Abstract
Blockade of undesired neutrophil migration to sites of inflammation remains an area of substantial pharmaceutical interest. To effect this blockade, a validated therapeutic target is antagonism of the chemokine receptor CXCR2. Herein we report the discovery of 6-(2-boronic acid-5-trifluoromethoxy-benzylsulfanyl)-N-(4-fluoro-phenyl)-nicotinamide 6, an antagonist with activity at both CXCR1 and CXCR2 receptors (IC50 values 31 and 21 nM, respectively). Compound 6 exhibited potent inhibition of neutrophil influx in a rat model of pulmonary inflammation, and is hypothesized to interact with a unique intracellular binding site on CXCR2. Compound 6 (SX-576) is undergoing further investigation as a potential therapy for pulmonary inflammation.
Keywords: CXCR2, Antagonist, Thionicotinamide, COPD, Asthma
Pulmonary inflammation by a predominantly neutrophil (polymorphonuclear leukocyte, PMN) infiltrate in response to chronic lung injury is a pathophysiologic mechanism common to several pulmonary diseases including severe asthma and chronic obstructive pulmonary disease (COPD),1 PMNs are large phagocytic cells whose primary function is to release an arsenal of degradative enzymes and NADPH-dependent oxidases at sites of injury or inflammation. Chronic ongoing extracellular release of cytotoxic enzymes permanently damages host tissues, playing a pivotal role in the pathogenesis of severe asthma and COPD. Due to their prominent role in numerous inflammatory diseases, one strategy to prevent or mitigate the severity of disease progression is to block the migration of PMNs to sites of inflammation.2
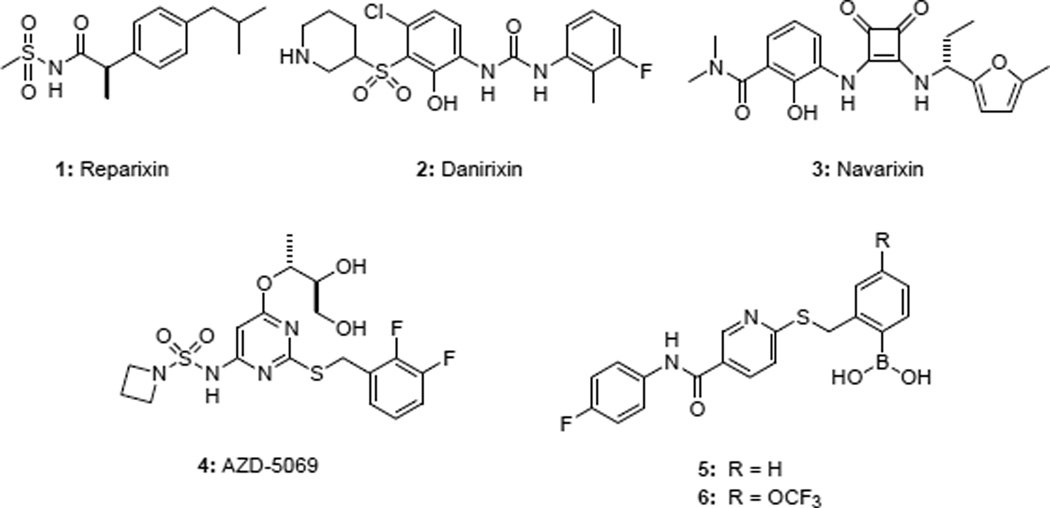
The ability of PMNs to migrate towards sites of injury or inflammation is known as chemotaxis, and is directed in large part by the “Cys-Xaa-Cys” (CXC) chemokine receptors CXCR1 and CXCR2. The endogenous ligands for these G-protein coupled receptors (GPCRs) include growth-related oncogene α (GROα, or CXCL1) and interleukin-8 (IL8, or CXCL8).3 Development of small molecule antagonists of CXCR2 is a major focus of contemporary pharmaceutical research.4,5 Reparixin 1 (Figure 1) is a ketoprofen derivative being investigated in trials for the prevention and treatment of delayed graft function and pancreatic islet transplantation.6,7 In 1998, the first small molecule CXCR2 antagonist based on the diaryl urea pharmacophore was reported.8
Figure 1.
