Abstract
Hepatitis C virus (HCV) infection is a leading cause of liver cirrhosis and cancer1. Cell entry of HCV2 and other pathogens3-5 is mediated by tight junction (TJ) proteins, but successful therapeutic targeting of TJ proteins has not been reported yet. Using a human liver-chimeric mouse model6 we show that a monoclonal antibody specific for TJ protein claudin-17 eliminates chronic HCV infection without detectable toxicity. This antibody inhibits HCV entry, cell-cell transmission and virus-induced signaling events. Antibody treatment reduces the number of HCV-infected hepatocytes in vivo, highlighting the need for de novo infection via host entry factors to maintain chronic infection. In summary, we demonstrate that an antibody targeting a virus receptor can cure chronic viral infection and uncover TJ proteins as targets for antiviral therapy.
The tight junction (TJ) protein claudin-1 (CLDN1) mediates hepatitis C virus (HCV) entry into host cells2. TJs are also implicated in the entry of other pathogens including dengue virus5, adenovirus3, coxsackievirus3 and shigella4. However, the role of TJ proteins in viral pathogenesis and as antiviral targets is unknown. To address this question we used a recently developed inhibitory CLDN1-specific monoclonal antibody (mAb)7 and the human chimeric uPA-SCID mouse model6. Owing to discontinuation of research in chimpanzees for ethical reasons, this mouse model is the only available model that supports robust long-term chronic HCV infection. Although these mice lack a functional immune system precluding the study of immune-mediated events, they have substantially contributed to our understanding of viral pathogenesis and the development of antivirals8-11.
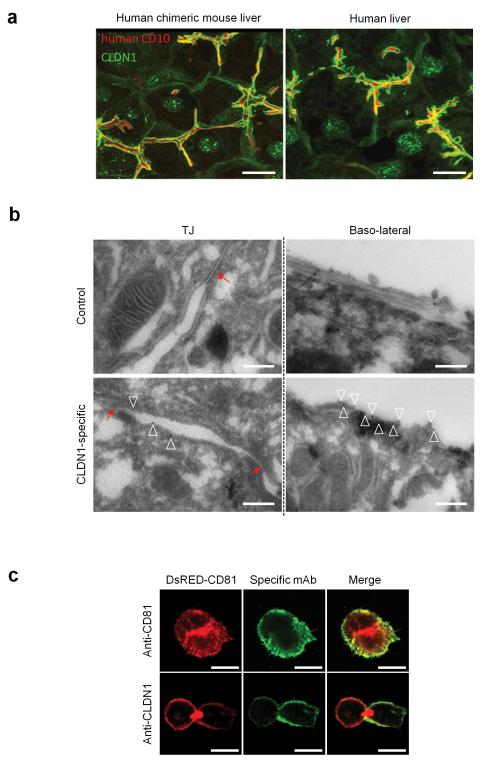
First, we analyzed CLDN1 expression in the chimeric liver using a commercially available mAb that recognizes human and mouse CLDN1 (Supplementary Fig.1). Confocal imaging demonstrated that the majority of CLDN1 on human hepatocytes co-localized with apical marker CD10 demonstrating the formation of bile canalicular structures (Fig.1a). A minor pool of protein was detected at the basolateral membrane as identified with cytokeratin-8 staining (Fig.1a, data not shown and 12). Comparative staining of normal human liver tissue demonstrated comparable subcellular localization (Fig.1a). Transmission electron microscopic (TEM) analysis confirmed that hepatocytes in the chimeric mouse liver form TJs that were structurally indistinguishable from those in human liver tissue (Supplementary Fig.2a-b). The similar localization of hepatocellular CLDN1 and hepatocyte architecture suggests that the uPA-chimeric mouse is a relevant model to evaluate CLDN1 as a therapeutic target.
Figure 1. Human CLDN1 expression and tight junction ultrastructure in the livers of human chimeric mice.
(a) Human CLDN1 expression in non-infected chimeric livers of uPA-SCID mice (left panel) as well as non-infected human livers (right panel) was assessed by confocal microscopy as described in Methods. Representative 3D composite images show human CLDN1 (green) co-stained with apical membrane marker human CD10 (red). Scale bars – 20 μm. (b) Binding of CLDN1-specific mAb to hepatocyte ultrastructures was assessed by transmission electron microscopy analyses and immunogold labeling of tissue sections of chimeric human mouse livers from mAb-treated mice. Images show TJ area (left panels) or basolateral membranes of hepatocytes (right panels). Red arrows indicate TJ, empty triangles indicate immunogold staining. Scale bars – 500nm. (c) Confocal microscopy of polarized HepG2-CD81 HCV permissive cells stained with CLDN1-specific antibody. HepG2 DsRED-CD81 cells were stained live with CD81- (upper panels) or CLDN1-specific (lower panels) antibodies and visualized by confocal microscopy. CLDN-1 specific staining is predominantly limited to the basolateral membrane of polarized cells. Scale bars – 20μm.
Next, we characterized the subcellular localization of CLDN1 recognized by the inhibitory CLDN1-specific mAb (OM-7D3-B3) when administered intraperitoneally in vivo. TEM and immunogold labeling of the CLDN1 mAb showed reactivity with basolateral and sinusoidal hepatocyte membranes in vivo (Fig.1b), whereas we failed to detect staining of TJs (Fig.1b). Basolateral membrane staining was confirmed using live cell imaging of polarized hepatoma HepG2 cells (Fig.1c)13. Collectively, these data suggest that the CLDN1-specific mAb predominantly binds to non-junctional pools of CLDN1 on the hepatocyte basolateral membrane in vivo.
