Abstract
The homozygous repeated epilation (Er/Er) mouse mutant of the gene encoding 14-3-3σ displays an epidermal phenotype characterized by hyperproliferative keratinocytes and undifferentiated epidermis. Heterozygous Er/+ mice develop spontaneous skin tumors and are highly sensitive to tumor-promoting DMBA/TPA induction. The molecular mechanisms underlying 14-3-3σ regulation of epidermal proliferation, differentiation, and tumor formation have not been well elucidated. In the present study, we found that Er/Er keratinocytes failed to sequester Yap1 in the cytoplasm, leading to its nuclear localization during epidermal development in vivo and under differentiation-inducing culture conditions in vitro. In addition, enhanced Yap1 nuclear localization was also evident in DMBA/TPA-induced tumors from Er/+ skin. Furthermore, shRNA knockdown of Yap1 expression in Er/Er keratinocytes inhibited their proliferation, suggesting that YAP1 functions as a downstream effector of 14-3-3σ controlling epidermal proliferation. We then demonstrated that keratinocytes express all seven 14-3-3 protein isoforms, some of which form heterodimers with 14-3-3σ, either full-length WT or the mutant form found in Er/Er mice. However Er 14-3-3σ does not interact with Yap1, as demonstrated by co-immunoprecipitation. We conclude that Er 14-3-3σ disrupts the interaction between 14-3-3 and Yap1, thus fails to block Yap1 nuclear transcriptional function, causing continued progenitor expansion and inhibition of differentiation in Er/Er epidermis.
Introduction
Epithelial differentiation has been intensively studied, and the process can be separated into two general steps: 1) the generation of proliferating progenitors from resident stem cells and 2) the subsequent differentiation of these progenitors into terminally differentiated epithelial cells. The balance between cell proliferation and differentiation must be tightly controlled, and the timing for initiation of progenitor proliferation is critical to precisely defining the number of cells and the topology of the tissue. Signals for proliferation arrest and differentiation are transmitted through cell-cell contacts that sense the cellular density or tension when the progenitor expansion covers a specific field and reach to a distinct density. The Hippo pathway plays a key role in controlling tissue size through regulation of cell proliferation by sensing and responding to cell density. Most of the signaling components in the Hippo pathway were initially identified by genetic screens in Drosophila (Pan, 2007) that were expanded to mammalian cells thereafter (Zhao et al., 2010). The Mst/Lats kinase cascade has been shown to relay membrane-bound signaling to nuclear transcriptional activity by phosphorylating Yap/Taz proteins, which are retained in the cytoplasm by binding to 14-3-3 proteins, which blocks them from nuclear translocation and transcriptional coactivity (Basu et al., 2003; Kanai et al., 2000). As a downstream effector of the Hippo kinase cascade and a transcriptional co-activator, Yap1 has been implicated in cell proliferation and the control of organ size. Yap1 is expressed in many tissues and cell types and interacts with multiple DNA-binding transcription factor partners, such as p73, Runx2, and TEDA. Genetic studies have demonstrated that Yap1 increases organ size and expands the progenitor cell population in liver, skin, and heart (Camargo et al., 2007; Schlegelmilch et al., 2011; Zhang et al., 2011; Zhou et al., 2009). The cell density-dependent activation of Hippo cascades and the resulting inactivation of Yap1/Taz activity are required for cell–cell contact inhibition in culture. Yap1 is critical for skin development and proliferation (Schlegelmilch et al., 2011; Zhang et al., 2011), and expression of a constitutively active Yap1 in epidermis induced an expansion of undifferentiated progenitor cells, accompanied by the disappearance of most differentiation makers. Transplantation of Yap1 transgenic skin into nude mice developed tumor types resembling squamous cell carcinoma (Schlegelmilch et al., 2011). By contrast, deletion of Yap1 driven by an epidermis-specific promoter diminished skin expansion (Schlegelmilch et al., 2011; Zhang et al., 2011). A recent study has suggested that Yap1 acts as a signaling molecule downstream of α-catenin, which is associated with a membrane sensing mechanism for cell density, suggesting that cell density controls the progenitor population and tissue expansion by modulating Yap1 function in epidermis (Schlegelmilch et al., 2011).