Chemokine antagonists
Danirixin 2 is a diaryl urea CXCR2 antagonist being developed for the treatment of pulmonary diseases, including COPD.9 The central urea motif in the diarylureas was later replaced with the cyclic urea bioisostere 3,4-diaminocyclobut-3-ene-1,2-dione to provide potent analogues as represented by navarixin 3.10 In recent clinical evaluation, navarixin inhibited ozone inhalation-induced sputum PMN recruitment in healthy subjects.11 AZD-5069 4 is a CXCR2 antagonist whose structure was only recently disclosed.12 AZD-5069 is being developed for the treatment of moderate to severe COPD.13 Recent publications detailing the development of novel antagonists from the Neamati group14,15 and Novartis16,17 as well as continued activity in the patent literature,18 underscore the continued interest in developing CXCR2 antagonists for inflammatory diseases.
Based on the known roles of CXCR1/2 in PMN chemotaxis and function, we hypothesized that dual blockade of CXCR1 and CXCR2 would provide critical therapeutic benefit to patients suffering from pulmonary inflammatory diseases and began a discovery program to identify and develop dual CXCR1/2 antagonists. The discovery and evaluation of the first reported boronic acid containing CXCR1/2 antagonist 5 (SX-517) was previously reported.19 Compound 5 belongs to the nicotinamide class of allosteric CXCR1/2 antagonists, which act via an intracellular mechanism of action20 and are unable to displace IL8 binding.19 Although 5 exhibited anti-inflammatory activity in vivo, further preclinical development was hindered by its metabolic instability. A focused SAR effort to increase metabolic stability was then undertaken. A major product of metabolic degradation was the result of oxidative deboronylation of 5 to yield the corresponding 2-hydroxy derivative. It was hypothesized that appropriate derivitization would hinder oxidative cleavage of the boronic acid, thereby increasing both metabolic stability and systemic exposure upon administration. From these efforts, the chemokine antagonist 6 was discovered. Herein we report the SAR studies that led to the discovery of compound 6, a thionicotinamide derivative that exhibits increased metabolic stability while retaining potent activity at both CXCR1 and CXCR2 receptors. Compound 6 was further evaluated in a rat model of pulmonary inflammation, and simulated receptor docking studies were performed to further understand the mechanism of action for this unique class of allosteric CXCR1/2 antagonists.
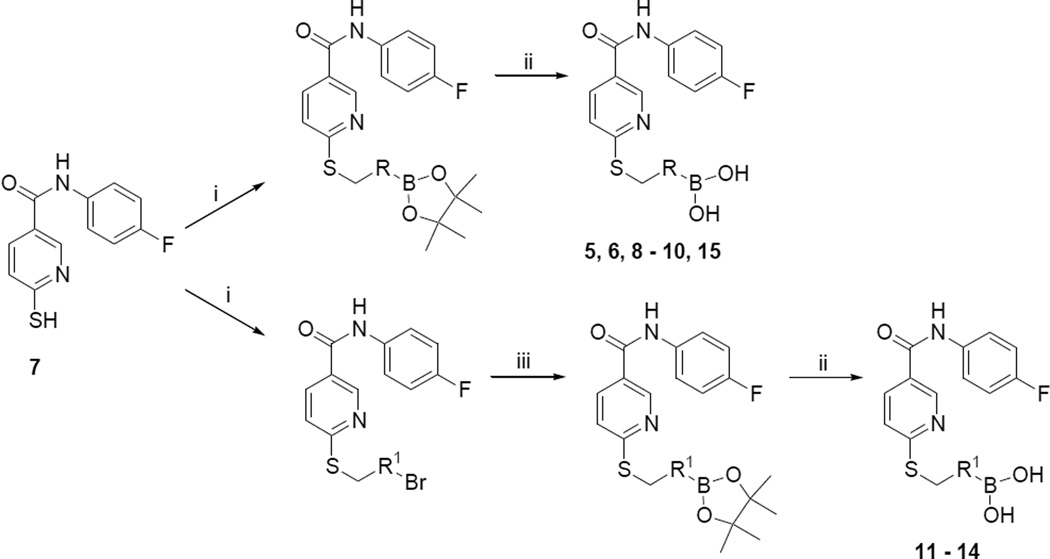
Synthesis of the evaluated compounds was achieved as shown in Scheme 1. Thionicotinamide 719 (1 eq.) and the corresponding bromomethyl derivative (1 eq.) were dissolved in anhydrous dimethylformamide (2 ml/mmol) in an oven dried round bottom flask. To the solution, triethylamine (1 eq) was added, and the reaction was allowed to proceed at room temperature. The reaction progress was monitored by either TLC or LC-MS until complete. The crude products were then precipitated out of solution by the addition of water (50 ml/mmol), filtered, washed with deionized water, and dried under vacuum. This facile filtration workup resulted in compound purities suitable for further advancement without the need for