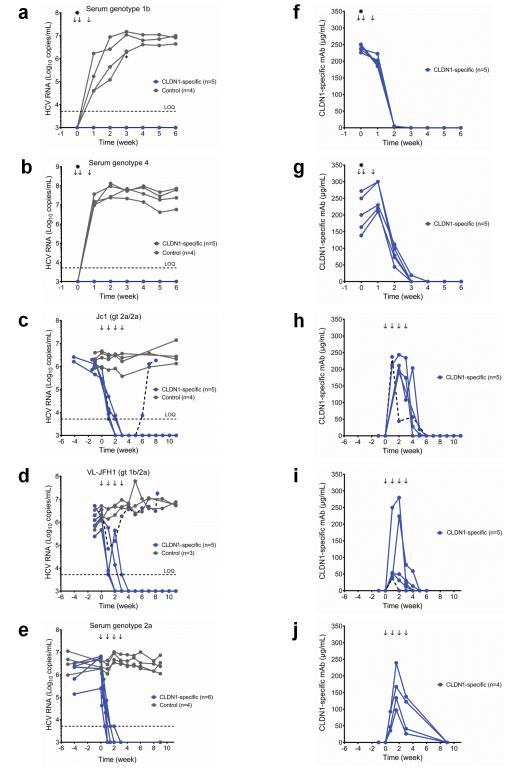
To investigate the antiviral effect of this CLDN1-specific mAb, chimeric uPA-SCID mice received CLDN1-specific (n=5) or control isotype-matched irrelevant mAb (n=4) one day prior to inoculation, and 1 and 5 days post-infection with a primary HCV genotype 1b serum. The CLDN1-specific mAb provided a complete and sustained protection from infection as evidenced by undetectable HCV RNA in the sera for up to 6 weeks post-infection (Fig.2a, Supplementary Table 1). In contrast, all control mAb-treated mice became infected (Fig.2a). Similar results were observed following infection with a HCV of the difficult-to-treat genotype 4 (isolate ED4314, Fig.2b, Supplementary Table 1) demonstrating a significant level of anti-viral activity (p< 0.0002, pooled data from experiments of both genotypes, Mann-Whitney test). Pharmacokinetic studies showed that CLDN1-specific mAb levels declined rapidly and were undetectable 2 weeks after administration (Fig.2f-g). Collectively, these data show that short-term administration of a CLDN1-specific mAb prevents de novo HCV infection and suggests that it might be used to prevent HCV re-infection during liver transplantation.
Figure 2. Prevention and clearance of chronic HCV infection using a CLDN1-specific mAb in vivo.
(a, b) Prevention studies. Chimeric uPA-SCID mice received 500 μg CLDN1-specific (n=5) or control mAb (n=4) on days −1, 1 and 5 (arrows) of inoculation (star) with genotype 1b (a) or genotype 4 (b) HCV-containing serum. One infected mouse injected with control mAb died on day 26 (a, cross). (c-e) Treatment studies. Chimeric uPA-SCID mice were chronically infected with HCVcc Jc115 (genotype 2a/2a) (c), HCVcc VL-JFH117 (genotype 1b/2a) (d) or serum of genotype 2a (e). Twenty-four to 50 days following inoculation, the animals received 500 μg control (c, d, e; n=4, 3 and 4 respectively) or CLDN1-specific mAb (c, d, e; n=5, 5 and 6 respectively) each week (arrows) for 4 weeks. Two CLDN1-specific mAb treated mice were sacrificed following viral clearance for FISH studies (e). (f-j) Serum concentration of CLDN1-specific mAb from (a-e respectively) was determined by ELISA. Serum viral load (a-e) was quantified by the clinically licensed Abbott RealTime™ HCV assay. The horizontal dashed line indicates the limit of quantification (LOQ). (c, d, h, i) The viral load and antibody concentration of the mice exhibiting a relapse is indicated by a dashed line. Values of individual animals are shown.
To investigate the effect of the CLDN1-specific mAb on chronic HCV infection we utilized mice persistently infected with the cell culture-derived HCV (HCVcc) strain Jc1 (genotype 2a/2a chimera)15, a JFH1-derived16 prototype strain. Persistently infected mice (24 to 50 days post-infection) were given 4 doses total (one in each week of the study period) of CLDN1-specific (n=5) or control mAb (n=4). All CLDN1-specific mAb-treated mice showed undetectable HCV RNA levels after 2 to 4 injections (Fig.2c, Supplementary Table 1) and, except for one case, animals remained virus-free until the end of the study period. The relapse mouse showed a low antibody serum concentration during the early treatment phase (Fig.2h). However, low levels of mAbs did not always correlate with relapse (Fig.2i-j). Sequencing and functional analysis of viral envelope glycoproteins from antibody-treated mice in HCV pseudoparticles (HCVpp) and HCVcc (Supplementary Fig.3a-e) suggested that the relapse was unlikely to be explained by the presence of resistant variants that escaped the CLDN1-specific mAb (Supplementary Fig.3c-d) or to CLDN1-independent cell entry (Supplementary Fig.3e). Noteworthy, human hepatocytes engrafted into uPA-SCID mice expressed lower CLDN6 levels than Huh7.5.1 cells used for escape studies (Supplementary Fig.4a-d). The mAb concentration peaked during the treatment period and declined rapidly following discontinuation of mAb injection (Fig.2h). Similar results were observed in 4 additional HCV Jc1-infected CLDN1-specific mAb-treated mice with follow-up of 3 weeks (data not shown).
CLDN1 mAb also induced viral clearance from mice chronically infected with HCVcc VL-JFH1 (genotype 1b/2a chimera) (n=5), a well-characterized strain17 encoding the structural proteins from a highly infectious HCV neutralization escape variant isolated from a patient undergoing liver transplantation (Fig.2d, Supplementary Table 1). One mouse showing a partial viral response (Fig.2d, Supplementary Table 1) was characterized by undetectable mAb concentration after the second injection (Fig.2i). The therapeutic effect was robust (HCV RNA decline > 2 logs), sustained (until the end of the study period corresponding to 11 weeks) and significant (p< 0.0007, pooled data for experiments of both genotypes, Mann-Whitney test). Furthermore, a similar effect was observed in mice chronically infected with a HCV genotype 2a serum (Fig.2e, Supplementary Table 1). Mice infected with primary isolates of HCV genotype 4 (n=4) also showed a marked and time-dependent decrease in HCV RNA levels (Supplementary Fig.5a) with no viral rebound and stable function of the engrafted human hepatocytes as shown by serum albumin levels (Supplementary Fig.5b). In summary, these data show that a CLDN1-specific mAb can clear chronic HCV infection in vivo and exhibits antiviral activity against different viral genotypes. These findings suggest CLDN1 as a therapeutic target in a clinically relevant animal model.
