingly, the boundaries for the two domains correspond with two proteins of roughly 800 residues and 300 residues encoded in the biosynthetic clusters of thiopeptides13, suggesting they carry out aminoacylation and elimination, respectively, to achieve the dehydration of their substrate peptides.
RNA binding proteins often have basic patches which mediate binding of their cognate RNAs via electrostatic interactions14. Mapping the electrostatic potential onto the surface of NisB reveals the presence of a densely basic cavity that maps to the inner face of the cleft-like structure (Extended Data Fig. 4). The dimensions of the basic cavity are suitable to accommodate the characteristic L-shape of polyanionic tRNA. Based on limited structural homology of this NisB region to the X. laevis dsRNA binding protein A complexed to its cognate RNA (PDB ID: 1DI2) and using a structure of bacterial tRNA from T. thermophilus (PDB ID: 1N78), a docking pose was derived for NisB binding to tRNAGlu (Extended Data Fig. 4). This model results in placement of the 3’ end of the tRNA in close proximity to the pocket defined by the NisB residues that are critical for glutamylation activity, providing additional support for the model.
Structure-based similarity searches against the Protein Data Bank failed to identify obvious homologous folds within the NisB structure. However, searches using smaller domains of NisB identified several structural homologs (Extended Data Fig. 4). A small domain spanning residues Phe136–Ile216 resembles the amino-terminal domain of TruD, a heterocyclase involved in the biosynthesis of cyanobactins15, another class of ribosomally synthesized and post-translationally modified peptides (RiPPs) that is made in a leader peptide-dependent manner16. A second region within the carboxy-terminus of NisB (Phe734–Tyr820) is similar to the LsrG protein that carries out the epimerization of activated quorum sensing molecules17. However, despite the structural homology and similar chemistry (abstraction of a relatively acidic proton adjacent to a carbonyl group), residues important for LsrG catalysis17 are not conserved in NisB. Instead residues important for glutamate elimination in NisB are located between the LsrG-like region and a second subdomain of the glutamate elimination site (Extended Data Fig. 5). Indeed, the crystal structure of NisB allowed us to identify an additional conserved positively charged residue in this pocket, Arg784. As anticipated, the NisB-R784A mutant resulted in the accumulation of glutamylated NisA intermediates (Extended Data Fig. 5), providing additional support for our assignment of this region as the glutamate elimination active site.
Clear and continuous electron density was observed for the NisA leader sequence spanning residues Lys–9 through Lys–20 (Fig. 2d). Beyond Lys–9, moving towards the C-terminal core region, the electron density diminishes probably due to NisA conformational flexibility. The NisA leader peptide binds to NisB residues Leu166–Asn175 as an anti-parallel β-strand. The site of binding combined with the extended conformation of the leader peptide ensures for the core region to reach the glutamylation active site. NisA, like most class I lantibiotic precursor peptides, contains a F-D/N-L-N/D conserved motif within the leader peptide important for substrate recognition by NisB18–20. The structure reveals that Phe–18 of NisA in this motif binds to NisB within a hydrophobic cage composed of Val176, Val198, Tyr202, Leu209, and Tyr213. Similarly, Leu–16 of NisA is clustered inside a NisB pocket consisting of residues Ile171, Tyr213, and Leu217. Lastly, Asp–15 of NisA is within hydrogen bonding distance of Arg154 (Fig. 2d). The interactions observed in the crystal structure provide a rationale for why mutations within the NisA FNLD sequence greatly reduce NisB binding19,20. Notably, NisA binds to the region in NisB homologous to the amino terminal domain of TruD (Extended Data Fig. 4), suggesting that some of the different classes of biosynthetic enzymes involved in the processing of RiPPs likely use similar structural elements for substrate binding.
The presence of the leader peptide-binding site on the N-terminal domain raised the question whether the glutamate elimination activity requires the leader peptide. To gain further insight, we tested NisB-catalyzed glutamate elimination activity on purified glutamylated NisA core peptide in the absence of the leader peptide. MALDI-TOF MS analysis after incubation with NisB revealed the presence of dehydrated NisA core peptide (Extended Data Fig. 6). Hence, the leader peptide does not appear to be critical for glutamate elimination, suggesting the local structure of glutamylated Ser/Thr is sufficient for substrate recognition by the elimination domain.
The data presented herein provide explanations for several long-standing questions. NisB substrate recognition is mediated by hydrophobic interactions between NisB and the FNLD motif in NisA. The presence of this motif in many class I lantibiotic precursor peptides suggests a similar recognition mechanism employed by their respective lantibiotic dehydratases. In addition, the NisB-catalyzed dehydration of NisA has been shown to proceed with general N-to-C terminal directionality21,22. This observation, combined with the demonstration that NisB dehydrates NisA in a distributive manner (Extended Data Fig. 1–3) and with the leader peptide binding site identified in NisB, suggests that directionality is achieved by the distance between the leader peptide binding site and the glutamylation active site. Residues closer to the leader peptide (N-terminus) have a higher likelihood to access the glutamylation active site than residues located at the C-terminus. The presence of a distinct leader peptide binding site also explains why Ser/Thr residues within the leader peptide are never dehydrated and why there is a minimal distance requirement from the F-D/N-L-N/D motif to the first dehydrated amino acid in class I lantibiotics19,23 (i.e. distance required to reach the glutamylation site). After glutamylation, the flexible core region reaches the glutamate elimination active site where the glutamate adduct is then eliminated. Such flexibility can account for the diminished electron density observed for the NisA C-terminal portion.