14-3-3 proteins are highly conserved throughout all eukaryotic organisms and modulate cellular function by interacting with more than 200 proteins (van Heusden, 2005). They share similar structures, can form homodimers or heterodimers, and are often functionally complementary to each other by binding to the same interacting partners, mostly phosphorylated proteins. Among seven isoforms in mammalian cells, 14-3-3σ appears to play a unique role in regulating epithelial cell proliferation and differentiation, due to its restricted expression, primarily in stratified epithelium cells (Lodygin and Hermeking, 2006; Wilker et al., 2005). We and others previously demonstrated that the 14-3-3σ gene is mutated in repeated epilation (Er) mice, and this mutation causes a complete block of epidermal differentiation (Herron et al., 2005; Li et al., 2005). Homozygous Er/Er pups died soon after birth and were characterized by severe dehydration due to lack of skin barrier protection (Herron et al., 2005; Li et al., 2005). Heterozygous Er/+ mice have a normal life span, but display semi-dominant phenotypes such as abnormal hair follicles, corneal epithelium plaque, and spontaneous skin tumors (Li et al., 2011; Lu et al., 2011; Xin et al., 2010). At the molecular level, the mutated 14-3-3σ protein in Er mice retains an intact functional N-terminal dimerization domain but lacks the C-terminal ligand-binding domain (Herron et al., 2005; Li et al., 2005). Such a domain structure, which remains in the truncated 14-3-3σ mutant protein, is considered critical for the phenotypes observed in Er/Er and Er/+ mice, since the mutant protein can potentially form stable heterodimers with other members of the 14-3-3 protein family and block their functions (Li et al., 2005; Xin et al., 2010).
Regulation of Yap1 cellular localization by 14-3-3 has been demonstrated (Hammond et al., 2012; Zhao et al., 2007), but whether Yap1 functions as a downstream target of 14-3-3σ in the regulation of epidermal proliferation and differentiation remains unknown. To fill this gap, we examined the relationship between Yap1 nuclear distribution and the proliferation phenotypes in Er/Er skin and keratinocytes and found enhanced nuclear localization of the Yap1 protein in the 14-3-3σ Er mutant keratinocytes. The unrestricted keratinocyte proliferation and expansion of Er/Er keratinocytes could be reversed by knocking down Yap1 mRNA using a shRNA lentiviral vector. We further demonstrated that the 14-3-3σ isoform could interact with Yap1 and other 14-3-3 protein isoforms, whereas the Er-truncated isoform of 14-3-3σ not only failed to interact with Yap1 and but also disrupted the interaction of other normal 14-3-3 isoforms with Yap1. These observations suggest that 14-3-3σ-mediated Yap1 cellular localization and function are essential for limiting epidermal progenitor cell growth and proliferation.
Results
Increased nuclear Yap1 localization in Er/Er epidermis and chemical-induced tumors in Er/+ mice
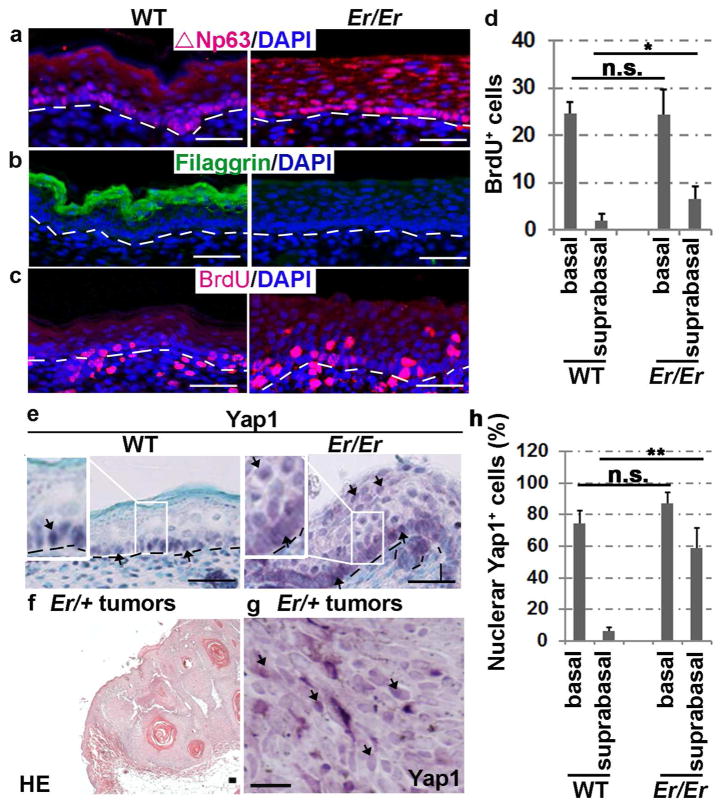