To assess antibody safety, we analyzed the histopathology of chimeric uPA-SCID mouse livers. Human hepatocyte-specific staining demonstrated similar repopulation and structure of human hepatocytes in HCV Jc1-infected mice treated with control or CLDN1-specific mAb (Supplementary Fig.6a). TEM analysis of chimeric infected livers of treated mice showed no detectable alteration in hepatocyte morphology or TJ ultra-structure (data not shown). Human albumin, transaminases (ALT, AST) and total bilirubin levels remained stable following antibody administration and were similar in control and CLDN1-specific mAb-treated mice at all time points tested (Supplementary Fig.6b-e).
To assess the functional integrity of human hepatocytes in CLDN1 mAb-treated mice we challenged the mice with HCV of a different strain and genotype. CLDN1-specific mAb-treated animals previously protected from HCV infection (Fig.2a) supported viral infection following mAb elimination (Supplementary Fig.6f). These functional data corroborate the presence of fully viable and functional hepatocytes following anti-CLDN1 mAb treatment and exclude adverse effects on hepatocyte function in vivo.
To address potential side effects of the CLDN1-specific mAb in other organs and tissues we assessed antibody binding to human and murine CLDN1 (Supplementary Fig.7a-g). The antibody bound primary mouse hepatocytes and 293T cells expressing murine CLDN1 with an apparent Kd of 83.6 ± 14.1 nM compared to the apparent Kd for human CLDN1 expressed in 293T cells (17.05 ± 1.14 nM) (Supplementary Fig.7g). Moreover, the latter was in a similar range as the apparent Kd of CLDN1-specific mAb binding to human hepatocytes7. Next we measured in vivo biodistribution in Balb/c and observed enrichment in skin, kidneys, lungs, intestines and liver (Supplementary Fig.8ab). Toxicity studies in Balb/c mice including clinical (data not shown), biochemical and hematological parameters as well as histopathological analyses did not reveal any toxicity (Supplementary Fig.9, Supplementary Tables 2, 3). Since TJs play a key role in intestinal paracellular permeability, we investigated the effect of the CLDN1-specific mAb on intestinal barrier function in vivo. The CLDN1-specific mAb had no effect on intestinal paracellular permeability (Supplementary Fig.8c) or total intestinal transit time (Supplementary Fig.8d), in contrast to irradiation which was used as positive control for intestinal injury. Additional ex vivo studies showed that the CLDN1-specific antibody had no detectable effects on paracellular permeability (Supplementary Fig.8e) or tissue conductance in the small intestine or colon (Supplementary Fig.8f). Moreover, the mAb had no effect on the trans-epithelial electrical resistance of intestinal Caco-2 cells (Supplementary Fig.8g) that express abundant CLDN1 levels (Supplementary Fig.8h) nor on TJ function in hepatoma HepG2 cells18. While CLDN1 knock-down significantly reduced TJ integrity (p< 0.001, t-test) in polarized HepG2 cells (Supplementary Fig.10a-b), the CLDN1-specific mAb had no impact (Supplementary Fig.10c). Furthermore, flow cytometry experiments measuring CLDN1 cell surface expression suggested that the CLDN1-specific mAb did not promote cellular CLDN1 internalization (Supplementary Fig.10d). Finally, a detailed analysis of cytokine mRNA expression in antibody-treated mice did not reveal evidence for antibody-induced inflammatory changes in the mouse jejunum (data not shown). Collectively, these data exclude major toxicity or side effects induced by the CLDN1-specific mAb in mice. Due to the absent interaction of the rat antibody with the mouse Fc receptor, immune-mediated adverse effects, e.g. ADCC, cannot be assessed in this model. Humanization of the antibody using an IgG4 backbone – a well established strategy to minimize ADCC19 – is underway for its clinical development.
To explore the antiviral mechanism of the CLDN1-specific mAb, we performed both in vitro and in vivo studies. Using protein association studies and a Förster resonance energy transfer (FRET)-based assay, we found that the CLDN1-specific mAb interfered with CD81-CLDN1 co-receptor complex formation (Supplementary Fig.11a) -a key step required for HCV entry20. Co-receptor perturbation most likely occurs at the hepatocyte basolateral membrane as shown by immunofluorescence studies of polarized HepG2 cells or immunogold affinity labeling of hepatocytes (Fig.1b-c). Furthermore, we demonstrate that the CLDN1-specific mAb significantly inhibits cell-free and cell-cell routes (p< 0.01, t-test) of transmission (Supplementary Fig.11b-e). Of note, inhibition of cell-cell transmission led to a marked decrease of virus spread in hepatoma cells when the CLDN1-specific mAb was added 48h post-infection (Supplementary Fig.11e). The ability of the antibody to inhibit cell-free and cell-cell infection may explain, at least partially, the antiviral effects of the CLDN1-specific mAb to cure persistent HCV infection in vivo. Since this mAb neither impaired HCV replication nor assembly/release (Supplementary Fig.11f-g), we conclude that the mAb acts predominantly by interfering with HCV entry.
Because CLDNs can modulate signaling pathways21, we investigated whether the antibody modulated the miRNA and kinome profiles of persistent HCV-infected Huh7.5.1 cells. Consistent with previous observations our deep sequencing studies confirmed that HCV infection modulates miRNA profiles in Huh7.5.1 cells. However, the CLDN1-specific mAb had no detectable effect on miRNA expression (Supplementary Fig.12).