additional purification procedures. Aryl bromide derivatives required the introduction of the boronic acid moiety, and this was accomplished thorough the use of a palladium catalyst21 in the presence of bispinacolato diboron. The aryl bromide (1 eq), PdCl2(CH3CN)2 (0.04 eq), and SPhos (0.16 eq) were placed in an oven dried pressure bottle, and anhydrous 1,4-dioxane added. Under argon, triethylamine (6 eq) and bispinacolato diboron (3 eq) were added, and the vessel sealed and heated to 110 °C for 24 hours. Upon completion, the mixture was cooled and filtered through celite. The resulting crude mixture was then purified using preparative HPLC. Yields for this boronylation reaction were typically low (< 20%), potentially due to catalyst poisoning by sulfur. While disappointing, these low yields were somewhat mitigated by the fact that the aryl bromide starting materials were largely able to be recovered following chromatography. The aryl pinacol boronate esters were deprotected through the use of KHF2, followed by hydrolysis with TMS-Cl/H2O22 to yield the boronic acid derivatives.
Scheme 1.
Reagents and conditions: (i) bromomethyl derivative, triethylamine, DMF, rt; (ii) KHF2, then TMS-Cl/H2O; (iii) PdCl2(CH3CN)2, bispinacolato diboron, triethylamine, dioxane, 110 °C, 24 hours.
The synthesized compounds were then screened for their ability to inhibit GROα-mediated intracellular calcium release in isolated human PMNs, as previously described (Table 1). With regards to lead compound 5, placement of the boronic acid at the 3-position (compound 8) of the phenyl ring resulted in a significant 30-fold drop in the ability to inhibit GROα-mediated intracellular calcium release in isolated human PMNs. Substitution at the 4-position (compound 9) was better tolerated, exhibiting an IC50 of 90 nM. The alkyl boronic acid derivative 10 resulted in an 18-fold drop in activity, suggesting that the aryl boronic acid is a necessary pharmacophore for activity. Compound 11 included the addition of a methylene unit between the phenyl boronic acid moiety and the nicotinamide core resulted in a 10-fold drop in activity, suggesting that a single methylene unit spacer provided the optimal geometry for activity. Both dimethoxy and dioxymethylene substitution at the 4,5 positions (compounds 12 and 13, respectively) resulted in a complete lack of activity in the PMN assay. Mono-substitutions were better tolerated, with methoxy derivatization at the 5-position (compound 14) resulting in a 2-fold drop in activity, and 5-fluoro substitution (compound 15) resulting in an equipotent compound as compared to compound 5. Substitution with a trifluoromethoxy group at the 5 position resulted in compound 6,23 which exhibited a 2-fold greater activity than 5 in the PMN assay. In RBL cells stably transfected with either CXCR1 or CXCR2 receptors, compound 6 exhibited significant antagonist activity at both receptors, suggesting that compound 6 is relatively non-selective. In PathHunter β-arrestin assays (DiscoveRx, Fremont CA) utilizing cells expressing CXCR2, compound 6 is a full antagonist with activity approximately equipotent to reference compound SB225002 (IC50 = 90 nM). In this assay, compound 6 exhibited almost 4-fold greater activity than compound 5.
Table 1.
