Abstract
Reactive oxygen species (ROS) are toxic molecules utilized by the immune system to combat invading pathogens. Recent evidence suggests that inefficiencies in ATP production or usage can lead to increased endogenous ROS production and sensitivity to oxidative stress in bacteria. With this as inspiration, and knowledge that ATP is required for a number of DNA repair mechanisms, we hypothesized that futile cycling would be an effective way to increase sensitivity to oxidative stress. We developed a mixed integer linear optimization framework to identify experimentally-tractable futile cycles, and confirmed metabolic modeling predictions that futile cycling depresses growth rate, and increases both O2 consumption and ROS production per biomass generated. Further, intracellular ATP was decreased and sensitivity to oxidative stress increased in all actively cycling strains compared to their catalytically inactive controls. This research establishes a fundamental connection between ATP metabolism, endogenous ROS production, and tolerance toward oxidative stress in bacteria.
Keywords: reactive oxygen species, oxidative stress, futile cycle, hydrogen peroxide, metabolism
1. Introduction
After phagocytic cells ingest pathogens, NADPH oxidase is recruited to the phagosome to generate an “oxidative burst” of O2−(Diacovich and Gorvel, 2010). The O2−generated can be dismutated to H2O2 and further processed to HOCl by myeloperoxidase (Hampton et al., 1998), or directly react with NO• to form OONO− (Fang, 2004). These ROS can damage a wide range of biomolecules, including iron-sulfur clusters (Jang and Imlay, 2007), lipids (Rubbo et al., 1994), cysteine residues (Kim et al., 2000; Storkey et al., 2014), methionine residues (Davies, 2005; Luo and Levine, 2009; Storkey et al., 2014), and tryptophan residues (Jerlich et al., 2000). To prevent or cope with these deleterious molecules, pathogens employ a number of detoxification and repair systems, as well as in some cases methods to inhibit NADPH oxidase and the originating oxidative burst. The importance of ROS to immunity is highlighted by the numerous pathogens that require such mechanisms to establish or sustain an infection [Bacillus anthracis (Crawford et al., 2006), Coxiella burnetii (Siemsen et al., 2009), Chlamydia trachomatis (Tauber et al., 1989), Salmonella Typhimurium (Hebrard et al., 2009), Mycobacterium tuberculosis (Li et al., 1998; Manca et al., 1999), Staphylococcus aureus (Cosgrove et al., 2007), Helicobacter pylori (Harris et al., 2003), Streptococcus pyogenes (Brenot et al., 2004), and Enterococcus faecalis (La Carbona et al., 2007)], and the shortened life expectancy of patients with chronic granulomatous disease (CGD), who suffer from recurring infections due to mutational defects in NADPH oxidase (Fang, 2004).
This strong dependency of virulence on oxidative stress suggests that bacterial tolerance to ROS may represent an effective target for anti-infective therapies. With that rationale, much research has been devoted to identifying the molecular mediators of oxidative stress responses in microbes. Inhibition of detoxification or repair systems (Ananthaswamy and Eisenstark, 1977; Bakshi et al., 2006; Cosgrove et al., 2007; De Groote et al., 1997; Hebrard et al., 2009; Imlay and Linn, 1986), as well as boosting ROS production from microbes themselves (Brynildsen et al., 2013), has proven effective at increasing sensitivity to exogenous oxidative stress. These results inspired us to explore the possibility of targeting two ROS tolerance determinants with a single metabolic perturbation. Specifically, numerous oxidative damage repair systems are ATP-dependent (Galletto et al., 2006; Heinze et al., 2009; Orren and Sancar, 1989; Selby and Sancar, 1995; Voloshin et al., 2003), and previous work suggested that inefficiency in ATP usage or production increases endogenous ROS generation and decreases tolerance to exogenous oxidants (Brynildsen et al., 2013). Futile cycles are an effective means to reduce efficiency in ATP usage, and previous research has demonstrated that active cycling reduces intracellular ATP levels (Izallalen et al., 2008; Koebmann et al., 2002). Therefore, we hypothesized that the stimulation of futile cycles would be an effective way to increase sensitivity to oxidants, through their combined effect on ATP levels and potential to increase endogenous ROS production.
Here, we explore the effects of three futile cycles in different areas of metabolism on E. coli K-12 MG1655. We experimentally validated computational predictions that cycling would decrease growth rate, and increase O2 consumption and ROS production relative to biomass produced. The anticipated decrease in ATP concentration (Koebmann et al., 2002) with futile cycling was also confirmed experimentally, and further, we discovered that futile cycling led to a significant increase in sensitivity to H2O2, consistent with our hypothesis. We found this result to not be explainable by growth rate alone, since a nutrient-deprived culture exhibited a significant increase in tolerance to H2O2 in comparison to its nutrient-replete control. Additionally, we found that futile cycling did not decrease cellular detoxification of H2O2, which indicated that the reduction in oxidant tolerance was the result of enhanced damage or defective repair. We anticipate that this fundamental knowledge connecting ATP metabolism to oxidant sensitivity could be leveraged to develop new anti-infective therapies for pathogens that rely on their oxidative stress tolerance to sustain an infection.
2. Materials and Methods
2.1 Flux balance analysis
To simulate ROS production, we used a previously developed ensemble modeling approach (Brynildsen et al., 2013). We note that although several reactions in E. coli are known to generate O2− and H2O2 (Korshunov and Imlay, 2010), the sources of the majority of ROS produced during aerobic growth remain ill-defined (Brynildsen et al., 2013; Seaver and Imlay, 2004). However, this uncertainty is constrained by the fact that enzymes with the potential to generate ROS use flavin, quinone, and/or transition metal electron carriers. The ensemble modeling approach leverages this knowledge to generate an ensemble of metabolic models where the total ROS production from each network is equivalent, but the quantitative contribution of each potential ROS-generating enzyme is distinct. To illustrate, Figure S1 depicts the total H2O2 generated by 100 distinct models (Fig. S1A), as well as the H2O2 generated by a single ROS generating reaction (NADH dehydrogenase I) (Fig. S1B). General trends obtained from the ensemble can then be used to formulate hypotheses that can be experimentally assessed, as was done here with respect to the impact of futile cycling on ROS metabolism. This ensemble modeling approach uses the iAF1260 genome-scale metabolic model of E. coli (Feist et al., 2007), and it has been shown previously to correctly predict the qualitative change in ROS production for single gene deletion mutants in approximately 90% of the tested cases (Brynildsen et al., 2013).
Flux balance analysis was performed using the COBRA Toolbox v2.0 (Schellenberger et al., 2011) in MATLAB while optimizing for biomass. The glucose uptake rate was set to 11 mmol/gDW/h (Brynildsen et al., 2013). Transcriptional regulation was applied (Covert and Palsson, 2002) with regulatory rule changes in b2458 (eutD), b3942 (katG), b0506 (allR), and b2087(gatR) (Brynildsen et al., 2013). Additional changes to the model include: correcting the electron acceptor of protoporphyrinogen oxidase from oxygen to ubiquinone (Mobius et al., 2010); unconstraining erroneous bounds on formate hydrogen lyase, catalase, and superoxide dismutase (Brynildsen et al., 2013); removing anaerobic pyridoxal-5-phosphate oxidase activity due to a lack of experimental evidence that it occurs in vivo (Feist et al., 2007); and only allowing oxygen as an electron acceptor for aspartate oxidase because it has been found to be the predominant electron acceptor under aerobic conditions (Korshunov and Imlay, 2010).
