Abstract
A widespread and complex distribution of vitamin requirements exists over the entire tree of life, with many species having evolved vitamin dependence, both within and between different lineages. Vitamin availability has been proposed to drive selection for vitamin dependence, in a process that links an organism's metabolism to the environment, but this has never been demonstrated directly. Moreover, understanding the physiological processes and evolutionary dynamics that influence metabolic demand for these important micronutrients has significant implications in terms of nutrient acquisition and, in microbial organisms, can affect community composition and metabolic exchange between coexisting species. Here we investigate the origins of vitamin dependence, using an experimental evolution approach with the vitamin B12-independent model green alga Chlamydomonas reinhardtii. In fewer than 500 generations of growth in the presence of vitamin B12, we observe the evolution of a B12-dependent clone that rapidly displaces its ancestor. Genetic characterization of this line reveals a type-II Gulliver-related transposable element integrated into the B12-independent methionine synthase gene (METE), knocking out gene function and fundamentally altering the physiology of the alga.
Introduction
All organisms must balance the cost of maintaining metabolic independence with the risk of restricting their niche by depending on environmental sources of enzyme cofactors. These cofactors perform essential metabolic functions and, when supplied externally, are known as vitamins. Animals obtain vitamins from their diet and are thus described as vitamin auxotrophs. Some organisms avoid the need for external sources of vitamins, because they synthesize the cofactors themselves. However, vitamin biosynthesis can be metabolically expensive, and as these compounds are required in only trace quantities, outsourcing production could be selected for if an exogenous vitamin supply is available. The loss of vitamin synthesis has happened frequently in both prokaryotes and eukaryotes (Helliwell et al., 2013), suggesting that the conditions for evolutionary shifts in vitamin requirements commonly occur in space and time. One well-known example of this is vitamin C auxotrophy, which arose independently in primates, guinea pigs, teleost fish and certain bat species as the result of loss of the final enzyme in the biosynthetic pathway, L-gulonolactone oxidase (Nishikimi et al., 1994; Drouin et al., 2011). As the lineages that can no longer synthesize this vitamin have a vitamin C-rich diet, it has been hypothesized that diet may have led to the evolution of this trait (Drouin et al., 2011).
Vitamin dependence is not, however, confined to animal taxa (Helliwell et al., 2013). For instance, the requirement for biotin (vitamin B7) varies between strains of the yeast Saccharomyces cerevisiae. Genomic evidence has revealed a partial pathway for biosynthesis of this vitamin in the strain S. cerevisiae S288c, suggesting that the ability to synthesize this cofactor has been lost recently (Hall and Dietrich, 2007). Among algae—taxonomically diverse photosynthetic eukaryotes—vitamin auxotrophy is also a highly variable trait. Over 50% of species surveyed require vitamin B12 (cobalamin), approximately 21% B1 (thiamine), and 5% B7 (biotin) (Croft et al., 2006), and the distribution of requirement does not follow phylogenetic lines. Unlike other B vitamins, vitamin B12 is synthesized only by prokaryotes (Warren et al., 2002). In aquatic ecosystems, ambient concentrations of B12 are extremely low (Sañudo-Wilhelmy et al., 2012), and it has been proposed that the availability of this factor may exert significant constraints on the distribution, taxonomic composition and primary productivity of algal communities (Gobler et al., 2007; Sañudo-Wilhelmy et al., 2012; Bertrand et al., 2012a). However, the prevalence of algal vitamin B12 requirers in nature implies that there is a readily available/common niche for auxotrophic algae to occupy. Current understanding suggests that B12 requirers may obtain a source of vitamin B12 through: (i) direct interactions with heterotrophic bacteria (Croft et al., 2005; Wagner-Döbler et al., 2010; Kazamia et al, 2012) and/or (ii) uptake from the dissolved vitamin pool, in patches of elevated microbial activity—that is, non-specific interactions with prokaryote producers (Karl, 2002; Azam and Malfatti, 2007). Based on genome analyses, prokaryotic taxa implicated in cobalamin synthesis include members of the Alphaproteobacteria, Gammaproteobacteria, Cyanobacteria and Bacteroidetes (Sañudo-Wilhelmy et al., 2014). A more recent study also revealed a globally significant role for the Archaea (Thaumarchaeota) in vitamin B12 production in aquatic ecosystems (Doxey et al., 2014).
Insights into the molecular basis underlying the vitamin requirements of algae have also been gained using available genome sequences. Unlike for other vitamins, where possession of the biosynthetic pathway means an organism does not require an external supply of the compound, vitamin B12 independence is conferred by the presence of an enzyme that does not need a cobalamin cofactor (Croft et al., 2005, 2006). Three B12-requiring enzymes are known in eukaryotes: (i) methylmalonyl-CoA mutase, used for odd chain-fatty-acid metabolism, (ii) type II ribonucleotide reductase involved in deoxyribose biosynthesis, and (iii) methionine synthase (METH), which catalyses the biosynthesis of methionine (Marsh, 1999). A B12-independent form of methionine synthase (METE) is found in land plants and fungi, and therefore these organisms do not require vitamin B12. A survey of algal genomes showed that algal B12 independence correlates with the presence of a functional copy of METE (Croft et al., 2005; Helliwell et al., 2011; Bertrand and Allen 2012b). The model green alga Chlamydomonas reinhardtii does not require vitamin B12 and possesses both isoforms of methionine synthase, whereas METE has been lost in other closely related B12-dependent species (Helliwell et al., 2011).
Determining and testing the selective pressures contributing to the evolution of vitamin dependence is a key component in understanding the evolution of a species niche and its biotic interactions with co-occurring species. Although comparative analyses can show which environmental conditions correlate with the evolution of vitamin dependencies, only experimentation can test definitively whether particular drivers, such as a shift in diet/environment, are sufficient to cause such major metabolic changes. A reliable and abundant external source of B12 may lead to the deterioration of METE through relaxed selection (Helliwell et al., 2011), whereby the negative regulatory effect of B12 on METE expression could facilitate this process (Helliwell et al., 2013, 2014). Here we adopt an experimental evolution approach using the fast growing alga C. reinhardtii to study the processes shaping the metabolic demand for vitamin B12. We focus on identifying the genetic changes involved, as previous work has suggested that the presence/absence of a single gene METE is a sufficient predictor of B12 auxotrophy in algae (Croft et al., 2005; Helliwell et al., 2011). Linking environmental conditions to evolutionary changes in basic metabolism in phytoplankton is vital to understand better ecosystem function and biogeochemical cycling in dynamic aquatic environments.