Inhibition of GROα-mediated intracellular calcium release in isolated human PMNs.
| Cpd | R | IC50 (nM) a | Cpd | R | IC50 (nM) |
|---|---|---|---|---|---|
| 5 | 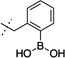 |
38 ± 3 | 8 | 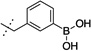 |
1220 ± 750 |
| 9 | 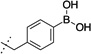 |
90 ± 15 | 10 | 689 ± 239 | |
| 11 | 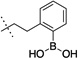 |
387 ± 125 | 12 | 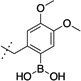 |
>10,000 |
| 13 | 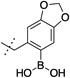 |
>10,000 | 14 | 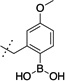 |
80 ± 8 |
| 15 | 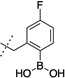 |
52 ± 6 | 6 | 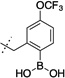 |
22 ± 3 |
Experiments performed in triplicate, reported as mean ± SE.
Further preclinical evaluation of compound 5 was hindered by rapid metabolism. It was of interest to determine whether the structural modifications that led to the discovery of compound 6 would result in increased metabolic stability. Isolated rat and monkey liver microsomes were used to evaluate metabolic stability, as they represent two species commonly used for toxicology experiments. Compounds 5 and 6 were incubated separately with either rat or monkey liver microsomes at a concentration of 1 μM for 60 min at 37°C. Samples were analyzed by LC-MS at 0 and 60 minutes. As shown in Table 2, 5 was extensively metabolized by both rat and monkey liver microsomes after an incubation period of 60 min (37 and 25%, respectively). In contrast, compound 6 exhibited significant stability against metabolic degradation in both rat and monkey liver microsomes (>90% remaining after 60 min).
Table 2.
Comparison of compounds 5 and 6.
| compound | RBL cell line IC50 (nM) a | CXCR2 PathHunter cell line (nM) b |
Microsome stability (% remaining @ 60 min) |
Rat PK AUC (μmol-hr/L) |
|||
|---|---|---|---|---|---|---|---|
| CXCR1 c | CXCR2 c | IC50 β-arrestin c | rat | monkey | IV | Oral | |
| 5 | 903 ± 361 | 356 ± 219 | 730 ± 72 | 37 | 25 | 0.39 | <LLOQ |
| 6 | 31 ± 4 | 21 ± 6 | 130 ± 6 | 92 | 95 | 5.40 | <LLOQ |
Experiments performed in triplicate, reported as mean ± SE;
Experiments performed in quadruplicate, reported as mean ± SE;
IL8 used as agonist.
In order to evaluate whether the increased metabolic stability of compound 6 would translate to improved systemic exposure in vivo, the pharmacokinetics of both compounds 5 and 6 were evaluated in the rat (Table 2). For both compounds, a dose of 1 mg/kg was administered intravenously. Plasma concentrations of both compounds 5 and 6 were determined by LC-MS/MS analysis, and calculated using a calibration curve of test compounds spiked in rat plasma run concurrently with the plasma samples. Following intravenous administration, compound 6 exhibited greater systemic exposure than compound 5 as evidenced by a larger area under the curve (AUC) in the PK graph of plasma concentration vs. time (5.40 vs. 0.39 μmol-hr/L, respectively). While systemic exposure levels rely on a number of factors, the increased metabolic stability of compound 6 may be an important factor that results in its larger AUC following intravenous administration. To establish oral bioavailability, both compounds 5 and 6 were administered via oral gavage to rats at a test concentration of 1 mg/kg. Due to their insolubility in aqueous systems, the compounds necessitated formulation in neat Labrasol (Gattefosse, Saint-Priest, Cedex France), which is a self-emulsifying drug delivery system (SEDDS).24 Despite the use of the SEDDS, plasma levels of both compounds 5 and 6 were below the lower limit of quantification (LLOQ) for our system, which was approximately 20 nM.
