Abstract
Advancements in photolithography have enabled us to spatially encode biochemical cues in biocompatible platforms such as synthetic hydrogels. Conventional patterning works through photo-activated chemical reactions on inert polymer networks. However, these techniques cannot be directly applied to protein hydrogels without chemically altering the protein scaffolds. To this end, we developed a non-covalent photo-patterning strategy for gelatin (denatured collagen) hydrogels utilizing a caged collagen mimetic peptide (caged CMP) which binds to gelatin strands through UV activated, triple helix hybridization. Here we present 2D and 3D photo-patterning of gelatin hydrogels enabled by the caged CMPs as well as creation of concentration gradients of CMPs. We show that photo-patterning of PEG-conjugated caged CMPs can be used to spatially control cell adhesion on gelatin films. CMP’s specificity for binding to gelatin allows patterning of almost any synthetic or natural gelatin-containing matrix, such as zymograms, gelatin-methacrylate hydrogels, and even a corneal tissue. Since the CMP is a chemically and biologically inert peptide which is proven to be an ideal carrier for bioactive molecules, our patterning method provides a radically new tool for immobilizing drugs to natural tissues and for functionalizing scaffolds for complex tissue formation.
Keywords: hydrogel, microenvironment, spatial control, tissue engineering, triple helix
1. Introduction
Native tissues exhibit complex architectural features ranging from micro to millimeter length scale. Such complex features are managed by cells in response to spatio-temporally dynamic microenvironment in the form of soluble cues (e.g., growth factors and hormones), as well as insoluble cues such as cell- and extracellular matrix (ECM)-bound signaling molecules. Controlling the interactions between cells and their microenvironment is crucial for guiding cells into formation of complex tissue constructs.[1] Recent advancements in micropatterning technology have enhanced our ability to spatially encode these biochemical signals in the cell microenvironment within biocompatible platforms. Many research groups have reported the use of photo-activated chemical reactions to pattern biomolecules onto hydrogels comprised of simple synthetic and natural polymers, such as poly(ethylene glycol) (PEG) and agarose.[2-14] Although such simple and inert polymer networks are easy to pattern by photo-chemistry, they are generally not ideal for cell culture because they are not adhesive to cells and/or cannot be degraded by cells. This lack of cell-interactive elements in synthetic scaffolds greatly limits the ability of cells to proliferate, migrate and grow into organized structures.[15] Bioactivity of such hydrogels can be improved to some extent by incorporation of basic cell interactive components (commonly derived from ECM), such as cell binding,[9] and matrix metalloproteinase (MMP)-degradable domains.[2] Although these patterned synthetic hydrogels are great systems to recapitulate and investigate the role of spatiotemporal cues in vitro,[16] they are not ideal for engineering complex tissues.
Conventional hydrogel patterning techniques use photo-activated reactions to conjugate biomolecules to chemically modified matrices;[2-14,17] in contrast, in natural tissues, many signaling molecules bind to ECM via non-covalent interactions (e.g., growth factor-ECM binding).[18] This inspired us to seek a natural ECM patterning technique based on non-covalent binding interactions. We envisioned that the non-covalent patterning of natural ECM would maintain the native chemical composition of the ECM and that such a patterning approach will have immediate translational applications in tissue engineering and regenerative medicine.
Gelatin is one of the most widely used biocompatible platforms for tissue engineering and drug delivery. Gelatin, which is an unfolded collagen denatured by heat or by fragmentation of protein chains, can be derived from a variety of sources by inexpensive means. Gelatin solution spontaneously forms a transparent hydrogel upon cooling from high temperature, and as a natural ECM protein, it inherently contains cell binding motifs, such as the RGD and GFOGER sequences,[19] as well as protease-cleavable sites, making it an ideal substrate for tissue culture. Gelatin is frequently used to coat cell culture plates to improve attachment of cells, and gelatin hydrogels have been used as scaffolds in delivering chondrocytes and stem cells for osteochondral tissue repair.[20,21] It is also a popular matrix to deliver various types of growth factors for tissue regeneration in vivo.[22-24]
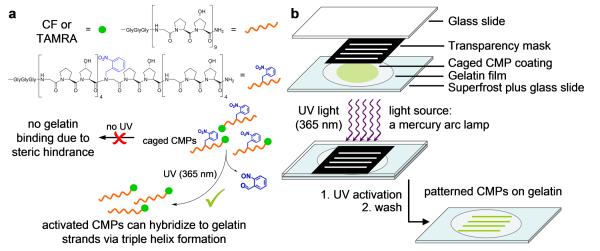
Previously, we discovered that a collagen mimetic peptide (CMP) [sequence: (GPO)n, n = 6–10, O: hydroxyproline] with strong propensity to fold into collagen-like triple helix can specifically hybridize to unfolded gelatin chains.[25-34] This binding is primarily driven by the triple helical hybridization between monomeric CMPs and the gelatin chains, which is similar to small DNA fragments binding to complimentary DNA strands. To enable photo-triggered gelatin binding, we incorporated a photo-labile nitrobenzyl protective group into the CMP backbone and developed a caged collagen mimetic peptide [sequence: (GPO)4NBGPO(GPO)4, designated as NB(GPO)9, NBG: N-o-nitrobenzyl-glycine] whose folding and gelatin binding are activated by UV light.[31,35] The caged CMP cannot hybridize to gelatin due to the steric hindrance caused by the NB cage group, yet removal of the cage group by UV light immediately triggers the peptide to hybridize with the gelatin chains (Figure 1).[31]
Figure 1.
Schematics of the photo-triggered activation of the caged CMP (a) and the experimental setup and process of photo-patterning gelatin films with caged CMPs.
The CMP, comprised of GlyProHyp repeats, is an excellent carrier for bioactive molecules.[25] The peptide is made entirely of neutral and hydrophilic amino acids which make them highly inert with respect to protein adsorption and enzymatic degradation. In addition, its simple chemical composition allows for easy modification and conjugation to other peptides and bioactive molecules, and even to inorganic and polymeric nanoparticles.[33,36] For example, CMPs have been conjugated to growth factor mimetics, cell binding motifs, antibody Fab regions, monosaccharides, fatty acids and a number of optical probes.[31,37-42] Such synthetic versatility, in conjunction with the ability to specifically hybridize with denatured collagen strand, makes CMP a truly unique tool for a wide range of targeting and delivery applications.
In this report, we present an in depth investigation into the photo-patterning (Figure 1) of 2D films and 3D hydrogels with caged CMPs and PEG-conjugated caged CMPs, as well as the cell adhesion properties of the patterned films. In addition, we show the photo-patterning of caged CMPs on other synthetic and natural gelatin-based materials, such as the gelatin-methacrylate hydrogel and bovine corneal tissue, which demonstrates the versatility of the method in patterning almost any gelatin-containing material for potential applications in drug immobilization and tissue scaffold engineering.