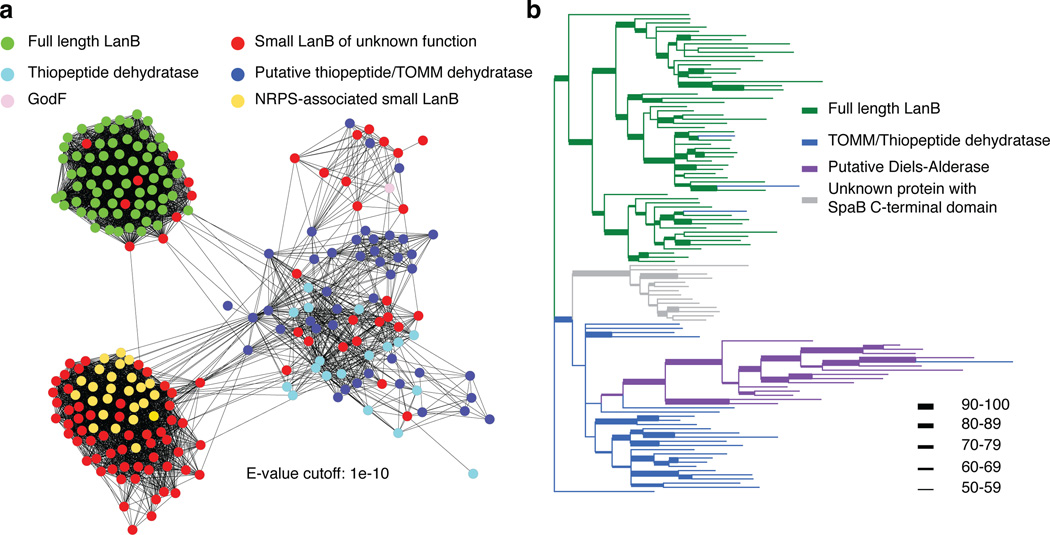
Lantibiotic dehydratases (LanB) are widely distributed among Gram-positive bacteria. A protein similarity map of the glutamylation domain of LanB proteins (Supplementary Table 1) in the databases identified several sub-families (Fig. 3a). One large group is formed predominantly by sequences that contain the glutamylation and elimination domains in one polypeptide (full length LanB, green). Another less well-defined group is formed by dehydratases involved in the biosynthesis of thiopeptides (cyan) and goadsporin (pink). In these LanBs the glutamylation and elimination domains are in separate polypeptides. Whether these enzymes indeed use glutamate or perhaps another amino acid for activation of Ser/Thr remains to be determined. Perhaps the most intriguing group is formed by shorter uncharacterized sequences (small LanBs, red) that lack the elimination domain altogether and for which no open reading frame encoding a stand-alone elimination protein is found within nearby genes. These small LanBs are often part of non-ribosomal peptide synthesis (NRPS) clusters (Fig. 3a, yellow), and the observation that they lack the elimination domain suggests they may add amino acids to a growing peptide in a tRNA-dependent manner.
Fig. 3. Protein similarity map and phylogenetic tree analysis of various lantibiotic dehydratases and homologs.
(a) Protein similarity map of the glutamylation domain (Lant_dehyd_C, PF04738) present in various lantibiotic dehydratases and homologs. Each node in the network represents a protein sequence and each edge represents sequences with BLASTP e-values below the indicated cutoff. (b) Phylogenetic tree analysis of the glutamate elimination domain (SpaB_C, PF14028) present in various lantibiotic dehydratases and homologs. Several clades can be identified based on the proposed function and natural product class. Only splits with a posterior probability higher than 50% are shown.
Further phylogenetic analysis also revealed distinct clades for the elimination domain (Fig. 3b). Some of the enzymes that have been connected to a putative Diels-Alder reaction involved in the biosynthesis of thiopeptides24 contain a domain that belongs to the same protein family (Pfam) as the LanB elimination domain (PF14028, also called SpaB_C) and clustered together as a separate clade (Fig. 3b, purple). Hence, despite their limited sequence similarity to the LanB elimination domain (Extended Data Fig. 6), these putative Diels-Alderases will likely contain a similar fold as the one presented herein.
Our observations predict that aminoacylated tRNA may find much broader participation in natural product biosynthesis than currently appreciated25–28. The demonstrated use of glutamylated tRNA by NisB to transiently affect glutamylation of Ser/Thr side chains in NisA is a new addition to a growing number of uses of aminoacylated tRNA for functions other than ribosomal protein synthesis29,30.
Methods
Oligonucleotides were purchased from Integrated DNA Technologies Inc. (Coralville, IA) (Extended Data Table 2). Reagents used for molecular biology were purchased from New England BioLabs (Ipswich, MA), Thermo Fisher Scientific (Waltham, MA), or Gold Biotechnology Inc. (St. Louis, MO). Chemicals were purchased from Sigma-Aldrich (St. Louis, MO). L-[14C(U)]-glutamic acid (0.1 µCi µL−1, 260 mCi mmol−1) was purchased from PerkinElmer (Waltham, MA). Escherichia coli DH5α and BL21 (DE3) strains were used for plasmid maintenance and protein overexpression, respectively. Cloning inserts were sequenced at ACGT Inc. (Wheeling, IL). MALDI-TOF MS analysis was performed using a Bruker UltrafleXtreme MALDI TOF-TOF (Billerica, MA) in reflective mode at the University of Illinois Mass Spectrometry Facility. Mass spectrometry data was analyzed, smoothed and baseline corrected using Bruker Daltonics Software flexAnalysis package. Data was exported, normalized and plotted using MS Excel. Reflective mode was used to resolve mass peaks differing by 18 Da within the 7–8 kDa size range. The small shoulder peaks appearing to the left of unmodified NisA (M) correspond to the laser-induced deamination of the parent peak. Deamination occurs as a consequence of analyzing large peptides using reflective mode (Extended Data Fig. 6). All biochemical assays were performed in duplicate with hexa-histidine tagged substrates and enzymes.
Expression and purification of His6-NisA, glutamylated His6-NisA, and His6-NisB
The nisA gene was cloned previously into pET15b resulting in a peptide with an N-terminal His6-tag28. E. coli BL21 (DE3) cells (50 µL) were electroporated with the plasmid his6-nisA-pET15b (50 ng), cells were plated on LB agar plates supplemented with ampicillin (amp, 100 µg mL−1) and grown at 37 °C for 12–15 h. A single colony was used to inoculate 50 mL of LB broth supplemented with amp, grown for 12–15 h at 37 °C, and the culture was used to inoculate 4 L of terrific broth (TB) media supplemented with amp, to an OD600 of 0.025. Cultures were grown at 37 °C to a final OD600 of 1.0. Peptide expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and cultures were grown at 18 °C for 18 h. Peptide purification was performed following a previously described method28. The cloning, expression, and purification of His6-NisB and glutamylated His6-NisA is described elsewhere3.