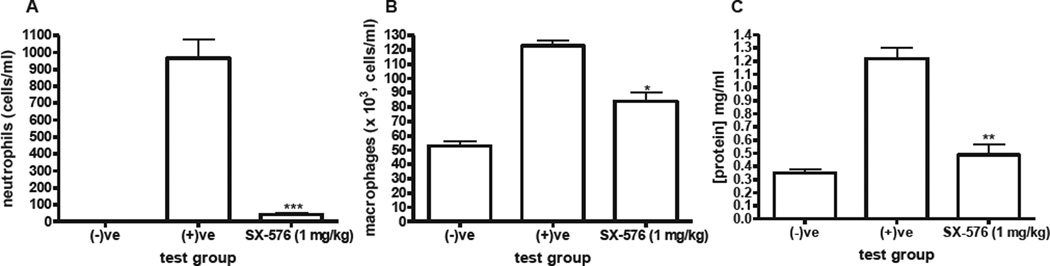
To establish efficacy, compound 6 was then evaluated in the ozone rat model of pulmonary inflammation (Figure 2). In this model, Sprague-Dawley rats (n = 4 per cohort) were dosed intravenously at t = 0 with either vehicle (negative and positive groups) or compound 6 (1 mg/kg). The rats were then placed in either air (negative) or 1 ppm ozone (positive and compound 6 groups) for 4 hours. In order to ensure that plasma concentrations of compound 6 remain within therapeutic levels, the rats were dosed again at t = 5 and 9 hours. The rats were then sacrificed at t = 24 hours, and the bronchoalveolar lavage fluid (BALF) collected. The cells were spun down, stained with Wright-Giemsa and counted. In the negative group, no PMNs were observed when stained (Figure 2A). Protein levels were quantified in BALF by Bradford protein assay analysis. Intravenous administration of compound 6 significantly decreased PMN influx to the lungs to near undetectable levels (Figure 2A). Administration of compound 6 also significantly decreased both macrophage influx and ozone-induced lung leakage (Figures 2B and 2C).
Figure 2.
Ozone rat model of pulmonary inflammation. *p < 0.05; **p < 0.01, ***p < 0.001, t-test vs. positive control.
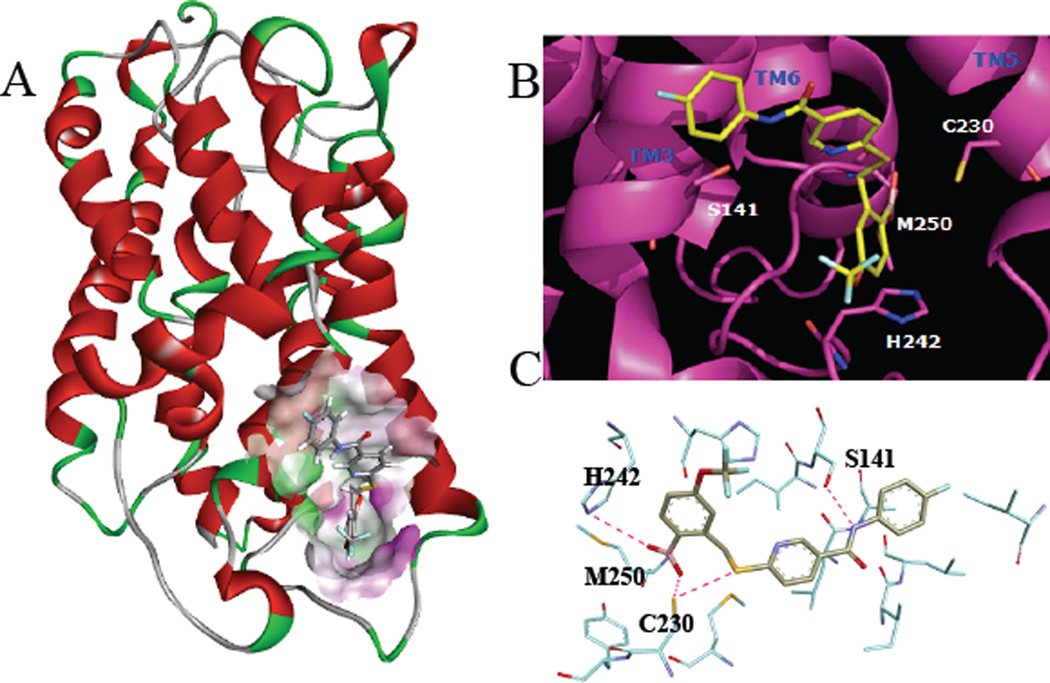
To further understand the mechanism of action of this novel class of allosteric antagonists, simulated receptor docking studies were performed (Figure 3). The 3D NMR structure of CXCR1 (PDB: 2LNL)25 was refined with molecular dynamic (MD) simulation26 using Discovery Studio 3.5 (DS3.5, BIOVIA, San Diego, CA). The 3D structure of CXCR2 is not yet reported, but due to the high level of homology between CXCR1 and CXCR2 (76% amino acid identity), the 3D structure of CXCR2 was constructed using the refined CXCR1 3D structure as a template utilizing the MODELER interface within DS3.5, and further refined with MD simulation. An intracellular allosteric binding site near intracellular loop 3 (ICL3) was identified using the grid search and eraser algorithms in DS3.5. The allosteric binding site also involves amino acids situated on transmembrane regions 3 (TM3), 5 (TM5) and 6 (TM6), as seen in Figures 3A and 3B. 3D coordinates for compound 6 were generated using the LigPrep and Epik programs in DS3.5, and docking studies were then performed using the GLIDE algorithm in DS3.5. Simulated docking with CXCR2 and compound 6 revealed six major interactions: 1) the boronic acid interacts with the imidazole ring in His242 ((B)OH--N, 3.02 Å); 2) the boronic acid oxygen interacts with the main chain NH of Met250 ((B) O--NH, 2.25 Å); 3) the other boronic acid oxygen also interacts with the main chain NH of Met250 ((B) O--NH, 3.00 Å) 4) the boronic acid oxygen interacts with the side chain SH of Cys230 ((B)O--SH, 2.43); 5) the amide nitrogen interacts with the side chain SH of Cys230 (N--SH, 2.57 Å); and 6) the amide interacts with the side chain OH of Ser141 (NH--OH, 2.24 Å). These interactions are illustrated in Figure 3C. The number of proposed interactions attributed to the boronic acid in this receptor binding model may explain