Abstract
The initiation and maintenance of a malignant phenotype requires complex and synergistic interactions of multiple oncogenic signals. The Hedgehog (HH)/GLI pathway has been implicated in a variety of cancer entities and targeted pathway inhibition is of therapeutic relevance. Signal cross-talk with other cancer pathways including PI3K/AKT modulates HH/GLI signal strength and its oncogenicity. In this study, we addressed the role of HH/GLI and its putative interaction with the PI3K/AKT cascade in the initiation and maintenance of chronic lymphocytic leukemia (CLL). Using transgenic mouse models, we show that B-cell-specific constitutive activation of HH/GLI signaling either at the level of the HH effector and drug target Smoothened or at the level of the GLI transcription factors does not suffice to initiate a CLL-like phenotype characterized by the accumulation of CD5+ B cells in the lymphatic system and peripheral blood. Furthermore, Hh/Gli activation in Pten-deficient B cells with activated Pi3K/Akt signaling failed to enhance the expansion of leukemic CD5+ B cells, suggesting that genetic or epigenetic alterations leading to aberrant HH/GLI signaling in B cells do not suffice to elicit a CLL-like phenotype in mice. By contrast, we identify a critical role of GLI and PI3K signaling for the survival of human primary CLL cells. We show that combined targeting of GLI and PI3K/AKT/mTOR signaling can have a synergistic therapeutic effect in cells from a subgroup of CLL patients, thereby providing a basis for the evaluation of future combination therapies targeting HH/GLI and PI3K signaling in this common hematopoietic malignancy.
Introduction
Hedgehog (HH)/GLI signaling has multiple etiologic roles in the initiation and progression of a variety of human cancers by regulating critical oncogenic traits such as cell proliferation, survival, metastasis and cancer stem cell fate.1, 2, 3, 4, 5
Activation and regulation of HH/GLI signaling is a complex molecular process. Control of pathway activity occurs at multiple levels within the signal cascade and frequently also involves cross-talk and signal integration with other pathways, thereby modifying the output of HH signaling (reviewed in refs 6, 7, 8, 9, 10, 11, 12, 13, 14).
Canonical HH signaling is activated upon binding of HH protein (either Sonic, Indian or Desert Hedgehog) to its receptor Patched (Ptch), a 12-transmembrane domain protein actively repressing the pathway in the absence of ligand by preventing the activation of the essential HH effector Smoothened (Smo). Binding of Hh to Ptch results in translocation of Smo into the primary cilium, followed by Smo activation and downstream signaling. Active ciliary Smo induces HH target gene expression by promoting the formation of the activator forms of the GLI zinc-finger transcription factors GLI3, GLI2 and GLI1 (for review see Hui et al.15 and Ruiz i Altaba et al.16 and references therein).
In addition to classical SMO-dependent regulation of GLI, SMO-independent control of GLI activity has recently emerged as crucial regulatory step in several human malignancies. For instance, GLI activation can be directly controlled by atypical protein kinase C in basal cell carcinoma, TGFβ/SMAD in pancreatic cancer or by the EWS-FLI1 oncogene in Ewing's Sarcoma.17, 18, 19, 20 Furthermore, activation of receptor tyrosine kinases (for example, EGFR and PDGFRA) and their downstream effectors including RAS/RAF/MEK/ERK and PI3K/AKT signaling enhance GLI activity by promoting GLI stability, nuclear import and transcriptional activity.21, 22, 23, 24, 25, 26, 27, 28, 29 Importantly, the identification of signals involved in Smo-independent GLI regulation has guided the development of rationale-based treatments also targeting GLI-promoting signals including atypical protein kinase C, PI3K/AKT and EGFR/PDGFR. This strategy proved particularly efficient in preclinical models of basal cell carcinoma, medulloblastoma, melanoma, pancreatic and esophageal cancer, even for SMO-inhibitor-resistant cancers.17, 21, 24, 28, 29, 30, 31
The therapeutic relevance and role of HH/GLI in the development and growth of various hematological malignancies has been demonstrated in several studies.32, 33, 34, 35, 36, 37, 38, 39, 40 However, the precise molecular–pathologic role of HH/GLI and its cross-talk with other oncogenic signals in entities such as chronic lymphocytic leukemia (CLL) is less well understood and partially controversial.6, 33, 41, 42 CLL is an abundant B-cell malignancy in the Western population, which typically manifests as an accumulation of CD5+ leukemic B cells in the peripheral blood (PB), bone marrow (BM) and secondary lymphoid organs.43, 44 Of note, survival of CLL cells critically depends on a complex molecular signal cross-talk between the leukemic cells with accessory immune and stromal cells as well as on B-cell receptor signaling (reviewed in Burger et al.45and Stevenson et al.46). These interactions trigger multiple signaling cascades within the CLL cells including Protein Kinase C and PI3K/AKT signaling, the latter of which having a crucial role in the survival of CLL cells.41 Of note, idelalisib, a first-in class oral PI3K delta inhibitor has recently been approved for the treatment of relapsed CLL patients.47, 48
As for the implication of HH/GLI in CLL Hegde et al.42 suggested a classical SMO-dependent HH/GLI signaling mechanism where stromal cells produce HH ligand that acts on leukemic cells in a paracrine manner. Inhibition of stroma-derived HH signaling using SMO inhibitors reduced survival of CLL cells.42 Our own studies also provided evidence for a pro-survival role of HH/GLI in CLL, although we found SMO-independent activation of GLI transcription factors to be critical for the survival of CLL cells.41 A more detailed analysis of CLL patients with distinct genetic aberrations revealed a critical role for SMO-dependent HH/GLI signaling only in CLL patients harboring the genomic aberration trisomy 12,33 which characterizes a CLL subgroup with high cell proliferation, enriched Notch1 mutations and an increased frequency to the aggressive Richter's transformation.49
Despite some discrepancies in the mechanisms by which HH/GLI contributes to CLL, the pro-survival role of the pathway in CLL maintenance is generally well accepted. By contrast, it is unclear whether aberrant oncogenic HH/GLI signaling is able and sufficient to also initiate a CLL-like phenotype and whether cross-talk of HH/GLI and PI3K signaling, a highly promising drug target in CLL therapy,47, 50 does have a role in CLL. As model of choice to address whether aberrant HH/GLI activation is sufficient for the initiation of a CLL-like phenotype, we took a genetic approach using conditional transgenic mouse models to activate Hh/Gli signaling in mouse CD19+ B cells either at the level of Smo or at the level of Gli. In addition, we addressed whether Hh/Gli and constitutive Pi3k/Akt signaling interact and synergize to drive a leukemic phenotype. Analysis of transgenic mice harboring single Hh/Gli or combined Hh/Gli–Pi3k/Akt-activating mutations in CD19+ B cells did not reveal a role for Hh/Gli in the initiation process of a CLL-like phenotype. However, we show that GLI and PI3K functions are required for survival of established human CLL cells, thereby providing a basis for the future evaluation of rational combination treatments simultaneously targeting GLI and PI3K/AKT in CLL patients.