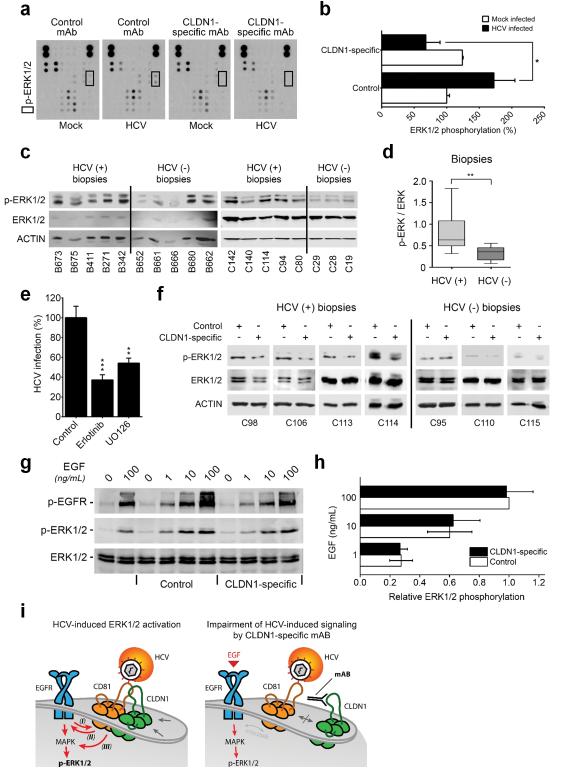
HCV binding to hepatocellular CD81 and associated receptors activates EGFR and mitogen-activated protein kinase (MAPK) signaling22,23. HCV Jc1 infection increased ERK1/2 phosphorylation and this was reversed by the CLDN1-specific mAb (Fig.3a-b). The significance of HCV-induced MAPK signaling was supported by analyzing liver biopsies from HCV-infected patients (Fig.3c-d, Supplementary Fig.13a, Supplementary Table 4, p< 0.01, Mann-Whitney test). We observed an antiviral effect of small molecule inhibitors targeting MAPK signaling (Fig.3e) at non-toxic concentrations9,24 (Supplementary Table 5). To confirm the effect of the CLDN1-specific mAb on HCV-induced signaling in liver tissue from HCV-infected patients (Supplementary Table 4), freshly isolated liver biopsies were treated ex vivo with CLDN1-specific or control mAb and we observed a reduction of ERK1/2 phosphorylation compared to control-treated biopsies (Fig.3f, Supplementary Fig.13b). These studies show that HCV infection activates MAPK signaling and CLDN1-specific mAb partially reverses this virus-dependant signaling event without impairing ERK activation by the physiological EGFR ligand EGF (Fig.3g-h, Supplementary Fig.13c). These data suggest a model in which the anti-CLDN1 mAb prevents HCV-induced cross-activation of EGFR and ERK by interfering with CD81-CLDN1 association (Fig.3i). Since MAPK signaling has been shown to be relevant for the HCV life cycle, including viral entry24,25, it is likely that this interference contributes to the ability of the antibody to clear an established infection.
Figure 3. CLDN1-specific mAb impairs HCV-induced host cell signaling.
(a) Detection of kinase phosphorylation in chronically HCV Jc1-infected Huh7.5.1 cells treated with control or CLDN1-specific mAbs (100 μg/mL; 24h) using human phosphokinase arrays. (b) p-ERK1/2 highlighted by black squares in (a) was quantified using Image J software (NIH). Results are shown as mean ± s.e.m. of integrated dot blot densities from 2 independent experiments performed in duplicate. (c) ERK1/2 phosphorylation is elevated in liver tissue of patients with chronic HCV infection. Expression of p-ERK1/2, total ERK1/2 and ACTIN was revealed by Western blotting (representative full length blots are presented in Supplementary Fig.13a). (d) Quantification of signal intensities of p-ERK1/2 in (c) normalized to total ERK1/2 expression. The quantification of p-ERK1/2 (normalized to ACTIN expression) relative to total ERK1/2 expression (normalized to ACTIN expression) is shown. Results are shown as the interquartiles (box) with the min and max values (whiskers). The line within the box indicates the median value. (e) Inhibition of the MAPK pathway inhibits persistent HCV infection in Huh7.5.1 cells. Luc-Jc1-infected Huh7.5.1 cells were treated 3 days after inoculation with erlotinib or UO126 (each at 10 μM) for 3 days. HCV replication was assessed by luciferase assay. Results are presented as mean ± s.e.m of 6 independent experiments performed in triplicate. (f) CLDN1-specific mAb reduces ERK1/2 phosphorylation in liver tissue from patients with chronic HCV infection. Fresh liver biopsies were divided in two equal pieces and maintained in DMEM medium supplemented with 10% foetal calf serum and 100 μg/mL control or CLDN1-specific mAb for 4h, respectively. Expression of p-ERK1/2, total ERK1/2 and ACTIN was determined by Western blotting. Information on liver biopsies is provided in Supplementary Table 4 (representative full length blots are presented in Supplementary Fig.13b). (g) CLDN1-specific mAb does not inhibit EGF-induced ERK1/2 phosphorylation. Huh7.5.1 cells were serum starved for 4h prior 1h incubation with 100 μg/mL control or CLDN1-specific mAb and 15 min incubation with increasing concentrations of EGF. P-EGFR, p-ERK1/2 and total ERK1/2 were quantified by Western blotting (full length blots are presented in Supplementary Fig.13c). (h) Quantification of EGF-induced ERK phosphorylation in the presence of CLDN1-specific or control mAb. P-ERK1/2 and total ERK1/2 Western blot signals described in (g) were quantified using a Typhoon Trio laser scanner and ImageQuant Software (GE Healthcare). Values are expressed as relative ratio of p-ERK1/2 to total ERK1/2 densities (two independent experiments ± s.e.m.). (i) Model of HCV-induced signal transduction during cell entry (left panel) and its impairment by CLDN1-specific mAb (right panel). (Left panel) (I) HCV entry is dependent on EGFR signaling which promotes CD81-CLDN1 co-receptor interactions according to Zona et al.24 and Lupberger et al.9 (II) At the same time viral binding to the target cell induces EGFR phosphorylation and signaling via CD81-EGFR interactions as shown by Diao et al.23 and (III) MAPK signaling according to Brazzoli et al. by cross-talk22. (Right panel) Combining these observations and results shown in panels (a-h) and Supplementary Fig.10, our findings are consistent with a model that the CLDN1-specific mAb impairs virus-induced MAPK/ERK1/2 signaling by interfering with CD81/CLDN1-MAPK crosstalk without impairing direct EGF-induced ERK1/2 phosphorylation. *p<0.05, **p<0.01, ***p<0.0001 (Student’s t-test (b, e), Mann-Whitney test (d)).