Given that 14-3-3 proteins are critical for excluding phosphorylated Yap1 from nuclei, we examined the cellular localization of Yap1 in Er/Er mouse skin to ask whether the Er 14-3-3σ mutation failed to retain Yap1 in the cytoplasm. E18.5 Er/Er embryos developed a multilayered, thicker epithelium lacking the cornified layer compared with well-differentiated WT skin (Li et al., 2005). Immunostaining with the antibody that specifically recognizes the C-terminus of 14-3-3σ, which was deleted in the Er mutant, showed that 14-3-3σ is mainly expressed in the differentiated suprabasal layers of the WT epidermis, but not in the Er/Er mutant epidermis (Li et al., 2005). The skin progenitor cells, as immunostained with ΔNp63α, were confined to the basal layer in the control epidermis but strongly expanded into the suprabasal layers in the mutant skin (Fig. 1a). A lack of the differentiation marker filaggrin was clearly evident in the mutant epidermis (Fig. 1b). The expansion of progenitor cells was associated with increased BrdU incorporation in the mutant suprabasal layers (Fig. 1c and 1d). Notably, Yap1 protein was clearly present in the nuclei of the mutant suprabasal layer, in contrast to the negative Yap1 nuclear staining in the WT suprabasel layer, although it was immunostained positively in the basal cell nuclei in both genotypes (Fig. 1e and 1h). Collectively, these results indicate that Yap1 is continuously localized in the nuclei from basal to suprabasal cells in the Er/Er epidermis, and this is correlated with an increase in the epidermal progenitor cell population and a lack of terminal differentiation.
Figure 1. Enriched nuclear localization of Yap1 in Er/Er epidermis and induced skin tumors.
(a–c) Immunofluorescence staining for ΔNp63 (a), filaggrin (b) and BrdU (c) in the E18.5 Er/Er epidermis (right) compared with the WT control (left). (d) Quantification of BrdU+ cells. The number of BrdU+ cells per 500 μm of fixed length parallel to the epidermis surface was counted. Two-tail t-test: *p<0.05, N=5 embryos. n.s., not significant. (e) Immunohistochemical staining of Yap1. H&E staining (f) and Yap1 cellular localization (brown) detected by immunohistochemical staining (g) of DMBA/TPA-induced tumors derived from Er/+ mice. The scale bars represent 50 μm in (a–f) and 150 μm in (g). The arrows indicate Yap1-positive cells in (e) and (g). (h) Quantification of nuclear Yap1-positive cells. Two-tail t-test: **p<0.01, N=3 embryos.
We have previously shown that Er/+ mice exhibit increased formation of skin papillomas and squamous cell carcinomas upon induction with 7,12-dimethylbenzanthracene (DMBA)/12-O-tetradecanoyl-phorbol-13-acetate (TPA) (Li et al., 2011). 14-3-3σ is therefore considered to have a tumor-suppressor function. Given that Yap1 is well known for its oncogenic function, and 14-3-3 proteins inhibit Yap1 transcriptional functions by retaining it in the cytoplasm, we then asked whether the skin tumors induced in Er mice have increased Yap1 nuclear localization. This is exactly what we observed: Yap1 was dramatically enriched in the nuclei of most tumor cells, suggesting a strong correlation and a likely contribution of Yap1 nuclear activity to the tumor cell growth seen in Er/+ mice (Fig. 1f–g).