Results
Constitutive Hh/Gli pathway activation in CD19+ B cells does not initiate a CLL-like phenotype in mice
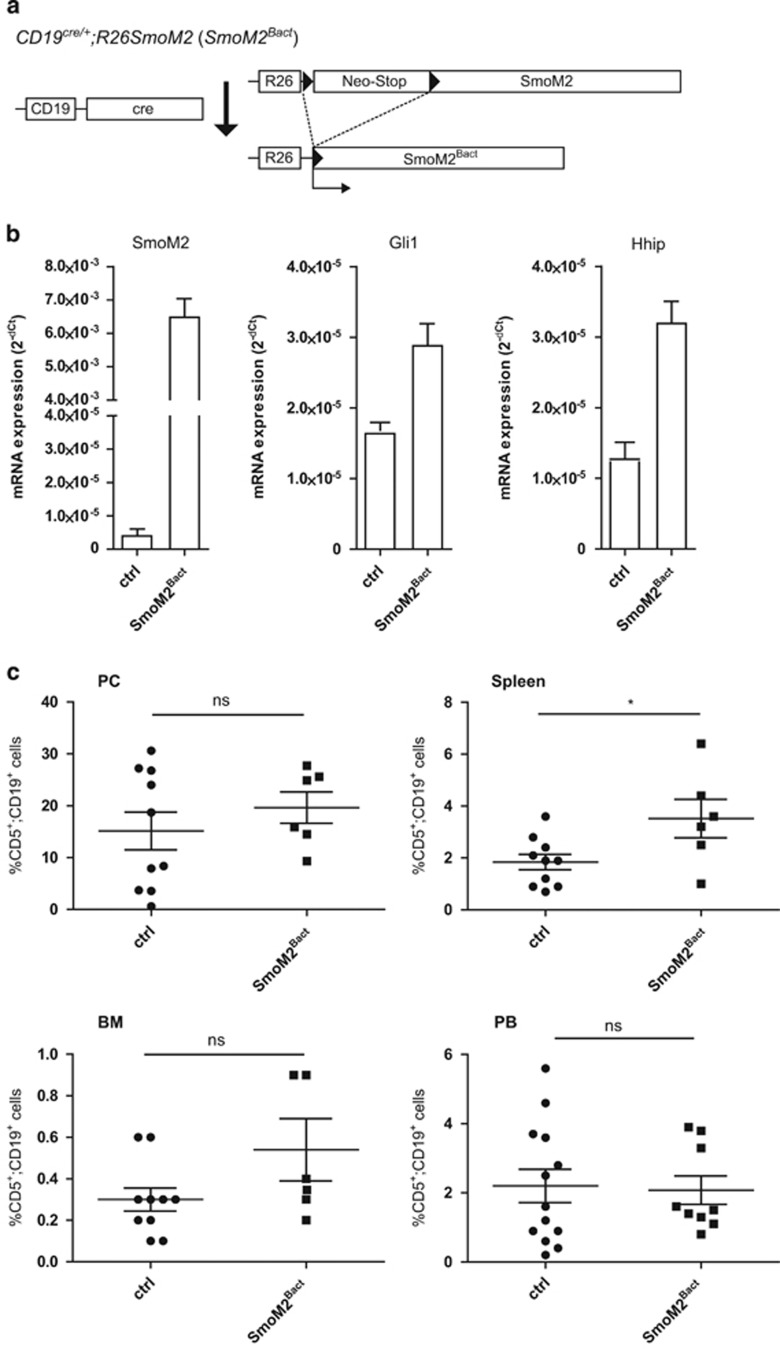
For B-cell-specific activation of Hh/Gli signaling, we crossed the B-cell-specific cre recombinase deleter strain CD19cre with R26SmoM2 mice carrying a conditional oncogenic Smoothened allele (SmoM2; Figure 1a). CD19cre/+;R26SmoM2 mice, hitherto referred to as SmoM2Bact, express in CD19+ B cells high levels of the SmoM2 transgene transcript and, importantly, also elevated levels of the Hh target genes Gli1 and Hhip, indicating B-cell-specific Hh pathway activation (Figure 1b).
Figure 1.
B-cell-specific expression of oncogenic Smo (SmoM2) does not lead to a systemic increase in leukemic CD5+ B cells. (a) Cre/loxP strategy for the activation of oncogenic SmoM2 expression in CD19+ B cells by the generation of CD19cre/+;R26SmoM2 (SmoM2Bact) mice. (b) Activation of Hh/Gli signaling in CD19-positive B cells by conditional SmoM2 expression. mRNA expression of transgenic SmoM2 and endogenous Hh target genes Gli1 and Hhip was measured by quantitative PCR (qPCR). ctrl: CD19-positive B cells from CD19cre/+ mice (n=6); SmoM2Bact: CD19-positive B cells from CD19cre/+;R26SmoM2 mice (n=6). (c) Percentage of CD5+/CD19+ cells in the PC, spleen, BM and PB of 10-month-old CD19cre/+ control mice (ctrl) and 10-month-old SmoM2Bact mice. Scatter dot plot with each dot representing an individual mouse. The mean value is indicated as bar, whiskers represent the s.e.m. ns: P>0.05; *P<0.05.
Accumulation of CD5+/CD19+ cells in the lymphatic system and PB is characteristic of a CLL-like phenotype. In Eμ-Tcl1 transgenic mice, a well-established mouse model of CLL,51 the increase in CD5+/CD19+ cells occurs first in the PC as early as 2 months after birth with an initial establishment of the leukemic phenotype at the age of ~8–10 months.51 To address whether persistent B-cell-specific activation of Hh/Gli alone is able to trigger a CLL-like phenotype, we analyzed by flow cytometry the number of CD5+/CD19+ cells in CD19cre/+ control mice and SmoM2Bact mice at the age of 10 months and also at earlier time points (that is, 12 weeks and 5 months after birth; Supplementary Figure S1A–F).
As shown in Figure 1c, B-cell-specific activation of oncogenic SmoM2 signaling is not sufficient to induce a CLL-like phenotype. We did not detect any significant difference in the amount of CD5+/CD19+ cells in the peritoneal cavity (PC), BM and PB of control (ctrl) and SmoM2Bact mice. Only in the spleen of 10-month-old (Figure 1c; 1.8% in control versus 3.5% in SmoM2Bact mice) and the PC of 5-month-old SmoM2Bact mice (Supplementary Figure S1B) did we detect a subtle increase in CD5+/CD19+ cells. We also analyzed CD19+ B2 cells in the BM, spleen and PB of CD19cre/+;R26SmoM2 mice but did not detect any significant changes in the amount of B2 cells compared with CD19cre/+ control mice (data not shown).
Given the lack of substantial CD5+ B-cell accumulation as described for other murine CLL models, we conclude that constitutive Hh/Gli signaling induced by B-cell-specific SmoM2 expression is insufficient for the initiation of a full-blown CLL-like phenotype in mice.
An alternative explanation for the inability of SmoM2 to expand CD5+/CD19+ cells may be its moderate activity as HH pathway activator.52 In addition, Smo signaling strictly depends on the presence of a functional primary cilium, an antenna-like organelle protruding from the cell surface and acting as critical organizing center of classical Smo-dependent Hh/Gli signaling.53, 54, 55 The primary cilium represents a feature characteristic mainly of adherent cell types, whereas cells of the hematopoietic system are typically considered to lack a primary cilium (reviewed in Finetti et al.56).