Preparation of Escherichia coli BL21 (DE3) cell extract
The cell extract of E. coli BL21 (DE3) was prepared following a previously described method29 with minor modifications. E. coli BL21 (DE3) cells were inoculated in 2 L of TB media and grown at 37 °C to a final OD600 of 1.0. Cells were harvested (4,650 × g, 10 min, 4 °C) and the pellet was washed three times by resuspending it in S30 buffer (10 mM Tris-acetate buffer pH 8.2, 14 mM Mg(OAc)2, 60 mM KOAc, 1 mM DTT, 0.3 mM EDTA, 0.3 mM MgCl2) followed by centrifugation (4,650 × g, 10 min, 4 °C). Cells were then resuspended in 1 mL of S30 buffer per 1 g of wet cells and lysed using a MultiFlex C3 Homogenizer (Avestin). The cell lysate was centrifuged twice (22,789 × g, 30 min, 4 °C) and the supernatant was dialyzed 4 times against 50 volumes of S30 buffer (without DTT) using a dialysis cassette with a molecular weight cutoff (MWCO) of 10 kDa. The cell extract was then centrifuged (4,000 × g, 10 min, 4 °C) and the supernatant was frozen and stored in 1 mL aliquots at −80 °C for future use.
Anion exchange fractionation of E. coli cell extract
An aliquot of 1 mL of cell extract was loaded onto a 1 mL Hi Trap DEAE FF (GE Healthcare) column equilibrated with 5 column volumes (CV) of Start Buffer (10 mM Tris-HCl pH 8.0) using an AKTA FPLC system (Amersham Pharmacia Biosystems). The column was washed with 10 CV of Start Buffer and the sample was eluted using a gradient of 0–100 % (v/v) of Elution Buffer (10 mM Tris-HCl, 1 M NaCl pH 8.0) in Start Buffer over 20 CV. Eluent was detected by absorbance at 280 nm (Extended Data Fig. 1). Collected fractions were desalted by washing five times with 500 µL of S30 buffer (without DTT) using a 10 kDa MWCO Amicon centrifugal filter (Millipore) (14,000 × g, 10 min, 4 °C). Fractions were stored at −80 °C.
Cloning, expression, and purification of Escherichia coli BL21 (DE3) GluRS
The gene gltX (WP_ 024237069) was cloned into pET28a resulting in a protein with an N-terminal hexahistidine tag. gDNA from E. coli BL21 (DE3) cells was extracted using an UltraClean Microbial DNA isolation kit (MoBio) following the manufacturer’s protocol. The gltX gene was PCR amplified using the following conditions: Phusion HF buffer (1×), dNTP (0.2 mM), primers (1 µM each, Extended Data Table 2), gDNA template (10 ng), DMSO (3% v/v), and Phusion polymerase (0.04 U µL−1) in a total volume of 50 µL. The amplification was performed in 20 cycles: denaturation (98 °C for 60 s), annealing (67 °C for 60 s), and amplification (72 °C for 42 s). The PCR product was purified by gel extraction on a 1% (w/v) agarose gel using the QIAquick Gel Extraction Kit (QIAGEN). The vector pET28a and gltX PCR fragment were digested using NdeI and XhoI (NEB) restriction endonucleases, purified by gel extraction as described above, and the resulting fragments were ligated in 20 µL using T4 DNA ligase (NEB) at 16 °C for 18 h. An aliquot of 10 µL from the ligation reaction was used to transform E. coli DH5α (DE3) cells using the heat shock method. The cells were plated on LB plates supplemented with kanamycin (kan, 50 µg mL−1), and the plates were incubated at 37 °C for 12–15 h. Single colonies were picked and grown in LB supplemented with kan at 37 °C for 12–15 h, and the plasmid his6-gltX-pET28a was isolated using a QIA prep Spin Miniprep Kit (QIAGEN). Insert integrity was verified by sequencing the plasmid with the appropriate primers.
E. coli BL21 (DE3) cells (50 µL) were electroporated with his6-gltX-pET28a (50 ng), plated on LB agar plates supplemented with kan, and grown at 37 °C for 12–15 h. A single colony was used to inoculate 50 mL of LB broth supplemented with kan, grown for 12–15 h at 37 °C, and the culture was used to inoculate 3 L of TB media supplemented with kan to an OD600 of 0.025. Cultures were grown at 37 °C to an OD600 of 0.5. Protein expression was induced by the addition of IPTG to a final concentration of 0.2 mM, and cultures were grown at 18 °C for 20 h. Protein purification was performed as described for His6-NisB3. After elution from a HisTrap HP 5 mL column (GE Healthcare), the protein was desalted using a PD-10 desalting column (GE Healthcare), concentrated using a 30 kDa MWCO Amicon centrifugal filter (Millipore) (4,000 × g, 20 min, 4 °C) and stored in Storage Buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10% glycerol) at –80 °C.
Cloning, expression and purification of Lactococcus lactis HP GluRS
The gene gltX (WP_011836000) from L. lactis HP was cloned into pET28a resulting in a protein with an N-terminal hexahistidine tag. gDNA from L. lactis HP cells was extracted using an UltraClean Microbial DNA isolation kit (MoBio) following the manufacturer’s protocol. The gltX gene was amplified by PCR using similar conditions as described above with appropriate primers (Extended Data Table 2). The amplification was performed in 20 cycles: denaturation (98 °C for 60 s), annealing (65 °C for 60 s), and amplification (72 °C for 60 s). The PCR purification, ligation, expression of L. lactis GluRS and purification were performed as described above for E. coli GluRS.
Purification of Escherichia coli tRNAGlu and glutamyl-tRNAGlu
Primers for E. coli tRNAGlu were designed according to a previously described method30 (Extended Data Table 2). The tRNAGlu dsDNA template for in vitro transcription was prepared by filling 5’ overhangs using the following conditions: NEB Buffer 2 (1×), primers (4 µM each), dNTP (33 µM), and NEB DNA pol I (large Klenow fragment) (1 U µg−1 DNA) in a final volume of 50 µL. The reaction was incubated at 25 °C for 15 min, quenched with EDTA (10 mM), incubated at 75 °C for 25 min, and dsDNA tRNAGlu template was precipitated with EtOH. In vitro transcription was performed using a previously described method31.