2. Results and Discussion
2.1. Photo-Activation of Caged CMP
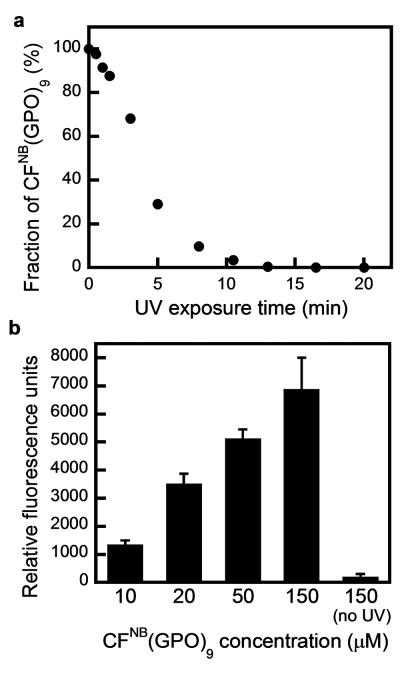
After synthesizing the fluorescently labeled caged CMPs, CFNB(GPO)9 and TAMRANB(GPO)9 (CF: carboxyfluorescein, TAMRA: carboxytetramethylrhodamine, see experimental section), we first studied the photo-cleavage efficiency of the nitrobenzyl (NB) group on the CMP backbone. A series of CFNB(GPO)9 solutions were irradiated with UV light (365 nm, 15.5 mW/cm2) for a range of time periods, and analyzed by high performance liquid chromatography (HPLC). Peak integration was used to calculate the fraction of caged CFNB(GPO)9 in each sample. Figure 2a shows an linear decrease of the CFNB(GPO)9 fraction overtime (R2 = 0.9736 for the linear fitting of the data) and a steady photo-decaging rate; 2.5 nmole of caged CMP was fully deprotected within 10 min. This photo-activation efficiency was used to estimate the UV exposure time in the subsequent binding and patterning experiments.
Figure 2.
Photo-triggered binding of caged CMP CFNB(GPO)9 on gelatin. (a) UV decaging efficiency estimated from a set of 50 μM CFNB(GPO)9 solutions exposed to UV light (365 nm, 15.5 mW/cm2) with variation in exposure time followed by HPLC analyses. The percentage of caged CFNB(GPO)9 remaining in each sample was calculated from HPLC peak integration. (b) Dose-dependent binding of photo-triggered CFNB(GPO)9 on gelatin films measured by fluorescence. Without UV activation, the caged CFNB(GPO)9 exhibited negligible affinity to gelatin even at the highest concentration tested. The binding assay was performed in triplicate, and error bars represent ±SD.
2.2. Photo-Triggered, Dose-Dependent Binding of CMP to Gelatin Matrix
To vary the concentration of photo-patterned CMP on gelatin substrates, we first investigated CMP’s dose-dependent binding. We performed a binding assay on thin films of gelatin hydrogels, where CFNB(GPO)9 solutions of various concentrations were applied and activated by a benchtop UV lamp for predetermined time periods. The fluorescence levels of the gelatin-bound CF(GPO)9 clearly indicated a dose-dependent binding of CMP to gelatin (Figure 2b). Without UV exposure, the caged CFNB(GPO)9 exhibited only negligible affinity to gelatin even at the highest concentration tested (150 μM, no UV), because the steric clash of the bulky NB cage group prevents triple helical hybridization. This drastic change in the caged CMP’s ability to bind to gelatin is the basis for its ability to be photo-patterned onto gelatin substrates. The results show that the local concentration of the patterned peptides can be readily controlled by both CMP dosage and UV exposure time.
2.3. Photo-Patterning of 2D Gelatin Films with Caged CMP
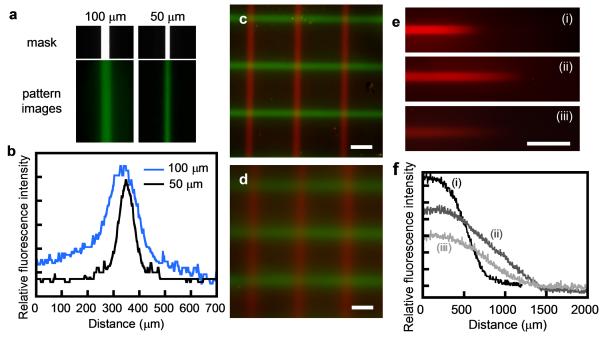
To demonstrate photo-patterning of CMPs on gelatin substrates, thin films of gelatin hydrogels (10% w/v) of approximately 50 μm thickness were fabricated and crosslinked by EDC/NHS. Crosslinking of gelatin is not necessary for the patterning of CMPs since the same CMP patterns can be created on uncrosslinked gelatin films using our method (data not shown), but it is important for the use of the gelatin material in cell and tissue culture because uncrosslinked gelatin gels dissolve in water at 37 °C.[43] We were able to pattern crosslinked gelatin films using a UV lamp (same light source used in the binding assay) and a transparency mask printed with desired patterns. Figure 3a shows the feature size of the patterns that were created on 2D gelatin films. For transparency line widths of 50 μm and 100 μm, the widths of the patterned CMP lines are below 60 μm and 120 μm, respectively, as estimated by the relative fluorescence intensity profiles (Figure 3b). This demonstrates great spatial fidelity which is comparable to other established covalent patterning methods.[4,9]
Figure 3.
Photo-patterning of fluorescent CMPs on thin gelatin films. (a) Fluorescence micrographs of the photo-patterned gelatin films along with photographs of the transparency masks showing the line patterns with 100 μm and 50 μm widths. (b) Relative fluorescence intensity profiles of the line patterns shown in (a). (c) A fluorescence image of a gelatin film sequentially patterned with two fluorescent CMPs [CF(GPO)9 in green, TAMRA(GPO)9 in red]. (d) A fluorescence image of a gelatin film patterned with the same method used in (c) but using a wet gelatin film, which results in diffusive gradients of patterned CMPs. (e) Fluorescence images of three photo-patterned lines of TAMRA(GPO)9 with different linear concentration gradients (i-iii) generated by continuous variation of UV exposure time using a movable cover. (f) Relative fluorescence intensity profiles of the line patterns (i, ii, iii) shown in (e). Scale bars: 300 μm in (c, d), 500 mm in (e).
Multiple types of CMP can be immobilized in distinct patterns on the same gelatin film by sequential application of the patterning process. Figure 3c shows a film patterned with horizontal lines of CFNB(GPO)9 and vertical lines of TAMRANB(GPO)9. By employing CMPs conjugated to bioactive molecules,[37] this approach could be used to display multiple bioactive cues to cells on gelatin scaffolds in a spatially controlled fashion. Therefore, the caged-CMP mediated photo patterning method can potentially provide new library screening platforms for identifying desired interactions between cells and matrix-bound factors.