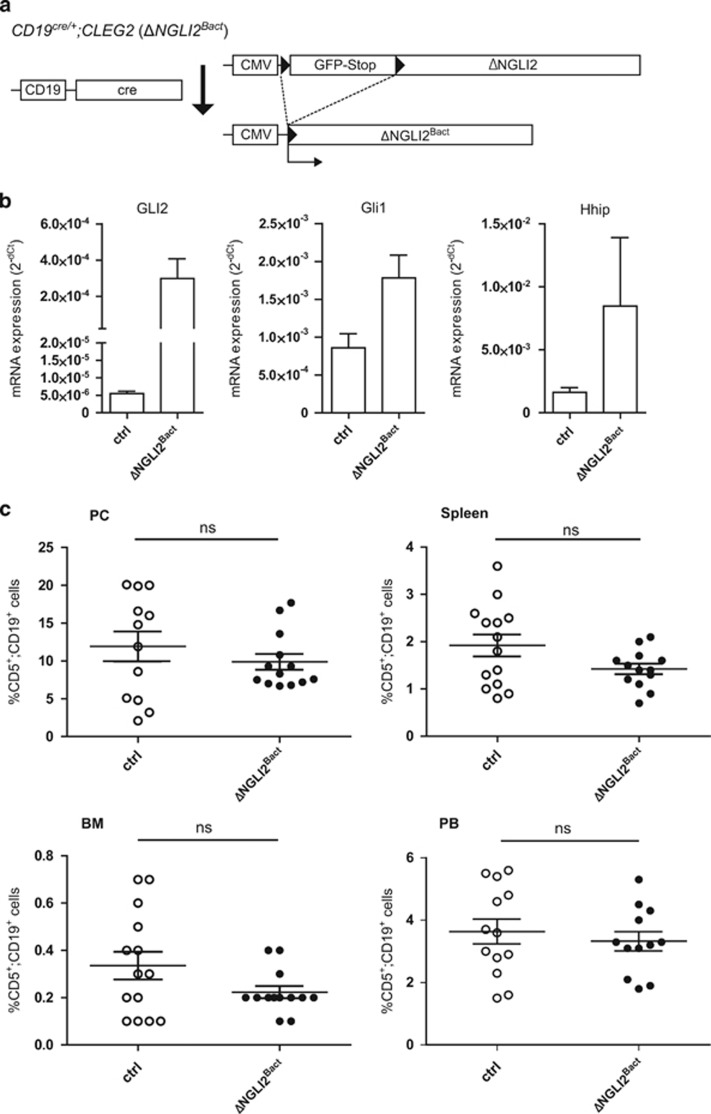
We therefore deployed an alternative genetic model, allowing us to activate in B cells Smo-independent high-level Hh signaling at the level of the Gli transcription factors by expression of a dominant active version of the Gli2 transcriptional activator referred to as ΔNGli2. For this purpose, we crossed CD19cre/+ mice with Cleg2 mice carrying a conditional ΔNGli2 allele57 (Figure 2a), yielding a progeny with B-cell-specific activation of Gli2 activator expression (ΔNGli2Bact mice). CD19+ B cells of ΔNGli2Bact mice express high levels of ΔNGli2 mRNA and increased levels of the Hh/Gli targets Gli1 and Hhip when compared with CD19+ cells of CD19cre/+ control mice (ctrl), demonstrating Smo-independent Hh/Gli pathway activation at the level of the Gli transcription factors (Figure 2b).
Figure 2.
B-cell-specific expression of Gli2 activator (ΔNGli2) does not lead to a systemic increase in leukemic CD5+ B cells. (a) Cre/loxP strategy for the activation of dominant active GLI2 expression in CD19+ B cells by the generation of CD19cre/+;CLEG2 (ΔNGli2Bact) mice. (b) qPCR analysis of transgenic GLI2 and endogenous Gli1 and Hhip mRNA expression in B cells of CD19cre/+ control mice (ctrl; n=6) and CD19cre/+;CLEG2 mice (ΔNGli2Bact; n=6). (c) Percentage of CD5+/CD19+ B cells in the PC, spleen, BM and PB of 10-month-old CD19cre/+ control mice (ctrl) and 10-month-old ΔNGli2Bact mice. Scatter dot plot with each dot representing an individual mouse. The mean value is indicated as bar, whiskers represent the s.e.m. ns: P>0.05.
Analysis of CD5+/CD19+ cells in ΔNGli2Bact mice revealed a similar phenotype as described for SmoM2Bact mice (Figure 2c). We did not observe a significant change of CD5+ B cells in ΔNGli2Bact mice in the PC with 11.9% in the control and 9.9% in the ΔNGli2Bact mice. Similarly, the number of CD5+ B cells in the BM, the spleen and the PB did not change significantly (Figure 2c). In addition, the weight of spleens from SmoM2Bact and ΔNGli2Bact mice was comparable to CD19cre/+ control mice and the number of B1b cells and B2 cells remained constant in SmoM2Bact and ΔNGli2Bact mice (data not shown). We conclude that mere activation of constitutive HH signaling at the level of Smo or at the level of Gli is insufficient to initiate a CLL-like phenotype in mice.
Pten deletion and simultaneous Hh/Gli activation do not cooperate in the initiation of a CLL-like phenotype in mice
PI3K/AKT signaling constitutes a critical signaling axis with relevance to CLL therapy.46, 47, 50 Of note, mice with B-cell-specific activation of Pi3k/Akt by genetic deletion of Pten showed increased numbers of CD5+-positive B cells, although they did not progress to a full-blown disease, suggesting that additional genetic/epigenetic alterations are required for malignant transformation.58 As PI3K/AKT has been shown to enhance the oncogenic HH/GLI signal strength,26, 28, 29, 59 we hypothesized that combined B-cell-specific activation of Pi3k/Akt and Hh/Gli may result in transformation of B cells into a CLL-like disease in mice.
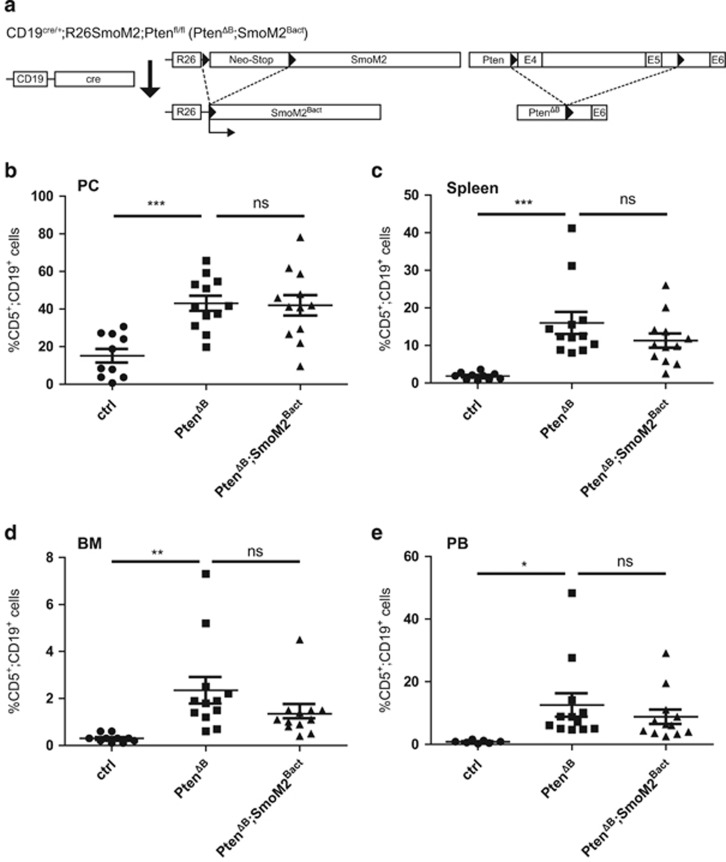
To genetically test this hypothesis, we crossed SmoM2Bactmice or ΔNGli2Bact mice to Ptenflox/flox mice, yielding mice with combined B-cell-specific activation of either SmoM2 and Pi3k/Akt (PtenΔB;SmoM2Bact; Figure 3a) or ΔNGli2 and Pi3k/Akt signaling (PtenΔB; ΔNGli2Bact; Figure 4a). Activation of Pi3k/Akt signaling in response to CD19cre-mediated Pten deletion was verified by phospho-Akt (pS473) analysis in CD19+ cells of compound transgenic mice (data not shown).
Figure 3.
Oncogenic SmoM2 signaling and PTEN deletion in B cells do not synergize in the initiation of a CLL-like phenotype in mice. (a) Cre/loxP strategy for the combined activation of oncogenic SmoM2 and Pten-deletion-mediated Pi3k/Akt signaling in CD19+ B cells by the generation of CD19cre/+;R26SmoM2;Ptenfl/fl (PTENΔB;SmoM2Bact) mice. (b–e) Flow cytometry analysis of CD5+/CD19+ cells in PC (b), spleen (c), BM (d) and PB (e) of 8-month-old CD19cre/+ control mice (ctrl), CD19cre/+;Ptenfl/fl (PtenΔB) and CD19cre/+;Ptenfl/fl;R26SmoM2 (PtenΔB;SmoM2Bact) mice. B-cell-specific Pi3k/Akt activation by Pten deletion significantly increases the amount of CD5+/CD19+ cells. Concomitant SmoM2 activation does not affect the CD5+/CD19+ population. Scatter dot plot with each dot representing an individual mouse. The mean value is indicated as bar, whiskers represent the s.e.m. ns: P>0.05; *P<0.05; **P<0.01; ***P<0.001.