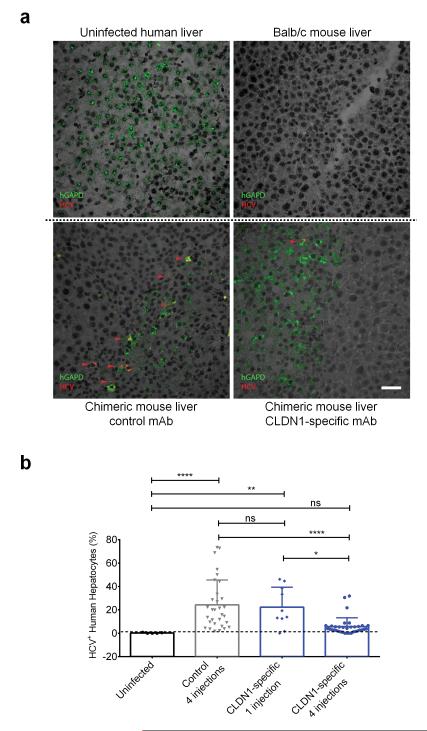
Finally, we performed mechanistic studies in chimeric mice to study the consequences of the observed mechanistic findings for viral clearance in vivo. Using FISH with HCV- and human GAPDH-RNA specific probes (Fig.4a) we demonstrate that the CLDN1-specific mAb results in a dose- and time-dependent decrease in the number of HCV-infected cells leading to their final elimination (Fig.4b). The decline of HCV-infected hepatocytes paralleled the decline in peripheral HCV RNA levels, showing a monophasic decline (Fig.4b, Supplementary Fig.14). Noteworthy, the baseline frequency of HCV-infected hepatocytes (Fig. 4b) in the chimeric mouse liver were comparable to those reported in chronically infected human liver26.
Figure 4. CLDN1-specific mAb leads to elimination of HCV-infected cells from the livers of human chimeric mice in a dose and time-dependant manner.
(a) Detection of human hepatocytes and HCV RNA with probes specific for human GAPDH mRNA (green) or HCV Jc1 RNA (red) using FISH. The specificity of the HCV- and GAPDH probes are shown by specific staining of HCV-infected human hepatocytes and absent staining of uninfected human liver (left top panel) and Balb/c mouse liver (right top panel). FISH analyses were then performed in livers of control (left bottom panel) or CLDN1-specific mAb (right bottom panel) treated chimeric uPA-SCID mice. Red arrows indicate HCV positive cells. Scale bar – 50μm. (b) Quantification of the percentage of infected cells in the liver of uninfected chimeric mice (black circles) or HCV Jc1-infected mice treated either four times with control mAb (grey triangles), or once (blue diamonds) or four times (blue circles) with CLDN1-specific mAb. Livers were collected 1 week after the last antibody injection. The quantification was performed after confocal microscopy acquisition. *p<0.05, **p<0.01, ****p<0.0001 (Kruskal-Wallis test).
Assuming an inhibition of entry (as shown in Supplementary Fig.11) and a negligible effect on viral replication/assembly (Supplementary Fig.11f-g), mathematic modeling predicts a time-dependent elimination of virus-infected hepatocytes with concomitant absent de novo-infection. Applying this model, we estimated the half-life of infected cells to be ~ 1.3 days for HCV genotype 2a and 5.4 days for HCV genotype 4 which is in the range of hepatocyte half-lives reported in patients27. Indeed, HCV has been reported to promote apoptosis (Supplementary Fig.15 and 28). During serum HCV RNA decline, virus infected hepatocytes are constantly replaced by non-infected hepatocytes resulting in viral control and clearance. This model is supported by both FISH analyses of liver tissue sections (Fig.4) and in vitro studies where the CLDN1-specific mAb eliminates HCV-infected cells (Supplementary Fig.15c-d). The different half-lives of HCV-infected cells and the magnitude of virus-induced cell death may explain the varying magnitude of the decline in viral load for diverse HCV strains (Supplementary Figs.5a, 14)27.
Collectively, we demonstrate that a host-targeting entry inhibitor clears chronic viral infection (Fig.2c-e). Our results show that viral entry and spread are required for persistent infection in vivo and that CLDN1 is a valid target to prevent and treat chronic HCV infection. The interaction of the mAb with non-junctional pools of CLDN1 (Fig.1b-c) that complex with the viral co-receptor CD81 (Supplementary Fig.11a), may explain the non-detectable toxicity (Supplementary Figs.6a-f, 8c-g, 9, Supplementary Tables 2, 3).
In summary, therapeutic administration of a CLDN1-specific mAb demonstrates activity against several HCV genotypes (Fig.2, Supplementary Fig.5, 11b) without detectable toxicity (Supplementary Figs.6, 8, 9, Supplementary Tables 2, 3). The absence of detectable viral resistance (Supplementary Fig.3) suggests that CLDN1 is an antiviral target with a high genetic barrier for resistance. While strain-dependent use of CLDN1 and 6 has been described in hepatoma cells29, our data suggest that CLDN1 is essential for HCV infection in vivo and low expression levels of CLDN6 and 9 in human hepatocytes (Supplementary Fig.4 and 29,30) appear to preclude a relevant entry function of these molecules in vivo.
In contrast to direct-acting antivirals, the CLDN1-specific mAb eliminates viral infection in monotherapy in the state-of-the-art animal model (Fig.2, Supplementary Fig.5a) without detectable resistance (Supplementary Fig.3). These features are likely due to its unique mechanism of action targeting a host factor (Supplementary Fig.11a) that is essential for viral persistence in vivo as well as modulating signaling of virus-infected cells (Fig.3) likely contributing to clearance. Our data suggest that antibodies or small molecules targeting CLDN1 are promising candidates to prevent liver graft infection and to eliminate chronic HCV infection. Finally, similar antimicrobial strategies may be designed for a broad range of pathogens that utilize TJ proteins as host factors including dengue virus, coxackievirus, adenovirus and shigella3-5.
ONLINE METHODS
Research experiments on live vertebrates
In vivo experiments were performed in the Inserm Unit 1110 animal facility according to local laws and ethical committee approval (CREMEAS, project numbers AL/02/19/08/12 and AL/01/18/08/12).
Human subjects
Human material including liver biopsies and liver tissue from patients undergoing surgical resection was obtained with informed consent from all patients. The respective protocols were approved by the Ethics Committee of the University Hospital of Basel, Switzerland (EKBB 13 December 2004), University of Strasbourg Hospitals (CPP 10-17), and the University of Birmingham (LREC, Birmingham, West Midlands, project 04/Q2708/40).