Glutamylation of tRNAGlu was performed using the following conditions: tRNAGlu (50 µM), L-glutamate (10 mM), ATP (10 mM), E. coli GluRS (5 µM), thermostable inorganic pyrophosphatase (TIPP) (New England BioLabs) (0.04 U µL−1), HEPES pH 7.5 (100 mM), KCl (10 mM), MgCl2 (20 mM) and DTT (1 mM) in a final volume of 500 µL. The reaction was incubated at 37 °C for 1 h and desalted by washing ten times with 500 µL HEPES pH 7.5 (100 mM) using a 10 kDa MWCO Amicon centrifugal filter (Millipore) (14,000 × g, 10 min, 4 °C). Glutamyl-tRNAGlu was then purified by acidic phenol extraction using a previously described method32.
To assess the extent of aminoacylation on in vitro transcribed E. coli tRNAGlu, glutamyl-tRNAGlu was analyzed by denaturing acidic polyacrylamide gel using a previously described procedure32,33. tRNA aminoacylation assays were performed using L-[14C(U)]-glutamic acid (0.1 µCi µL−1, 260 mCi mmol−1) under the following conditions: in vitro transcribed E. coli tRNAGlu (3 µM), L-[14C(U)]-glutamic acid (50 µM), HEPES pH 7.5 (100 mM), ATP (5 mM), TCEP (1 mM), KCl (10 mM), MgCl2 (20 mM), TIPP (0.02 U µL−1) and purified E. coli GluRS (1 µM) in a final volume of 50 µL. The reaction was incubated at 37 °C for 1 h and quenched with NaOAc pH 5.6 (300 mM). Glutamyl-tRNAGlu was precipitated by standard EtOH precipitation. The resulting pellet was dissolved in 10 µL of acidic loading buffer (100 mM NaOAc pH 5.6, 7 M urea, 0.05% bromophenol blue) and the sample was loaded onto a 7% acidic polyacrylamide gel (100 mM NaOAc, 8 M urea). Electrophoresis was performed at 100 V (constant voltage) in an ice bath for 120 min using 100 mM NaOAc pH 5.6 as running buffer. To visualize tRNA, the gel was stained with staining solution (500 mM NaOAc pH 5.6, 0.06% methylene blue) for 30 min and destained in water. After drying the gel overnight using a gel drying kit (Promega) following the manufacturer’s procedure, glutamyl-tRNAGlu was visualized by exposing the dried gel onto a phosphorimager cassette for 4 days. The film was then visualized using a STORM 840 Phosphorimager Scanner (Amersham Biosciences) (Extended Data Fig. 2).
RNA extraction from Lactococcus lactis HP
Lactococcus lactis HP cells were grown in 2 L of M17 media supplemented with 0.5% (w/v) glucose for 16 h at 30 °C. Cells were washed three times with S30 Buffer and cell extract was prepared as described above for E. coli cells. RNA was extracted from the cell extract by acidic phenol extraction. Briefly, an aliquot of 500 µL of cell extract was mixed with 500 µL of acidic phenol (pH ~4.5) and the resulting emulsion was vortexed for 1 min. The sample was centrifuged (14,000 × g, 5 min, 4 °C) and the aqueous phase was collected for a second acidic phenol extraction. After centrifugation, the aqueous layer was extracted twice with 500 µL of chloroform:isoamyl alcohol (24:1). The sample was vortexed for 1 min and centrifuged (14,000 × g, 5 min, 4 °C). Total RNA was precipitated by standard EtOH precipitation and dissolved in RNase-free distilled water. To remove any aminoacylated tRNA, total RNA (0.66 µg µL−1) was incubated for 1 h at 37 °C in EDTA (0.6 mM) and Tris-HCl pH 9.0 (66 mM). The sample was acidified with NaOAc (300 mM) and RNA was extracted by standard EtOH precipitation33.
In vitro dehydration assays
The following reaction conditions were used for dehydration assays: E. coli cell extract/fraction (10 µL), HEPES pH 7.5 (100 mM), DTT (1 mM), L-glutamate (10 mM), NisA (200 µM), MgCl2 (10 mM), KCl (10 mM), NisB (5 µM), TIPP (0.02 U µL−1) and ATP (5 mM) in a final volume of 30 µL. The assay was incubated at 30 °C for 5 h, centrifuged to remove insoluble material (14,000 × g, 5 min, 25 °C), and desalted using a C-18 ZipTip concentrator (Millipore). The sample was mixed in a 1:1 ratio with sinapinic acid matrix and spotted on a Bruker MALDI-TOF plate. When noted, the cell extract (10 µL) was substituted with E. coli GluRS (5 µM) and E. coli tRNAGlu (50 µM). In addition when noted, the cell extract (10 µL) was treated with either RNase (10 U) or RNase-free DNase (10 U) in the presence of CaCl2 (100 µM) and incubated at 25 °C for 1 h prior to dehydration assays.
For dehydration assays using glutamyl-tRNAGlu the following conditions were used: HEPES pH 7.5 (100 mM), DTT (500 µM), MgCl2 (10 mM), KCl (10 mM), His6-NisA (10 µM), Glu-tRNAGlu (~100 µM), and His6-NisB (5 µM) in a final volume of 30 µL. The assay was incubated at 30 °C for 12 h and prepared as described above for analysis by MALDI-TOF MS. In a separate experiment, ATP (1 mM) was added to the reaction mixture.
For dehydration assays using L. lactis HP total RNA the following conditions were used: L. lactis RNA (1 µg µL−1), glutamate (10 mM), MgCl2 (10 mM), KCl (10 mM), HEPES pH 7.5 (50 mM), L. lactis GluRS (10 µM), NisB (5 µM), NisA (100 µM), ATP (5 mM), TIPP (0.02 U µL−1), and TCEP (1 mM). The assay was incubated at 30 °C for 3 h and prepared as described above for analysis by MALDI-TOF MS.