Unlike conventional photo-patterning systems, in which hydrogel scaffolds are photo-activated to covalently capture soluble peptides,[2,4,7,9,17,44] our method is prone to blurring of the photo-pattern due to the UV-decaged CMP’s ability to diffuse away from the activation site before binding to the gelatin substrate. We were able to minimize this blurring by concentrating and drying the CMP solution on the gelatin film prior to UV exposure (see experimental section and Figure 3a,c). In a near-dry state, the diffusion of CMP is greatly limited which allows for patterning with high spatial fidelity. The hydrogel film quickly re-hydrates during the wash step after UV exposure. Because the film is thin, unbound CMPs were removed quickly during washing, leaving sharp 2D patterns (Figure 3a-c). In contrast, when wet films were used, wider lines and blurry edges with concentration gradients were produced (Figure 3d). Therefore, by controlling the wetness of the gelatin substrate, the caged CMP could be used to create either clear-cut photo-patterns or diffusion-mediated concentration gradients, both of which could be useful for advanced cell culturing systems.
2.4. Creation of Concentration Gradients of Patterned CMPs on Gelatin Films
Signaling molecules, such as growth factors, are often presented to cells in the form of concentration gradients, as they diffuse through tissues. The concentration gradients are believed to control cell differentiation and tissue morphogenesis by inducing and maintaining the expression of target genes at distinct concentration thresholds. Although many researchers have created concentration gradients of soluble cues using microfluidic devices,[45,46] it remains a challenge to generate gradients of matrix-bound bioactive molecules, particularly in natural tissue scaffolds. To this end, we developed a method of creating concentration gradients of the patterned CMPs by continual change of UV exposure time. This was accomplished by placing a cover on top of the transparency mask to partially block the UV beam during decaging of the TAMRANB(GPO)9 on the gelatin film. The cover was moved at a predetermined speed, creating a patterned area in which the UV exposure time was increased continuously from one end to the other. The experiment produced patterned lines with linear concentration gradients of immobilized TAMRA(GPO)9 as seen Figure 3e and 3f. This result demonstrated the high fidelity of the decaging kinetics also seen in Figure 2a. Using this technique, we were able to control all three key features of the gradients- the total concentration of the bound peptides, and the slope and length of the gradients, simply by varying the caged CMP concentration, UV exposure time, and the rate at which the cover moved across the gelatin surface. This experiment showcases an effective way to produce patterned gradients on gelatin at a sub-millimeter length scale. We believe that more complex gradient features can be achieved by the use of sophisticated light barrier, or by using an advanced programmable light source.[2,7]
2.5. Cell Adhesion Studies of PEG-CMP Patterned Gelatin Films
To evaluate the potential of the patterning technique in modulating cellular behavior, we prepared a conjugate of a PEG polymer and caged CMPs which can be photo-patterned on gelatin and reduce local cell adhesiveness. As a natural ECM, gelatin and collagen are highly adhesive substrates for cell attachment, and their adhesive nature is the main cause for abnormal tissue adhesion after surgery.[47,48] Modifying exposed collagen with a large hydrophilic PEG polymer could block cell binding sites, making them cell-repelling.[27,28,38,44] Caged CMPs featuring a cysteine residue at the N-terminus [sequence: AcCK(CF)Ahx(GPO)4NBGPO(GPO)4AhxY, Ac = acetylation, Ahx = aminohexanoic acid linker] were attached to a 40 kDa 8arm-PEG-maleimide polymer via the Michael addition reaction.[15,49] Each caged CMP was labeled with a CF fluorophore to allow easy visualization of the photo-pattern. The conjugated product, named 8arm-PEG-CFNB(GPO)9, was successfully patterned on an EDC-crosslinked thin gelatin film, and fibroblasts were seeded afterwards. Shortly after seeding (~5 h), cells only attached to the unmodified areas of the gelatin film, and even after three to four days of incubation, the areas patterned with PEG-CMP were devoid of cells. In contrast, a confluent cell layer formed in untreated areas (Figure 4). The results show the ability of this technique to control the organization of cells in gelatin matrices and the potential to spatially immobilize CMP-conjugated therapeutic drugs and bioactive molecules. In addition, this approach could be used to prevent abnormal tissue adhesion after surgical procedure.
Figure 4.
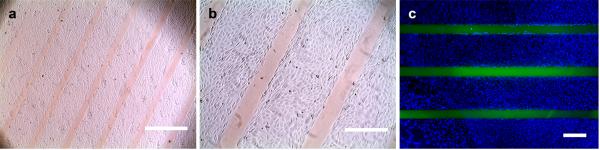
Optical photographs (a, b) and a fluorescence image (c) of fibroblasts cultured on a gelatin film that was photo-patterned with 8arm-PEG-CF(GPO)9. Cells exclusively attached to the unpatterned areas and formed a confluent monolayer. The immobilized CF-labeled PEG-CMP conjugate appears as clear orange (a, b) or green lines (c); the fibroblasts were fixed and stained with DAPI to visualize the nuclei (c). Scale bars: 1 mm in (a), 400 mm in (b), and 380 mm in (c).
2.6. Three Dimensional Patterning of Caged CMPs in Gelatin Hydrogels
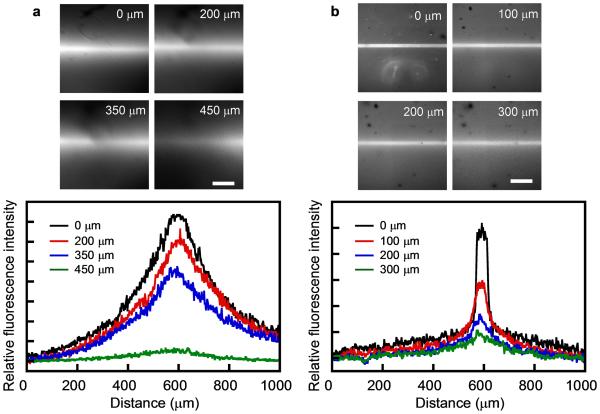
In vivo, cells interact dynamically with a variety of bioactive cues presented within a 3D microenvironment. Adding the 3rd dimension of interactions between ECM and cells can bring cellular behavior closer to natural tissues which is not easily recapitulated on 2D flat surfaces.[1] To test the CMP patterning technique in a 3D setting, we prepared a gelatin hydrogel of 1 mm thickness soaked in either CFNB(GPO)9 or 8arm-PEG-CFNB(GPO)9. To prevent the blurring/expansion of the photo-pattern within the bulk hydrogel due to unparallel UV light, the hydrogel attached to the transparency mask was irradiated by a laser beam through an infinity-focused objective lens under a fluorescence microscope (DAPI channel, ~10 mW/cm2) instead of the simple mercury lamp. After photo-patterning and extensive washing, the x-y cross-section micrographs at different z depths were recorded by confocal microscopy, and the fluorescence intensity profiles of the CMP and the PEG-CMP patterned hydrogels were acquired. Figure 5 shows the fluorescence signals of the patterned CMP and PEG-CMP, with the patterned lines fading away approximately 500 μm below the irradiated top surfaces. This attenuation is likely due to the peptide (labeled with carboxyfluorescein and caged with nitrobenzyl) absorbing the UV beam as it travels down through the thick gel, resulting in a decrease of light intensity and corresponding reduction in concentration of decaged CMPs.[4] The feature of CMP pattern seems diffusive compared to the mask (Figure 5a), presumably because the decaged CMP molecules diffused away from the photo-activation site prior to binding to the gelatin matrix. The diffusion simultaneously created a concentration gradient on both sides of the patterned line, and the widths of the diffusive gradients seemed consistent at all depths despite the decrease in activated CMP concentration (Figure 5a). In contrast, the pattern of 8arm-PEG-CFNB(GPO)9 showed much higher spatial fidelity than the CFNB(GPO)9, with little line expansion from the pattern of the photo-mask (Figure 5b). This, we hypothesize, is the result of two main factors: (i) slower diffusion due to the large molecular mass of the PEG-CMP conjugate (~70K Da, which is over 20 times the mass of the CMP) and (ii) possibly the multi-ligand effect of the eight CMP arms giving stronger gelatin binding. Many matrix-associated signaling molecules (e.g., growth factors) are proteins of relatively large molecular mass. Therefore, according to our results, CMP conjugated to such proteins might benefit from their high molecular mass and therefore be immobilized on 3D gelatin scaffolds with good spatial fidelity. Overall, the results demonstrate the feasibility of creating patterns and concentration gradients of biomolecules in a 3D gelatin hydrogel through photo-patterning caged CMPs. The limitation regarding the patterning depth and line diffusion could be overcome by using more light-sensitive cage groups[5,50] and multiphoton confocal microscopy, both of which are currently under development in our lab.