Calcium-induced cytoplasmic retention of Yap1 protein is impaired in Er/Er keratinocytes
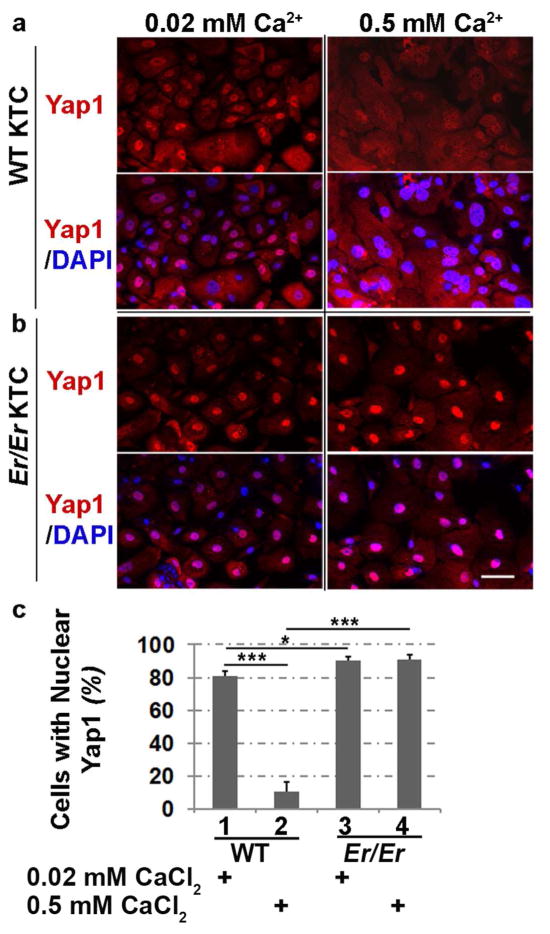
The enhanced Yap1 nuclear localization in Er/Er epidermis led us to further analyze Yap1 expression in primary cultured keratinocytes isolated from E18.5 WT or Er/Er embryos. In primary WT control keratinocytes, Yap1 was distributed throughout both cytoplasm and nucleus but was preferentially concentrated in the nucleus in the undifferentiated cells under low-calcium culture conditions, whereas in the differentiated cells, upon induction with a high concentration of calcium, nuclear Yap1 staining signal was lost, which was accompanied by enhanced cytoplasmic staining (Fig. 2a and 2c). Compared with the Yap1 staining pattern in WT cells, Yap1 was mainly concentrated in the mutant nuclei, negligibly staining the cytoplasm, and such strong nuclear localization in the Er/Er keratinocytes remained predominant throughout the entire differentiation induction period in high-calcium culture (Fig. 2b and 2c). This data indicates that Yap1 cytoplasmic retention can’t be induced under high-calcium differentiation culture conditions in Er/Er keratinocytes that harbor the truncated 14-3-3σ mutation.
Figure 2. Increased nuclear localization of Yap1 in Er/Er keratinocytes.
Yap1 cellular localization (red) was shown by immunostaining in primary cultured keratinocytes isolated from E18.5 WT (a) and Er/Er (b) skin. The keratinocytes were cultured for 48 h in SFM medium containing 0.02 mM Ca2+ (maintenance medium) or 0.5 mM Ca2+ (differentiation medium) before immunostaining. DAPI (blue) was used for nuclear counterstaining. The scale bars represent 50 μm. (c) Quantification of the number of Yap1-positive cells. The cells were immunostained with Yap1-specific antibody and calculated as percentage of total DAPI-labeled cells. The error bars represent the standard deviation from three independent experiments. The statistical significance was analyzed by two-tailed Student’s t-test. *p≤0.05 and ***p≤0.005, n=3.
Yap1 function is correlated with its nuclear localization
To investigate whether Yap1 nuclear enrichment in Er/Er keratinocytes led to elevated expression of its target genes, we performed qPCR to assay the mRNA levels of the genes Cyr61, Ctgf, Zeb1, and Snai2, which were previously shown to be Yap1 transcriptional targets (Heallen et al., 2011; Shao et al., 2014). They exhibited reduced expression in differentiating WT keratinocytes (Fig. 3a), which was correlated with the Yap1 cytoplasmic retention pattern shown above. By contrast, the high calcium-induced reduction in their expression level did not occur in the Er/Er keratinocytes, consistent with the fact that high calcium-induced cytoplasmic retention of Yap1 protein was impaired due to the absence of functional 14-3-3σ in Er mutant cells (Fig. 3a). In addition, Cyr61, Zeb1, and Snai2 expression was significantly higher in Er/Er keratinocytes than in WT keratinocytes. To exclude the possibility that the increased transcription of Yap1 target genes was caused by increased Yap1 expression, we performed quantification by qPCR for its mRNA level (Fig. 3a) and by Western blotting for its protein level (Fig. 3b). Both showed indistinguishable Yap1 expression between high-and low-calcium culture conditions or between WT and Er/Er keratinocytes, suggesting that Yap1 activity is mainly regulated through its cellular localization. Furthermore, we examined phospho-Yap1 and detected only slightly reduction of Yap1Ser127 in the high calcium induced Er/Er keratinocytes compared to WT keratinocytes under the same conditions (Fig. 3b).
Figure 3. Enhanced expression of Yap1 target genes in Er/Er keratinocytes.