Antibodies
The human CLDN1-specific (OM-7D3-B3, IgG2b, rat7), CLDN6-specific (WU-9E1-G2, IgG2b, rat30), CLDN9-specific (YD-4E9-A2, IgG2b, rat30) mAbs have been described previously and were produced by Aldevron Freiburg. HCV E2-specific AP33 (mouse)7, E1-specific IGH433 (human)7 E2-specific CHB-23 (human)17 mAbs and patient-derived HCV-specific IgG7 have been described. Control mAb (rat IgG2b clone LTF-2, Bio X Cell), human CLDN1-specific (Life Technologies, rabbit; R&D, rat polyclonal), human CD10-specific (clone 56C6, Vector Laboratories, mouse), human CD81-specific (clone 2s131, mouse), human CK18-specific (clone VP-C414, Vector Laboratories, mouse), Alexa-fluor 488 conjugated IgG rabbit-specific, Alexa-fluor 488 conjugated IgG mouse-specifc, Alexa-fluor 488 conjugated IgG rat-specific and Alexa-fluor 594 conjugated IgG mouse-specific Abs (Invitrogen, goat) were used for fluorescent imaging as described31. ERK1/2 (rabbit, Cell Signaling), pERK1/2 (Thr202, Tyr204, mouse, clone E10, Cell Signaling) and beta-actin (clone AC-15, mouse, Sigma)-specific antibodies and alkaline–phosphatase (AP)–labeled (goat, GE Healthcare) as well as IRDye 800CW-conjugated goat anti-rabbit and IRDye 680-conjugated goat anti-mouse (Li-Cor Biosciences) secondary antibodies were used for immunoblot analyses. Rabbit anti-rat bridging antibody (Cappel #55704) was used for immunogold labeling.
Cell lines
Huh7.5.1, Huh7.5-GFP, Huh7.5 shCD81.1, 293T, HepG2 DsRed-CD81 and Caco-2 cells were cultured as described9,32-35.
Imaging of CLDN1 in human and chimeric livers
Chimeric mouse and human liver samples were formalin fixed and paraffin embedded. Human liver tissue was obtained from patients undergoing liver transplantation due to HCV or from normal livers taken from surplus donor tissue used for reduced size transplantation. Informed consent from each patient was obtained with regional ethics committee approval (project number 04/Q2708/40). Sections were subjected to low temperature antigen retrieval pre-treatment, followed by fluorescent detection of CLDN1, CD10 or CK18 as previously described31. Labeled sections were visualized using laser scanning confocal microscopy (Zeiss LSM 780, 100 x Plan Apochromat 1.4NA oil immersion objective). Background and autofluorescence of tissue samples were corrected throughout and 3D composite images were generated using the Zeiss Zen analysis software.
Transmission electron microscopy and immunogold labeling of CLDN1 in vivo
Human liver-chimeric mice were injected with 500 μg control or CLDN1-specific mAb and sacrificed 5h later. Mice were perfused with PBS for 3 min through portal vein infusion followed by 5 min perfusion with 4% electron microscopy (EM)-grade paraformaldehyde (PFA). Livers were harvested, minced in 3 mm pieces and fixed in 4% PFA at room temperature for 24h. Subsequently liver pieces were placed in PBS and kept at 4°C before processing for EM. Prior to freezing in liquid nitrogen the tissue were infiltrated with 2.3M sucrose in PBS containing 0.2M glycin for 15 min. Frozen samples were sectioned at −120°C, the sections were transferred to formvar-carbon coated copper grids. Grids were floated on PBS at room temperature (RT) until the immunogold labeling was carried out at RT on a piece of parafilm using control or CLDN1-specific mAb, rabbit anti-rat bridging antibody and 10 nm (for CLDN1-specific mAb) or 15 nm (for isotype control mAb) protein-A gold. Contrasting/embedding of the labeled grids was carried out on ice in 0.3% uranyl acetete in 2% methyl cellulose for 10 min. Grids were examined in a JEOL 1200EX Transmission electron microscope and images were recorded with an AMT 2k CCD camera.
CLDN1 staining on polarized HepG2 cells
HepG2 DsRED-CD81 cells were grown on 13 mm diameter borosilicate glass coverslips at 4×104 cells/coverslip (Fisher Scientific, UK). Laser scanning confocal microscopy (LSCM) was performed on a Zeiss Meta Head Confocal Microscope with a 63x water immersion objective. For live cell receptor staining of CD81 and CLDN1 in HepG2 DsRED-CD81 cells, cells were washed twice with PBS before blocking in PBS-BSA-0.01% sodium azide (Sigma, UK) for 15 min. Cells were incubated with the following primary antibodies: anti-CD81 (1μg/mL) and anti-CLDN1 (1:200) in PBS-BSA-azide for 30 min at 37oC. Cells were washed 3x in PBS before fixing with 3.6% PFA for 30 min. Cells were washed 3x with PBS before addition of a goat anti-mouse secondary Ab (Alexa 488, Invitrogen) at a 1:1000 dilution in PBS-BSA-azide or a goat anti-rat secondary Ab (Alexa 488, Invitrogen) for 30 min at 37oC. Cells were then washed with PBS before mounting and visualization.
Human liver-chimeric mice
Human liver-chimeric mice were produced by transplanting cryopreserved human hepatocytes into the spleen of 3 week-old homozygous urokinase-type plasminogen activator-transgenic Severe Combined Immuno-Deficiency-beige of both sex (uPA-SCID-bg) mice6,36. Human hepatocytes were purchased from Becton Dickinson or isolated as described7, 9.
In vivo experimentation
For prevention studies, chimeric uPA-SCID mice were intraperitoneally injected with 500 μg CLDN1-specific or control mAb at day −1 before and days 1 and 5 after inoculation of a genotype 1b or 4 HCV-infected serum. HCV infection was performed as previously described6. For treatment studies, chimeric uPA-SCID mice were chronically infected with HCVcc of prototype strain Jc115, a genotype 1b/2a chimeric VL-JFH117 or genotype 2a and 4 serum. Chronically infected mice received 500 μg CLDN1-specific or control mAb once a week for 4 weeks, except for genotype 4 where mice received 500μg twice a week for 3 weeks followed by 200μg twice a week up to the end of the study period. Blood was harvested by retro-orbital puncture under general anesthesia. Mice were randomly assigned to the different experimental groups by a blinded technician. Experiments were performed in the Inserm Unit 1110 animal facility according to local laws and ethical committee approval (AL/02/19/08/12 and AL/01/18/08/12).