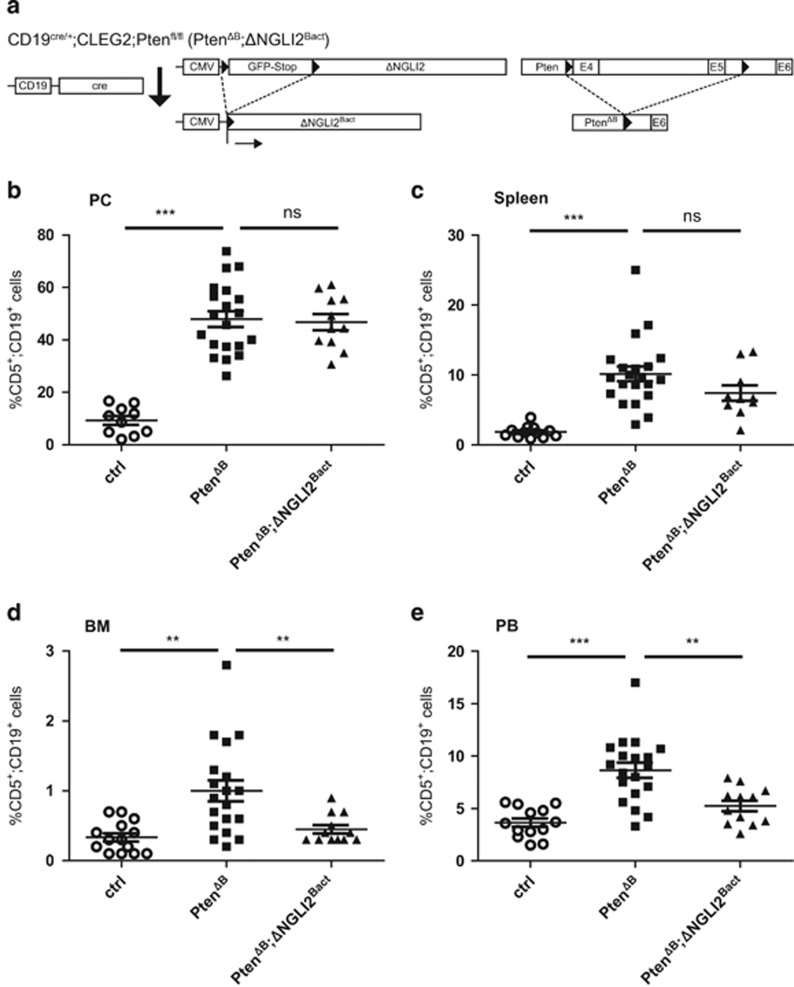
Figure 4.
Combined in vivo activation of Gli2 and Pi3K/Akt signaling in murine B cells does not initiate a CLL-like phenotype. (a) Cre/loxP strategy for the combined activation of dominant active GLI2 and Pten-deletion-mediated Pi3k/Akt signaling in CD19+ B cells by the generation of CD19cre/+;CLEG2;Ptenfl/fl (PtenΔB; ΔNGli2Bact) mice. (b–e) Flow cytometry analysis of CD5+/CD19+ cells in PC (b), spleen (c), BM (d) and PB (e) of 8-month-old CD19cre/+ control mice (ctrl), CD19cre/+;Ptenfl/fl (PtenΔB) and CD19cre/+;Ptenfl/fl;CLEG2 (PtenΔB; ΔNGli2Bact) mice. B-cell-specific deletion of Pten increases CD5+/CD19+ cells in all organs analyzed. Simultaneous activation of Gli2 expression reverses this effect in BM and PB. Scatter dot plot with each dot representing an individual mouse. The mean value is indicated as bar, whiskers represent the s.e.m. ns: P>0.05; **P<0.01; ***P<0.001.
In line with previous data,58 10-month-old PtenΔB mice showed a significant increase in the number of CD5+ B cells in the PC (15% in control CD19cre/+ mice (ctrl) versus 43% in PtenΔB mice), spleen (1.8% versus 16%), BM (0.4% versus 1.3%) and PB (0.6% versus 12% Figure 3b–e). We also confirm previous data showing that B-cell-specific loss of Pten results in an increase in marginal zone B cells and splenomegaly (data not shown).58 Of note, B-cell-specific deletion of Pten did not significantly affect Gli1 or Hhip expression (Supplementary Figure S2), suggesting that loss of Pten does not enhance Hh signaling in B cells.
PtenΔB mice with concomitant activation of SmoM2 expression (PtenΔB;SmoM2Bact) did not show an enhanced CD5+/CD19+ phenotype in any of the organs analyzed (Figure 3b–e), nor did PtenΔB; SmoM2Bact display any obvious adverse symptoms or reduced viability compared with PtenΔB (data not shown). We conclude that simultaneous activation of SmoM2 and Pi3k/Akt signaling does not synergize in the initiation of a full-blown CLL-like phenotype in mice.
Pi3k/Akt signaling has been shown to directly affect and enhance the transcriptional activity of Gli proteins by promoting Gli protein stability, nuclear import and/or by releasing Gli from its negative regulator suppressor of fused via mTOR/S6K activation.26, 28, 29 We therefore also tested whether combined Pi3k/Akt signaling and Gli activator expression cooperate in the transformation of CD19+ B cells. For this, we generated PtenΔB; ΔNGli2Bact mice with B-cell-specific deletion of Pten and concomitant expression of dominant active Gli2 (Figure 4a). We could again confirm the increase in CD5+/CD19+ cells in PtenΔB mice (Figure 4b–e). However, and similar to results of PtenΔB;SmoM2Bact mice, Gli2 activation in combination with Pten deletion did not further enhance the amount of CD5+/CD19+ cells in the PC and spleen compared with CD19cre/+ control mice (Figure 4b and c). Surprisingly, however, Gli2 activator expression reversed the increase in CD5+/CD19+ cells in the BM and PB of PtenΔB mice (Figure 4d and e), pointing to an unexplained negative impact of Gli2 activator on Pi3k/Akt in CD5+/CD19+ cells.