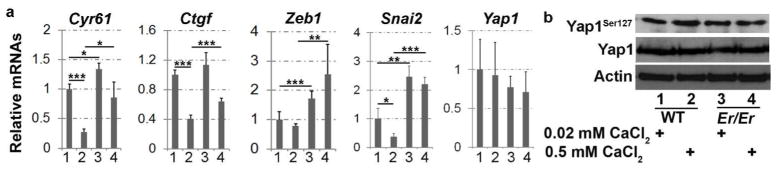
(a) qPCR analysis of the transcription of Yap1 and Yap1 transcriptional targets. Both WT and Er/Er keratinocytes were cultured for 48 h in medium containing 0.02 mM Ca2+ or 0.5 Ca2+ mM before isolation of RNA for quantification of Cyr61, Ctgf, Zeb1, Snai2, and Yap1 expression. The comparative threshold cycle (CT) methods normalized to Gapdh was used to analyze relative changes in gene expression. Data are expressed as mean±SD for three independent experiments. The statistical significance of the differences between means was assessed by a two-tailed Student’s t-test. *P≤0.05, **P≤0.01, and ***P≤0.005, n=3. (b) Yap1Ser127, Yap1, and actin protein expressions in keratinocyte samples shown by Western blotting. Data are representative of three independent experiments.
Proliferation phenotype in Er/Er keratinocytes depends on Yap1 activity
Yap1 mediates cell survival and expansion by its nuclear transcriptional activity. To investigate the functional role of Yap1 in promoting continuous cell proliferation of Er/Er keratinocytes under differentiation-induction conditions, we transduced Er/Er keratinocytes with Yap1 shRNA-expressing lentivirus to knockdown Yap1 expression and function. Primary WT and Er/Er keratinocytes were transduced with a scrambled shRNA or Yap1 shRNA for 2 days prior to culturing in 0.02 mM or 0.5 mM calcium-containing medium for an additional 2 days. Puromycin (2.5 μg/ml) was added to the medium to select for cells that stably express shRNA. Transduction of keratinocytes with Yap1 shRNA, but not control lentivirus, resulted in a dramatic reduction in Yap1 protein, as detected by immunostaining and Western blotting using a Yap1-specific antibody (Fig. 4a and 4c). The reduction in Yap1 protein was due to decreased Yap1 RNA expression (Fig. 4d). To investigate the effect of Yap1 knockdown on keratinocyte proliferation, we examined BrdU incorporation into the cells at 4 days after Yap1 shRNA lentivirus transduction. The proliferating cells were labeled by BrdU incorporation for 90 minutes before adding 4% PFA fixative. Blocking Yap1 expression dramatically reduced cell proliferation in both WT and Er/Er keratinocytes (Fig. 4b and 4e), which suggests that overgrowth of the Er mutant keratinocytes depends on Yap1 activity.
Figure 4. Inhibition of cell proliferation in both WT and Er/Er keratinocytes transduced with Yap1-specific shRNA lentivirus.
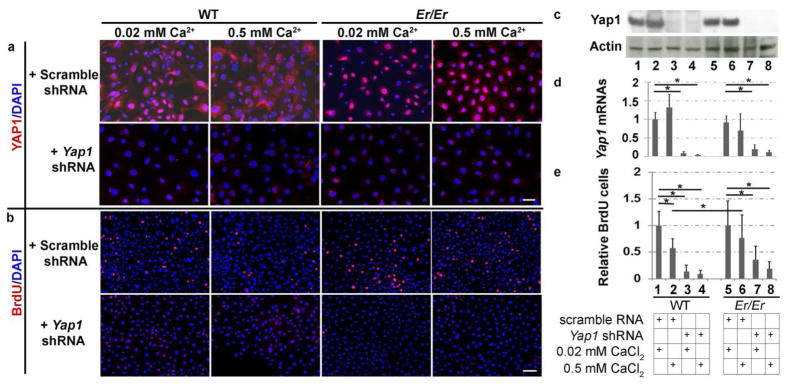
Immunofluorescence staining for Yap1 (a) or BrdU (b) were performed and counterstained with DAPI in the WT and Er/Er keratinocytes. Yap1 knockdown was confirmed by Western blotting with β-actin as loading control (c) and qPCR analysis with the CT method normalized to Actb (d). (e) Quantification of BrdU+ cells as shown in (b). Values are relative to the number of BrdU+ cells in keratinocytes transduced with scrambled shRNA lentivirus and cultured in 0.02 mM calcium-containing medium. Data are expressed as mean±SD, n=3. Two-tailed Student’s t-test was performed. *p < 0.005. The scale bars represent 50 μm in (a) and 100 μm in (b).