Glutamate elimination assays
The glutamylated precursor peptide NisA was incubated with endoprotease ArgC under the following conditions: glutamylated NisA (500 µM), Tris-HCl (20 mM), CaCl2 (10 mM), EDTA (0.5 mM), DTT (5 mM), and ArgC (0.005 µg µL−1). The reaction was incubated for 4 h at 37 °C. Glutamylated core and leader peptide were purified by analytical high performance liquid chromatography (HPLC) on a Phenomenex C18 column (Luna, 250 mm × 4.60 mm, 10 µ, 100 Å) connected to an Agilent 1260 infinity liquid chromatography system. Peptides were separated using a linear gradient of 2% (v/v) Solvent B (80% (v/v) Acetonitrile (MeCN), 20% (v/v) H2O, 0.086% (v/v) trifluoroacetic acid (TFA)) in Solvent A (0.1% (v/v) TFA in H2O) to 100% (v/v) Solvent B over 60 min at a flow rate of 1 mL min−1. Peptide elution was monitored using a 220 nm wavelength. Desired fractions were lyophilized and stored at −20 °C.
The following reaction conditions were used for the glutamate elimination assays: HEPES pH 7.5 (100 mM), glutamylated His6-NisA core peptide (~ 100 µM), NisB (5 µM), MgCl2 (10 mM), KCl (10 mM), and TCEP (1 mM). The reaction was performed in a 15 µL volume, incubated for 60 min at 30 °C and analyzed by MALDI-TOF MS as described above.
Crystallization and structure determination of NisB
His6-tagged NisB was coexpressed with untagged NisA and purified as described previously elsewhere3. Prior to crystallization, samples of NisB were methylated using a previously described method34 and purified again by size exclusion chromatography (HiLoad 16/600 Superdex 75 pg column, GE Healthcare) into a buffer of HEPES pH 7.5 (20 mM) and KCl (300 mM). Crystals of methylated NisB (5 mg mL−1) were grown using the vapor diffusion method at 9 °C using a precipitant of 100 mM Bicine (pH = 8.5), and 15–25% poly(ethylene) glycol 6000. Macro- and micro-seeding facilitated the formation of crystals suitable for diffraction data collection. SeMet NisB was expressed, purified, and crystallized in a similar manner. Crystallographic phases were determined by single wavelength anomalous diffraction methods from data collected on crystals of SeMet NisB to a resolution limit of 3.2 Å. Native and SeMet data were collected at Sector 21 ID (LS-CAT, Advanced Photon Source, Argonne National Labs, IL) and data were integrated and scaled using HKL200035 or XDS36. Heavy atom sites were located using the SHELX suite of programs37 and refinement of heavy atom parameters in SHARP38 yielded an initial figure of merit of 0.276. Density modification and twofold symmetry averaging produced experimental maps that allowed for most secondary structural elements to be manually assigned using COOT39. Further model building utilized the 2.9 Å resolution data collected on crystals of native NisB. While obvious electron density corresponding to the NisA peptide (co-purified from the coexpression with NisB) can be observed in experimentally phased maps, the peptide was only built into the model once the free R factor for the structure had dropped below 0.30. The validity of all models were routinely determined using MOLPROBITY and by using the free R factor to monitor improvements during building and crystallographic refinement.
Coexpression of NisB-R784A with NisA
Site directed mutagenesis was performed on his6-nisA/nisB pRSF Duet-13 to replace the conserved Arg784 in NisB to Ala using a previously described method40 with appropriate primers (Extended Data Table 2). The resultant plasmid was used for coexpression of His6-NisA with NisB-R784A and modified His6-NisA was purified following a previously described method3.
Construction of the LanB protein sequence similarity map
The LanB sequences were obtained by iterative Psi-Blast search, and the hits were grouped into full-length LanB and short LanB based on whether they had the SpaB-C terminal domain. Thiopeptide dehydratase sequences were obtained from the ThioBase database24. Network analysis was performed by BLASTP searches comparing each sequence against each other. A Matlab script was written to remove all duplicate comparisons, and the result was imported into the Cytoscape software package. Each node in the network represents a protein sequence, and each edge represents the pairwise connection between two sequences with BLASTP e-value lower than the cutoff value. The nodes were arranged using the yFiles organic layout provided with Cytoscape version 2.8.3. Accession numbers are listed in the Supplementary Table 1
Phylogenetic tree construction
Sequences for the SpaB C-terminal domain containing proteins were obtained by searching the NCBI assembled bacterial genomes database with the SpaB_C PFAM41 hidden Markov model using HMMER342. Putative Diels-Alderase sequences were identified constructing a hidden Markov model from an alignment of NosO orthologs obtained from the Thiobase24 database. This hidden Markov model was used in the same manner as the SpaB_C hidden Markov model. The portions of protein sequences that aligned to the SpaB_C PFAM or the putative Diels-Alderase hidden Markov model were combined and clustered using CD-HIT43, identifying representative sequences that shared no more than 50% identity. The genomic contexts of these representative sequences were examined to determine the type of protein and biosynthetic cluster in which these domains resided. Then, the sequences were aligned using PRALINE44. The alignment was then manually adjusted to remove poorly aligning regions and subsequently used to construct a phylogenetic tree using Mr. Bayes45. A portion of invariable and gamma-distributed (4 categories) rates, the WAG amino acid substitution model, and random starting trees were utilized to calculate the likelihood over 7,500,000 generations of 2 runs with 4 chains each. The final tree was generated using a 25% relative burn in. Accession numbers are listed in Supplementary Table 1.
Representative sequence alignment
Representative sequences for thiopeptide dehydratases and putative Diels-Alderases were selected from ThioBase24. The SpaB C-terminal domains of these proteins were identified by HMMER as described above and aligned with the SpaB C-terminal domains of characterized full length LanB proteins using MUSCLE46.
Extended Data
Extended Data Figure 1. RNA-dependent dehydration of His6-NisA.