Figure 5.
Photo-patterning of the caged CMP in 1 mm thick 3D gelatin hydrogels. Top: representative x-y cross-section images of the fluorescent lines of patterned CF(GPO)9 (a) or 8arm-PEG-CF(GPO)9 (b) at the indicated z depths from the UV-irradiated hydrogel surface, shown in grey scale. Scale bar: 300 μm. Bottom: corresponding relative fluorescence intensity profiles across the patterned lines at the indicated z depths.
2.7. Patterning CMPs on Hybrid Gelatin Materials and Natural Tissues
Gelatin is a popular biopolymer used in a variety of biomaterials and drug delivery systems. Due to its high water solubility, it is commonly used in crosslinked forms and/or combined with other synthetic materials to limit the dissolution process and enhance cell adhesiveness.[17,51-53] We envisioned that the CMP photo-patterning method can be applied to these gelatin-based hydrogel systems to give spatially-encoded functionalities. We tested the versatility of CMP patterning on a gelatin zymogram gel and a gelatin-methacrylate hydrogel. The zymogram gel is a polyacrylamide based hydrogel (SDS-PAGE gel) containing 0.1% gelatin which is added as a substrate for studying collagenases activity.[54] Although not a traditional scaffold for tissue engineering, gelatin zymogram is a readily available gelatin-based hybrid material, and similar polyacrylamide hydrogels have been used as matrices to direct stem cell differentiation.[55] Gelatin-methacrylate hydrogel (GelMA) is a photopolymerizable synthetic hydrogel formed from gelatin whose amine-containing side chains are modified with methacrylate groups. GelMA has become a promising hydrogels for tissue engineering and microfluidic applications because it can not only interact with cells (i.e., cell binding and matrix degradation) but can also be easily fabricated into a variety of shapes and configurations similar to photo curable synthetic polymers (e.g., PEG-diacrylate). For examples, the Khademhosseini group and others have developed bioprinted microchannels and hydrogel microarrays based on GelMA.[56-59]
As shown in Figure 6a & b, both the zymogram and the GelMA hydrogels were successfully photo-patterned with CMPs using our technique. The patterning resulted in clear fluorescent lines and a letter “U” shape with line sizes on the order of hundreds of micrometers. Because the CMP-gelatin binding affinity comes from the triple helical propensity of the gelatin chains and not from any specific epitope, the patterning efficiency was not compromised although GelMA is a chemically modified and highly crosslinked version of natural gelatin.[17] GelMA hydrogel contains photo-polymerizable groups and can be photo-patterned with acrylate-containing molecules.[55] However, since the hydrogel itself crosslinks via the same patterning chemistry, it is difficult to decouple the patterning of the gel from further crosslinking of the gel which strongly affects the gel’s mechanical property that critically dictates the behavior of seeded cells.[1] The caged CMP technique provides a patterning tool that is independent from the crosslinking of the GelMA hydrogel, and therefore can be utilized as an alternative patterning method or in combination with the chemical patterning of the GelMA scaffolds.
Figure 6.

Photo-pattering of caged CMPs on various gelatin containing matrices. (a) A photograph of lines of CF(GPO)9 patterned into a gelatin zymogram polyacrylamide hydrogel shown together with the image of the mask (0.4 mm transparent lines with 4 mm wide spacing). (b) A letter U created by TAMRA(GPO)9 patterned on a film of photo-crosslinked gelatin methacrylate hydrogel (scale bar: 500 μm). (c) A fluorescence image of lines of CF(GPO)9 (arrows) patterned on the stroma of a bovine corneal tissue (scale bar = 4 mm).
One unique advantage of the CMP patterning is that it could be directly applied to natural tissues with high collagen/gelatin content. High levels of denatured collagen are found in many pathological or damaged tissues (e.g., arthritis and wounds) since high level of ECM remodeling occurs during wound healing. Therefore, photo-immobilization of CMP derivatives on these abnormal tissues could be developed into a new therapy in wound healing. Previously, our group and others have shown that CMP can readily bind to denatured collagen strands through triple helical hybridization both ex vivo and in vivo.[31,32,60] In this study, we demonstrated the ability of caged CMPs to bind to natural tissues through photo-patterning corneal stroma. The corneal stroma was chosen since it is primarily composed of collagen and because it has an ideal laminar geometry which can be laid flat for patterning. More importantly, corneal collagen damage and denaturation are closely associated with many ophthalmologic procedures and disorders such as laser eye surgeries and keratoconus.[61-65] After removal of the epithelium, a fresh bovine corneal tissue was treated with hot water for 10 s to mimic eye injury. This procedure presumably generated large amount of denatured collagen strands. The tissue was then soaked in the CFNB(GPO)9 solution and exposed to UV light through a mask as described above. After patterning and washing, regular line patterns of approximately 400 μm in width and 4 mm in length were clearly seen from the fluorescence image (Figure 6c). As a proof-of-concept, this experiment demonstrates the great potential of the CMP patterning technique in spatially immobilizing exogenous molecules on native tissues that contain high levels of denatured collagen, which are extremely challenging to pattern with conventional synthetic photo sensitive materials. We believe that this technique can lead to new strategies in drug application particularly for tissues that can readily be exposed to light such as eyes and skin.