Combined targeting of GLI and PI3K/AKT reduces survival of human CLL cells
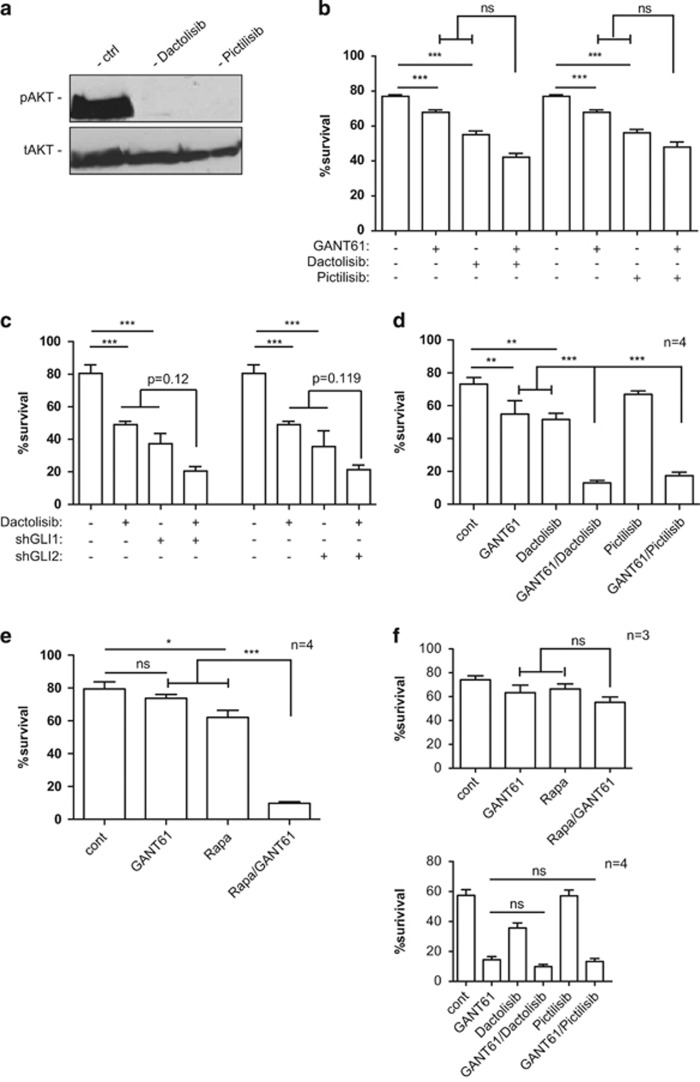
Although we did not find evidence for Hh/Gli–Pi3k/Akt cooperation in the establishment of a CLL-like disease in mice, pathway interaction may well occur in established CLL. We have previously uncovered a critical SMO-independent role of GLI activator for the survival of human CLL cells.41 Likewise, PI3K/AKT signaling is frequently active in CLL and targeting PI3K in CLL patients currently emerges as a novel targeted therapy with unprecedented efficacy.47, 50 We therefore hypothesized that GLI and PI3K/AKT may cooperate in established CLL cells to synergistically promote survival, which would open new avenues for rational combination therapy. To test for GLI–PI3K interactions, we performed single and combined chemical and genetic inhibition experiments of GLI and PI3K signaling, followed by monitoring the effects on CLL cell survival via flow cytometry of Annexin V/7AAD-stained cells. As cellular model for assessing putative GLI–PI3K/AKT interactions, we first used the CLL-derived cell line MEC1,60 which displays both GLI and PI3K/AKT activation Desch et al.41(Figure 5a). To inhibit PI3K activity, we treated MEC1 cells with the pan-PI3K inhibitors dactolisib or pictilisib.61, 62 Both compound effectively inhibited PI3K signaling in MEC1 CLL cells as evidenced by efficient abrogation of AKT phosphorylation (Figure 5a). We tested for synergistic reduction of CLL survival by combining PI3K inhibitors with the GLI antagonist GANT61.63 In single treatment settings, PI3K inhibition reduced CLL cell survival by ~30%. Treatment with GANT61 had a less pronounced, yet significant, negative effect on CLL survival, consistent with our previous data.41 Combined inhibition of PI3K and GLI enhanced the single treatment effect to up to 50% reduction of survival (combination GANT61/dactolisib). However, statistical testing for synergistic interactions of GANT61 and dactolisib did not reach significance. In line with enhanced reduction of MEC1 survival by targeting GLI and PI3K, combined GANT61/dactolisib treatment was more efficient in inhibiting GLI1 mRNA expression than single treatments (Supplementary Figure S3). We also evaluated the possible therapeutic efficacy of combined PI3K and GLI targeting by lentiviral short hairpin RNA (shRNA) knockdown of GLI1 and GLI2, either alone or in combination with dactolisib. In line with our previous data,41 inhibition of GLI1 or GLI2 significantly reduced CLL survival. Inhibition of PI3K signaling in combination with GIL1 or GLI2 knockdown further reduced CLL survival, although again synergy did not reach statistical significance.
Figure 5.
Efficient inhibition of CLL survival by combined targeting of GLI and PI3K/AKT signaling. (a) Human MEC1 CLL cells display activated PI3K/AKT signaling, as evidenced by expression of phopho-AKT (pAKT; pS473) in control-treated cells (solvent only). Inhibition of PI3K signaling either by dactolisib (1 μM) or pictilisib (1 μM) both abrogated AKT activation. (b) Single and combined GLI and PI3K/AKT targeting reduces CLL survival. Statistical analysis of single versus combined treatments did not reveal significant synergistic interactions of the GLI and PI3K inhibitor drugs. CLL cells were treated for 72 h with 10 μM GANT61 and/or 1 μM dactosilib/1 μM pictilisib before 7AAD/Annexin V staining and multicolor flow cytometry analysis. (c) Genetic perturbation of GLI1 and GLI2 in MEC1 CLL cells by stable lentiviral shRNA both strongly reduced survival. Control cells were transduced with nontarget control shRNA. Dactosilib treatment of GLI1 and GLI2 knockdown cells led to a further decrease in the number of viable cells, yet synergistic interaction of GLI1/2 depletion and PI3K targeting did not reach statistical significance. Data represent the mean values of three independent biological replicate experiments, error bars indicate the s.e.m. (d) Primary CLL cell survival in response to single and combined targeting of GLI (GANT61) and PI3K (dactolisib or pictilisib). Only patient samples sensitive to combined targeting are shown (that is, ID 7643, 7644, 7889 and 7890). (e) Primary CLL cell survival in response to single and combined targeting of GLI (GANT61) and mTOR (Rapa) signaling. Only patient samples sensitive to combined targeting are shown (that is, ID 1753, 7579, 7643 and 7644). (f) Upper panel: survival of three primary CLL cell samples (ID 544, 7538 and 7632) in response to single or combined targeting of GLI (GANT61) and mTOR (Rapa); no significant reduction of CLL survival by single or combined GLI-mTOR targeting; lower panel: primary CLL cell survival of four patients (ID 7530, 7913, 7914 and 7915) preferentially sensitive to GLI targeting; combined treatment with GANT61 and dactolisib or pictilisib did not further enhance apoptosis; n=number of patient samples. Error bars represent s.e.m; ns: P>0.05; *P<0.05; **P<0.01; ***P<0.001.
As MEC1 cells do not fully represent the molecular and genetic features of CLL cells, we tested the clinical relevance of our findings by analyzing the survival of primary human CLL cells in response to single or combined targeting of GLI and PI3K signaling. In total, we analyzed the effect of single and combined GLI–PI3K targeting on primary CD19+/CD5+ leukemic cell survival of a total of 14 CLL patients (for summary, see Supplemenatry Tables S1 and S2). To inhibit PI3K activity, we treated cultured primary CLL cells with dactolisib, pictilisib or wortmannin64 either alone or in combination with GANT61. We also analyzed the effect of combined treatment with GANT61 and rapamycin, a potent inhibitor of the PI3K effector mTOR.65 Notably, six patient samples were highly sensitive to combined GLI–PI3K and/or GLI-mTOR targeting, but only moderately to single treatments (that is, 1753, 7579, 7643, 7644, 7889 and 7890 (Figure 5d and e); also see Supplemenatry Table S1). As shown in Figure 5d, combination treatment of four primary CLL samples (patient ID 7643, 7644, 7889 and 7890) with GANT61/dactolisib or GANT61/pictilisib synergistically enhanced CLL apoptosis by more than 65%, whereas single treatment protocols reduced survival by only less than 30% compared with controls. Further, combination treatment with GANT61 and rapamycin (Rapa) synergistically reduced survival of four primary CLL samples (Figure 5e, patient ID 1753, 7579, 7643 and 7644), two of which were also highly sensitive to GLI/PI3K targeting (ID 7643 and 7644; see Supplementary Table S1). The heterogeneity of treatment responses is further shown in Figure 5f. CLL cells of three patients (ID 544, 7538 and 7632) were essentially resistant to single and combined GLI/mTOR targeting, whereas five patient samples were mainly sensitive to GLI targeting, with no further enhancement of apoptosis by concomitant PI3K inhibition (patient ID 7530, 7577, 7913, 7914 and 7915, also see Supplemenatry Table S1).