(a) Anion exchange chromatogram of E. coli cell extract. Fractionation was monitored at 280 nm (blue) and sample was eluted using a NaCl gradient (red). Two peaks were observed with a retention volume of 7.3 mL and 10.3 mL, respectively. (b–f) MALDI-TOF mass spectra of His6-NisAafter in vitro reaction with His6-NisB, ATP, Glu in the presence of (b) the 10.3 mL fraction, (c) the 7.3 mL fraction, (d)E. coli cell extract, and E. coli cell extract treated with (e) DNase or (f) RNase. Numbers above the peaks correspond to the number of dehydrations of His6-NisA after incubation with His6-NisB (panels b–f) or the elution fraction (panel a). (M) Unmodified (7992 m/z, calc. 7996 m/z), (3) three-fold dehydrated (7941 m/z, calc. 7942 m/z), (5) five-fold dehydrated (7905 m/z, calc. 7906 m/z), and (7) seven-fold dehydrated (7870 m/z, calc. 7870 m/z) His6-NisA.
Extended Data Figure 2. Glutamyl-tRNA dependent dehydration of His6-NisA.
(a) Glutamylation of in vitro transcribed E. coli tRNAGlu by purified E. coli GluRS using L-[14C(U)]-glutamic acid was analyzed by gel electrophoresis. The gel was stained with methylene blue (top), dried, exposed to a phosphorimager screen, and scanned (bottom). Thus, recombinant purified GluRS is capable of aminoacylating E. coli tRNAGlu lacking post-transcriptional modifications. The gel was cropped for visualization purposes. MALDI-TOF mass spectra of the precursor peptide His6-NisA after incubation with (b) His6-NisB, glutamyl-tRNAGlu and ATP, (c) His6-NisB and glutamyl-tRNAGlu, and (d) glutamyl-tRNAGlu in the absence of His6-NisB. Numbers above the mass spectra correspond to the number of dehydrations of His6-NisAafter incubation with His6-NisB. (M) Unmodified(7995 m/z, calc. 7996 m/z), (2) two-fold dehydrated (7959 m/z, calc. 7960 m/z), and (4) four-fold dehydrated (7924 m/z, calc. 7924 m/z) His6-NisA.
Extended Data Fig. 3. His6-NisB-catalyzed dehydration of His6-NisA in the presence of Lactococcus lactis HPRNA and GluRS.
MALDI-TOF MS analysis of His6-NisA after incubation with (a) His6-NisB, L. lactis His6-GluRS, RNA isolated from L. lactis, glutamate, and ATP, (b) His6-NisB, L. lactis His6-GluRS and RNA, and ATP, (c) His6-NisB, L. lactis His6-GluRS, Glu, and ATP, (d) His6-NisB, L. lactis RNA, Glu, and ATP, and (e) L. lactis His6-GluRS and RNA, Glu, and ATP. Numbers above the mass spectra correspond to the number of dehydrations of His6-NisA after incubation with His6-NisB. (M) Unmodified (7997 m/z, calc. 7996 m/z), (5) five-fold dehydrated (7906 m/z, calc. 7906 m/z), (7) seven-fold dehydrated (7870 m/z, calc. 7870 m/z), and (8) eight-fold dehydrated (7853 m/z, calc. 7852 m/z)His6-NisA.
Extended Data Figure 4. Surface representation, structural homology, and model for tRNA engagement by NisB.
(a) The NisB homodimer is shown with one monomer colored in gold (glutamylation domain) and blue (glutamate elimination domain) while the other monomer is colored pink (glutamylation domain) and green (glutamate elimination domain). The NisA peptide is shown as spheres. (b) Surface cartoon representation showing the clustering of the residues important for glutamylation (in the pink domain) and glutamate elimination (in the green domain).(c)Calculated electrostatic potential mapped onto the NisB surface showing the basic patch (blue) that likely engages the glutamyl-tRNAGlu. The NisA peptide is shown in yellow.(d) tRNAGlu-NisB binding model with the NisB glutamylation domain in pink and elimination domain in green. The leader peptide is shown in a yellow ball-and-stick representation. The dsRNA binding protein A complexed with its cognate RNA (PDB ID: 1DI2) was used to derive a NisB docking pose for binding to bacterial tRNAGlu(T. thermophilus tRNA taken from 1N78). The model results in the placement of the aminoacylated CCA terminus in the vicinity of residues that have been shown to be important for glutamylation activity.(e) Domains within the overall structure of NisB that share notable homology are shown for the leader peptide binding site (gold), and the site for glutamate elimination (cyan). Structures that are related by homology are shown adjacent to the respective domains.
Extended Data Figure 5. Structure analysis of the glutamate elimination domain in NisB.