2.2 Bacterial strains
E. coli K-12 MG1655 was used in all experiments. ΔkatE, ΔkatG, and ΔptsI mutations were transduced into MG1655 from their respective strains in the Keio collection (Baba et al., 2006) by the P1 phage method. The ΔahpCF mutation was generously provided by Michael Kohanski and transduced into MG1655 using the P1 phage method. In generating the Hpx– strain (ΔkatE ΔkatG ΔahpCF), antibiotic markers were cured out using pCP20 before subsequent P1 transductions (Datsenko and Wanner, 2000), and the known antioxidant pyruvate was included in the LB agar plates to prevent a toxic build-up of H2O2 (Nath et al., 1995). Deletions were PCR checked for proper chromosomal integration with a forward primer external to the gene and reverse primer within the kanamycin resistance cassette (kanR). Internal primers were used to check for gene duplication. In cured strains, external primers before and after the gene were used to check for proper scar size. All PCR primers are listed in Table S1. Hpx– retains no catalase or peroxidase activity, which was confirmed through H2O2 clearance assays (Fig. S13).
2.3 Plasmid construction
Genes were amplified off of purified E. coli K-12 MG1655 genomic DNA using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Inc.). All restriction enzymes were purchased from New England Biolabs, Inc., and digestions were carried out according to manufacturer instructions. Ligations were done using the NEB Quick Ligation Kit, and ligated plasmids were transformed into Z-competent (Zymo Research) E. coli XL1-Blue (Stratagene). Transformed cells were immediately plated on LB-agar with 100 μg/mL ampicillin (amp). Inserts of all plasmids (active and inactive) were confirmed by sequencing through GENEWIZ, Inc.
Overexpression plasmids were generated using pQE80 (Qiagen). Primers used for amplifying genes off of MG1655 genomic DNA are listed in Table S2. Forward primers consist of a GC-clamp followed by a restriction site, spacer, ribosomal binding site, and the genomic sequence beginning 8 base pairs upstream of the start codon. The EcoRI cut site on pQE80 was used to remove the 6xHis-tag coding sequence, with either HindIII (pQE80-acs and pQE80-pck) or BamHI (pQE80-atp) on the other side of the gene. For the pQE80-atp (atpAGD) insert, MfeI, which has compatible cohesive ends with EcoRI, was used in the forward primer because EcoRI was present within the insert. Inactive enzymes were generated by site-directed mutagenesis using the overlap extension method (Ho et al., 1989) with primers listed in Table S2. Inactivations were suggested by literature: D268N for Pck (Jabalquinto et al., 2002), K609A for Acs (Starai and Escalante-Semerena, 2004), and βR246A for AtpD (Ahmad and Senior, 2004).
2.4 Growth rates
Optical densities at 600 nm (OD600) were measured on a Synergy H1 Microplate Reader (BioTek Instruments, Inc.). Growth rates were determined in MOPS minimal 10 mM glucose medium and MOPS EZ-rich 10 mM glucose medium, both purchased from Teknova. EZ-rich medium is supplemented with all 20 amino acids, adenine, cytosine, uracil, guanine, and the vitamins thiamine, pantothenate, p-aminobenzoic acid, p-hydroxybenzoic acid, and 2,3-dihydroxybenzoic acid (Neidhardt et al., 1974). Overnight cultures were inoculated from −80°C stocks. From a 20 hour 1 mL LB + 100 μg/mL amp overnight culture, 20 mL of media (MOPS 10 mM glucose medium or EZ-rich 10 mM glucose medium) + 100 μg/mL amp was inoculated to an OD600 of 0.005 in a 250 mL baffled flask. The culture was grown at 37°C with 250 rpm shaking to an OD600 of 0.04 before inducing with 1 mM IPTG. OD600 was measured every hour for minimal media cultures, and every 20 minutes for cultures in rich media. Growth rate was calculated from OD600 measurements 2 to 4 hours after induction in minimal media cultures, and 40 minutes to 1 hour 40 minutes after induction in rich media.
2.5 Oxygen measurements
From a 20 hour LB + 100 μg/mL amp overnight culture originating from a −80°C stock, 20 mL of media (MOPS 10 mM glucose medium, or MOPS EZ-rich 10 mM glucose medium) + 100 μg/mL amp was inoculated to an OD600 of 0.005 in a 250 mL baffled flask. The culture was grown at 37°C with 250 rpm shaking to an OD600 of 0.1 before inducing with 1 mM IPTG (0.1 mM IPTG for fcatp/* in minimal glucose medium to allow for measurable growth during oxygen measurement). After two hours of induction in minimal media or one hour in rich media, 12 mL of culture was removed and centrifuged at 37°C and 4,000 rpm for ten minutes. 10.8 mL of the spent media was removed. The cell pellet was resuspended in the remaining media, and 1 mL was transferred to a 1.5 mL microcentrifuge tube and centrifuged at 14,000 rpm for 2 minutes. 980 μL of the spent media was removed, and the cell pellet was resuspended with 980 μL of pre-warmed fresh media + amp + IPTG (same concentrations used in the growth in flasks). The resuspended cells in fresh media were used to inoculate 10 mL of pre-warmed fresh media + amp + IPTG to an OD600 of 0.1 in a 50 mL Falcon tube containing a sterile magnetic stirring bar. The Falcon tube was immersed in a stirred water bath at 37°C. For the fcacs/* cycle, 3.5 mL instead of 10 mL was used in the Falcon tube to enhance cycling, as determined by depressed growth rate of fcacs compared to fcacs*.
OD600 measurements were taken every 10 minutes for strains with inactive enzymes and 20 minutes with active enzymes in rich media, and every 20 minutes for strains with inactive enzymes and hourly with active enzymes in minimal media. Dissolved O2 concentration was continuously measured (one reading per second) using the FireStingO2 fiber-optic O2 meter with the OXROB10-CL2 robust oxygen miniprobe (PyroScience, GmbH). The effect of fluctuations in temperature was automatically compensated for using the TDIP15 temperature sensor (PyroScience GmbH) and the FireSting Logger Software. For details of how the FireSting data was used to calculate the total O2 consumed, please see section S.7 of the Supplementary material.
2.6 ATP measurement
For cycling assays, cells were grown, induced, and washed as described for the oxygen experiments. All cycles were fully induced (1 mM IPTG). The washed cells were used to inoculate pre-warmed fresh media + amp + IPTG to an OD600 of 0.2. For measuring MG1655 and MG ΔptsI ATP levels, 20 h LB (+ 30 μg/mL kan for MG ΔptsI) overnight cultures were washed with MOPS 10 mM glucose medium to remove trace LB components and then used to inoculate 20 mL MOPS 10 mM glucose medium in a 250 mL baffled flask to an OD600 of 0.05. A higher cell density was necessary for ΔptsI experiments because the ΔptsI mutant will not grow and the cultures were washed and concentrated to an OD600 of 0.2. Cultures were grown for 5 hours at 37°C with 250 rpm shaking (one doubling after leaving lag phase for MG1655), then washed and adjusted to an OD600 of 0.2 as described above. ATP was measured using the BacTiter-Glo Microbial Cell Viability Assay Kit (Promega) according to the manufacturer's instructions. Briefly, 100 μL of the OD600 0.2 culture was mixed with 100 μL BacTiter-Glo reagent in an opaque 96-well plate. After five minutes at room temperature, luminescence was read on a Synergy H1 Microplate Reader (BioTek Instruments, Inc.). An ATP standard curve was used to calculate ATP concentrations.