Together, these results suggest that PI3K/mTOR signaling and GLI proteins can cooperate in a fraction of CLL patients to promote CLL cell survival.
Discussion
Oncogenic HH/GLI signaling accounts for the development and progression of a number of human cancers (for review see Teglund and Toftgard5 and references therein). Its clinical relevance has been convincingly evidenced by the striking therapeutic efficacy of HH pathway inhibitors targeting SMO for the treatment of patients suffering from HH-driven basal cell carcinoma or medulloblastoma.66, 67, 68, 69, 70 However, the rapid development of drug resistance30, 31, 66, 71 as well as noncanonical SMO-independent activation of GLI call for a detailed molecular analysis of HH/GLI and interacting oncogenic pathways to develop better treatment strategies.
The causal involvement of the HH/GLI pathway in malignancies of the hematopoietic system such as B-cell malignancies including CLL has previously been reported,33, 34, 41, 42 although its precise role is not fully understood, partly also because of the lack of sophisticated mouse models mimicking oncogenic Hh/Gli and signal cross-talk specifically in B cells. We therefore generated and analyzed transgenic mice with conditional B-cell-specific activation of Hh/Gli signaling alone or in combination with oncogenic PI3K signaling, an emerging and highly promising therapeutic target in CLL patients known to promote GLI activity.47, 50
Several groups have addressed the role of HH/GLI in CLL, yet its involvement in CLL is poorly defined. CLL cell survival critically depends on signals derived from the surrounding tumor stroma. Hegde et al.42 showed that stroma-derived Sonic HH protein is likely to protect CLL cells from apoptosis, suggesting paracrine and SMO-dependent activation of HH signaling in CLL cells. By contrast, our own previous study identified non-canonical, SMO-independent activation of GLI transcription factors in CLL cells as a crucial pro-survival signal,41 a finding in line with another report on the absence of HH expression in the lymph node stroma of CLL patients.72 The controversial results may also be because of the highly diverse expression of HH pathway members in CLL. Indeed, sensitivity to SMO inhibitors correlates only with high HH target gene levels and a trisomy 12 karyotype.33 In essence and despite these discrepancies, the results clearly support a critical role of HH pathway effectors particularly of GLI in established CLL.
Whether HH/GLI signaling does have a role in the initiation of CLL has not been addressed so far. The results of our present study do not support a critical contribution of Hh/Gli in triggering a CLL-like phenotype in mice, as evidenced by the lack of a substantial and systemic increase in CD5+/CD19+ B cells in response to pathway activation. We also propose that the apparent lack of a significant response to Hh/Gli activation in B cells is not simply because of the absence of a primary cilium on hematopoietic cells, on which SMO critically depends.53, 56 First, we detected activation of Hh/Gli target genes in B cells expressing constitutively active SmoM2. Second, we did not observe significant changes in CD5+ B-cell numbers in mice with B-cell-specific expression of dominant active Gli2, activating Hh/Gli downstream of Smo independently of a primary cilium. That said, expression of active SMO led to a subtle increase in CD5+ B cells in the spleen, with a similar yet non-significant trend in the BM. This may point to a possible involvement of HH/GLI in the proliferative niches of CLL cells, which may be boosted by additional driver mutations. We therefore tested for a possible interaction of HH/GLI and PI3K/AKT signaling, as (i) PI3K/AKT has been shown to cooperate with HH/GLI in cancer,28, 30, 31, 59 (ii) PI3K is frequently activated in CLL41 and PI3K targeting is highly effective in the treatment of CLL patients47, 50 and (iii) mice with Pten-deficient B cells display a systemic expansion of CD5+ B cells without progressing to a full-blown CLL-like phenotype as Eμ-Tcl1 transgenic mice do.58 Furthermore, transgenic expression of the AKT enhancer TCL1 in immature and mature murine B cells induces a CLL-like phenotype that presents with an expansion of CD5+ B cells in the PC, and later on the accumulation of leukemic cells in the BM, spleen and PB.51, 73
We found that activation of Hh/Gli in B cells with Pi3K/Akt activation does not promote the expansion of CD5+ B cells, again suggesting that Hh/Gli does not have a major role in the early initiation phase of a CLL-like phenotype in mice. In summary, we propose that B-cell-specific activation of Hh/Gli signaling, even in combination with Pi3k/Akt, does not suffice to elicit a malignant B-cell phenotype in mice. It may, however, be possible that Hh/Gli provides a permissive signal being required for the establishment of a malignant CLL-like phenotype. This will be an important question to be addressed in future studies and will require genetic and pharmacological inactivation of Hh/Gli signaling in sophisticated in vivo CLL models such as Eμ-Tcl1 transgenic mice.51
In contrast to the initiation and onset of CLL, PI3K/AKT and GLI activation is crucial in established CLL, mainly supporting the survival of malignant cells.41, 74, 75 We therefore addressed the possible cooperation of GLI and PI3K/AKT in CLL survival. We first confirm the importance of GLI activator forms (GLI1/GLI2) and PI3K/AKT signaling for CLL survival and, second, show that combined GLI and PI3K/AKT inhibition in primary CLL cells strongly reduces CLL survival in an undefined subgroup of CLL patients. This may open up new opportunities for future rationale-based combination treatments relying on simultaneous targeting of GLI and PI3K. Such a strategy may even further enhance and/or prolong the striking therapeutic benefit of PI3K targeting.47, 50 In this context it is noteworthy that arsenic trioxide—an efficient therapeutic for the treatment of acute promyelocytic leukemia76—induces apoptosis of CLL cells even from patients with poor prognosis.77 As arsenic trioxide is able to inhibit both PI3K/AKT signaling and GLI activator forms,78, 79, 80, 81 its efficacy in triggering apoptosis may be the result of combined targeting of PI3K/AKT and GLI.
In summary, our present study suggests a critical role of PI3K/AKT and GLI activator forms in the survival of established CLL rather than in the initiation of the disease phenotype and warrants further investigation into combined targeting of GLI–PI3K using recently validated and promising GLI inhibitors such as arsenic trioxide or atypical protein kinase C antagonists.17, 78
Materials and methods
Transgenic mice and genotyping
All animal experiments were performed in compliance with the national requirements. All transgenic lines were maintained on a C57BL/6 N background for at least 10 generations. CD19cre mice82 with a knock-in allele of cre recombinase in the CD19 gene locus and R26SmoM2 mice83 carrying a conditional SmoM2 allele under control of the Rosa26 promoter were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) (JAX strains B6.129P2(C)-Cd19tm1(cre)Cgn/J and Gt(ROSA)26Sortm1(Smo/EYFP)Amc/J). Conditional PTENflox/flox mice were kindly provided by Professors Tak Mak and Josef Penninger.84 Cleg2 mice carry a conditional transgenic allele of a dominant active version of GLI2.57
PCR genotyping of CD19cre mice was performed using the following primers: forward 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and reverse 5′-GTGAAACAGCATTGCTGTCACTT-3′. Genotyping of floxed PTEN mice was carried out using forward 5′-GTCACCAGGATGCTTCTGAC-3′ and reverse 5′-GAAACGGCCTTAACGACGTAG-3′ primers, for wild-type controls we used forward 5′-GTCACCAGGATGCTTCTGAC-3′ and reverse 5′-GTGACATCAACATGCAACACTG-3′ primers. Genotyping of the SmoM2 allele was accomplished using the following oligonucleotide primers: forward 5′-CTGACCCTGAAGTTCATCTGC-3′ and reverse 5′-GTGCGCTCCTGGACGTAG-3′. The wild-type was genotyped with forward 5′-CGTGATCTGCAACTCCAGTC-3′ and reverse 5′-GGAGCGGGAGAAATGGATATG-3′ primers.
Transgenic GLI2 was detected using the following primers: forward 5′-CCCGCCTGGAGAACCTGAAGACAC-3′ and reverse 5′-CCCCGGGGCTGGACTGACAC-3′.