4. Conclusions
The ability to micropattern hydrogels has provided new tools to manipulate cell behaviors. Most efforts have focused on conventional patterning processes on synthetic polymers or chemically altered forms of naturally derived biopolymers. Such artificial systems are generally inferior to natural scaffolds for in vivo use and for engineering tissue constructs to be used in tissue replacement therapies. Since conventional photo-patterning methods cannot be easily applied to scaffolds based on natural ECM proteins, we developed a new photo-patterning strategy for gelatin hydrogels that relies on non-covalent hybridization interactions between gelatin strands and collagen mimetic peptides. By employing photo-masks and the caged CMP that can be photo-triggered to bind to gelatin, CMPs and 8arm-PEG-CMP conjugates were patterned onto 2D gelatin films and 3D gelatin hydrogels. Depending on the patterning conditions, a range of photo-patterns were achieved, from clear-cut line patterns which are tens of micrometers in width, to highly diffusive concentration gradients over hundreds of micrometers. The study showing patterning of 8arm-PEG-CMP can confine growth of fibroblasts to localized areas on a gelatin surface suggests the feasibility to spatially control cell behaviors using the patterning technique and CMP-conjugates, and due to the high molar mass and multivalency, the 8arm-PEG-CMP can pattern even a thick 3D hydrogel with high spatial fidelity. This ability to photo-pattern the caged CMP is highly versatile as demonstrated by patterning of various gelatin-containing hydrogels and a corneal tissue. To our knowledge, this is the first ex vivo study that shows direct photo-patterning of a natural tissue via peptide binding, and clearly demonstrates the unique application enabled by this technique. With this first proof-of-concept work, we envision that the CMP-mediated photo-patterning strategy will provide unpresented tools for engineering spatially complex tissue constructs, especially those based on natural ECM scaffolds, and also facilitate translation of conventional photo-patterning technologies to in vivo therapeutic applications.
4. Experimental Section
4.1. Materials
Gelatin from porcine skin (gel strength ~300 g Bloom) was obtained from Sigma-Aldrich (St Louis, MO). For peptide synthesis and labeling, Fmoc-Gly-OH, Fmoc-Pro-OH, Fmoc-Ahx-OH, H-Gly-OH, fluorenylmethyloxy chloroformate (Fmoc-Cl), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), and hydroxybenzotriazole (HOBt) were purchased from Advanced ChemTech (Louisville, KY). O-(Benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HBTU) was purchased from AAPPTec (Louisville, KY), N,N-diisopropylethylamine (DIPEA) from Acros (Geel, Belgium), and dimethylformamide (DMF), N-Methyl-2-pyrrolidone (NMP), and trifluoroacetic acid (TFA) were purchased from Fisher (Pittsburgh, PA) and used without further purification. Fmoc-Hyp(tBu)-OH, Fmoc-Cys(trt)-OH, Fmoc-Lys(Dde)-OH and bromotripyrrolidinophosphonium hexafluorophosphate (PyBroP) were purchased from EMD millipore (Temecula, CA). TentaGel R RAM resin was purchased from Peptides International (Louisville, KY). 5(6)-carboxytetramethylrhodamine (TAMRA) was purchased from Life Technologies (Grand Island, NY). Piperidine, (7-azabenzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyAOP), nitrobenzaldehyde, 5(6)-carboxyfluorescein (CF), triisopropylsilane (TIS), N-hydroxysuccinimide (NHS), methacrylic anhydride (MA) and all other unspecified chemicals were purchased from Sigma-Aldrich. 8arm-PEG-maleimide was purchased from JenKem Technology USA (Plano, TX). MES [2-(N-morpholino)ethanesulfonic acid] buffered saline was purchased from Thermo Pierce (Rockford, IL). DAPI (4',6-diamidino-2-phenylindole) was obtained from Roche Applied Science (Indianapolis, IN). Cell culture supplies were purchased from Life Technologies.
4.2. Synthesis and Characterization of Fluorescently Labeled Caged CMPs and PEG Conjugated Caged CMPs
Standard peptide synthesis was carried out using an automated peptide synthesizer (model 433A) from Applied BioSystems on TentaGel R RAM resin (reactive site density: 0.18 mmol/g), as reported previously.[66] The caged CMP NB(GPO)9 was prepared by introducing Fmoc(N-o-nitrobenzyl)Gly-OH (synthesized according to Tatsu et al.[31,67]) in the middle of the standard peptide synthesis.[31] The Hyp residue following the NBGly was conjugated for over 24 h using 9 molar equiv of Fmoc-Hyp(tBu)-OH, 8.8 molar equiv of PyBroP, and 20 molar equiv of DIPEA to overcome the low reactivity of the NBGly. Synthesis of the remaining sequence including the GGG spacer was completed by HBTU chemistry, followed by on-resin labeling with 6 molar equivalent of CF or TAMRA activated by 6 molar equivalent of PyAOP in NMP for over 24 h.[68-70] The full length fluorescent CMPs were cleaved from the resin by treating the resin with TFA/TIS/H2O (95:2.5:2.5) for 3 h, and the cleaved peptide was purified by reverse phase HPLC on a semipreparative Vydac C18 column using a linear gradient mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) (5-45% acetonitrile gradient in 40 min, flow rate: 4 mL/min). The purified peptides were analyzed by matrix assisted laser desorption ionization time-of-flight mass spectroscopy (MALDI-TOF MS, UltrafleXtreme, Bruker Daltonics): m/z calculated 3110.2 [M + Na]+ for CFNB(GPO)9, found 3109.4 [M + Na]+; m/z calculated 3142.3 [M + H]+ for TAMRANB(GPO)9, found 3141.4 [M + H]+.
To prepare PEG-conjugated caged CMP, AcCK(Dde)Ahx(GPO)4NBGPO(GPO)4AhxY which contains Dde protected Lys residue was synthesized on solid support and peptide’s N-terminus was capped on-resin by acetylation (Ac) reaction. The Dde group was removed by treating the peptide-resin with 3% hydrazine monohydrate in 10 mL NMP for 5 min, and CF was subsequently coupled onto the deprotected Lys side chain on resin using PyAOP. The CF labeled caged peptide AcCK(CF)Ahx(GPO)4NBGPO(GPO)4AhxY was cleaved from the resin with TFA/DTT/H2O/TIS (88:5:5:2) and purified by HPLC. MALDI-TOF MS analysis: m/z calculated 3579.8 [M + H]+ for the purified peptide, found 3577.4 [M + H]+. The purified CF-labeled caged CMP (5 mg, 12 molar equiv) was dissolved in a mixture of 280 μL of DMSO and 520 μL of 1×PBS (phosphate buffered saline), and 4.66 mg of 8arm-PEG-maleimide (1 molar equiv) in 50 μL of 1×PBS was added to the peptide solution in a drop-wise fashion. The thiol-maleimide reaction was allowed to take place overnight at room temperature and the final product of 8arm-PEG-CFNB(GPO)9 conjugate was purified by HPLC on a semipreparative Vydac C18 column using a linear gradient mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) (20-50% acetonitrile gradient in 30 min, flow rate: 4 mL/min).