2.7 H2O2 production
H2O2 production was measured using an Amplex Red H2O2 quantification assay. An E. coli strain deficient in catalase and alkyl hydroperoxidase (MG1655 ΔkatE ΔkatG ΔahpCF, denoted Hpx–) was necessary for these assays because wild-type MG1655 does not release measureable amounts of H2O2 (Seaver and Imlay, 2001). Different but comparable media was used in this assay, as it was found that MOPS, cysteine, lysine, methionine, and EZ-rich vitamins all increased background fluorescence in the presence of Amplex Red. Minimal media assays were performed in M9 10 mM glucose medium, while rich media assays were performed in M9 10 mM glucose medium supplemented with the concentrations of adenine, cytosine, uracil, guanine, and amino acids except cysteine, lysine, and methionine present in EZ-rich defined medium (Neidhardt et al., 1974). In Hpx– strains, catalase was added to prevent a toxic buildup of H2O2. Catalase from bovine liver (2,000-5,000 units/mg protein: Sigma Aldrich) was used, and the concentrations for overnight cultures were chosen based on tests that found addition of more catalase no longer led to an increase in terminal cell density. For growth in flasks, catalase concentrations were chosen based on tests that found the addition of more catalase no longer increased the growth rate.
For cycling assays, overnight cultures were inoculated from −80°C stocks and grown for 20 hours in 1 mL LB + 100 μg/mL amp. For Hpx– strains, 30-75 U/mL of catalase was added to prevent a toxic buildup of H2O2 prior to beginning the assay. From overnight cultures, 20 mL of minimal or rich media in a 250 mL baffled flask was inoculated to an OD600 of 0.005. Hpx– strains were supplemented with 30-75 U/mL catalase in minimal media, or 93-233 U/mL in rich media. When cultures reached an OD600 of 0.1, they were induced with 1 mM IPTG (rich media assays) or 0.1 mM IPTG in the case of fcatp in minimal glucose medium, which had failed to grow at 1mM IPTG due to excessive futile cycling.
After one hour of induction in rich media, or two hours in minimal, 12 mL was removed to a pre-warmed 15 mL Falcon tube and centrifuged at 37 °C and 4,000 rpm for 10 minutes. 10.8 mL of spent media was removed, the cell pellet resuspended, and 1 mL transferred to a warm 1.5 mL microcentrifuge tube. Cells were washed four times to remove all catalase. Washes consisted of spinning down at 14,000 rpm for 2 minutes, removing 980 μL of media, and resuspending the cell pellet with 980 μL fresh media + amp + IPTG. Washed cells were adjusted to an OD600 of 0.1 in warm, fresh media + amp + IPTG.
For MG1655 and MG ΔptsI experiments, 20 h LB (+ 30 μg/mL kan for MG ΔptsI) overnight cultures were washed in M9 10 mM glucose medium and then used to inoculate 20 mL of M9 10 mM glucose medium in a 250 mL baffled flask to an OD600 of 0.05. After MG1655 doubled once (6 hours), cultures were washed and adjusted to an OD600 of 0.1 in warm, fresh media as described for the cycling strains, except no amp or IPTG was included.
In a black clear-bottom 96-well plate, 50 μL of the OD600 0.1 culture was added to 50 μL media + 100 μM Amplex Red reagent (Life Technologies) + 0.2 U/mL horseradish peroxidase (Life Technologies), with appropriate concentrations of amp and IPTG for the cycling assays. The plate was mixed in a dark shaker at 37°C with 250 rpm shaking for 2 minutes before reading the initial (time = 0) OD600 and fluorescence (550/590 nm) on a Synergy H1 Microplate Reader (BioTek Instruments, Inc.). OD600 and fluorescence were measured every 30 min in rich media and every hour in minimal. While in the shaker, the plate was covered with Breathe-Easy film (Diversified Biotech), which is O2 permeable. Results of this assay are presented as fluorescence versus time, where fluorescence is proportional to cumulative H2O2 production. This method was used rather than measuring H2O2 concentration at specific time points, because the assay conditions for quantifying H2O2 concentrations from Hpx– led to significant growth defects due to the accumulation of H2O2 in the media. By continuously incubating the cells with Amplex Red and horseradish peroxidase, we were able to measure H2O2 released with the added benefit of providing an extracellular sink for H2O2 that allows Hpx– growth to more closely resemble that of wild-type.
2.8 Sensitivity assays
For cycling assays, scavenging proficient wild-type cells were grown, induced, and washed as described for the oxygen experiments and ATP measurements. The washed cells were used to inoculate pre-warmed fresh media + amp + IPTG to an OD600 of 0.2. For treatment with H2O2, 1 mL of the OD600 0.2 culture was placed in a warm test tube and treated with H2O2. Due to the different media used and different level of cycling between systems, H2O2 concentrations were chosen such that bacterial killing in excess of 90% of the population would be observed for at least one strain (strain harboring the active or inactive enzyme). These concentrations were 4 mM H2O2 for fcpck/* in MOPS 10 mM glucose medium, 5 mM H2O2 for fcatp/* in MOPS 10 mM glucose medium, 2 mM H2O2 for fcatp/* in EZ-rich 10 mM glucose medium, and 1.5 mM H2O2 for fcacs/* in EZ-rich 10 mM glucose medium. For MG1655 and MG ΔptsI, cells were grown, washed, and adjusted to an OD600 of 0.2 as described in the MG1655 and MG ΔptsI ATP measurement protocol. They were treated with 12 mM H2O2, a concentration at which at least 90% killing was observed in wild-type or ΔptsI.
Initial cells were plated prior to the addition of H2O2. At each time point, 200 μL was removed to a 1.5 mL microcentrifuge tube and spun down at 15,000 rpm for 3 minutes. Cells were washed by removing 180 μL of media, resuspending with 980 μL PBS, spinning down again at 15,000 rpm for 3 minutes, removing 980 μL, and resuspending a final time in 80 μL PBS. Plating was done on LB agar using the serial dilution method, and plates were incubated for 16 hours at 37°C prior to counting colonies.
2.9 H2O2 detoxification assays
Samples were prepared and treated identically to the sensitivity assays. Washed induced cells were used to inoculate pre-warmed fresh media + 100 μg/ml amp + 1 mM IPTG to an OD600 of 0.2, 1 mL of which was placed in a warm test tube and treated with H2O2. Initial H2O2 concentrations matched those used in the sensitivity assays. At each time point, 200 μL was removed to a microcentrifuge tube and centrifuged at 15,000 rpm for 3 minutes. To remove the cells, 150 μL of the supernatant was removed to a clean microcentrifuge tube. The initial time points were obtained by adding an identical amount of H2O2 to a tube containing only MOPS 10 mM glucose medium + 100 μg/ml amp + 1 mM IPTG. H2O2 concentration was measured in the cell-free samples using an Amplex Red Hydrogen Peroxide/Peroxidase kit (Life Technologies) according to the manufacturer's instructions. A standard curve was used to convert measured fluorescence values to H2O2 concentration.
2.10 Statistical analysis
Statistical significance was assessed using two-tailed t-tests with unequal variances. Two values were considered statistically different if their respective p-value was less than 0.05. For experiments involving survival fractions, values were log transformed prior to statistical analysis. For untreated sensitivity assay controls, a one-sample t-test was used to compare the survival fractions to 1. Again, log transformed values were used when determining significance of survival fractions.