The truncated Er mutant 14-3-3σ interacts with other 14-3-3 isoforms and blocks interaction of other 14-3-3 members and Yap1
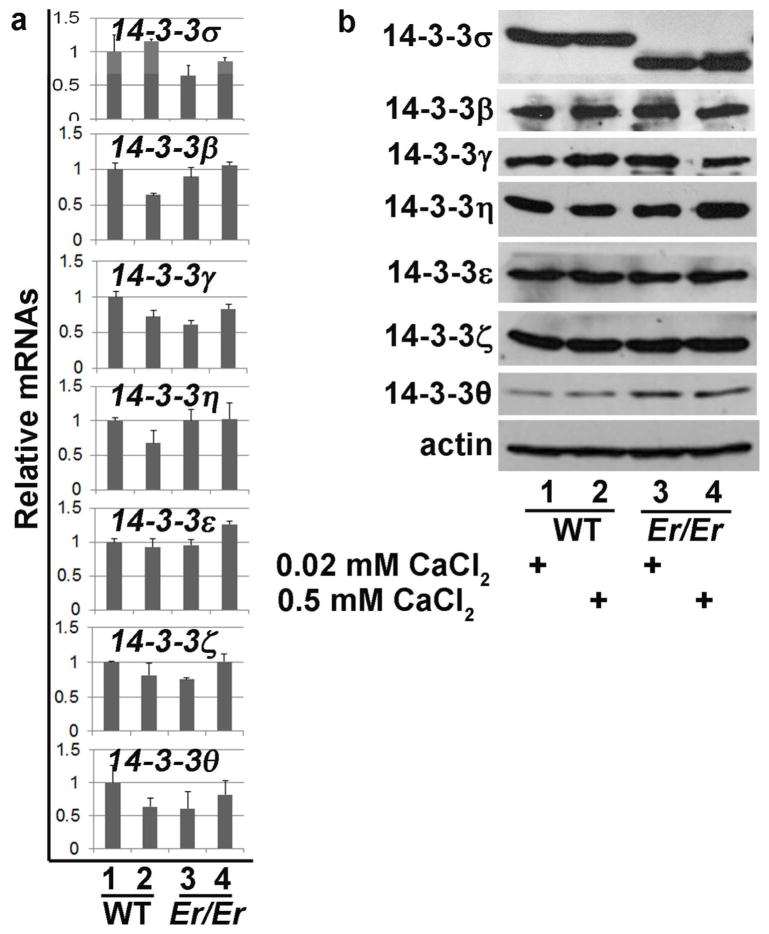
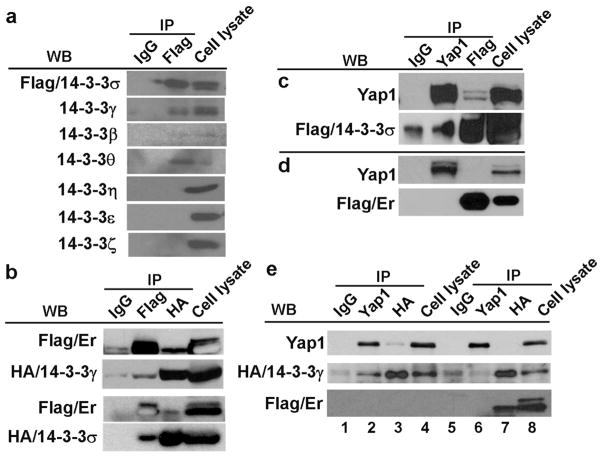
It was reported that 14-3-3σ knockout mice had a normal development and life span (Su et al., 2011), a distinct phenotype from what was seen in Er/Er mice, which died at birth due to severe dehydration caused by undifferentiated skin (Li et al., 2005). The remarkable difference in the phenotypes between Er/Er mice and 14-3-3σ knockout mice reflects the differences in the deletions of its functional domains. The truncated form of 14-3-3σ protein in Er/Er mice contains a functional dimerization domain that interacts and dimerizes with other 14-3-3 isoforms and consequently interferes with their functions, while 14-3-3σ protein in knockout mice was completely deleted (Su et al., 2011), leaving other 14-3-3 isoforms unaffected. We hypothesized that the lack of a skin phenotype in the 14-3-3σ KO mice was due to the presence of other 14-3-3 isoforms that compensated for the loss of 14-3-3σ. To test this hypothesis, we examined the keratinocyte expression of other isoforms in the 14-3-3 family by qPCR and Western blotting analyses. The qPCR results showed that all seven 14-3-3 isoforms were expressed by keratinocytes, and the mutation in 14-3-3σ in Er/Er keratinocytes did not cause a significant change in the mRNA levels of the other six 14-3-3 isoforms (Fig. 5a). Western blotting also detected all seven 14-3-3 isoforms in keratinocytes using the antibodies specific for each isoform, although a truncated Er form of 14-3-3σ was detected in the Er/Er samples (Fig. 5b). We further hypothesized that the truncated 14-3-3σ isoform in the Er mutant had a dominant-negative effect and suppressed the functions of other 14-3-3 isoforms in Er/Er cells. To test for heterodimerization of 14-3-3σ with other 14-3-3 isoforms in keratinocytes, we overexpressed flag-tagged 14-3-3σ and performed serial co-immunoprecipitation assays. 14-3-3σ and its associated proteins were pulled down from the transduced keratinocytes using anti-flag antibody, and the co-immunoprecipitated complex was subjected to Western blotting. Although all seven isoforms of 14-3-3 proteins were detected in the keratinocyte lysates (Fig. 5a), only the 14-3-3γ, 14-3-3β, and 14-3-3θ isoforms were co-immunoprecipitated with overexpressed 14-3-3σ and not the 14-3-3η, 14-3-3ε, and 14-3-3ζ isoforms (Fig. 6a). This result indicates that 14-3-3σ can form heterodimers with 14-3-3γ, 