4.3. Study of Photo-Triggered CMP Binding on Gelatin Films
Except for the 3D gel patterning, a mercury arc lamp (1447T17 UV lamp, McMaster-Carr) was used as the UV light source in all the following CMP binding and photo-patterning experiments. To investigate the photo-activation kinetics of the caged CMP, a series of CFNB(GPO)9 solutions (50 μM in 50 μL 1×PBS) were exposed to 365 nm UV light (15.5 mW/cm2) in a 96-well plate for designated time periods (0-20 min), followed by HPLC analysis on a Vydac C18 analytical RP-HPLC column using a linear gradient mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) (13-43% acetonitrile gradient in 30 min, flow rate: 1 mL/min, UV detection: 275 nm). For each sample, the faction of caged CMP was calculated from the ratio of the peak area between CFNB(GPO)9 and others corresponding to the photo-cleaved CMP.
To test the dose-dependent binding of CMP on gelatin films, wells in a 96-well black/clear-bottom plate (Costar) were coated with warm gelatin solutions (10% w/v in 1×PBS) and the solutions were allowed to gel and form thin films at 4 °C. PBS solutions containing CFNB(GPO)9 of various concentrations (5-150 μM) were added onto the gelatin films and exposed to 365 nm UV light (15.5 mW/cm2). This UV intensity was arbitrarily chosen in this study; however, the intensity and the UV exposure time could be adjusted to avoid affecting cell viability in future studies which may involve cell encapsulation. The duration of the UV exposure time was proportional to the CMP concentrations. Wells for the UV-negative control groups were covered with aluminum foil. After incubation at room temperature for 2 h, the unbound materials were removed by rinsing with PBS buffer. The fluorescence levels of the films were measured with a SpectraMax Gemini XPS microplate reader (Molecular Devices, ex: 489 nm, em: 533 nm). Each binding experiment was conducted in triplicate.
4.4. Photo-Patterning Gelatin Films with Caged CMPs
The line patterns were prepared using CorelDraw and printed onto transparencies by CAD/Art Services (Bandon, OR). A solution of 10% w/v gelatin in 1×PBS was prepared and heated to 50 °C. Twenty-five μL of the heated gelatin solution was added to the surface of a superfrost plus microscope slide (Fisher) and covered with a Permanox plastic slide. The sandwiched gelatin solution was allowed to gel at 4 °C for 10 min, followed by careful removal of the upper plastic slide. The gelatin film (size: ~5 cm2, thickness: ~50 μm) was crosslinked overnight in a MES solution (pH 4.7) containing EDC (10 mM) and NHS (2 mM),[43] and washed with PBS and water. A solution (150 μL) of CFNB(GPO)9 or TAMRANB(GPO)9 (50 μM) was applied to the film surface and allowed to fully diffuse into the thin gel for 10 min, and the excess solution was removed from the surface of the gel by pipetting. The film was air-dried in dark. The transparency mask was placed directly on the dry film with the printed side facing down, and another microscope slide was placed on top of the mask to ensure flatness of the whole system. The film was exposed to UV light (from a mercury arc lamp, 365 nm, 15.5 mW/cm2) through the mask for 4 min. After removing the mask, the film was washed in a staining jar with 100 mL of PBS for 5 min three times to remove the unbound peptides. The patterned gelatin films were imaged using a Nikon Eclipse E600 microscope. All fluorescence images were analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
4.5. CMP Concentration Gradient on Gelatin Films
As described above, a transparency mask with line pattern was placed on top of a dry EDC-fixed gelatin film containing TAMRANB(GPO)9 on a microscope slide. A piece of aluminum foil placed directly on top of the mask was attached to the moving block of a syringe pump (Harvard Apparatus, Holliston, MA). The syringe pump was programmed to move the aluminum cover approximately 500 to 1400 μm at a speed ranging from 1.4 to 7.8 μm/s across the mask during the UV exposure time. The gelatin film was washed with PBS and imaged as described above.
4.6. Cell Adhesion Studies of PEG-CMP Patterned Gelatin Films
A thin gelatin film (size: ~2.5 cm2, thickness: ~50 μm) was prepared on a petri dish (Nunc) and crosslinked by EDC/NHS. A solution (50 μL) of 8arm-PEG-CFNB(GPO)9 (31.25 μM) in water was carefully applied to the film surface and allowed to dry completely. The film was exposed to UV light (from a mercury arc lamp, 365 nm, 27 mW/cm2) for 15 min through a transparency mask directly placed on the film surface, followed by three runs of 5 min wash with 1×PBS solution at 37 °C. The patterned petri dish was sterilized with 70% ethanol in water for 20 min and washed with sterile 1×PBS solution. NIH 3T3 fibroblasts (5×104 cells/mL) in 10 mL of Dulbecco's modified eagle medium (supplemented with 10% fetal bovine serum and 1% mixture of penicillin-streptomycin antibiotics) were added to the petri dish, and incubated at 37 °C in a 5% CO2 atmosphere. The growth media were exchanged every 3 days. Cell attachment was monitored and recorded with an EVOS light microscope (Life Technologies). When confluent, the cells were fixed with PBS buffered 4% formaldehyde solution for 3 h and stained by DAPI (1.65 μg/mL in PBS) for 1 min. After washing, the cells and patterned lines of fluorescent PEG-CMPs were imaged using a Nikon Eclipse E600 microscope.
4.7. Patterning 3D Gelatin Hydrogels
After detaching the media chamber from the silicone gasket of an 8 well Permanox chamber slide (Nunc, Thermo Scientific), 100 μL of heated gelatin solution (10% w/v, 70 °C) was added to a well, and a glass slide was placed on top of the solution and gasket. The solution was cooled to gel at 4 °C for 10 min before the gasket and plastic slide were removed, leaving a gelatin hydrogel of 1 mm thickness. The gel was crosslinked (EDC: 10 mM, NHS: 2 mM, in MES buffer) at room temperature overnight. After washing in 1×PBS for 2 h, the hydrogel was soaked in 150 μL of CFNB(GPO)9 (20 μM) or 8arm-PEG-CFNB(GPO)9 (2.5 μM) for over 4 h with gentle shaking. The transparency mask was directly placed on the gel surface with the printed side facing down, and the gel was irradiated for 6 min with the Nikon Eclipse E600 microscope (DAPI channel) through a 4× infinity corrected objective lens (beam size: 6 mm in diameter, ~10 mW/cm2). Unbound CMPs were removed by washing the gel in PBS extensively overnight (or for 2 days at 37 °C for PEG-CMP). CMP (or PEG-CMP) distribution in the hydrogel was imaged using a laser scanning confocal microscope (FV1000, Olympus) and analyzed by ImageJ.
4.8. Patterning Zymogram Gel and Corneal Tissue
A piece of 10% zymogram (gelatin) gel (Life Technologies, 1 mm thickness) was soaked in a 5 μM CFNB(GPO)9 solution overnight followed by photo-patterning as described above. The gel was washed with PBS and water overnight, and imaged by a digital camera. Corneal tissues were harvested from fresh bovine eyeballs purchased from local slaughterhouse. The corneal epithelium was carefully removed by surgical tools. The exposed corneal stroma was treated with 1 mL of hot water (75 °C) for 10 s and rinsed with cold PBS. The tissue was soaked in 200 μL of CFNB(GPO)9 solution (50 μM), and covered with a transparency mask and exposed to UV light for 10 min. The sample was incubated at 4 °C for 30 min and washed with PBS, followed by fluorescence imaging using the Xenogen IVIS Spectrum optical imaging device (Caliper Life Sciences, excitation: 500 nm, emission: 540 nm). The fluorescence signals were analyzed by Living Image Software (Caliper Life Sciences).