3. Results
3.1 Flux balance analysis predictions
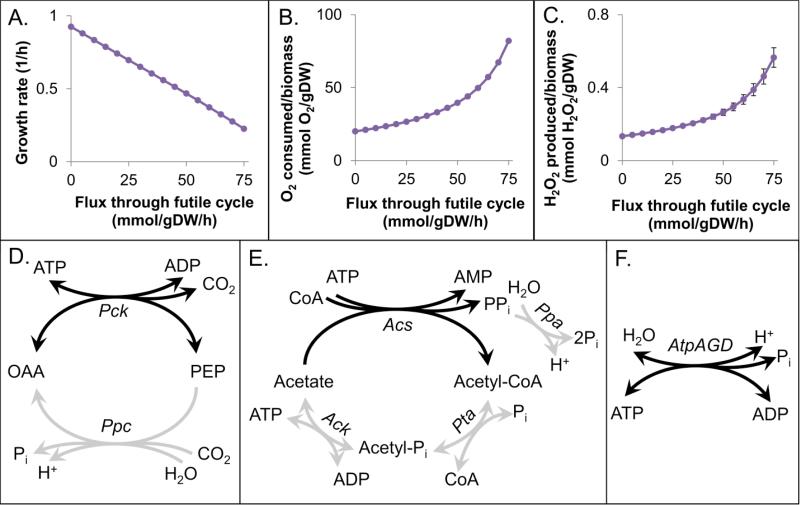
To explore the impact of futile cycling on ROS production, we performed in silico experiments using an ensemble approach, which was required due to the indeterminacy associated with endogenous sources of O2− and H2O2 (Methods). We observed that increasing flux through a futile cycle led to a decrease in growth rate (Fig. 1A), an increase in O2 consumed relative to biomass produced (Fig. 1B), as well as an increase in ROS production per biomass produced (Fig. 1C), consistent with the notion of decreased productive output for the same energy invested. An analogous plot for O2− production per biomas produced can be found in the Supplementary material (Fig. S2). These effects were independent of the specific futile cycle theat was functioning, which was expected since the net effect of flux through a futile cycle is ATP hydrolysis. To confirm these predictions experimentally, we sought to measure growth rate, O2 consumption, and H2O2 production from strains experiencing futile cycling in different areas of metabolism. To accomplish this, we needed to identify experimentally tractable futile cycles.
Figure 1. Futile cycle effects and cycles to be experimentally assessed.
Predictions on the effect of futile cycling on growth rate (A), O2 consumption (B), and H2O2 production (C). Points represent the mean prediction of an ensemble of 100 models, and error bars show the standard deviation. The O2− and H2O2 stoichiometric coefficients used in these models can be found in Table S4. Depictions of the Ppc-Pck, acetate, and context independent ATP synthase futile cycles are shown in D, E, and F, respectively. Enzymes present during growth in minimal glucose medium are shown in gray, and the reaction required for cycling is highilighted in black.
3.2 Identification of theoretical and experimental cycles
To identify futile cycles throughout E. coli metabolism, we sought sets of reactions within the iAF1260 genome-scale metabolic model that resulted in ATP hydrolysis. Using mixed integer linear optimization (MILO), we identified futile cycles with the smallest number of reactions (Supplementary material). This process identified a total of 111 “theoretical” futile cycles in E. coli that include only one, two, or three reactions. However, these cycles were not equally accessible experimentally. For example, glycerol kinase and glycerol-3-phosphatase form a two-step cycle that would require the overexpression of both enzymes in minimal glucose medium, whereas the cycle created by phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase requires the overexpression of only one protein (Pck). To identify futile cycles that were the most straight-forward to engage experimentally (requiring the synthetic expression of only a single enzyme), we modified the MILO problem to weight only enzymatic reactions that would require proteins to be synthetically expressed (Supplementary material). This process identified 25 futile cycles that required only one enzymatic reaction to be expressed in E. coli during growth on minimal glucose medium. For comparative purposes, we also identified all cycles that would require synthetic expression of two or three enzymatic reactions, and these are presented in the Supplementary material along with the theoretical cycles. A complete list of all one-, two-, and three-step theoretical and experimental cycles can be found in Table S5.
The ATP hydrolyzing cycles we chose to explore are shown in Fig. 1. The Ppc-Pck futile cycle, fcpck, could be engaged by expression of Pck (Ppc is utilized during growth on minimal glucose medium), whereas the acetate cycle, fcacs, requires the expression of Acs (Ack and Pta are expressed during overflow metabolism and acetate excretion, whereas a basal level of Ppa expression is required for growth (Chen et al., 1990)). We note that these cycles are both dependent on media conditions, and for that reason, we chose to also look at an ATP synthase cycle, fcatp, although it required expression of three proteins (AtpA, AtpG, and AtpD), because it was context independent (Koebmann et al., 2002). These cycles are located in different parts of metabolism, with the only common feature being ATP hydrolysis. We chose to analyze all three systems to provide generality to the results of this study.
3.3 Verification of futile cycling
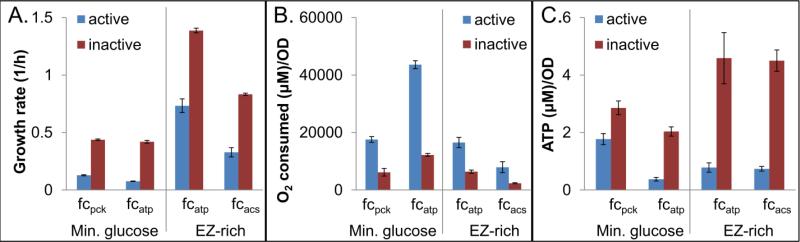
To verify that futile cycling was occurring upon expression of pck, acs, and atpAGD, we sought to confirm the FBA predictions that in the presence of futile cycling growth rate would decrease (Fig. 1A) and O2 consumption per biomass would increase (Fig. 1B). Previous studies analyzing the impacts of futile cycling have used similar measures to establish the presence of an active cycle (Chao and Liao, 1994; Izallalen et al., 2008; Patnaik et al., 1992). We also expected that futile cycling would decrease the cellular ATP concentration based on previous work in which expression of the F1 subunits from ATP synthase was found to cause uncoupled ATPase activity within the cytoplasm (Koebmann et al., 2002). Therefore, we measured growth rate, relative oxygen consumption, and ATP in strains expressing the futile cycle enabling enzymes, and compared those to strains expressing catalytically inactive mutants of each enzyme. The inactivating mutations were suggested by literature: D268N for Pck (Jabalquinto et al., 2002), K609A for Acs (Starai and Escalante-Semerena, 2004), and R246A for AtpD (Ahmad and Senior, 2004), and enzyme assays were performed to verify successful removal of catalytic activity (Fig. S4).
Growth rates were first measured in MOPS 10 mM glucose medium for all cycles (Fig. 2A and S9). We observed that the growth rate was not significantly suppressed for fcacs in minimal glucose medium (Fig. S9). The fcacs requires Ack and Pta to produce acetate to enable cycling, so increased flux to acetate was predicted to result in more significant cycling. We note that acetate production increases with increasing growth rate (Eiteman and Altman, 2006), and this inspired us to test fcacs under rich media conditions. When we expressed acs in EZ-rich 10 mM glucose medium, we observed a significant depression of growth rate (Fig. 2A). In addition, since fcatp was predicted to cycle independently of media conditions, we also engaged this cycle in the rich media and observed a significant depression in growth rate (Fig. 2A). When O2 consumption relative to biomass production was assayed for these four experimental scenarios, we found that strains expressing cycle-enabling enzymes consumed significantly more O2 per biomass produced than strains carrying their respective catalytically inactive mutants (Fig. 2B, Fig.S12). To further confirm the presence of futile cycling, we measured ATP and observed that ATP concentrations were significantly reduced in all cycling strains (Fig. 2C). These experiments confirm that we were able to stimulate futile cycling in E. coli with different enzymes under different environmental conditions. Effects on growth rate, relative oxygen consumption, and ATP levels match predictions based on FBA and information from the literature.