Materials and methods
Selection experiment
Selection was carried out in 24-well plates containing 2 ml of TAP medium (Gorman and Levine, 1965) at 25 °C in continuous light (20 μmol m−2 sec−1) with shaking (140 r.p.m.). Forty-six independent populations were founded from a single colony of the ancestral line (AL) C. reinhardtii strain 12, derived from wild-type strain 137c. Cells were transferred every Monday, Wednesday and Friday, with growth periods approximately 51, 53 and 64 h, respectively. Optical density (OD730) was measured after every transfer, which determined the subsequent transfer volume to obtain ∼8000 cells per inoculum. As such, cells never exceeded a cell density of ∼3 × 106 cells ml−1. Stock-points were taken after 13, 25, 40, 50, 60 and 70 transfers and maintained on 2% TAP agar in 24-well plates in the dark.
Pure culture growth rates
Pure culture growth assays were measured in 24-well plates in the presence of vitamin B12 (1000 ng l−1) in the same growth chamber and conditions as used for the selection experiment (described above). Ten independent S-type (B12-dependent), H-type (B12-independent) and R-type (B12-independent, derived from S-type clones following loss of the transposon from METE) clones from population E8+ at transfer T70, alongside 10 AL clones were isolated from single colonies grown on 2% TAP agar and allowed to recover for 3–6 days. Prior to the growth assay, cultures were acclimated to the growth assay conditions (with 1000 ng l−1 B12 supplementation) for 4 days and then diluted to a cell density of 4000 cells ml−1 (that is, an 8000-cell inoculum). The number of cells ml−1 was subsequently measured every 12 h over a 96-h time period using the Duel Threshold Beckman Coulter (Z2) Particle Counter and Size Analyzer (Brea, CA, USA) with a 70-μm diameter aperture, counting between 3 μm (Tl) and 9 μm (Tu). Values given are means of 10 independent replicates.
Molecular methods
DNA/RNA were extracted, and PCR/reverse transcriptase-PCR experiments were performed as described by Helliwell et al. (2011). Primers used are listed in Supplementary Table S1.
Southern blotting
Extracted DNA, 1.5 μg from each sample, was digested with NaeI and BamHI (NEB, Hichin, UK), separated by agarose-gel electrophoresis and then transferred to Hybond-N+ (GE-Healthcare, Chalfont St Giles, UK) membranes. A 339-bp probe (Supplementary Table S1) was amplified using PCR and labelled with [α-32 P]dCTP using Ready-to-Go DNA-labelling beads (GE-Healthcare). The blots were prehybridized overnight at 65 °C in Church buffer (Church and Gilbert, 1984). The probe was denatured by heating at 100° C for 10 min and added to the hybridization tubes. Hybridization was carried out overnight at 65 °C. Filters were washed at 65 °C in increasingly stringent buffers (2 × sodium chloride/sodium citrate (SSC), 0.1% sodium dodecyl sulfate (SDS) to 0.2 × SSC, 0.1% SDS) until counts were ∼1000 c.p.m.
Western blotting
Total protein was extracted, and western blotting experiments were performed as described by Helliwell et al. (2014). To verify adequate transfer and equal loading, the membrane was stained in Ponceau stain (0.2% (w/v) Ponceau-S, 3% (w/v) TCA) (Romero-Calvo et al., 2010).
Results
Rapid evolution of a vitamin B12-dependent line of C. reinhardtii
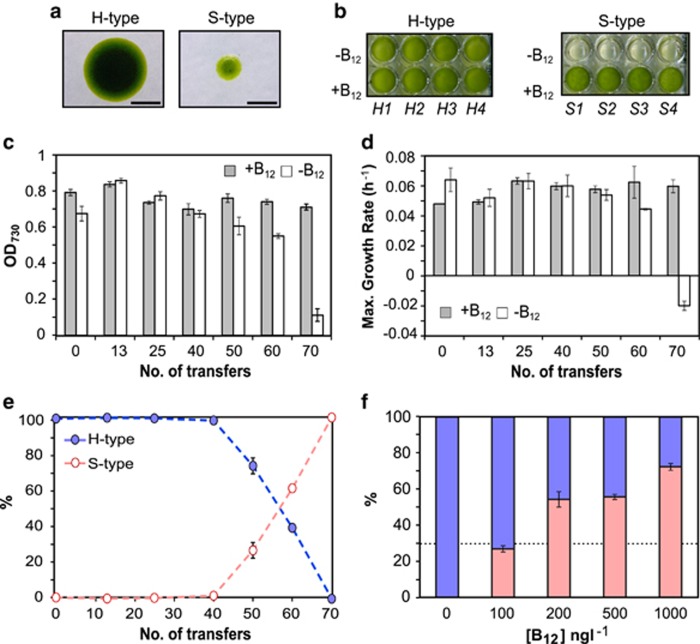
To investigate whether an exogenous supply of vitamin B12 could lead to auxotrophy, we established an evolution experiment where 46 independent populations of C. reinhardtii, were founded from a single clone (the AL). Half the populations were grown without B12 on TAP medium (in Materials and methods section) and the other half with 1000 ng l−1 vitamin B12, an amount that exceeds the growth requirements of B12-requiring algae (Croft et al., 2005). The populations were sub-cultured into fresh medium at regular intervals, with the maximum cell density reaching ∼3 × 106 cells ml−1. Populations were scored for B12 dependence every 10 transfers (T). At T60 (∼600 generations), one of the populations supplemented with B12 (evolved line, E8+) had impaired ability to grow without the vitamin. When E8+ cells were plated on solid media so that colonies could grow from single cells, in the absence of B12 two colony morphologies were evident: healthy (H-type) normal-sized colonies, and smaller (S-type) colonies impaired in growth (Figure 1a); on B12-containing medium all colonies appeared normal-sized. Growth assays in liquid culture revealed that cells isolated from H-type colonies were vitamin B12 independent, while S-type cells were dependent on the vitamin for growth in liquid culture during a 72-h cultivation window (Figure 1b). We found no evidence of S-type cells in any of the other replicate populations, when cells were plated out on TAP media in the absence of B12.