4.9. Patterning Gelatin Methacrylate Hydrogels
The methacrylated gelatin was synthesized according to Nichol and coworkers.[17] Briefly, 0.25 g of gelatin (porcine skin) dissolved in 2.5 mL of 1×PBS buffer in a 60 °C water bath was mixed with 31.25 μL (1.25% v/v) of methacrylic anhydride (Alfa Aesar, Ward Hill, MA). Reaction was allowed to proceed for 1 h followed by quenching by diluting the solution with 7.5 mL of warm (50 °C) 1×PBS buffer. The diluted solution was dialyzed against deionized water using a 10 kDa cutoff dialysis cassette (Pierce) at 50 °C for 5 days. The solution was lyophilized, and the gelatin powder was dissolved in deionized water to produce a 10% w/v gelatin-methacrylate solution. Fifty μL/mL of 10% w/v 2-hydroxy-1-(4-(hydroxyethoxy)phenyl)-2-methyl-1-propanone (Irgacure 2959, Sigma-Aldrich) in 50:50 DMSO and water was added to a solution of 10% GelMA at 50 °C. Approximately 18 μL of the warm GelMA/Igracure solution was polymerized via exposure to UV light (from a mercury arc lamp, 365 nm, 15.5 mW/cm2) for 5 min between a superfrost plus microscope slide and a silanized glass coverslip separated by thin spacers. The upper coverslip was removed in water to expose the crosslinked GelMA film, and the film was patterned with CFNB(GPO)9 or TAMRANB(GPO)9 as described.
Acknowledgements
We thank Dr. Patrick Tresco (University of Utah) for access to the microscopes and the kind gift of the NIH 3T3 fibroblasts. This work was supported by grants from NIAMS/NIH (R01-AR060484 and R21-AR065124) and DOD (W81XWH-12-1-0555) awarded to S.M.Y., by the National Eye Institute Grant (P30EY001765, J.H. and Q.X.), and by the College of Engineering Wayne Brown Fellowship (University of Utah) awarded to J.L.K.
References
- [1].Underhill GH, Peter G, Chen CS, N. Bhatia S. Annu. Rev. Cell Dev. Biol. 2012;28:385. doi: 10.1146/annurev-cellbio-101011-155709. [DOI] [PubMed] [Google Scholar]
- [2].DeForest CA, Polizzotti BD, Anseth KS. Nature Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].DeForest CA, Anseth KS. Nature Chem. 2011;3:925. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Luo Y, Shoichet MS. Nature Mater. 2004;3:249. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- [5].Wosnick JH, Shoichet MS. Chem. Mater. 2007;20:55. [Google Scholar]
- [6].Wylie RG, Shoichet MS. J. Mater. Chem. 2008;18:2716. [Google Scholar]
- [7].Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Nature Mater. 2011;10:799. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- [8].Hahn MS, Miller JS, West JL. Adv. Mater. 2005;17:2939. [Google Scholar]
- [9].Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- [10].Hahn MS, Miller JS, West JL. Adv. Mater. 2006;18:2679. [Google Scholar]
- [11].Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. FASEB J. 2007;21:790. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- [12].Lee S-H, Moon JJ, West JL. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].West JL. Nature Mater. 2011;10:727. doi: 10.1038/nmat3132. [DOI] [PubMed] [Google Scholar]
- [14].Culver JC, Hoffmann JC, Poché RA, Slater JH, West JL, Dickinson ME. Adv. Mater. 2012;24:2344. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rubert Pérez CM, Panitch A, Chmielewski J. Macromol. Biosci. 2011;11:1426. doi: 10.1002/mabi.201100230. [DOI] [PubMed] [Google Scholar]
- [16].Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Park JE, Keller GA, Ferrara N. Mol. Biol. Cell. 1993;4:1317. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Knight CG, Morton LF, Peachey AR, Tuckwell DS, Farndale RW, Barnes MJ. J. Biol. Chem. 2000;275:35. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- [20].Hoshikawa A, Nakayama Y, Matsuda T, Oda H, Nakamura K, Mabuchi K. Tissue Eng. 2006;12:2333. doi: 10.1089/ten.2006.12.2333. [DOI] [PubMed] [Google Scholar]
- [21].Mazaki T, Shiozaki Y, Yamane K, Yoshida A, Nakamura M, Yoshida Y, Zhou D, Kitajima T, Tanaka M, Ito Y, Ozaki T, Matsukawa A. Sci. Rep. 2014:4. doi: 10.1038/srep04457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Patel ZS, Young S, Tabata Y, Jansen JA, Wong MEK, Mikos AG. Bone. 2008;43:931. doi: 10.1016/j.bone.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu X, Ma L, Wang C, Gao C. Macromol. Biosci. 2009;9:1194. doi: 10.1002/mabi.200900275. [DOI] [PubMed] [Google Scholar]
- [24].Chen F-M, Chen R, Wang X-J, Sun H-H, Wu Z-F. Biomaterials. 2009;30:5215. doi: 10.1016/j.biomaterials.2009.06.009. [DOI] [PubMed] [Google Scholar]
- [25].Li Y, Yu SM. Curr. Opin. Chem. Biol. 2013;17:968. doi: 10.1016/j.cbpa.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yu SM, Li Y, Kim D. Soft Matter. 2011;7:7927. doi: 10.1039/C1SM05329A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang AY, Mo X, Chen CS, Yu SM. J. Am. Chem. Soc. 2005;127:4130. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- [28].Wang AY, Foss CA, Leong S, Mo X, Pomper MG, Yu SM. Biomacromolecules. 2008;9:1755. doi: 10.1021/bm701378k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang AY, Leong S, Liang YC, Huang RCC, Chen CS, Yu SM. Biomacromolecules. 2008;9:2929. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- [30].Lee HJ, Lee J-S, Chansakul T, Yu C, Elisseeff JH, Yu SM. Biomaterials. 2006;27:5268. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [31].Li Y, Foss CA, Summerfield DD, Doyle JJ, Torok CM, Dietz HC, Pomper MG, Yu SM. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14767. doi: 10.1073/pnas.1209721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li Y, Ho D, Meng H, Chan TR, An B, Yu H, Brodsky B, Jun AS, Michael Yu S. Bioconjug. Chem. 2013;24:9. doi: 10.1021/bc3005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mo X, An YJ, Yun CS, Yu SM. Angew. Chem. Int. Ed. 2006;45:2267. doi: 10.1002/anie.200504529. [DOI] [PubMed] [Google Scholar]