Figure 2. Experimental measurement of growth rate, O2 consumption, and ATP concentration.
Effect of expression of catalytically active or inactive Pck, AtpAGD, or Acs on growth rate (A), O2 consumption relative to biomass produced (B), and ATP concentration (C). All bars represent at least three biological replicates, and error bars represent standard error of the mean. In each case, the active and inactive cycles are statistically different (p<0.05). In the absence of induction, strains with plasmids encoding the active and inactive enzymes were indistinguishable with regard to growth rate and ATP measurements (Fig. S10). Cells/ml for a given OD600 were found to be equivalent for the active and inactive strains (Fig. S5), and further, the cells were found to be approximately the same size (Fig. S6 and S7).
3.4 Increased H2O2 production in futile cycling strains
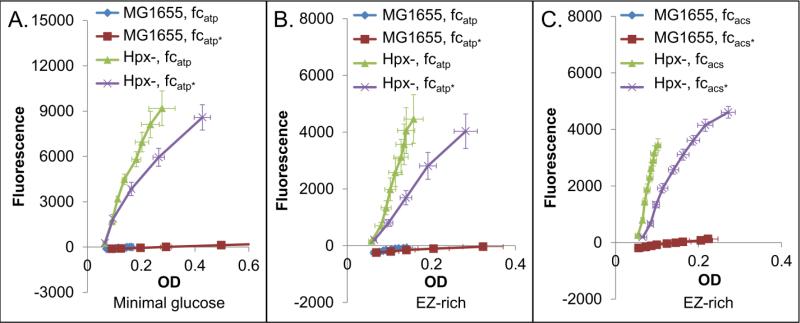
Simulations also predicted an increase in H2O2 produced per biomass produced under conditions of active futile cycling (Fig 1C). H2O2 represents a good approximation of cumulative ROS because there is approximately a 3:1 ratio of H2O2 to O2− produced (Imlay and Fridovich, 1991; Seaver and Imlay, 2001), and most O2− is dismutated to H2O2 (Imlay and Fridovich, 1991). To explore H2O2 production experimentally, we performed Amplex Red based experiments in an Hpx– strain (ΔkatE ΔkatG ΔahpCF). The Hpx– strain is necessary because wild-type MG1655 does not accumulate measurable amounts of H2O2 in the media (Seaver and Imlay, 2001). In the Amplex Red assay, H2O2 reacts with Amplex Red Reagent in the presence of horseradish peroxidase to produce resorufin, a fluorescent product. Using this assay we confirmed that our Hpx– strains accumulated H2O2 in the media, whereas wild-type did not (Fig. 3). Additionally, complementation of Hpx– with ahpCF prevented accumulation (Fig. S14). All experiments were performed with cell-free controls, which were used as blanks for OD600 and fluorescence measurements.
Figure 3. Endogenous H2O2 production in cycling strains.
H2O2 production in fcatp in minimal media (A), fcatp in rich media (B), and fcacs in rich media (C), approximated by fluorescence of resorufin vs. OD600 in an Hpx– strain. Presence of alkyl hydroperoxidase and catalases (MG1655) eliminates fluorescence in all cases. Each graph represents at least three biological replicates, and error bars show the standard error of the mean. We note that the scavenger-proficient wild-type MG1655 is able to detoxify H2O2 generated by salt-catalyzed glucose oxidation in the media (Seaver and Imlay, 2001), so the fluorescence of this strain was sometimes below that of the blank. An asterisk (*) indicates a strain expressing the catalytically inactive form of the cycle-enabling enzyme. Cells/ml for a given OD600 were found to be equivalent for the active and inactive strains (Fig. S5), and further, the cells were found to be approximately the same size (Fig. S6 and S7).
When strains expressing cycle-enabling enzymes and their catalytically inactive controls were assayed, we observed an increase in H2O2 produced per OD600 for fcatp in minimal media (Fig. 3A), fcatp in rich media (Fig. 3B), and fcacs in rich media (Fig. 3C). Interestingly, we were unable to perform the assay for fcpck because cycling was not observed in the Hpx– strain, as determined by growth rate (Fig. S15). Although we did not explore the cause of this effect, it is likely due to the rewiring of metabolism that occurs in Hpx– due to the constitutive oxidative stress it encounters. Failure of Pck to produce a futile cycle could be due to the close proximity of the futile cycle to pyruvate and oxaloacetate, which are both a-ketoacids known to react with H2O2 (Nath et al., 1995). If metabolites in or near the cycle were being consumed by direct reaction with H2O2, one would expect reduced flux through this cycle in Hpx–. We note that all Hpx– strains where cycling was observed demonstrated an increase in H2O2 production per biomass (Fig. 3), suggesting the effect is independent of the cycle.
3.5 Increased oxidant sensitivity in futile cycling strains
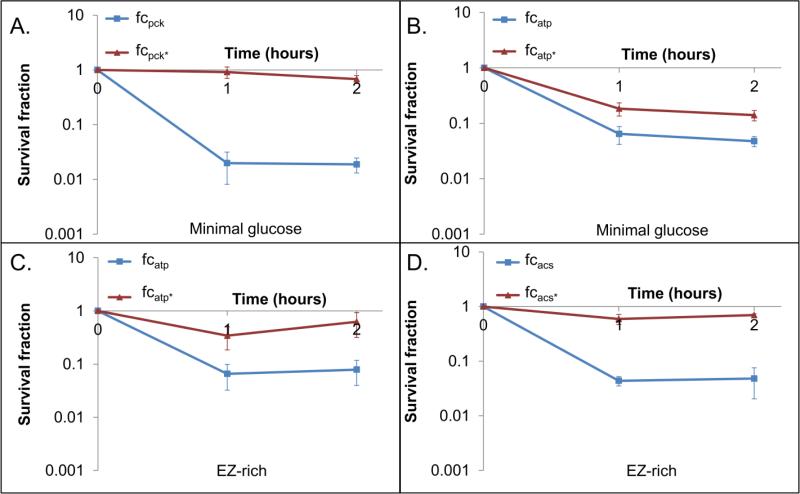
Based on previous studies, we had hypothesized that futile cycling would be an effective means to increase sensitivity toward oxidants. One study showed that inefficiencies in ATP production or usage resulted in increased endogenous production of ROS and increased sensitivity to oxidants (Brynildsen et al., 2013). By increasing the basal H2O2 production, we suspect the cells are closer to their threshold tolerance, beyond which they are unable to detoxify oxidants or repair the damage they cause. Another study demonstrated that futile cycling reduced intracellular ATP (Koebmann et al., 2002), and with knowledge that a number of DNA repair systems that process oxidative damage rely on ATP [e.g. DinG (Voloshin et al., 2003), Mfd (Selby and Sancar, 1995), RecA (Galletto et al., 2006), Uvr (Orren and Sancar, 1989), Vsr (Heinze et al., 2009)], we rationalized that reduced ATP could also yield sensitivity to oxidative stress. Upon confirming that the endogenous H2O2 production increased and ATP levels decreased in cycling strains, we sought to test whether oxidant sensitivity was affected by cycling. To do this, we treated cells with the model oxidant H2O2 (Diaz-Acosta et al., 2006; Martinez and Kolter, 1997; Tamarit et al., 1998; Tkachenko et al., 2001), and measured survival as a function of time. As depicted in Figure 4, we observed significantly increased sensitivity for all strains with active cycling in comparison to their controls. Given that different media were used and different areas of metabolism were targeted for cycling, these data suggest a highly general relationship between oxidative stress tolerance and futile cycling that is independent of the local metabolic network in which the cycle is acting. The increased oxidant sensitivity would likely originate from phenotypic changes shared by the different systems, which suggests that depressed growth, increased ROS production, and/or reduced ATP levels would be responsible. In addition, more generally, increased H2O2 sensitivity suggests impaired H2O2 detoxification capabilities, enhanced oxidative damage, and/or impaired repair capabilities.