Viral load
Viral load was quantified by a blinded technician using Abbott RealTime™ HCV assay (Abbott). The linear range of the assay is 12 IU/mL to 108 IU/mL, the limit of quantification (LOQ) of HCV RNA is 12 IU/mL. Given a mouse serum dilution of 1:100 in PBS, LOQ is 1200 UI/mL, i.e. 5160 copies/mL.
Analysis of CLDN1-specific mAb serum concentrations
Serum antibody concentrations were quantified using a rat IgG2b antibody-specific ELISA (ref. E110-111, Bethyl Laboratories, Euromedex) following the recommendations of the supplier.
Liver biopsies
Eligible patients were identified by a systematic review of patient charts at the Hepatology outpatient clinic of the University Hospital of Basel, Switzerland. The protocol was approved by the Ethics Committee of the University Hospital of Basel, Switzerland. Written informed consent was obtained from all patients. Histopathological grading and staging of the HCV liver biopsies according to the Metavir classification system was performed at the Pathology Institute of the University Hospital Basel and are summarized in Supplementary Table 3. All patients that donated liver tissue were male between 27 and 61 years old and female between 39 and 62 years old (Supplementary Table 4). Liver biopsy tissues from patients were analyzed as described37. In brief, liver tissue was lysed in 100 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 0.1 % Triton X-100, 10 mM NaF, 1 mM PMSF, and 1 mM sodium ortho-vanadate during mechanic homogenization followed by 20 min incubation on ice. Lysates were cleared by centrifugation at 4 °C and 21,000 × g for 5 min prior to immunoblot analysis of 5 μg protein per sample. For ex vivo stimulation, fresh liver biopsies were divided in two equal pieces and incubated for 4h at 37°C in DMEM medium supplemented with 10 % fetal calf serum and 100 μg/mL control or CLDN1-specific mAb, respectively.
Analysis of protein expression and phosphorylation
Immunoblots of cell lysates using ERK1/2-specific antibodies were performed using Hybond–P membranes and visualized using ECF substrate (GE Healthcare) according to the manufacturer’s protocol. Fluorescence emission was detected using Typhoon Trio high performance fluorescence scanner (GE Healthcare) and quantified using Image Quant TL software (GE Healthcare). Phospho-array analysis was performed using Proteome Profiler Human Phospho-kinase Array (R&D Systems) as described by the manufacturer. For imaging, blots were incubated with ECL (GE Healthcare) and exposed to ECL Hyperfilm (GE Healthcare). Phospho-kinase array results were quantified by integrating the dot blot densities using Image J software (NIH). Immunoblots from biopsies were analyzed as described24.
Inhibition of HCV infection by protein kinase inhibitors
Huh7.5.1. cells were plated in 96 well-plates (2 × 103 cells/well) and infected 24h later for 3 days with HCVcc Luc-Jc1. Medium was then removed and replaced with medium containing 10 μM erlotinib (LC Laboratories) or UO126 (Calbiochem) and final concentration of 1% DMSO. Control medium contains only 1% DMSO. Three days later, HCV replication was quantified by luciferase activity measurement.
EGF-induced MAPK activation
Huh7.5.1 cells were serum starved for 4h prior 1h incubation with 100 μg/mL control or CLDN1-specific mAb and 15 min incubation with increasing concentrations (1, 10 and 100 ng/mL) of EGF. P-EGFR, p-ERK1/2 and total ERK were detected by Western blotting. Quantification was performed using a Typhoon Trio laser scanner and ImageQuant Software (GE Healthcare).
FISH analyzes of human chimeric livers
Liver samples from mice were collected during necropsy, immediately embedded in optimal cutting temperature compound (OCT) and frozen in liquid nitrogen chilled 2-methylbutane. Tissues were then stored at −80°C until use. Sections (10 μm) were cut at cryostat (Leica), fixed overnight in 4% formaldehyde at 4°C and hybridized, as previously described26, with the following modifications. Briefly, tissue sections were pre-treated by boiling (90°-95°C) in pre-treatment solution (Affymetrix – Panomics) for 1 min, followed by a protease QF (Affymetrix – Panomics) digestion for 10 min at 40°C. Hybridization was performed using probe sets against JFH-1 HCV RNA (target region 4513-6253) and against human GAPDH mRNA (VA6-12786-06, Affymetrix-Panomics). Pre-amplification, amplification and detection were performed according to provider’s protocol. Images were acquired with a LSCM (LSM710, Carl Zeiss Microscopy) and Zen2 software, using same settings for all the tissues analyzed. Five to seven random fields were selected from each section, based only on the presence of human GAPDH signal to ensure the presence of human hepatocytes. Images were then acquired also including the channel for the HCV detection. Image analysis was performed using ImageJ and CellProfiler software, with a customized pipeline. Total number of cells, number of human hepatocytes (HH), number of HCV+ HH and HCV signal average intensity (and s.d) were evaluated.
Statistical analyzes
For in vivo and ex vivo data, the one-way ANOVA followed by Tukey’s Post-Hoc test or the Kruskal-Wallis followed by Dunn’s Post Hoc test were used after determination of distribution by the Shapiro-Wilk normality test. The Wilcoxon rank test as well as the two-tailed Mann-Whitney test were also used. The in vitro data are presented as the mean ± s.d. except where mean ± s.e.m. is indicated, and were analyzed by the unpaired Student’s t-test or the two-tailed Mann-Whitney test as indicated. A p-value ≤ 0.05 was considered significant. Statistical analyzes were performed with GraphPad Prism 6 software.