its necessity in this class of CXCR1/2 antagonists.19 The proposed binding model for compound 6 differs from previously reported binding models for small molecule CXCR2 antagonists.27–30 This is in accordance with previous findings that structurally distinct small molecule antagonists interact with the CXCR2 receptor via different binding sites.31 Further refinement of this binding model is currently underway through the use of NMR experiments and CXCR2 receptors in phospholipid bilayers.
Figure 3.
Simulated receptor docking of compound 6 to CXCR2. (A) ribbon diagram of CXCR2 with compound 6 docked to intracellular binding site. (B) Predicted binding mode of compound 6 to intracellular binding site of CXCR2. (C) Key receptor-ligand interactions.
In conclusion, compound 6 is an equipotent CXCR1 and CXCR2 antagonist discovered as a result of SAR experiments involving previous lead compound 5. Compound 6 exhibits increased potency in vitro, increased metabolic stability, and improved PK in the rat. In a rat ozone model of pulmonary inflammation, compound 6 exhibited potent inhibition of PMN recruitment upon intravenous administration. Compound 6 (now designated as SX-576) represents a possible therapeutic for the treatment of a variety of inflammatory disorders, and formulation efforts are currently underway to improve the oral bioavailability of this compound.
Acknowledgments
This work was supported by National Institutes of Health grant R44HL072614 (D.Y.M.) from the National Heart Lung and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Chapman RW, Phillips JE, Hipkin RW, Curran AK, Lundell D, Fine JS. Pharmacol. Ther. 2009;121:55. doi: 10.1016/j.pharmthera.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Boppana NB, Devarajan A, Gopal K, Barathan M, Bakar SA, Shankar EM, Ebrahim AS, Farooq SM. Exp. Biol. Med. 2014;239:509. doi: 10.1177/1535370213520110. [DOI] [PubMed] [Google Scholar]
- 3.Baggiolini M. J. Intern. Med. 2001;250:91. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 4.Busch-Petersen J. Curr. Top. Med. Chem. 2006;6:1345. doi: 10.2174/15680266106061345. [DOI] [PubMed] [Google Scholar]
- 5.Dwyer MP, Yu Y. Curr. Top. Med. Chem. 2014;14:1590. doi: 10.2174/1568026614666140827144615. [DOI] [PubMed] [Google Scholar]
- 6.Allegretti M, Bertini R, Cesta MC, Bizzarri C, Di Bitondo R, Di Cioccio V, Galliera E, Berdini V, Topai A, Zampella G, Russo V, Di Bello N, Nano G, Nicolini L, Locati M, Fantucci P, Florio S, Colotta F. J. Med. Chem. 2005;48:4312. doi: 10.1021/jm049082i. [DOI] [PubMed] [Google Scholar]
- 7.Bertini R, Allegretti M, Bizzarri C, Moriconi A, Locati M, Zampella G, Cervellera MN, Di Cioccio V, Cesta MC, Galliera E, Martinez FO, Di Bitondo R, Troiani G, Sabbatini V, D'Anniballe G, Anacardio R, Cutrin JC, Cavalieri B, Mainiero F, Strippoli R, Villa P, Di Girolamo M, Martin F, Gentile M, Santoni A, Corda D, Poli G, Mantovani A, Ghezzi P, Colotta F. Proc. Natl. Acad. Sci. U S A. 2004;101:11791. doi: 10.1073/pnas.0402090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White JR, Lee JM, Young PR, Hertzberg RP, Jurewicz AJ, Chaikin MA, Widdowson K, Foley JJ, Martin LD, Griswold DE, Sarau HM. J. Biol. Chem. 1998;273:10095. doi: 10.1074/jbc.273.17.10095. [DOI] [PubMed] [Google Scholar]