14-3-3β, or 14-3-3θ isoforms in keratinocytes. We next investigated whether the Er 14-3-3σ found in Er/Er mice could also interact with other isoforms of 14-3-3 or with WT 14-3-3σ. We performed co-immunoprecipitation by either using anti-flag antibody to pull down the flag-tagged truncated Er 14-3-3σ isoform and Western blotting with anti-HA antibody to detect the HA-tagged 14-3-3γ or WT form of 14-3-3σ, or using anti-HA antibody to pull-down complex followed by Western blot with flag antibody. We indeed detected the co-immunoprecipitated 14-3-3γ and 14-3-3σ (Fig. 6b), suggesting that Er 14-3-3σ can truly bind to other 14-3-3 protein isoforms. We further tested whether the WT or Er 14-3-3σ proteins could interact with Yap1 protein. Co-immunoprecipitaton using anti-flag antibody could pull down Yap1 with the full-length version of WT 14-3-3σ (Fig. 6c), but not with the truncated form of 14-3-3σ (Fig. 6d). Similarly, we also detected an interaction between the 14-3-3γ and Yap1 (Fig. 6e, 1–4), and this interaction was blocked by co-expression of Er 14-3-3σ (Fig. 6e, 5–8). On the other hand, we showed that over-expression of 14-3-3γ antagonized Er 14-3-3σ and retained Yap1 cytoplasmic localization, and therefore suppressed HaCat cell proliferation. We transduced HaCat cell with14-3-3γ or Er 14-3-3σ, or both. Yap1 nuclear localization was enhanced when cells over-expressed Er 14-3-3σ (Fig. S1c), but not GFP (Fig. S1a) and 14-3-3γ (Fig. S1b). Co-expression of 14-3-3γ neutralized the effects of Er 14-3-3σ on the default Yap1 nuclear localization (Fig. S1d). Consistently, co-expression of the 14-3-3γ suppressed the over-expression of Er 14-3-3σ induced cell proliferation (Fig. S1e).
Figure 5. Expression of all seven 14-3-3 isoforms in WT and Er/Er keratinocytes as detected by qPCR and Western blotting.
RNA samples or protein lysates were collected from primary cultured keratinocytes isolated from E18.5 WT or Er/Er epidermis. The cells were cultured for 48 h in medium containing 0.02 mM Ca2+ or 0.5 mM Ca2+. (a) Expression of 14-3-3 isoforms determined via qPCR. The CT method normalized to Actb was used to analyze relative changes in gene expression. The relative expression (arbitrary units) is expressed as a ratio. Data are expressed as mean±SD, n=3. (b) Western blot analysis of protein lysates using 14-3-3 isoform-specific antibodies. Actin was used as a loading control. Data were representative of three independent experiments.
Figure 6. The truncated Er mutant 14-3-3σ interacts with other 14-3-3 isoforms but not Yap1.