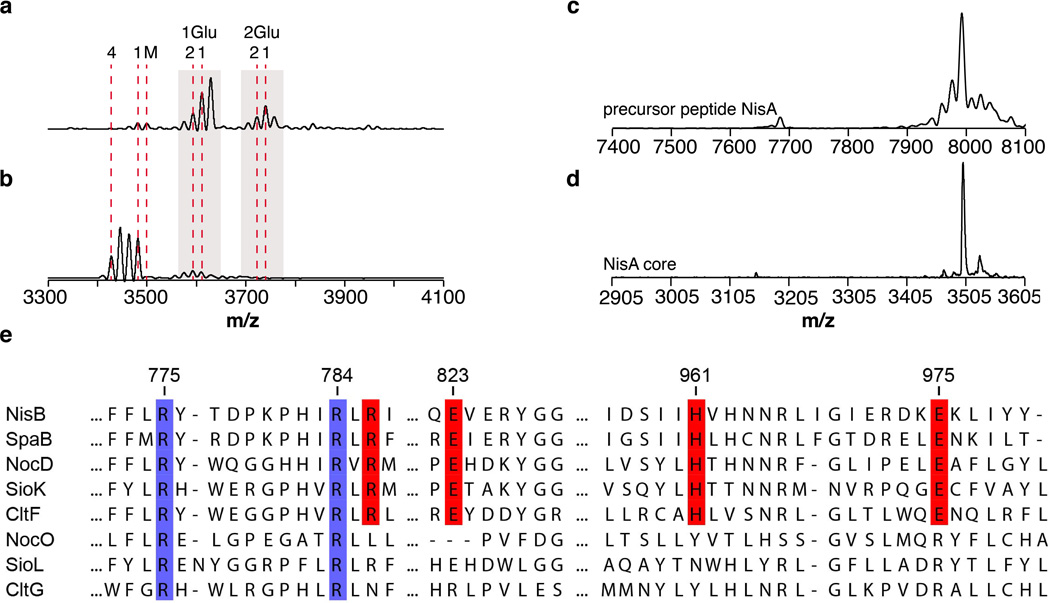
(a) Cartoon representation of the LsrG (PDB ID: 3QMQ) protein. Residues involved in LsrG catalysis are shown as sticks. The putative LsrG active site, located between the α helices and β sheet, was proposed based on the presence of an unidentified ligand17. Mutations of residues in the proposed active site of LsrG demonstrated their importance in activity but no functions were assigned to individual residues17. (b) Cartoon representation of the NisB glutamate elimination domain. Segment with structural homology to LsrG is colored light blue while the remainder of the elimination domain is colored yellow. Residues important for glutamate elimination or dehydration3 are shown as sticks. (c) Residues important for glutamate elimination and net dehydration delineate a putative glutamate elimination active site. Residues Arg786, Arg826, and His961 are important for glutamate elimination3 and Glu823has been previously shown to be partially important for dehydration3. The importance of Arg784 in the glutamate elimination step was determined in this study. (d) MALDI-TOF MS analysis of His6-NisA purified after coexpression with His6-NisB-R784A. The presence of multiple glutamylated intermediates demonstrates that Arg784 is important for glutamate elimination and not for glutamylation. The designation of 1Glu and 2Glu above the peaks indicates the number of glutamate adducts on the family of peaks, with the number shown below indicating the additional number of dehydrations for each peak.
Extended Data Figure 6. NisB-catalyzed glutamate elimination of glutamylated NisA core peptide, laser induced deamination of full-length precursor peptide NisA, and sequence analysis of the glutamate elimination domain in NisB.
MALDI-TOF MS analysis of glutamylated NisA core peptide(a) before, and (b) after incubation with His6-NisB. The data show that the leader peptide is not required for NisB-catalyzed glutamate elimination. Numbers above correspond to the number of glutamate adducts or dehydrations of NisA core peptide. (M) NisA core peptide (3499 m/z, calc. 3498 m/z), (1) one-fold dehydrated (3481 m/z, calc. 3480 m/z), and (4) four-fold dehydrated (3427 m/z, calc. 3426 m/z) NisA core peptide. (1Glu-1) NisA core peptide after formation of one glutamate adduct and one dehydration (3611 m/z, calc. 3609 m/z), (1Glu-2) NisA core peptide after formation of one glutamate adduct and two dehydrations (3593 m/z, calc. 3591 m/z), (2Glu-1) NisA core peptide after formation of two glutamate adducts and one dehydration (3740 m/z, calc. 3738 m/z), and (2Glu-2) NisA core peptide after formation of two glutamate adducts and two dehydrations (3722 m/z, calc. 3720 m/z). (c) MALDI-TOF MS in reflective mode of the precursor peptide NisA. (d) MALDI-TOF MS in reflective mode of NisA core peptide after treatment with the protease ArgC. ArgC cleaves after Arg–1 in the leader peptide of NisA (Fig. 1a). MALDI-TOF MS analysis of the precursor peptide NisA in reflective mode caused the appearance of smaller shoulder peaks next to the parent mass. These shoulder peaks correspond to laser induced deamination of the parent mass and are only observed for high molecular weight peptides (> 6 kDa). The shoulder peaks are not observed after proteolytic removal of the leader peptide, confirming that they were not the result of dehydrations. (e) Sequence alignment of selected elimination domains (SpaB_C, PF14028) present in full LanBs (NisB, SpaB), thiopeptide dehydratases (NocD, SioK, CltF), and putative24 thiopeptide Diels-Alderases (NocO, SioL, and CltG). Residues involved in glutamate elimination (red) are conserved in full LanBs and thiopeptide dehydratases but not in the putative Diels-Alderases.
Extended Data Table 1.
Data collection, phasing and refinement statistics for NisB.
| Native 1 | SeMet | |
|---|---|---|
| Data collection | ||
| a, b, c (Å) | 98.5, 107.3, 135.7 | 100.8, 108.5, 136.5 |
| β (°) | 109.9 | 110.1 |
| Resolution (Å)* | 50.00 −2.9 (3.0 - 2.9) | 50.00 - 3.2 (3.2 - 3.26) |
| Total reflections | 257,756 | 217,912 |
| Unique reflections | 58,894 | 45,467 |
| Rsym (%) | 10.2 (87.1) | 7.9 (82.5) |
| I/σ(I) | 10.6 (1.9) | 17.9 (2.0) |
| Completeness (%) | 99.6 (99.8) | 99.2 (98.7) |
| Redundancy | 4.4 (4.4) | 4.8 (4.8) |
| Phasing | ||
| Figure of Merit† | 0.276/0.76 | |
| Refinement | ||
| Resolution (Å) | 34.5 - 2.9 | |
| No. reflections used | 58,852 | |
| Rwork /Rfree‡ | 19.4/25.7 | |
| Number of atoms | ||
| Protein | 15,901 | |
| Peptide | 200 | |
| B-factors | ||
| Protein | 27.3 | |
| Peptide | 38.6 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles (°) | 1.36 |
Highest resolution shell is shown in parenthesis.
Figure of merit- Probability weighted averaged of the cosine of the phase error, before and after density modification.
R-factor =Σ(|Fobs|−k|Fcalc|)/Σ|Fobs| and R-free is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
Extended Data Table 2.