Figure 4. Effect of futile cycling on sensitivity to H2O2.
Induced cultures were treated with H2O2 concentrations that yield at least 90% reduction in CFUs in one strain (active or inactive cycle) within 2 hours. These concentrations were 4 mM H2O2 for fcpck in MOPS 10 mM glucose medium (A), 5 mM H2O2 for fcatp in MOPS 10 mM glucose medium (B), 2 mM H2O2 for fcatp in EZ-rich 10 mM glucose medium (C), and 1.5 mM H2O2 for fcacs in EZ-rich 10 mM glucose medium (D). These assays were performed in a wild-type MG1655 background. The p-value of the log transformed values was <0.05 by hour 2 for all cycles. Uninduced (Fig. S16) and untreated (Fig. S17) controls can be found in the Supplementary material.
3.6 Increased oxidant sensitivity is not due to decreased growth rate or detoxification capacity
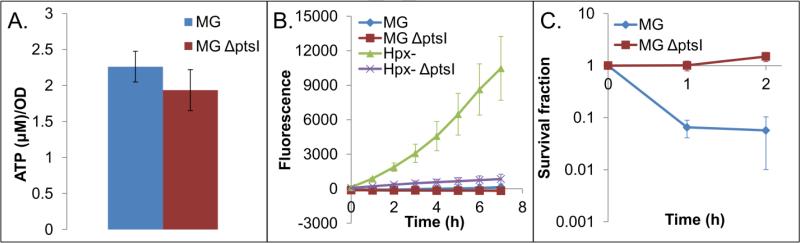
In an attempt to disentangle the origins of H2O2 sensitivity in futile-cycling strains, we first measured oxidative stress tolerance in a strain experiencing nutrient-dependent growth inhibition in comparison to an uninhibited control. Specifically, we explored the effects of nutrient deprivation on ATP levels, H2O2 production, and sensitivity to H2O2 using a ΔptsI mutant of MG1655 (Fig. 5). MG1655 lacking PTS enzyme I (ptsI) is unable to metabolize glucose (Patrick et al., 2007), so transferring this strain to minimal glucose medium will result in growth arrest by nutrient deprivation, rather than futile cycling. As depicted in Figure 5, we did not observe a significant difference in ATP concentration (Fig. 5A), and we did not observe significant H2O2 production in Hpx– ΔptsI when compared with its MG1655 control (Fig. 5B), which was anticipated due to the lack of glucose consumption. When ΔptsI and wild-type were assayed for H2O2 tolerance, we observed that ΔptsI was significantly more tolerant to H2O2. Interestingly, while the effects on growth rate qualitatively mirror those of futile cycling strains, the ATP depletion and endogenous H2O2 production are absent, and the effect on sensitivity is reversed (p<0.05).
Figure 5. EffMs of growth inhibition through nutrient deprivation.
ATP levels (A), H202 production (B), and sensitivity to oxidative stress in a nutrient deprived strain (MG ΔptsI mutant transferred to minimal glucose medium) (C) (p<0.05). In the sensitivity assay, cultures were treated with 12 mM H2O2, a concentration that led to at least a 90% reduction in CFUs for one of the strains. At least 3 biological replicates were performed for each experiment, and error bars represent the standard error of the mean. Untreated sensitivity assay controls can be found in Fig. S18.
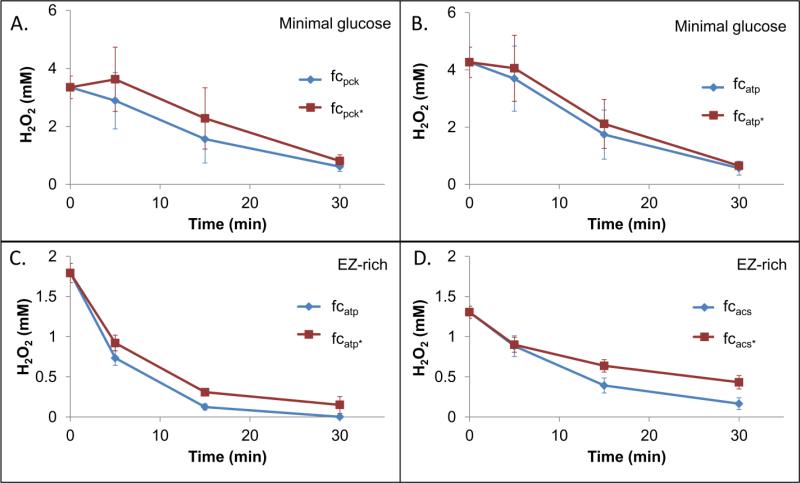
To test whether futile cycling enhanced H2O2 sensitivity through inhibition of detoxification capabilities, we prepared samples identically to the sensitivity assays and measured H2O2 concentration as a function of time. None of the actively cycling strains removed H2O2 more slowly than their inactive controls (Fig. 6). Collectively, these data show the increase in sensitivity to H2O2 is not attributable to growth inhibition or a reduced ability to detoxify H2O2. Rather, the data suggests that increased damage and/or impaired repair mechanisms caused the heightened oxidant sensitivity, which is consistent with the low ATP levels and enhanced endogenous ROS production that is present in these strains.
Figure 6. H2O2 removal by cells.
H2O2 concentration in the presence of cells (OD600 = 0.2) was measured at various time points. Initial concentrations were 4 mM H2O2 for fcpck in MOPS 10 mM glucose medium (A), 5 mM H2O2 for fcatp in MOPS 10 mM glucose medium (B), 2 mM H2O2 for fcatp in EZ-rich 10 mM glucose medium (C), and 1.5 mM H2O2 for fcacs in EZ-rich 10 mM glucose medium (D). Data points represent three biological replicates with error bars showing the standard error of the mean.
4. Discussion
Oxidative stress is a common challenge experienced by all microbes. O2− and H2O2 are generated as unintended byproducts of endogenous metabolism, but they are also used as weapons in interspecies competition. For example, ROS are a major defense used by phagocytic cells of the innate immune system to kill infecting pathogens (Flannagan et al., 2009), and they are also used by microbes during niche competitions, as is the case of Streptococcus pneumonia generating H2O2 to stimulate prophage induction and cell death in Staphylococcus aureus (Selva et al., 2009). To cope with oxidative stress, bacteria have developed sophisticated responses that include the induction of detoxification and repair systems such as catalase and alkyl hydroperoxidase (Cosgrove et al., 2007; Harris et al., 2003; Hebrard et al., 2009; La Carbona et al., 2007; Li et al., 1998; Manca et al., 1999; Yoshpe-Purer and Henis, 1976), superoxide dismutase (Bakshi et al., 2006), and DNA repair systems [e.g. repair of single-strand breaks (Ananthaswamy and Eisenstark, 1977; Carlsson and Carpenter, 1980), and base excision repair (Schalow et al., 2011)]. In addition, bacteria rewire their metabolism to provide precursors to synthesize detoxification and repair systems [e.g. amino acids, nucleotides], reducing power to drive detoxification systems [e.g., NADH (Seaver and Imlay, 2001)], and energy and material to repair or re-synthesize damaged biomolecules [e.g., ATP (Galletto et al., 2006; Heinze et al., 2009; Orren and Sancar, 1989; Selby and Sancar, 1995; Voloshin et al., 2003), amino acids, nucleotides (Schalow et al., 2011)]. These dependencies suggest that targeting metabolism would be an effective way to sensitize bacteria to oxidants.