Figure 1.
The evolution of vitamin B12 dependence in C. reinhardtii. (a) E8+ cells plated onto solid medium −B12 give rise to two colony morphologies: healthy (H-type) colonies, and smaller (S-type) colonies (as visualized under a dissecting microscope), scale bar: 1 mm. (b) Growth of four independent H- and S-type colonies +B12 (1000 ng l−1) and −B12 after 72 h (mean±s.e.m.) n=3. Mean optical density (OD)730 values for H- and S-type clones at this time point were: 0.78±0.08 s.e.m. (+B12) and 0.78± 0.04 (−B12) and 0.64±0.009 (+B12) and 0.04± 0.02 (−B12), respectively. (c) OD730 of stock-points cultures on liquid medium with (1000 ng l−1; grey) and without B12 after 72 h (mean±s.e.m.) n=3 and (d) maximal growth rate, μ (h−1) of stock-points cultures on liquid medium with (1000 ng l−1; grey) and without B12 as calculated from panel (c) (mean±s.e.m.) n=3. (e) Percentage of S- vs H-type colonies within the population at independent stock-points (mean±s.e.m.) n=3. (f) Percentage of S- (red) and H-type colonies (blue) after replaying selection from T50 (where S-type cells represent <30% of the population, broken black line) for 10 transfers at different concentrations of B12 (mean±s.e.m.) n=3.
Selective sweep of the novel B12-dependent clone
Stocks of independent populations were collected throughout the experiment at T13, 25, 40, 50, 60 and 70, and stored on solid medium. To identify the point at which the S-type cells arose, we grew each stock-point for the E8+ population in liquid medium with or without B12. Growth in the presence of B12 was comparable between stocks (Figures 1c and d). In contrast, on medium without the vitamin, the B12-dependent phenotype was more pronounced in the E8+ population with increasing transfers (Figures 1c and d). Plate assays to quantify the percentage of cells giving rise to S-type colonies on medium without B12 showed S-type cells increased in frequency within the population from 1.6% to 99.7% over 30 transfers (T40–T70) (Figure 1e). To define the level of B12 sufficient to produce this response, a ‘replay' experiment was conducted. We returned to stock-point T50 (where S-type cells comprised <30% of the population) and repeated the selective regime, at a range of concentrations of B12. After 10 transfers with ⩾200 ng l−1 (0.2 μM), the B12-dependent cells rose in frequency within the population (Figure 1f). Indeed, a B12-dose response confirms that S-type cells can grow unimpaired at this concentration (Supplementary Figure S1).
A transposition event underlies the B12-dependent phenotype
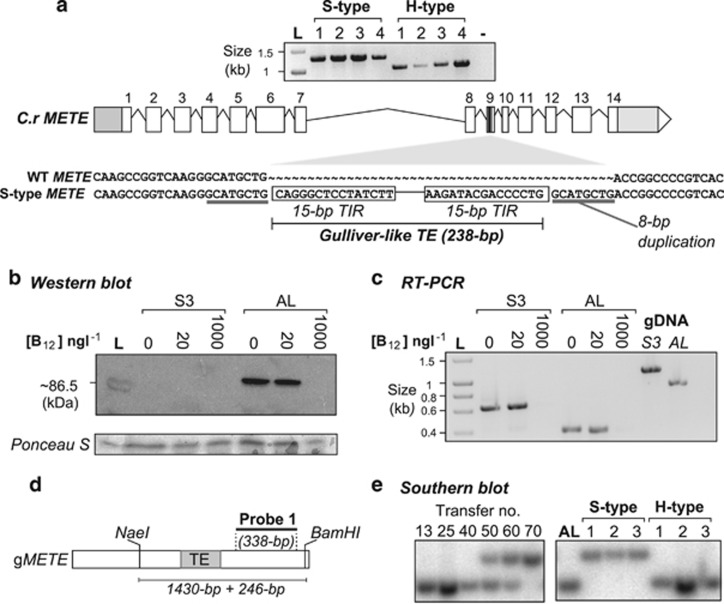
To characterize the genetic cause of the novel B12-dependent phenotype, we conducted a PCR-based analysis of the METE gene in S- and H-type clones. This approach revealed a size polymorphism between the different clone types, in the region corresponding to the ninth exon of the gene (Figure 2a). Sequencing and BLAST analysis revealed that a 238-bp class-II (‘cut-and-paste') Gulliver-related transposable element (GR-TE) had integrated into METE in S-type cells (Figure 2a, Supplementary Figures S2a and b). GR-TEs have been described previously in C. reinhardtii (Kim et al., 2005, 2006). Such elements belong to a family of >200 small, non-autonomous TEs and feature characteristic 15-bp imperfect terminal-inverted repeats that are also found in a larger transposon (∼12 kb) known as Gulliver, which is thought to activate mobilization of the GR-TE elements (Ferris, 1989; Kim et al., 2006). The transposition event described here causes an 8-bp duplication of the target-site in the gene (Figure 2a), characteristic of Gulliver elements (Ferris, 1989). Insertion of the GR-TE was into a highly conserved region of the protein and resulted in an in-frame stop codon that would be likely to cause premature termination of translation (Gonzalez et al., 1992; Pejchal and Ludwig, 2005) (Supplementary Figure S3). Indeed, western blotting analysis using a polyclonal antibody against C. reinhardtii METE (Schneider et al., 2008) detected a band of 86.5 kDa in AL cells but no cross-reacting polypeptide in an S-type clone from the E8+ population (Figure 2b). Nonetheless, the METE transcript remained expressed (at 0 and 20 ng l−1 B12), and repressed by B12 (1000 ng l−1), as is characteristic for wild-type C. reinhardtii METE (Figure 2c; Croft et al., 2005; Helliwell et al., 2011, 2014).