- 9.Miller BE, Smart K, Mistry S, Ambery CL, Bloomer JC, Connolly P, Sanderson D, Shreeves T, Smith R, Lazaar AL. Eur. J. Drug Metab. Pharmacokinet. 2014;39:173. doi: 10.1007/s13318-014-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwyer MP, Yu Y, Chao J, Aki C, Biju P, Girijavallabhan V, Rindgen D, Bond R, Mayer-Ezel R, Jakway J, Hipkin RW, Fossetta J, Gonsiorek W, Bian H, Fan X, Terminelli C, Fine J, Lundell D, Merritt JR, Rokosz LL, Kaiser B, Li G, Wang W, Stauffer T, Ozgur L, Baldwin J, Taveras AG. J. Med. Chem. 2006;49:7603. doi: 10.1021/jm0609622. [DOI] [PubMed] [Google Scholar]
- 11.Holz O, Khalilieh S, Ludwig-Sengpiel A, Watz H, Stryszak P, Soni P, Tsai M, Sadeh J, Magnussen H. Eur. Respir. J. 2010;35:564. doi: 10.1183/09031936.00048509. [DOI] [PubMed] [Google Scholar]
- 12.Nicholls DJ, Wiley K, Dainty I, MacIntosh F, Phillips C, Gaw A, Mardh CK. J. Pharmacol. Exp. Ther. 2015 doi: 10.1124/jpet.114.221358. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 13.Kirsten AM, Forster K, Radeczky E, Linnhoff A, Balint B, Watz H, Wray H, Salkeld L, Cullberg M, Larsson B. Pulm. Pharmacol. Ther. 2015 doi: 10.1016/j.pupt.2015.02.001. advance online publication. [DOI] [PubMed] [Google Scholar]
- 14.Ha H, Bensman T, Ho H, Beringer PM, Neamati N. Br. J. Pharmacol. 2014;171:1551. doi: 10.1111/bph.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha H, Neamati N. Mol. Pharmaceutics. 2014;11:2431. doi: 10.1021/mp500180e. [DOI] [PubMed] [Google Scholar]
- 16.Porter DW, Bradley M, Brown Z, Canova R, Charlton S, Cox B, Hunt P, Kolarik D, Lewis S, O'Connor D, Reilly J, Spanka C, Tedaldi L, Watson SJ, Wermuth R, Press NJ. Bioorg. Med. Chem. Lett. 2014;24:72. doi: 10.1016/j.bmcl.2013.11.074. [DOI] [PubMed] [Google Scholar]
- 17.Porter DW, Bradley M, Brown Z, Charlton SJ, Cox B, Hunt P, Janus D, Lewis S, Oakley P, O'Connor D, Reilly J, Smith N, Press NJ. Bioorg. Med. Chem. Lett. 2014;24:3285. doi: 10.1016/j.bmcl.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Dwyer MP, Yu YN. Expert Opin. Ther. Pat. 2014;24:519. doi: 10.1517/13543776.2014.887682. [DOI] [PubMed] [Google Scholar]
- 19.Maeda DY, Peck AM, Schuler AD, Quinn MT, Kirpotina LN, Wicomb WN, Fan GH, Zebala JA. J. Med. Chem. 2014;57:8378. doi: 10.1021/jm500827t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda DY, Quinn MT, Scheptekin IA, Kirpotina LN, Zebala JA. J. Pharmacol. Exp. Ther. 2009;332:145. doi: 10.1124/jpet.109.159020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billingsley KL, Buchwald SL. J. Org. Chem. 2008;73:5589. doi: 10.1021/jo800727s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuen AKL, Hutton CA. Tetrahedron Lett. 2005;46:7899. [Google Scholar]
- 23.Characterization data for compound 6: ESI-MS m/z = 467.2 [M+H]+. 1H NMR (300 MHz, MeOH-d4) d 9.0 (s, 1H), 8.1 (dd, 2H), 7.86 (s, 1H), 7.7 (q, 2H), 7.6 (d, 1H), 7.4 (d, 1H), 7.3 (d, 1H), 7.1 (t, 2H), 4.5 (s, 2H). Calcd. for C20H15BF4N2O4S: C, 51.52; H, 3.24; N, 6.01; S, 6.88; F, 16.3. Found: C, 51.27; H, 3.33; N, 5.95; S, 6.67; F, 16.45.