- [34].Lee HJ, Yu C, Chansakul T, Hwang NS, Varghese S, Yu SM, Elisseeff JH. Tissue Eng. Part A. 2008;14:1843. doi: 10.1089/ten.tea.2007.0204. [DOI] [PubMed] [Google Scholar]
- [35].Li Y, Foss CA, Pomper MG, Yu SM. J. Vis. Exp. 2014:e51052. doi: 10.3791/51052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kojima C, Suehiro T, Tada T, Sakamoto Y, Waku T, Tanaka N. Soft Matter. 2011;7:8991. [Google Scholar]
- [37].Chan TR, Stahl PJ, Yu SM. Adv. Funct. Mater. 2011;21:4252. doi: 10.1002/adfm.201101163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Stahl PJ, Yu SM. Soft Matter. 2012;8:10409. doi: 10.1039/C2SM25903F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fan CY, Huang CC, Chiu WC, Lai CC, Liou GG, Li HC, Chou MY. FASEB J. 2008;22:3795. doi: 10.1096/fj.08-111484. [DOI] [PubMed] [Google Scholar]
- [40].Pires MM, Przybyla DE, Chmielewski J. Angew. Chem. Int. Ed. 2009;48:7813. doi: 10.1002/anie.200902375. [DOI] [PubMed] [Google Scholar]
- [41].Okada T, Isobe C, Wada T, Ezaki S, Minoura N. Bioconjug. Chem. 2013;24:841. doi: 10.1021/bc3006013. [DOI] [PubMed] [Google Scholar]
- [42].Sarkar N, Banerjee J, Hanson AJ, Elegbede AI, Rosendahl T, Krueger AB, Banerjee AL, Tobwala S, Wang R, Lu X, Mallik S, Srivastava DK. Bioconjug. Chem. 2008;19:57. doi: 10.1021/bc070081p. [DOI] [PubMed] [Google Scholar]
- [43].Kuijpers AJ, Engbers GHM, Krijgsveld J, Zaat SAJ, Dankert J, Feijen J. J. Biomater. Sci., Polym. Ed. 2000;11:225. doi: 10.1163/156856200743670. [DOI] [PubMed] [Google Scholar]
- [44].Stahl PJ, Chan TR, Shen Y-I, Sun G, Gerecht S, Yu SM. Adv. Funct. Mater. 2014;24:3213. doi: 10.1002/adfm.201303217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim S, Kim HJ, Jeon NL. Integr. Biol. 2010;2:584. doi: 10.1039/c0ib00055h. [DOI] [PubMed] [Google Scholar]
- [46].Ostrovidov S, Annabi N, Seidi A, Ramalingam M, Dehghani F, Kaji H, Khademhosseini A. Anal. Chem. 2012;84:1302. doi: 10.1021/ac202256c. [DOI] [PubMed] [Google Scholar]
- [47].Schnüriger B, Barmparas G, Branco BC, Lustenberger T, Inaba K, Demetriades D. Am. J. Surg. 201:111. doi: 10.1016/j.amjsurg.2010.02.008. [DOI] [PubMed] [Google Scholar]
- [48].Hellebrekers BWJ, Trimbos-Kemper TCM, Trimbos JBMZ, Emeis JJ, Kooistra T. Fertil. Steril. 2000;74:203. doi: 10.1016/s0015-0282(00)00656-7. [DOI] [PubMed] [Google Scholar]
- [49].Rubert Perez CM, Rank LA, Chmielewski J. Chem. Commun. 2014 doi: 10.1039/c4cc03171g. [DOI] [PubMed] [Google Scholar]
- [50].Ellis-Davies GCR. Nat. Methods. 2007;4:619. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Paguirigan AL, Beebe DJ. Nat. Protoc. 2007;2:1782. doi: 10.1038/nprot.2007.256. [DOI] [PubMed] [Google Scholar]
- [52].Young S, Wong M, Tabata Y, Mikos AG. J. Controlled Release. 2005;109:256. doi: 10.1016/j.jconrel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- [53].Olsen D, Yang C, Bodo M, Chang R, Leigh S, Baez J, Carmichael D, Perälä M, Hämäläinen E-R, Jarvinen M, Polarek J. Adv. Drug Del. Rev. 2003;55:1547. doi: 10.1016/j.addr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- [54].Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G. Nat. Methods. 2013;10:211. doi: 10.1038/nmeth.2371. [DOI] [PubMed] [Google Scholar]
- [55].J. Engler A, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- [56].Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Lab on a Chip. 2014;14:2202. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gauvin R, Chen Y-C, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A. Biomaterials. 2012;33:3824. doi: 10.1016/j.biomaterials.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dolatshahi-Pirouz A, Nikkhah M, Gaharwar AK, Hashmi B, Guermani E, Aliabadi H, Camci-Unal G, Ferrante T, Foss M, Ingber DE, Khademhosseini A. Sci. Rep. 2014:4. doi: 10.1038/srep03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Eng G, Lee BW, Parsa H, Chin CD, Schneider J, Linkov G, Sia SK, Vunjak-Novakovic G. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4551. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chattopadhyay S, Murphy CJ, McAnulty JF, Raines RT. Organic & Biomolecular Chemistry. 2012;10:5892. doi: 10.1039/c2ob25190f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Asiyo-Vogel MN, Brinkmann R, Notbohm H, Eggers R, Lubatschowski H, Laqua H, Vogel A. J. Cataract Refract. Surg. 1997;23:515. doi: 10.1016/s0886-3350(97)80208-8. [DOI] [PubMed] [Google Scholar]
- [62].Matteini P, Cicchi R, Ratto F, Kapsokalyvas D, Rossi F, de Angelis M, Pavone FS, Pini R. Biophys. J. 2012;103:1179. doi: 10.1016/j.bpj.2012.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Theodossiou T, Rapti GS, Hovhannisyan V, Georgiou E, Politopoulos K, Yova D. Laser Med. Sci. 2002;17:34. doi: 10.1007/s10103-002-8264-7. [DOI] [PubMed] [Google Scholar]
- [64].Leonardi A, Brun P, Abatangelo G, Plebani M, Secchi AG. Invest. Ophthalmol. Vis. Sci. 2003;44:3052. doi: 10.1167/iovs.02-0766. [DOI] [PubMed] [Google Scholar]
- [65].Lema I, Durán JA. Ophthalmology. 2005;112:654. doi: 10.1016/j.ophtha.2004.11.050. [DOI] [PubMed] [Google Scholar]
- [66].Li Y, Mo X, Kim D, Yu SM. Biopolymers. 2011;95:94. doi: 10.1002/bip.21536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tatsu Y, Nishigaki T, Darszon A, Yumoto N. FEBS Lett. 2002;525:20. doi: 10.1016/s0014-5793(02)03000-4. [DOI] [PubMed] [Google Scholar]
- [68].Stahl PJ, Cruz JC, Li Y, Yu SM, Hristova K. Anal. Biochem. 2012;424:137. doi: 10.1016/j.ab.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fischer R, Mader O, Jung G, Brock R. Bioconjug. Chem. 2003;14:653. doi: 10.1021/bc025658b. [DOI] [PubMed] [Google Scholar]
- [70].Fernádez-Carneado J, Giralt E. Tetrahedron Lett. 2004;45:6079. [Google Scholar]