Figure 2.
Identification of a Gulliver-related transposable element (GR-TE) in the METE gene of E8+ S-type cells. (a) PCR on genomic DNA of four independent S- and H-type clones using primer pair F2b/R3b (amplifying a 1-kb region between 4.4 kb and 5.4 kb from the start codon) reveals an unexpectedly large product for S-type clones (expected product size for wild-type (WT) METE: 1003 bp). A BLAST search using the sequence from the S-type product revealed a strong (E-value: 8e−67) hit for C. reinhardtii METE (Supplementary Figure S2a). Another hit (E-value: 2e−87) 238 bp in size was identified as a class-II GR-TE (Kim et al., 2005, 2006; Supplementary Figure S2b). The schematic diagram shows an alignment between C. reinhardtii WT METE in this region compared with the ‘S-type' product sequence. A target-site duplication of METE (grey underline) flanks a 15-bp terminal-inverted-repeat (boxed). (b) Western blotting analysis on total protein of E8+ and AL cells using a polyclonal antibody against C. reinhardtii METE (∼86.5 kDa; Schneider et al., 2008; L: Ladder). To verify adequate transfer and equal loading, the membrane was stained in Ponceau stain (Ponceau S) (c) Reverse transcriptase-PCR reveals that METE is expressed and regulated by B12 in E8+. Expected products using primers Transcript_F1/R1: AL gDNA: 902 bp (+246 bp with TE+8-bp METE repeat, that is, 1148 bp), cDNA: 371 bp (+246 bp, that is, 617 bp). (d) Schematic diagram of probe used for Southern blotting. (e) Southern blotting analysis using the METE probe (probe 1) on genomic samples for stock-points and independent S- and H-type clones.
A Southern blotting analysis of genomic DNA prepared from each of the stock-points was carried out, using a probe (338 bp) to an internal region of METE (Figure 2d). This probe hybridized to a band of the expected size (1430 bp) in the AL and in all but the last stock-points (Figure 2e). However, a second, larger band appears at T50 (∼500 generations), which corresponds to the B12-dependent phenotype, likely to be the arrival of the TE. Both the large and small METE bands are evident between T50 and T60, until T70, where only the large band is detectable. We interpret these data to confirm that a B12-dependent phenotype of C. reinhardtii arose between T40 and T50, through transposition of a GR-TE into METE, correlating with growth experiments (Figures 1d and e). These B12-dependent cells remained in co-culture with their B12-independent predecessors for a further 20 transfers (<200 generations), until eventually the B12-dependent clones dominated the population (T70, <700 generations). Samples prepared from individual S- and H-type clones (Figure 1b) show only the larger and smaller products, respectively (Figure 2e).
Phenotypic plasticity in response to exogenous levels of vitamin B12
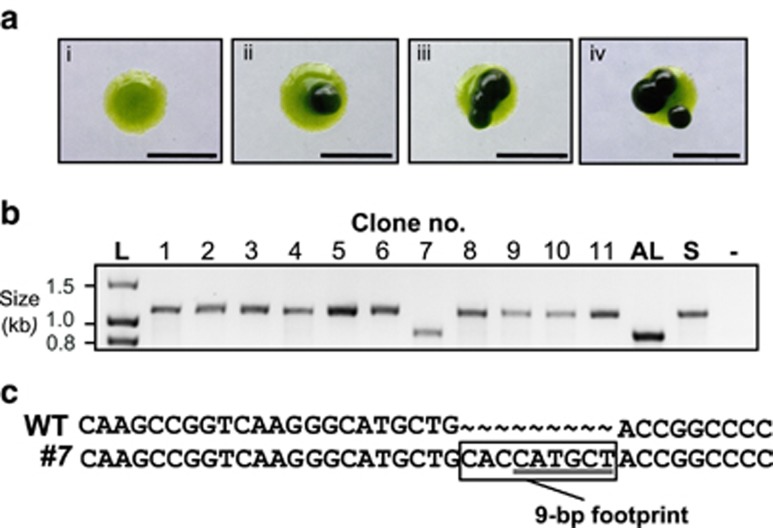
Reversion of mutant phenotypes by transposon excision is well documented, especially in conditions of physiological stress (McClintock, 1948; Maumus et al., 2009). We sought to investigate the occurrence of reversion in the evolved E8+ S-type cells in B12-deplete conditions. Eight days after plating on solid medium, S-type colonies were seen on plates lacking B12 (Supplementary Figure S4). However, after a further 3 days, darker bodies of cells appeared within the S-type colonies on the plates without B12 (Figure 3a; Supplementary Figure S4). As they grew after colonies on the control plate with B12 were already visible, we reasoned that they were likely to be revertants. Sequencing revealed complete excision of the transposon from the METE gene in such cells. We also screened 11 S-type colonies that showed no evidence of phenotypic reversion after 15 days on B12-deplete medium (Supplementary Figure S4). All 11 clones were confirmed to be vitamin B12 dependent, and using PCR with primers spanning the GR-TE the majority were also shown to have GR-TE (Figure 3b). However, one B12-dependent clone (clone no. 7) generated a PCR product with the size expected for wild-type METE (Figure 3b). Sequencing revealed that the GR-TE was absent except for a 9-bp footprint sequence (CACCATGCT), the latter 6 bp of which is a remnant of the METE repeat (Figure 2a; Figure 3c). This in-frame insertion leads to inclusion of three extra amino acids in a conserved region of the METE gene (Supplementary Figure S3) resulting in a stable vitamin B12-dependent mutant.
Figure 3.
Characterization of mutant phenotype revertants and isolation of a stable METE insertion mutant (a) A non-reverting colony (i) alongside three independent revertant colonies (ii–iv) visualized under a dissecting microscope, after 11 days on solid medium −B12. (b) PCR screen for the presence of GR-TE insertion in METE gene of clones using primers spanning GR-TE insertion site (METE_revert F1/R1). Clone no. 7 is vitamin B12 dependent yet lacks the GR-TE (expected product sizes: wild-type METE− 913 bp, and METE with GR-TE insertion 913 bp+246 bp=1159 bp). Sequencing revealed a 9-bp footprint (CACCATGCT) in this clone (c) the latter 6 bp of which (underlined grey) is a remnant of the METE repeat.