Flow cytometry and apoptosis assays
Peritoneal cells were isolated by peritoneal lavage. Spleen cells were purified by maceration through a 40-μm cell strainer (BD Biosciences, San Jose, CA, USA) and separation on a lymphocyte separation medium gradient (Sigma, St Louis, MO, USA). Samples were washed twice with phosphate-buffered saline before staining. PB cells were incubated with Red Blood Lysis Buffer and washed twice before staining. BM cells were macerated through a 40-μm cell strainer (BD Biosciences) and washed twice in phosphate-buffered saline. Peritoneal cells, spleen cells, PB and BM cells were stained with the following antibodies: CD5-PE, CD19-FITC (Immunotools, Friesoythe, Germany), IgM-APC (BD Biosciences), CD23-PerCP-Cy7 and/or CD11b-APC (Biolegends, San Diego, CA, USA). CD16/32 Antibody (Biolegends) was used to prevent unspecific binding to FcγIII and FcγII receptors. To analyze cell survival and apoptosis, respectively, CLL cells were stained with Annexin-V-FITC (Immunotools), 7-Aminoactinomycin D (7AAD, BD Biosciences) and the surface markers CD19-APC (Immunotools). All flow cytometry analyses were performed on a BD FACS Canto II and data were analyzed with the BD FACS Diva Software (BD Biosciences).
RNA isolation and real-time PCR
Total RNA was isolated using the Micro RNA Kit (Life Technologies, Vienna, Austria) according to the manufacturer's protocol. RNA from MEC cells were isolated using TRI Reagent (Molecular Research Center, Cincinnati, OH, USA) followed by a LiCl precipitation step for enrichment of highly pure RNA. Reverse transcription of RNA was performed using the Superscript II (Life Technologies) and quantitative real-time PCR was performed on a Rotor gene 3000 Cycler (Qiagen, Hilden, Germany) using Sybr Green (Bio-Rad, Hercules, CA, USA) as described previously.41
Cell culture and inhibitor treatments
All work with primary human material was performed in compliance with the national and local regulations. Primary CLL cells were isolated from PB and BM of CLL patients via Biocoll (Biochrom AG, Berlin, Germany) density gradient centrifugation and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin (62.5 μg/ml) and streptomycin (100 μg/ml) at a density of 1 × 106 cells/ml. MEC1 cells (ATCC, Manassas, VA, USA) were cultured in IMDM medium containing 10% fetal calf serum, penicillin and streptomycin. Cells were split twice a week at a density of 106 cells per ml medium. For lentiviral packaging, HEK293FT (ATCC) cells were grown in DMEM supplemented with 10% fetal bovine serum (Life Technologies), penicillin and streptomycin, L-glutamate (100x concentrate, Life Technologies) and essential amino acids (100 × concentrate, Life Technologies) and split three times a week in a ratio of 1:5. Dactolisib, pictilisib, rapamycin, wortmannin (LC Laboratories, Woburn, MA, USA) and GANT61 (Merck, Darmstadt, Germany) were dissolved in dimethylsulfoxide, stored as stock solution at −20 and diluted to final concentrations as indicated in the Results section.
RNA interference experiments
Lentiviral shRNA knockdown experiments in MEC1 cells were performed as described in Desch et al.41 and lentiviral production and transduction following the protocol published by Kasper et al.,85 except that packaging plasmid transfection was carried out using Metafectene Pro (Biontex GmbH, Planegg, Germany) according to the manufacturer's instructions. The following lentiviral shRNA constructs (Sigma-Aldrich mission TRC library, Sigma) were used: shGLI1 (TRCN0000020486), shGLI2 (TRCN0000238361) and nontarget control shRNA (shc002). Efficient knockdown of GLI1 and GLI2 was verified by quantitative PCR (data not shown).
Western blot analysis
Protein detection was performed using the SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific, Waltham, MA, USA) and Hyperfilm ECL films (GE Healthcare, Amersham, UK). Antibodies against Akt and phospho-Akt (pS473) were purchased from Cell Signaling Technology (Beverly, MA, USA).
Statistical analysis
Statistical analyses (two-way analysis of variance with post hoc Bonferroni or unpaired t-test for normally distributed data, respectively, Mann–Whitney test or Kruskal–Wallis test for non-normally distributed data) were performed using SPSS 22 (IBM Corp, Armonk, NY, USA). Effects were considered significant at P⩽0.05 with *P⩽0.05; **P⩽0.001; ***P⩽0.0001.
Acknowledgments
We are grateful to Professors Tak Mak and Josef Penninger for providing transgenic mouse strains, and Dr Sandrine Tonon and Professor Christoph Binder for help with B-cell analysis. Work of FA has been supported by the Austrian Science Fund FWF (project W1213), the Austrian Genome Project GEN-AU and the priority program Biosciences and Health of the Paris-Lodron University of Salzburg. Work of RG, TNH, SWH and PA was supported by the Austrian Science Fund FWF (SFB-P021 and W1213 to RG), the SCRI-LIMCR GmbH and the province of Salzburg. We dedicate this study to Gernot Achatz who tragically passed away during the course of this study.
The authors declare no conflict of interest
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- 1Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004; 432: 324–331. [DOI] [PubMed] [Google Scholar]
- 2Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008; 8: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Gulino A, Di Marcotullio L, Ferretti E, De Smaele E, Screpanti I. Hedgehog signaling pathway in neural development and disease. Psychoneuroendocrinology 2007; 32: S52–S56. [DOI] [PubMed] [Google Scholar]
- 4Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 2009; 30: 303–312. [DOI] [PubMed] [Google Scholar]
- 5Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta 2010; 1805: 181–208. [DOI] [PubMed] [Google Scholar]
- 6Aberger F, Kern D, Greil R, Hartmann TN. Canonical and noncanonical Hedgehog/GLI signaling in hematological malignancies. Vitam Horm 2012; 88: 25–54. [DOI] [PubMed] [Google Scholar]
- 7Aberger F, Ruiz i Altaba A. Context-dependent signal integration by the GLI code: the oncogenic load, pathways, modifiers and implications for cancer therapy. Semin Cell Dev Biol 2014; 33: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Brennan D, Chen X, Cheng L, Mahoney M, Riobo NA. Noncanonical Hedgehog signaling. Vitam Horm 2012; 88: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol 2005; 6: 306–317. [DOI] [PubMed] [Google Scholar]
- 10Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- 11Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 2011; 12: 393–406. [DOI] [PubMed] [Google Scholar]
- 12Mangelberger D, Kern D, Loipetzberger A, Eberl M, Aberger F. Cooperative Hedgehog-EGFR signaling. Front Biosci 2012; 17: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol 2007; 9: 1005–1009. [DOI] [PubMed] [Google Scholar]
- 14Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol 2010; 2: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol 2011; 27: 513–537. [DOI] [PubMed] [Google Scholar]
- 16Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol 2007; 17: 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature 2013; 494: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Beauchamp E, Bulut G, Abaan O, Chen K, Merchant A, Matsui W et al. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J Biol Chem 2009; 284: 9074–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Dennler S, Andre J, Alexaki I, Li A, Magnaldo T, ten Dijke P et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res 2007; 67: 6981–6986. [DOI] [PubMed] [Google Scholar]
- 20Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev 2009; 23: 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Eberl M, Klingler S, Mangelberger D, Loipetzberger A, Damhofer H, Zoidl K et al. Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol Med 2012; 4: 218–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol 2006; 26: 6283–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Lauth M, Bergstrom A, Toftgard R. Phorbol esters inhibit the Hedgehog signalling pathway downstream of Suppressor of Fused, but upstream of Gli. Oncogene 2007; 26: 5163–5168. [DOI] [PubMed] [Google Scholar]
- 24Pelczar P, Zibat A, van Dop WA, Heijmans J, Bleckmann A, Gruber W et al. Inactivation of Patched1 in mice leads to development of gastrointestinal stromal-like tumors that express Pdgfralpha but not kit. Gastroenterology 2013; 144: 134–144 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Riobo NA, Haines GM, Emerson CP Jr. Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res 2006; 66: 839–845. [DOI] [PubMed] [Google Scholar]