Supplementary Material
Acknowledgements
This work was supported by the European Union (ERC-2008-AdG-233130-HEPCENT, ERC-2010-StG-260767-ncRNAVIR) INTERREG-IV-Rhin Supérieur-FEDER-Hepato-Regio-Net 2009 and 2012), ANRS (ANRS 2009/183, 2009/136, 2011/132, 2012/239, 2013/108), ANR (Laboratoires d’excellence ANR-10-LABX-0028_HEPSYS and ANR-10-LABX-36 netRNA), Fondation ARC pour la recherche (NanoISI and TheraHCC IHUARC IHU201301187), Institut Hospitalo-Universitaire (IHU) Strasbourg, the Wilhelm Sander Foundation, Région Alsace, Institut National du Cancer, the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Université de Strasbourg, the Ghent University (GOA 01G01712) and the Research Foundation – Flanders (projects 1500910N and G052112N). We are grateful to Dr. S. Ito (Harvard Medical School) for electron microscopy studies, Dr. F.-L. Cosset (Inserm U1111, ENS Lyon, France) and Dr. J. Ball (University of Nottingham, Nottingham, UK) for retroviral vectors for HCVpp production, Dr. F. Chisari (The Scripps Research Institute, La Jolla, USA) for the gift of Huh7.5.1 cells, Dr. A. Patel (MRC Virology Unit, Glasgow, UK) for E2-specific mAb AP33 and Huh7.5-GFP cells, Dr. S. Foung (Stanford Blood Center, Palo Alto, USA) for E2-specific mAb CHB-23 and Drs. C. M. Rice and M. Evans (Rockefeller University and Mount Sinai School of Medicine, New York, NY) for providing human and mouse CLDN1 expression constructs as well as Huh7.5 cells. We acknowledge S. Durand, L. Heydmann, E. Soulier, Dr. J. Barths, N. Brignon, S. Pernot (Inserm U1110, Strasbourg), Drs. O. Wendling and N. Messadeq (ICS, Illkirch), C. Valencia (PCBIS, Illkirch), S. Kallis (University of Heidelberg, Germany) for technical work, Drs. F. Grunert and J. Thompson (Aldevron, Freiburg) for helpful discussions, Drs. H. Jacob and M.F. Champy (ICS, Illkirch) for histopathological, hematological and biochemical analyzes, Prof. P. Bachellier (Strasbourg University Hospitals) for providing liver resections for isolation of primary human hepatocytes, the Laboratoire Schuh – groupement Bio67, Strasbourg and the Plateau Technique de Microbiologie, Laboratoire de Virologie (Prof. S. Fafi-Kremer and Prof. F. Stoll-Keller), University Hospital Strasbourg for performing viral load analyses, and the IGBMC microarray and sequencing platform, member of the France Génomique program, for the sequencing of our libraries. Part of the animal experiments was carried out within the small animal exploration facility Cardiex (Nantes), which is supported by the GIS-IBiSA program.
Footnotes
Competing Financial Interests. The authors declare no competing financial interests. Inserm, the University of Strasbourg and Genovac/Aldevron Freiburg have filed a patent application on monoclonal anti-claudin 1 antibodies for the inhibition of hepatitis C virus infection (US Patent # 8,518,408; WO2010034812; PCT/EP 08 305 597 0). T. F. B. has served as scientific advisor for Gilead, Biotest and Vironexx.
REFERENCES
- 1.Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 3.Cohen CJ, et al. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci U S A. 2001;98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukumatsu M, et al. Shigella targets epithelial tricellular junctions and uses a noncanonical clathrin-dependent endocytic pathway to spread between cells. Cell Host Microbe. 2012;11:325–336. doi: 10.1016/j.chom.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Che P, Tang H, Li Q. The interaction between claudin-1 and dengue viral prM/M protein for its entry. Virology. 2013;446:303–313. doi: 10.1016/j.virol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Mercer DF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 7.Fofana I, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–964. e954. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 8.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 9.Lupberger J, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meuleman P, et al. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology. 2012;55:364–372. doi: 10.1002/hep.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi N, et al. Combination therapies with NS5A, NS3 and NS5B inhibitors on different genotypes of hepatitis C virus in human hepatocyte chimeric mice. Gut. 2013;62:1055–1061. doi: 10.1136/gutjnl-2012-302600. [DOI] [PubMed] [Google Scholar]
- 12.Harris HJ, Wilson GK, Hubscher SG, McKeating JA. Heterogeneous claudin-1 expression in human liver. Hepatology. 2013;57:854–855. doi: 10.1002/hep.25910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher NF, et al. Activated macrophages promote hepatitis C virus entry in a tumor necrosis factor-dependent manner. Hepatology. 2014;59:1320–1330. doi: 10.1002/hep.26911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukh J, et al. Challenge pools of hepatitis C virus genotypes 1-6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis. 2010;201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietschmann T, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fofana I, et al. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology. 2012;143:223–233. doi: 10.1053/j.gastro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger SE, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 19.Dübel S, Reichert JM, editors. Handbook of Therapeutic Antibodies. Wiley-Blackwell; 2014. p. 2544. [Google Scholar]
- 20.Harris HJ, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh Y, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32:4873–4882. doi: 10.1038/onc.2012.505. [DOI] [PubMed] [Google Scholar]
- 22.Brazzoli M, et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diao J, et al. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86:10935–10949. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zona L, et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Menzel N, et al. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 2012;8:e1002829. doi: 10.1371/journal.ppat.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieland S, et al. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2014;59:2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann AU, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 28.Lan L, et al. Hepatitis C virus infection sensitizes human hepatocytes to TRAIL-induced apoptosis in a caspase 9-dependent manner. J Immunol. 2008;181:4926–4935. doi: 10.4049/jimmunol.181.7.4926. [DOI] [PubMed] [Google Scholar]
- 29.Haid S, et al. Isolate-dependent use of Claudins for cell entry by hepatitis C virus. Hepatology. 2014;59:24–35. doi: 10.1002/hep.26567. [DOI] [PubMed] [Google Scholar]
- 30.Fofana I, et al. Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J Virol. 2013;87:10405–10410. doi: 10.1128/JVI.01691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds GM, et al. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology. 2008;47:418–427. doi: 10.1002/hep.22028. [DOI] [PubMed] [Google Scholar]
- 32.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris HJ, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol. 2008;82:5007–5020. doi: 10.1128/JVI.02286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piche T, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: involvement of soluble mediators. Gut. 2009;58:196–201. doi: 10.1136/gut.2007.140806. [DOI] [PubMed] [Google Scholar]
- 35.Zahid MN, et al. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology. 2013;57:492–504. doi: 10.1002/hep.26097. [DOI] [PubMed] [Google Scholar]
- 36.Petersen J, et al. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol. 2008;26:335–341. doi: 10.1038/nbt1389. [DOI] [PubMed] [Google Scholar]
- 37.Dill MT, et al. Interferon-gamma-stimulated genes, but not USP18, are expressed in livers of patients with acute hepatitis C. Gastroenterology. 2012;143:777–786. e771–776. doi: 10.1053/j.gastro.2012.05.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.