Comparison of growth rates of S, H, R and AL clones in pure culture
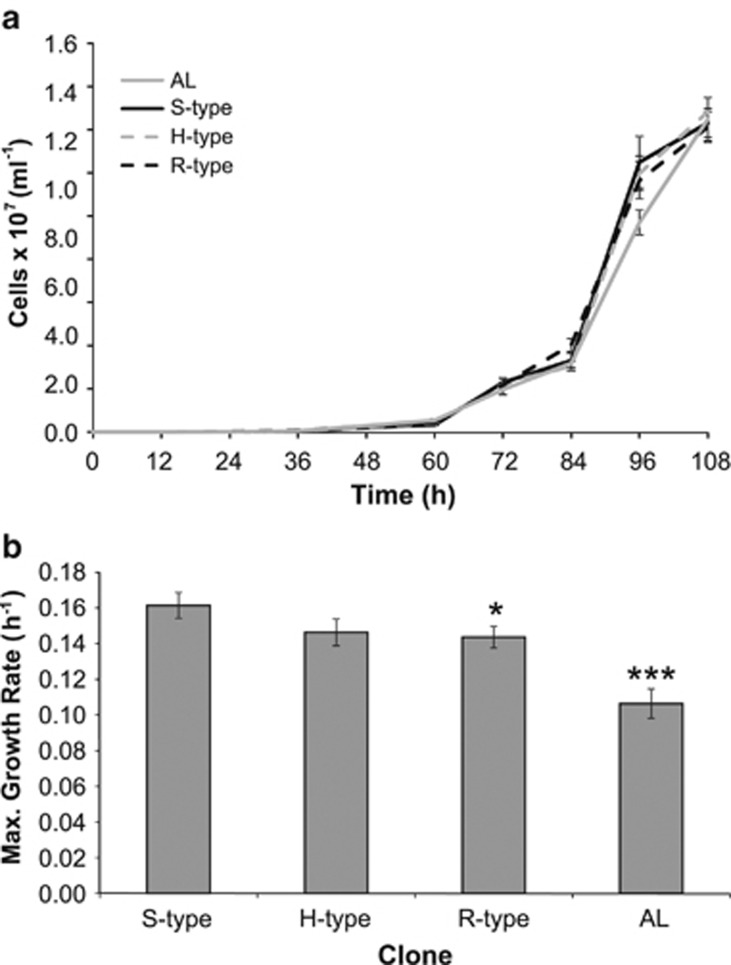
The selective sweep, which we observed in several independent experiments (including different B12 treatments), suggests that S-type cells have a growth advantage compared with their B12-independent counterparts in B12-replete conditions. Theoretical calculations (Table 1) illustrate that only a very minor increase in specific growth rate (∼4%) is required to cause the rise from 30% to 71% on hypothetical ‘strain B' within 10 transfers, similar to the population shifts we observe over this timescale with S-type cells in the replay experiment (Figure 1f). To investigate whether a growth advantage is detectable, a growth assay with pure cultures of 10 independently isolated S-type, H-type and AL clones was carried out. We also included within this analysis 10 independent R-type clones (that is, revertants derived from 10 different S-type colonies and thus representing independent reversion events) to investigate the link between fitness and METE presence/absence. We detected a ∼9% higher maximal growth rate (h−1) of S-type compared with H-type clones (Figures 4a and b); however, the difference was not statistically different using a Student's t test with a P-value of ⩽0.05. We did, however, observe a statistical difference in growth rate between S- and R-type cells (two-tailed Student's t-test P⩽0.05, n=10). The mean growth rates for H- and R-type cells were virtually identical (0.146±0.008 s.e.m. and 0.144±0.006). Moreover, all evolved lines (S-, H- and R-type) exhibited a faster maximal growth rate under the selective regime compared with the AL (P⩽0.001, n=10).
Table 1. Theoretical calculation of population shifts between two algal strains in co-culture after 24 days (10 transfers), assuming initial populations of 70% A: 30% B.
| Strain A μ (h−1) | Strain B μ (h−1) | Strain B divisions per day | Strain B % population (24 days) |
|---|---|---|---|
| 0.075 | 0.075 | 2.60 | 30 |
| 0.075 | 0.076 | 2.63 | 44 |
| 0.075 | 0.077 | 2.67 | 58 |
| 0.075 | 0.078 | 2.70 | 71 |
The calculations assume a constant specific growth rate (μ) of 0.075 h−1 in Strain A and are designed to mimic the conditions of the ‘selective sweep' experiment described in Figure 1f. The data demonstrate that only a minimal increase in specific growth rate in Strain B is required to observe a dramatic shift in the proportions of the respective populations over 24 days.
Figure 4.
Characterization of growth of S-type, H-type, R-type and AL cells (a). Growth over time of S-type, H-type, R-type and AL clones in the presence of vitamin B12 (1000 ng l−1) (mean±s.e.m.) n=10. (b) Mean maximal growth rate, μ (h−1) of S-type, H-type, R-type and AL clones as calculated from panel (a). *P⩽0.05, **P⩽0.001 compared with the S-type clones (two-tailed Student's t-test) (mean±s.e.m.) n=10.