- 26Riobo NA, Lu K, Ai X, Haines GM, Emerson CP Jr. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA 2006; 103: 4505–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res 2009; 69: 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 2007; 104: 5895–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY et al. The cross-talk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012; 21: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med 2010; 2: 51ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Gruber Filbin M, Dabral SK, Pazyra-Murphy MF, Ramkissoon S, Kung AL, Pak E et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med 2013; 19: 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Dao KH, Tyner JW. Next-generation medicine: combining BCR-ABL and Hedgehog-targeted therapies. Clin Cancer Res 2013; 19: 1309–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Decker S, Zirlik K, Djebatchie L, Hartmann D, Ihorst G, Schmitt-Graeff A et al. Trisomy 12 and elevated GLI1 and PTCH1 transcript levels are biomarkers for Hedgehog-inhibitor responsiveness in CLL. Blood 2012; 119: 997–1007. [DOI] [PubMed] [Google Scholar]
- 34Dierks C, Grbic J, Zirlik K, Beigi R, Englund NP, Guo GR et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nat Med 2007; 13: 944–951. [DOI] [PubMed] [Google Scholar]
- 35Dierks C, Beigi R, Guo GR, Zirlik K, Stegert MR, Manley P et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell 2008; 14: 238–249. [DOI] [PubMed] [Google Scholar]
- 36Katagiri S, Tauchi T, Okabe S, Minami Y, Kimura S, Maekawa T et al. Combination of ponatinib with Hedgehog antagonist vismodegib for therapy-resistant BCR-ABL1-positive leukemia. Clin Cancer Res 2013; 19: 1422–1432. [DOI] [PubMed] [Google Scholar]
- 37Merchant A, Joseph G, Wang Q, Brennan S, Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood 2010; 115: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA 2007; 104: 4048–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014; 511: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature 2009; 458: 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Desch P, Asslaber D, Kern D, Schnidar H, Mangelberger D, Alinger B et al. Inhibition of GLI, but not Smoothened, induces apoptosis in chronic lymphocytic leukemia cells. Oncogene 2010; 29: 4885–4895. [DOI] [PubMed] [Google Scholar]
- 42Hegde GV, Peterson KJ, Emanuel K, Mittal AK, Joshi AD, Dickinson JD et al. Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: a potential new therapeutic target. Mol Cancer Res 2008; 6: 1928–1936. [DOI] [PubMed] [Google Scholar]
- 43Pleyer L, Egle A, Hartmann TN, Greil R. Molecular and cellular mechanisms of CLL: novel therapeutic approaches. Nat Rev Clin Oncol 2009; 6: 405–418. [DOI] [PubMed] [Google Scholar]
- 44Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50. [DOI] [PubMed] [Google Scholar]
- 45Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 2009; 114: 3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood 2011; 118: 4313–4320. [DOI] [PubMed] [Google Scholar]
- 47Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 2014; 370: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Wu M, Akinleye A, Zhu X. Novel agents for chronic lymphocytic leukemia. J Hematol Oncol 2013; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Tsimberidou AM, Keating MJ. Richter syndrome: biology, incidence, and therapeutic strategies. Cancer 2005; 103: 216–228. [DOI] [PubMed] [Google Scholar]
- 50Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND et al. Idelalisib, an inhibitor of phosphatidylinositol 3 kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood 2014; 123: 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA 2002; 99: 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A et al. The magnitude of hedgehog signaling activity defines skin tumor phenotype. Embo J 2003; 22: 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature 2005; 437: 1018–1021. [DOI] [PubMed] [Google Scholar]
- 54Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA 2005; 102: 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007; 317: 372–376. [DOI] [PubMed] [Google Scholar]
- 56Finetti F, Paccani SR, Rosenbaum J, Baldari CT. Intraflagellar transport: a new player at the immune synapse. Trends Immunol 2011; 32: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev 2006; 20: 3161–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Suzuki A, Kaisho T, Ohishi M, Tsukio-Yamaguchi M, Tsubata T, Koni PA et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med 2003; 197: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Ju B, Spitsbergen J, Eden CJ, Taylor MR, Chen W. Co-activation of hedgehog and AKT pathways promote tumorigenesis in zebrafish. Mol Cancer 2009; 8: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res 1999; 23: 127–136. [DOI] [PubMed] [Google Scholar]
- 61Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G et al. The identification of 2-(1 H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem 2008; 51: 5522–5532. [DOI] [PubMed] [Google Scholar]
- 62Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther 2008; 7: 1851–1863. [DOI] [PubMed] [Google Scholar]
- 63Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA 2007; 104: 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J 1993; 296: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994; 369: 756–758. [DOI] [PubMed] [Google Scholar]
- 66Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med 2009; 361: 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 2012; 366: 2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68Skvara H, Kalthoff F, Meingassner JG, Wolff-Winiski B, Aschauer H, Kelleher JF et al. Topical treatment of Basal cell carcinomas in nevoid Basal cell carcinoma syndrome with a smoothened inhibitor. J Invest Dermatol 2011; 131: 1735–1744. [DOI] [PubMed] [Google Scholar]
- 69Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med 2012; 366: 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 2009; 361: 1164–1172. [DOI] [PubMed] [Google Scholar]
- 71Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 2009; 326: 572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72Kim JE, Singh RR, Cho-Vega JH, Drakos E, Davuluri Y, Khokhar FA et al. Sonic hedgehog signaling proteins and ATP-binding cassette G2 are aberrantly expressed in diffuse large B-cell lymphoma. Mod Pathol 2009; 22: 1312–1320. [DOI] [PubMed] [Google Scholar]
- 73Gold MR. Akt is TCL-ish: implications for B-cell lymphoma. Trend Immunol 2003; 24: 104–108. [DOI] [PubMed] [Google Scholar]
- 74de Frias M, Iglesias-Serret D, Cosialls AM, Coll-Mulet L, Santidrian AF, Gonzalez-Girones DM et al. Akt inhibitors induce apoptosis in chronic lymphocytic leukemia cells. Haematologica 2009; 94: 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood 2010; 116: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med 1998; 339: 1341–1348. [DOI] [PubMed] [Google Scholar]
- 77Merkel O, Heyder C, Asslaber D, Hamacher F, Tinhofer I, Holler C et al. Arsenic trioxide induces apoptosis preferentially in B-CLL cells of patients with unfavourable prognostic factors including del17p13. J Mol Med (Berl) 2008; 86: 541–552. [DOI] [PubMed] [Google Scholar]
- 78Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest 2011; 121: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79Kim J, Lee JJ, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci USA 2010; 107: 13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80Mann KK, Colombo M, Miller WH Jr.. Arsenic trioxide decreases AKT protein in a caspase-dependent manner. Mol Cancer Ther 2008; 7: 1680–1687. [DOI] [PubMed] [Google Scholar]
- 81Xue P, Hou Y, Zhang Q, Woods CG, Yarborough K, Liu H et al. Prolonged inorganic arsenite exposure suppresses insulin-stimulated AKT S473 phosphorylation and glucose uptake in 3T3-L1 adipocytes: involvement of the adaptive antioxidant response. Biochem Biophys Res Commun 2011; 407: 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acid Res 1997; 25: 1317–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 2004; 18: 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 2001; 14: 523–534. [DOI] [PubMed] [Google Scholar]
- 85Kasper M, Regl G, Eichberger T, Frischauf AM, Aberger F. Efficient manipulation of Hedgehog/GLI signaling using retroviral expression systems. Methods Mol Biol 2007; 397: 67–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.