- 24.Hu Z, Tawa R, Konishi T, Shibata N, Takada K. Life Sci. 2001;69:2899. doi: 10.1016/s0024-3205(01)01375-3. [DOI] [PubMed] [Google Scholar]
- 25.Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, Opella SJ. Nature. 2012;491:779. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chemistry at HARvard Macromolecular Mechanics (CHARMM) force field was utilized. A dipalmitoylphosphotidylcholine lipid bilayer was embedded during the MD simulation studies of CXCR1. The potential energy of CXCR1 was minimized using the steepest descent algorithm (5000 steps) followed by conjugated gradient method (2000 steps). The conjugate gradient method was applied until the RMS gradient of the structure reached below 0.0001 kcal/molÅ−1. The heating of CXCR1 was performed for 6000 steps, with a step size of 0.001 ps. The initial temperature for heating process was set to 50 K and the final target temperature to 300 K and the velocities are adjusted in every 50 steps. The system was equilibrated until average temperature and structure remained stable and the total energy converged. MD simulations were performed for 50,000 steps, with a step size of 0.0005 ps. The canonical production ensemble NVT was used for the production simulation using SHAKE algorithm for 20,000,000 steps, with a step size of 0.001 ps.
- 27.Nicholls DJ, Tomkinson NP, Wiley KE, Brammall A, Bowers L, Grahames C, Gaw A, Meghani P, Shelton P, Wright TJ, Mallinder PR. Mol. Pharmacol. 2008;74:1193. doi: 10.1124/mol.107.044610. [DOI] [PubMed] [Google Scholar]
- 28.Salchow K, Bond ME, Evans SC, Press NJ, Charlton SJ, Hunt PA, Bradley ME. Br. J. Pharmacol. 2010;159:1429. doi: 10.1111/j.1476-5381.2009.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Kruijf P, Lim HD, Roumen L, Renjaan VA, Zhao J, Webb ML, Auld DS, Wijkmans JC, Zaman GJ, Smit MJ, de Graaf C, Leurs R. Mol. Pharmacol. 2011;80:1108. doi: 10.1124/mol.111.073825. [DOI] [PubMed] [Google Scholar]
- 30.Bertini R, Barcelos LS, Beccari AR, Cavalieri B, Moriconi A, Bizzarri C, Di Benedetto P, Di Giacinto C, Gloaguen I, Galliera E, Corsi MM, Russo RC, Andrade SP, Cesta MC, Nano G, Aramini A, Cutrin JC, Locati M, Allegretti M, Teixeira MM. Br. J. Pharmacol. 2012;165:436. doi: 10.1111/j.1476-5381.2011.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Kruijf P, van Heteren J, Lim HD, Conti PG, van der Lee MM, Bosch L, Ho KK, Auld D, Ohlmeyer M, Smit MJ, Wijkmans JC, Zaman GJ, Smit MJ, Leurs R. J. Pharmacol. Exp. Ther. 2009;329:783. doi: 10.1124/jpet.108.148387. [DOI] [PubMed] [Google Scholar]