Vitamin B12-dependent growth is rescued by B12-synthesizing bacteria
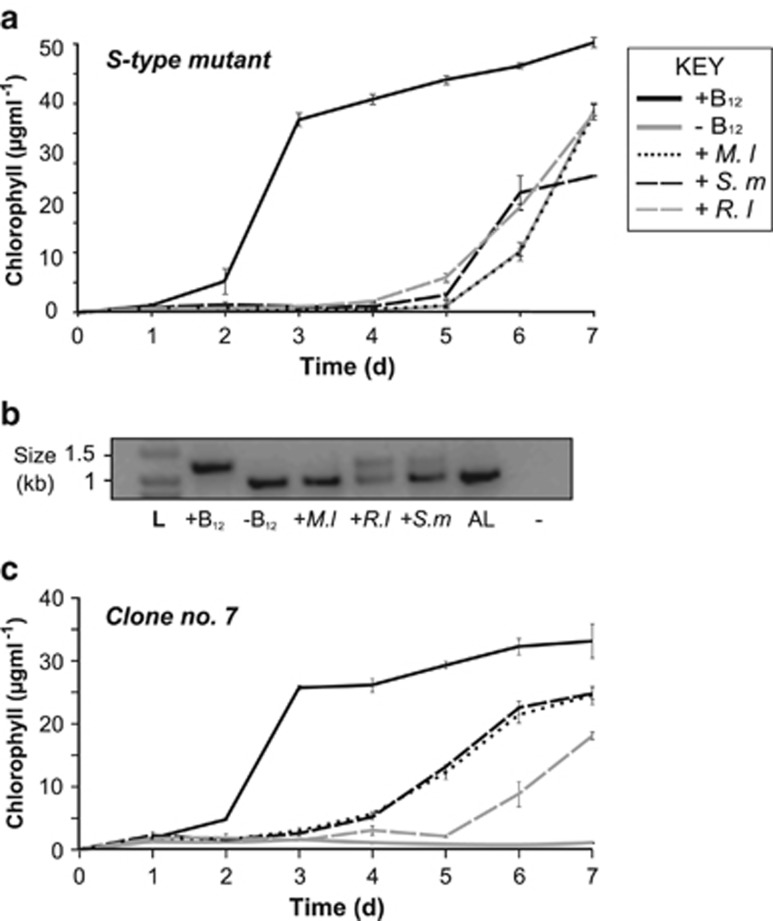
Vitamin B12 biosynthesis is confined to prokaryotes (Croft et al., 2005). The irreversible loss of METE, therefore, not only forces the evolution of vitamin auxotrophy but also an absolute dependency on a bacterial supply of the vitamin. Algal acquisition of vitamin B12 through direct mutualism with bacteria has been demonstrated previously by our laboratory (Kazamia et al., 2012), in which Lobomonas rostrata, a known B12 auxotroph, and a bacterial partner, Mesorhizobium loti, can grow stably for an indefinite period in co-culture in the absence of vitamin B12 or fixed carbon. This system has also been described mathematically (Grant et al., 2014). To test whether a similar exchange is able to support the growth of the newly evolved line, we set up co-cultures of the non-stable S-type line with one of the three B12-synthesizing rhizobial species of bacteria (M. loti (strain MAFF 303099) Rhizobium leguminosarum (RL3841) and Sinorhizobium meliloti (RM 1021)) in TAP medium lacking B12. Using chlorophyll concentration as a proxy for algal growth, we found that for the first 5 days there was no growth of the alga, except when exogenous B12 was present in the medium (Figure 5a). However, after 5 days all inocula grew well, even the control with no B12/bacterial supplementation. We interpreted that this was a result of B12-independent revertants rising to dominance within the population. Using PCR with primers spanning the GR-TE, a larger product in the +B12 treatment was identified (Figure 5b) indicating the presence of the GR-TE in the METE gene. However, for the –B12, and M. loti treatments the product was smaller, confirming excision of the transposon in these cultures. Interestingly, in the other co-cultures two products were amplified, revealing a mixed population of revertant and non-revertant clones (Figure 5b). The proportion of the two bands varied depending on which bacterial species was present, suggesting that different bacteria can support the alga to different levels and thus may dictate the frequency of B12-dependent vs independent algal clones within a population. We repeated this experiment with the stable B12-requiring clone no. 7. All three bacteria were able to support the mutant in the absence of B12, with no growth observed in the –B12 treatment (Figure 5c). Moreover, the algal–bacterial co-culture reached a lower carrying capacity compared with the +B12 treatment indicating a degree of regulation, as seen with the L. rostrata/M. loti co-culture (Kazamia et al., 2012; Grant et al., 2014). A similar result was observed in medium lacking an organic carbon source, so bacterial growth is in turn dependent on algal photosynthate (Supplementary Figure S5).
Figure 5.
Vitamin B12 dependence is rescued by three B12-synthesizing rhizobial species of bacteria. (a) Growth of S-type mutant (unstable) in different B12 regimes, including: (i) +B12 (1000 ng l−1), (ii) −B12, (iii) Mesorhizobium loti, (iv) Sinorhizobium meliloti and (v) Rhizobium leguminosarum. The latter three treatments were grown in the absence of B12 in TAP medium (mean±s.e.m.) n=3. (b) PCR with METE primers spanning the GR-TE from DNA extracted from the different conditions at day 7. (c) Growth of stable-METE-insertion mutant clone no. 7 in B12 regimes described in panel (a). This experiment was carried out in TAP medium (mean±s.e.m.) n=3.
Discussion
The evolution of vitamin dependence has been a recurrent event across the tree of life, with important implications for the basic physiology and ecology of all organisms. The processes underlying how species become dependent on an external source of these organic micronutrients are inherently difficult to test empirically. In this study, we explored directly whether a key factor hypothesized to drive the evolution of vitamin auxotrophy was able to do so. By adopting an experimental evolution approach, we found direct support for the hypothesis that an exogenous supply of vitamin B12 can lead to the evolution of B12 dependence (Figure 1). Additionally, we were able to define in detail the genetic mechanism (transposition), population dynamics (including phenotypic reversibility) and the environmental context in which this evolutionary event occurred. By establishing the genetic basis for the change in phenotype, we were able to pinpoint the precise timing of the change in genotype and characterize temporally the rise to dominance of the novel clone within the population.