Oligonucleotides used in this study.
| Primer Name | 5' Sequence 3' |
| NisB R784A F | GAT ATA CTG ATC CTA AAC CAC ATA TTG CAT TGC GTA TAA AAT GTT CAG ATT TAT TTT TA |
| NisB R784A R | AAT ATG TGG TTT AGG ATC AGT ATA TCT TAG GAA GAA TAG ATT TCC ACC CA |
| L. lactis gltX Ndel F | AAT ATA ATC ATA TGA ACA AAA AAA TCC GCG TC |
| L. lactis gltX Xhol R | AAT AAT ACT CGA GTT ATT TAT AAG CGG CAT CCA AG |
| E. coli gltX Ndel F | AAT CAA TCA TAT GAA AAT CAA AAC TCG CTT CG |
| E. coli gltX Xhol R | TTA CTA CTC GAG TTA CTG CTG ATT TTC GCG TTC AG |
| E. coli tRNAGlu F | AAT TCC TGC AGT AAT ACG ACT CAC TAT AGT CCC CTT CGT CTA GAG GCC CAG GAC ACC |
| E. coli tRNAGlu R* | mUmGG CGT CCC CTA GGG GAT TCG AAC CCC TGT TAC CGC CGT GAA AGG GCG GTG TCC TGG |
m stands for methylation at the 2’ of the ribose sugar
Acknowledgements
We thank Keith Brister and colleagues for facilitating data collection at LS-CAT (Argonne National Labs, IL). This work was supported by grants from the National Institutes of Health (R01 GM 058822 to W.A.v.d.D. and RO1 GM079038 to S.K.N.). M.A.O was supported partially by the NIGMS-NIH Chemistry-Biology Interface Training Grant (5T32-GM070421) and by the Ford Foundation. Y.H. was supported partially by a Lowell P. Hager fellowship from the Department of Biochemistry. The Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the National Institutes of Health (S10 RR027109 A). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or Ford Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at nature.com/nature.
Authors Contributions M.A.O. performed all biochemical assays, which were designed and analyzed by M.A.O. and W.A.V. Y.H. and S.K.N. performed and interpreted all structural studies. Q.Z. and M.C.W. performed bioinformatic analysis. M.A.O., S.K.N., and W.A.V. wrote the manuscript. M.A.O. and Y.H. contributed equally to this study.
Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Base (http://www.rcsb.org) under accession code (4WD9).
The authors declare no competing financial interest.
References and Notes
- 1.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 2.Sen AK, et al. Post-translational modification of nisin. The involvement of NisB in the dehydration process. Eur. J. Biochem. 1999;261:524–532. doi: 10.1046/j.1432-1327.1999.00303.x. [DOI] [PubMed] [Google Scholar]
- 3.Garg N, Salazar-Ocampo LM, van der Donk WA. In vitro activity of the nisin dehydratase NisB. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7258–7263. doi: 10.1073/pnas.1222488110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breukink E, et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 6.Wiedemann I, et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 7.Hasper HE, et al. A new mechanism of antibiotic action. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 8.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 9.Kaletta C, Entian KD. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J. Bacteriol. 1989;171:1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, et al. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 11.van der Meer JR, et al. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnell N, et al. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 14.Shazman S, Mandel-Gutfreund Y. Classifying RNA-binding proteins based on electrostatic properties. PLoS Comput. Biol. 2008;4:e1000146. doi: 10.1371/journal.pcbi.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehnke J, et al. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew. Chem. Int. Ed. 2013;52:13991–13996. doi: 10.1002/anie.201306302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition Sequences and Substrate Evolution in Cyanobactin Biosynthesis. ACS Synth. Biol. 2014 doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques JC, et al. Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): characterization of phospho-(S)-4,5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. J. Biol. Chem. 2011;286:18331–18343. doi: 10.1074/jbc.M111.230227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavaro A, et al. Substrate recognition and specificity of NisB, the lantibiotic dehydratase involved in nisin biosynthesis. J. Biol. Chem. 2011;286:30552–30560. doi: 10.1074/jbc.M111.263210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plat A, Kluskens LD, Kuipers A, Rink R, Moll GN. Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 2011;77:604–611. doi: 10.1128/AEM.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khusainov R, Heils R, Lubelski J, Moll GN, Kuipers OP. Determining sites of interaction between prenisin and its modification enzymes NisB and NisC. Mol. Microbiol. 2011;82:706–718. doi: 10.1111/j.1365-2958.2011.07846.x. [DOI] [PubMed] [Google Scholar]
- 21.Lubelski J, Khusainov R, Kuipers OP. Directionality and Coordination of Dehydration and Ring Formation during Biosynthesis of the Lantibiotic Nisin. J. Biol. Chem. 2009;284:25962–25972. doi: 10.1074/jbc.M109.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, et al. Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12031–12036. doi: 10.1073/pnas.1406418111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lubelski J, Overkamp W, Kluskens LD, Moll GN, Kuipers OP. Influence of shifting positions of Ser, Thr, and Cys residues in prenisin on the efficiency of modification reactions and on the antimicrobial activities of the modified prepeptides. Appl. Environ. Microbiol. 2008;74:4680–4685. doi: 10.1128/AEM.00112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, et al. ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS One. 2012;7:e45878. doi: 10.1371/journal.pone.0045878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Ntai I, Kelleher NL, Walsh CT. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12249–12253. doi: 10.1073/pnas.1109539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bougioukou DJ, Mukherjee S, van der Donk WA. Revisiting the biosynthesis of dehydrophos reveals a tRNA-dependent pathway. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10952–10957. doi: 10.1073/pnas.1303568110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 29.Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584:366–375. doi: 10.1016/j.febslet.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
References cited in Methods
- 28.Li B, Cooper LE, van der Donk WA. In vitro studies of lantibiotic biosynthesis. Methods Enzymol. 2009;458:533–558. doi: 10.1016/S0076-6879(09)04821-6. [DOI] [PubMed] [Google Scholar]
- 29.Kigawa T, et al. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 30.Sherlin LD, et al. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. Rna. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 31.Rio DC, Ares MJ, Hannon GJ, Nilsen TW. RNA: A laboratory Manual. Cold Spring Harbor Laboratory Press; 2011. pp. 216–219. [Google Scholar]
- 32.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssen BD, Diner EJ, Hayes CS. Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Methods. Mol. Biol. 2012;905:291–309. doi: 10.1007/978-1-61779-949-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. Sect. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorn A, Sheldrick GM. Extending molecular-replacement solutions with SHELXE. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:2251–2256. doi: 10.1107/S0907444913027534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]