Recently, we discovered that metabolism could be rationally modified to increase endogenous ROS production in E. coli, despite the fact that a high degree of uncertainty exists with regard to the enzymes responsible for ROS production under normal growth conditions (Brynildsen et al., 2013). This uncertainty was addressed using an ensemble modeling approach, and it was found that increased ROS production correlated with increased sensitivity toward oxidative stress as measured by bacterial killing (Brynildsen et al., 2013). An observation from that study was that mutations that caused inefficiencies in ATP production or usage increased ROS production and sensitivity to oxidative stress. Inspired by that result and recognizing that ATP depletion could reduce the activity of DNA repair systems (Galletto et al., 2006; Heinze et al., 2009; Orren and Sancar, 1989; Selby and Sancar, 1995; Voloshin et al., 2003), we hypothesized that futile cycling would result in a two-pronged attack on bacterial oxidant tolerance. With depleted ATP levels, the cell's ability to repair the damage caused by oxidative stress was likely to be compromised (Galletto et al., 2006; Heinze et al., 2009; Orren and Sancar, 1989; Selby and Sancar, 1995; Voloshin et al., 2003), whereas increased endogenous H2O2 production would reduce the cells’ ability to cope with additional oxidative stress.
To test our hypothesis, we used an ensemble modeling approach to predict that futile cycling would yield a decrease in growth rate, an increase in O2 consumption per biomass, and an increase in H2O2 production relative to biomass produced. Both the growth rate and O2 consumption predictions were consistent with previous simulations of futile cycles (Izallalen et al., 2008; Sun et al., 2009), whereas the prediction of increased H2O2 production was unique to the current study due to the use of an ROS model. We confirmed these predictions experimentally with four distinct futile cycling systems, which operate in different areas of metabolism and/or different media conditions. Consistent with previous investigations, futile cycling was found to depress growth rate (Chao and Liao, 1994; Izallalen et al., 2008; Koebmann et al., 2002), increase O2 consumption (Chao and Liao, 1994; Izallalen et al., 2008; Patnaik et al., 1992), and lower ATP concentrations (Izallalen et al., 2008; Koebmann et al., 2002). In addition, futile cycling was confirmed to yield more H2O2 production per cell division. Further, we found that our hypothesis that futile cycling's two-pronged effect would sensitize bacteria to oxidative stress was correct. We determined that this sensitivity was not attributable solely to growth-rate depression, since nutrient-deprived cells are more tolerant to H2O2 in comparison to their nutrient-supplied controls. Additionally, we found that futile cycling cells are not impaired in their ability to detoxify H2O2. Collectively, these results suggest that the increased oxidant sensitivity in futile cycling strains is caused by increased damage and/or impaired repair mechanisms.
Futile cycling is a process that organisms largely avoid through the use of transcriptional and allosteric regulation (Liao et al., 1994; Xu et al., 2012). Beyond the inherent loss of energy and resulting growth-rate depression, we have shown that futile cycling increases endogenous ROS production, and therefore, may facilitate mutagenesis through the accumulation of DNA damage. This fundamental knowledge on the relationship between metabolism and oxidative stress presented here could have implications for understanding host-pathogen interactions, as well as developing novel anti-infectives. With the ever increasing failure rate of antibiotics (Taubes, 2008), alternative approaches such as engineered bacteriophage have become increasingly attractive. This approach has been explored with an enzyme that disperses biofilms (Lu and Collins, 2007) and a repressor of genes involved in the SOS response (Lu and Collins, 2009). Analogously, bacteriophage engineered to stimulate futile cycles could be used to sensitize bacteria to oxidants, which are important antimicrobials of the immune system.
Supplementary Material
Highlights.
A method for identifying experimentally-tractable futile cycles is presented.
Futile cycling elevates endogenous reactive oxygen species production.
Futile cycling increases bacterial sensitivity to oxidative stress.
Acknowledgments
This work was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI105342, the National Science Foundation Division of Materials Research through the Princeton University MRSEC (NSF-DMR 0819860), and with start-up funds from Princeton University. We would like to acknowledge Sara Sukenik and Jeff Morell, who did preliminary work on the project, as well as Sophia Leonard, who performed the Hpx– clearance assays to demonstrate that Hpx– retained no catalase activity. We acknowledge the Lidow Senior Thesis Fund for allowing them to explore their projects. We thank Christina J. DeCoste and the Flow Cytometry Resource Facility for assistance with flow cytometry experiments. We thank Michael Kohanski for providing the ΔahpCF mutation required to generate the Hpx– strain. In addition, we thank Professors Christodoulos Floudas, Joshua Rabinowitz, and A. James Link for their suggestions on this project, and Jonathan Robinson for his comments on the manuscript.
Abbreviations
- Acs
acetyl-coA synthetase
- Pta
phosphate acetyltransferase
- Ack
acetate kinase
- Pck
phosphoenolpyruvate carboxykinase
- Ppc
phosphoenolpyruvate carboxylase
- fcacs
cycling engaged by overexpression of acetyl-coA synthetase
- fcacs*
catalytically inactive control for fcacs
- fcatp
cycling engaged by overexpression of the soluble subunits of ATP synthase
- fcatp*
catalytically inactive control for fcatp
- fcpck
cycling engaged by overexpression of phosphoenolpyruvate carboxykinase
- fcpck*
catalytically inactive control for fcpck
- ROS
reactive oxygen species
- Hpx–
Escherichia coli strain MG1655 ΔkatE ΔkatG ΔahpCF
- amp
ampicillin
- kan
kanamycin
- IPTG
isopropyl β-D-1-thiogalactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad Z, Senior AE. Mutagenesis of residue betaArg-246 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F1-ATPase. J Biol Chem. 2004;279:31505–13. doi: 10.1074/jbc.M404621200. [DOI] [PubMed] [Google Scholar]
- Ananthaswamy HN, Eisenstark A. Repair of hydrogen peroxide-induced single-strand breaks in Escherichia coli deoxyribonucleic acid. Journal of bacteriology. 1977;130:187–91. doi: 10.1128/jb.130.1.187-191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology. 2006;2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. Journal of bacteriology. 2006;188:6443–8. doi: 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun. 2004;72:408–13. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nature biotechnology. 2013;31:160–5. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, Carpenter VS. The Reca+ Gene-Product Is More Important Than Catalase and Superoxide-Dismutase in Protecting Escherichia-Coli against Hydrogen-Peroxide Toxicity. Journal of bacteriology. 1980;142:319–321. doi: 10.1128/jb.142.1.319-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YP, Liao JC. Metabolic responses to substrate futile cycling in Escherichia coli. J Biol Chem. 1994;269:5122–6. [PubMed] [Google Scholar]
- Chen J, Brevet A, Fromant M, Leveque F, Schmitter JM, Blanquet S, Plateau P. Pyrophosphatase is essential for growth of Escherichia coli. Journal of bacteriology. 1990;172:5686–9. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. Journal of bacteriology. 2007;189:1025–35. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Palsson BO. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J Biol Chem. 2002;277:28058–64. doi: 10.1074/jbc.M201691200. [DOI] [PubMed] [Google Scholar]
- Crawford MA, Aylott CV, Bourdeau RW, Bokoch GM. Bacillus anthracis toxins inhibit human neutrophil NADPH oxidase activity. J Immunol. 2006;176:7557–65. doi: 10.4049/jimmunol.176.12.7557. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MJ. The oxidative environment and protein damage. Bba-Proteins Proteom. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- De Groote MA, Ochsner UA, Shiloh MU, Nathan C, McCord JM, Dinauer MC, Libby SJ, Vazquez-Torres A, Xu Y, Fang FC. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13997–4001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nature reviews. Microbiology. 2010;8:117–28. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- Diaz-Acosta A, Sandoval ML, Delgado-Olivares L, Membrillo-Hernandez J. Effect of anaerobic and stationary phase growth conditions on the heat shock and oxidative stress responses in Escherichia coli K-12. Archives of microbiology. 2006;185:429–38. doi: 10.1007/s00203-006-0113-9. [DOI] [PubMed] [Google Scholar]
- Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends in biotechnology. 2006;24:530–6. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nature reviews. Microbiology. 2004;2:820–32. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular systems biology. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nature reviews. Microbiology. 2009;7:355–66. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–8. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92:3007–17. [PubMed] [Google Scholar]
- Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiol-Sgm. 2003;149:665–672. doi: 10.1099/mic.0.26012-0. [DOI] [PubMed] [Google Scholar]
- Hebrard M, Viala JPM, Meresse S, Barras F, Aussel L. Redundant Hydrogen Peroxide Scavengers Contribute to Salmonella Virulence and Oxidative Stress Resistance. Journal of bacteriology. 2009;191:4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze RJ, Giron-Monzon L, Solovyova A, Elliot SL, Geisler S, Cupples CG, Connolly BA, Friedhoff P. Physical and functional interactions between Escherichia coli MutL and the Vsr repair endonuclease. Nucleic Acids Res. 2009;37:4453–63. doi: 10.1093/nar/gkp380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–65. [PubMed] [Google Scholar]
- Imlay JA, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. Journal of bacteriology. 1986;166:519–27. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izallalen M, Mahadevan R, Burgard A, Postier B, Didonato R, Jr., Sun J, Schilling CH, Lovley DR. Geobacter sulfurreducens strain engineered for increased rates of respiration. Metabolic engineering. 2008;10:267–75. doi: 10.1016/j.ymben.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Jabalquinto AM, Laivenieks M, Gonzalez-Nilo FD, Yevenes A, Encinas MV, Zeikus JG, Cardemil E. Evaluation by site-directed mutagenesis of active site amino acid residues of Anaerobiospirillum succiniciproducens phosphoenolpyruvate carboxykinase. Journal of protein chemistry. 2002;21:393–400. doi: 10.1023/a:1021178432158. [DOI] [PubMed] [Google Scholar]
- Jang SJ, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerlich A, Hammel M, Nigon F, Chapman MJ, Schaur RJ. Kinetics of tryptophan oxidation in plasma lipoproteins by myeloperoxidase-generated HOCl. European journal of biochemistry / FEBS. 2000;267:4137–43. doi: 10.1046/j.1432-1327.2000.01449.x. [DOI] [PubMed] [Google Scholar]
- Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. Journal of bacteriology. 2002;184:3909–16. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov S, Imlay JA. Two sources of endogenous hydrogen peroxide in Escherichia coli. Molecular microbiology. 2010;75:1389–401. doi: 10.1111/j.1365-2958.2010.07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Molecular microbiology. 2007;66:1148–63. doi: 10.1111/j.1365-2958.2007.05987.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Kelley C, Collins F, Rouse D, Morris S. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. The Journal of infectious diseases. 1998;177:1030–5. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]
- Liao JC, Chao YP, Patnaik R. Alteration of the biochemical valves in the central metabolism of Escherichia coli. Annals of the New York Academy of Sciences. 1994;745:21–34. doi: 10.1111/j.1749-6632.1994.tb44361.x. [DOI] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4629–34. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Levine RL. Methionine in proteins defends against oxidative stress. Faseb J. 2009;23:464–72. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Paul S, Barry CE, Freedman VH, Kaplan G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infection and immunity. 1999;67:74–79. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. Journal of bacteriology. 1997;179:5188–94. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobius K, Arias-Cartin R, Breckau D, Hannig AL, Riedmann K, Biedendieck R, Schroder S, Becher D, Magalon A, Moser J, Jahn M, Jahn D. Heme biosynthesis is coupled to electron transport chains for energy generation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10436–10441. doi: 10.1073/pnas.1000956107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath KA, Ngo EO, Hebbel RP, Croatt AJ, Zhou B, Nutter LM. alpha-Ketoacids scavenge H2O2 in vitro and in vivo and reduce menadione-induced DNA injury and cytotoxicity. The American journal of physiology. 1995;268:C227–36. doi: 10.1152/ajpcell.1995.268.1.C227. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. Journal of bacteriology. 1974;119:736–47. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5237–41. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik R, Roof WD, Young RF, Liao JC. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. Journal of bacteriology. 1992;174:7527–32. doi: 10.1128/jb.174.23.7527-7532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Multicopy suppression underpins metabolic evolvability. Molecular biology and evolution. 2007;24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA. Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J Biol Chem. 1994;269:26066–75. [PubMed] [Google Scholar]
- Schalow BJ, Courcelle CT, Courcelle J. Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo. Journal of molecular biology. 2011;410:183–93. doi: 10.1016/j.jmb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BO. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nature protocols. 2011;6:1290–307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. Journal of bacteriology. 2001;183:7173–81. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J Biol Chem. 2004;279:48742–50. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–9. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, Novick RP, Penades JR. Killing niche competitors by remote-control bacteriophage induction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1234–8. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemsen DW, Kirpotina LN, Jutila MA, Quinn MT. Inhibition of the human neutrophil NADPH oxidase by Coxiella burnetii. Microbes and infection / Institut Pasteur. 2009;11:671–9. doi: 10.1016/j.micinf.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Acetyl-coenzyme A synthetase (AMP forming). Cellular and molecular life sciences : CMLS. 2004;61:2020–30. doi: 10.1007/s00018-004-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storkey C, Davies MJ, Pattison DI. Reevaluation of the rate constants for the reaction of hypochlorous acid (HOCl) with cysteine, methionine, and peptide derivatives using a new competition kinetic approach. Free radical biology & medicine. 2014 doi: 10.1016/j.freeradbiomed.2014.04.024. [DOI] [PubMed] [Google Scholar]
- Sun J, Sayyar B, Butler JE, Pharkya P, Fahland TR, Famili I, Schilling CH, Lovley DR, Mahadevan R. Genome-scale constraint-based modeling of Geobacter metallireducens. BMC systems biology. 2009;3:15. doi: 10.1186/1752-0509-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit J, Cabiscol E, Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–32. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- Tauber AI, Pavlotsky N, Lin JS, Rice PA. Inhibition of human neutrophil NADPH oxidase by Chlamydia serovars E, K, and L2. Infection and immunity. 1989;57:1108–12. doi: 10.1128/iai.57.4.1108-1112.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321:356–61. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Tkachenko A, Nesterova L, Pshenichnov M. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Archives of microbiology. 2001;176:155–7. doi: 10.1007/s002030100301. [DOI] [PubMed] [Google Scholar]
- Voloshin ON, Vanevski F, Khil PP, Camerini-Otero RD. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J Biol Chem. 2003;278:28284–93. doi: 10.1074/jbc.M301188200. [DOI] [PubMed] [Google Scholar]
- Xu YF, Amador-Noguez D, Reaves ML, Feng XJ, Rabinowitz JD. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nature chemical biology. 2012;8:562–8. doi: 10.1038/nchembio.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshpe-Purer Y, Henis Y. Factors affecting catalase level and sensitivity to hydrogen peroxide in Escherichia coli. Applied and environmental microbiology. 1976;32:465–9. doi: 10.1128/aem.32.4.465-469.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.