Experimental evolution has been used widely as a powerful approach for understanding microbial evolution—exploiting the fast generation time and large population size of these organisms (Elena and Lenski, 2003). It allows fundamental evolutionary principles to be tested directly, and with greater rigour than alternative approaches, such as specific genome manipulation. Moreover, this technique allows detection of subtle fitness differences that would otherwise be overlooked via standard growth assays (Collins, 2011). C. reinhardtii has the lowest spontaneous mutation rate described for any eukaryote (Ness et al., 2012), and yet previous artificial selection experiments with C. reinhardtii have observed major evolutionary novelties (likely encompassing multiple gene alterations) such as loss of regulation in the carbon-concentrating mechanism (Collins and Bell, 2004) and evolution of a two-stage life cycle (Ratcliff et al., 2013), after 1000 and 312 generations, respectively. Nonetheless, the underlying genetic components of these phenotypes were not determined, so the contributions of epigenetics, point mutations, transposition events and other genetic changes to adaptive phenotypes remain unknown. To our knowledge, this is the first study characterizing transposition in an experimentally evolved algal population. Indeed, although TEs have been studied extensively in animals, plants and fungi, little is known about their significance in algal evolution. Transposons have, however, been identified in the genomes of several algal species (Armbrust et al., 2004; Bowler et al., 2008; Cock et al., 2010; Read et al. 2013), and nutrient stress (nitrate limitation) activated transposition has been observed in the marine diatom Phaeodactylum tricornutum (Maumus et al., 2009), which has also been observed with our system. Moreover, differential insertion patterns among natural isolates of diatom species from different geographic locations have been observed (Maumus et al., 2009). Together these findings suggest that TEs may have an important role in naturally evolving algal populations. An exciting area of future research will be to elucidate the impact of TEs on genome evolution of individual members of complex microbial communities, in particular understanding the frequency of transposition events and whether certain gene classes are more prone to disruption.
The fact that the METE gene loss in E8+ that we observed was due to transposition (Figure 2) has further significance, as the re-excision of the transposon allows reversion to B12 independence in response to the absence of environmental B12 (Figure 3). This temporary ‘get out of jail free card' could thus facilitate evolutionary escape from a B12-dependent lifestyle before METE further deteriorates (Helliwell et al., 2011). If similar processes happened in other algal lineages, this may explain the differences in B12 requirements observed between closely related strains by allowing for rapid and reversible evolution in environments where levels of B12 may fluctuate. The observed selective sweep of the novel evolved line E8+ within the population shows that this clone has a selective advantage compared with its ancestor. However, we must consider the possibility that genetic changes other than that to the METE gene have contributed to this fitness advantage. Whole genome analyses will be important in the future to pinpoint whether/what other genome modifications may have occurred. Nonetheless, as multiple independent isolates exhibiting reversion of the METE transposition event have a reduced growth rate relative to S-type cells (Figure 4), the selective advantage appears to be associated specifically with the loss of METE.
Vitamin B12 auxotrophy is found in half of over 300 species surveyed (Croft et al., 2005; Tang et al., 2010), and evidence suggests that B12-dependent metabolism is beneficial in certain scenarios, if B12 is readily available. For instance, METH has a catalytic efficiency 100 times greater (Gonzalez et al., 1992) and exhibits enhanced thermal tolerance, in comparison to METE (Xie et al., 2013). Moreover, theoretical calculations estimate that utilization of METH in P. tricornutum is more resource efficient than B12-independent metabolism, as the use of METE was calculated to require 30±9 times more nitrogen and 42± 5 times more zinc than METH (Bertrand et al. 2013). B12-dependent growth that favours the use of METH could therefore offer an advantage when Zn/N are limited. However, as METE expression is repressed in the presence of B12 (Croft et al., 2005; Helliwell et al., 2011; Bertrand et al., 2012a; Bertrand and Allen, 2012b; Bertrand et al., 2013; Helliwell et al., 2014) how fitness maybe conferred from inactivating a gene that is already switched off remains a conundrum. One possibility is that, in habitats where levels of vitamin B12 fluctuate, algae that have both forms of the enzyme may benefit from maintaining a low level of the METE protein, to facilitate rapid response to environmental fluctuations of B12 levels. Indeed, some METE transcript/protein can be detected under B12-replete conditions (Xie et al., 2013; Helliwell et al., 2014). However, as the levels are so low, it is unclear whether complete loss of METE would confer a metabolic saving. It is possible that METE function, even at low protein abundance, may exert an as yet unidentified energetic cost beyond simply the composition of the protein.
Whatever the explanation for the observed selective advantage of the S-type line, this study validates the hypothesis that B12 availability in the environment can lead to the taxonomically variable presence and absence of METE. In this context, it is relevant to consider levels of B12 occurring in natural aquatic environments. Recent measurements have revealed vitamin B12 depletion in large areas of coastal ocean, and the vitamin is typically absent from the euphotic zone (Sañudo-Wilhelmy et al., 2012). Moreover, levels of B12 are reportedly <10 ng l−1 (∼10 pM) in some freshwater habitats (Kurata, 1986). However, as this molecule will likely be rapidly consumed as it becomes available within the water column, measurements of standing stock concentrations alone might not accurately reflect B12 availability. Moreover, vitamin levels will vary to some extent on the microscale, with discrete vitamin patches arising due to localized microbial activity and/or the presence of particulate matter (Azam and Malfatti 2007; Stocker, 2012; Yawata et al., 2014). Interestingly, a recent study found that microscale nutrient heterogeneity could drive ecological differentiation in nutrient-acquisition strategies in marine bacteria (Yawata et al., 2014). This raises interesting eco-evolutionary considerations with regards to algal vitamin-acquisition strategies and METE presence/absence. A comprehensive comparison of the geographic distribution of vitamin B12 auxotrophs vs non-requirers in aquatic environments in relation to B12 levels has not yet been attempted. However, it is known that B12 auxotrophs such as the picoeukaryote Ostreococcus tauri are represented in environments, where ambient concentrations of vitamins are extremely low (Sañudo-Wilhelmy et al., 2012). Evolutionary adaptations enabling B12 auxotrophs to be successful competitors in B12-deprived regions could include becoming specialized at nutrient patch exploitation—being able to migrate rapidly to new nutrient sources upon a temporal change in the nutrient landscape for instance. Or alternatively, these organisms may meet their vitamin demands though the establishment and maintenance of direct symbiotic interactions with other microbes (Croft et al., 2005; Wagner-Döbler et al., 2010; Kazamia et al., 2012). As algae in possession of both METE and METH may use B12 if it is available, loss of METE could be a plausible mechanism to cause sympatric populations to embark on different evolutionary trajectories, driving the evolution of symbiotic interactions and/or other specialist nutrient-acquisition strategies. A challenging question that remains to be answered is to what extent these different strategies are represented in the natural world.
Acknowledgments
This work was supported by BBSRC grant: BB/I013164/1. We thank Professor Roger Sloboda (Dartmouth College, USA) for the METE antibody. We also thank Dr Ian Henderson, Dr Mark Scaife, Ginnie Nguyen and Vaibhav Bhardwaj (University of Cambridge) for helpful discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE, et al. Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA. 2012;109:E1762–E1771. doi: 10.1073/pnas.1201731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, Allen AE. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol. 2012;3:375. doi: 10.3389/fmicb.2012.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, McIlvin MR, Hoffman JM, Allen AE, Saito MA. Methionine synthase interreplacement in diatom cultures and communities: implications for the persistence of B12 use by eukaryotic phytoplankton. Limnol Oceangr. 2013;58:1431–1450. [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Collins S. Competition limits adaptation and productivity in a photosynthetic alga at elevated CO2. Proc R Biol Soc B. 2011;278:247–255. doi: 10.1098/rspb.2010.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Bell G. Phenotypic consequences of 1000 generations of selection at elevated CO2 in a green alga. Nature. 2004;431:566–569. doi: 10.1038/nature02945. [DOI] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- Croft MT, Warren MJ, Smith AG. Algae need their vitamins. Eukaryot Cell. 2006;5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey AC, Kurtz AK, Lynch MDJ, Suader LA, Neufeld JD.2014Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production ISME Je-pub ahead of print 15 August 2014; doi: 10.1038/ismej.2014.142 [DOI] [PMC free article] [PubMed]
- Drouin G, Godin JR, Page B. The genetics of vitamin C loss in vertebrates. Curr Genomics. 2011;12:371–378. doi: 10.2174/138920211796429736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Ferris PJ. Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher-plants. Genetics. 1989;122:363–377. doi: 10.1093/genetics/122.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Norman C, Panzeca C, Taylor GT, Sanudo-Wilhelmy SA. Effect of B vitamins and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Microb Ecol. 2007;49:181–194. [Google Scholar]
- Gonzalez JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli—2 solutions to the same chemical problem. Biochemistry. 1992;31:6045–6056. doi: 10.1021/bi00141a013. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MAA, Kazamia E, Cicuta P, Smith AG. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal-bacterial cocultures. ISME J. 2014;8:1418–1427. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Dietrich FS. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics. 2007;177:2293–2307. doi: 10.1534/genetics.107.074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Scaife MA, Sasso S, Ulian Araujo A, Purton S, Smith AG. Unravelling vitamin B12-responsive gene regulation in algae. Plant Physiol. 2014;165:388–397. doi: 10.1104/pp.113.234369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B-12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Smith AG. Widespread decay of vitamin-related pathways: coincidence or consequence. Trends Genet. 2013;29:469–478. doi: 10.1016/j.tig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10:410–418. doi: 10.1016/s0966-842x(02)02430-7. [DOI] [PubMed] [Google Scholar]
- Kazamia E, Czesnick H, Nguyen TT, Croft MT, Sherwood E, Sasso S, et al. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol. 2012;14:1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- Kim KS, Feild E, King N, Yaoi T, Kustu S, Inwood W. Spontaneous mutations in the ammonium transport gene AMT4 of Chlamydomonas reinhardtii. Genetics. 2005;170:631–644. doi: 10.1534/genetics.105.041574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata A.1986Blooms of Uroglena americana in relation to concentrations of B group vitaminsIn Kristiansen J, Andersen RA (eds).Chrysophytes: Aspects and Problems Cambridge University Press: Cambridge, UK; 185–196. [Google Scholar]
- Kim KS, Kustu S, Inwood W. Natural history of transposition in the green alga Chlamydomonas reinhardtii: Use of the AMT4 locus as an experimental system. Genetics. 2006;173:2005–2019. doi: 10.1534/genetics.106.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh EN. Coenzyme B12 (cobalamin)-dependent enzymes. Essays Biochem. 1999;34:139–154. doi: 10.1042/bse0340139. [DOI] [PubMed] [Google Scholar]
- Maumus F, Allen AE, Mhiri C, Hu H, Jabbari K, Vardi A, et al. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics. 2009;10:624. doi: 10.1186/1471-2164-10-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Mutable loci in maize. Carnegie Institute of Washington Year Book. 1948;47:155–169. [Google Scholar]
- Ness RW, Morgan AD, Colegrave N, Keightley PD. An estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics. 2012;192:1447–1454. doi: 10.1534/genetics.112.145078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M, Fukuyama R, Minoshima S, Shimizu N, Yagi K. Cloning and chromosomal mapping of the human nonfunctional gene for L-gulono-gamma-lactone oxidase, the enzyme for L-ascorbic-acid biosynthesis missing in man. J Biol Chem. 1994;269:13685–13688. [PubMed] [Google Scholar]
- Pejchal R, Ludwig ML. Cobalamin-independent methionine synthase (MetE): A face-to-face double barrel that evolved by gene duplication. PLoS Biol. 2005;3:254–265. doi: 10.1371/journal.pbio.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Herron M, Howell K, Rosenzweig F, Travisano M. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat Comms. 2013;4:2742. doi: 10.1038/ncomms3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read B, Kegal J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499:209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- Romero-Calvo I, Ocón B, Martínez-Moya P, Suárez M, Zarzuelo A, Martínez-Agustin O, et al. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal Biochem. 2010;401:318–320. doi: 10.1016/j.ab.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109:14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci. 2014;6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Ulland M, Sloboda RD. A protein methylation pathway in Chlamydomonas flagella is active during flagellar resorption. Mol Biol Cell. 2008;19:4319–4327. doi: 10.1091/mbc.E08-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R. Marine microbes see a sea of gradients. Science. 2012;338:628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B-1 and B-12 auxotrophs. Proc Natl Acad Sci USA. 2010;107:20756–20761. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker's guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- Xie B, Bishop S, Stessman D, Wright D, Spalding MH, Halverson LJ. Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J. 2013;7:1544–1555. doi: 10.1038/ismej.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. A competition-dispersal trade-off ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA. 2014;111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.