(a) The primary WT keratinocytes were transduced with lentivirus expressing flag-14-3-3σ. Three days after transduction, cellular lysates were immunoprecipitated with mouse anti-flag antibody to pull down the flag-tagged 14-3-3σ and its associated proteins. Western blots were probed with antibodies specifically recognizing each 14-3-3 isoform. (b) The interactions between the 14-3-3σ Er mutant and 14-3-3σ or 14-3-3γ were detected when they were overexpressed in 293T cells. (c–d) Immunoprecipitation showed that flag-tagged WT (c) but not Er 14-3-3σ (d) interacted with Yap1 when they were overexpressed in 293T cells. Furthermore, (e) HA-tagged 14-3-3γ interacted with Yap1 when they were overexpressed in 293T cells and this interaction was abolished when co-expressing Er 14-3-3σ.
Discussion
The molecular mechanism underlying 14-3-3σ regulation of epithelial differentiation has not been fully elucidated. 14-3-3σ interacts with a wide variety of cellular proteins and mediates multiple signaling pathways, generally through a phosphoserine-containing motif. Finding the principal targets that are responsible for its effect on homeostatic regulation of progenitor cell proliferation and the differentiation of keratinocytes has been challenging. Regulation of Yap1 cellular localization by 14-3-3 and the similar defective epidermal phenotypes observed between the 14-3-3σ mutant and Yap1 gain-of-function mutant skins prompted us to further investigate whether Yap1 functions as a bona fide downstream effector of 14-3-3σ protein in mediating keratinocyte proliferation. Our data in the present study indicate that nuclear localization of Yap1 is significantly enhanced in both Er/Er epidermis in vivo and its derived keratinocytes in vitro. In cultured keratinocyte progenitors, a high calcium concentration that normally induces keratinocyte differentiation was not able to induce cytoplasmic retention and nuclear exclusion of Yap1 protein and, as consequence, failed to induce cell cycle arrest of the mutant progenitor cells, which is in contrast to its effect on WT control cells. Reduction of Yap1 expression with shRNA knockdown blocked keratinocyte proliferation of both WT and Er/Er keratinocytes, suggesting an essential role for Yap1 for keratinocyte proliferation. Cell cycle arrest is considered necessary for keratinocytes to commit to terminal differentiation. However we have not examined the differentiation of the Yap1 knockdown keratinocyte, since these cells are unhealthy and undergo apoptosis (data not shown).
Er/Er mice harboring a truncated form of 14-3-3σ protein showed lethal epidermal defects (Li et al., 2005). However, 14-3-3σ gene knockout mice exhibited normal epidermal development and epithelial differentiation (Su et al., 2011). This suggests that the truncated form of 14-3-3σ in Er/Er mice has additional effects on epidermal development that are missing from the homozygous recombinant 14-3-3σ protein in knockout mice. The Er 14-3-3σ mutant protein possesses a functional dimerization domain and has lost its N-terminal ligand-binding domain (Li et al., 2005). We hypothesized that the dimerization domain retained in the truncated Er 14-3-3σ protein might allow its interaction with other 14-3-3 protein isoforms and function as a dominant-negative partner to form non-functional heterodimers in Er mutant cells; on the other hand, the complete deletion of 14-3-3σ in knockout mice should not interfere with the dimerization and function of other 14-3-3 protein isoforms. Motivated to test this hypothesis, we found that keratinocytes did express all seven isoforms of known 14-3-3 proteins, and full-length 14-3-3σ was proven to bind at least three isoforms of 14-3-3 (14-3-3γ, 14-3-3β, and 14-3-3θ). Furthermore, Er 14-3-3σ was bound to full-length forms 14-3-3σ and 14-3-3γ, as demonstrated by co-immunoprecipitation, suggesting that mutant Er 14-3-3σ can form heterodimers with other 14-3-3 isoforms. Although 14-3-3σ is believed to exist mostly in the homodimeric form (Verdoodt et al., 2006; Yaffe et al., 1997), dimerization of 14-3-3σ with 14-3-3γ has also been found in colorectal cancer cells (Benzinger et al., 2005). In addition, Er 14-3-3σ disrupted the 14-3-3σ and 14-3-3γ interaction with Yap1, whereas over-expression of 14-3-3γ relieved the effects of Er 14-3-3σ on Yap1 cellular localization and cell proliferation.
Our results showed that Er mutant acts as dominant negative that inhibits interaction of other 14-3-3 members with Yap1. 14-3-3 recognized phospho-Yap1 through core consensus binding site and this interaction is required to retain phospho-Yap1 in the cytoplasm. In the absence of this cytoplasmic retention, Yap1 becomes constitutively nuclear, correlating with enhanced expression of its target genes regulating keratinocyte proliferation. Consequently, in the Er mutant epidermis, the keratinocytes progenitors continue proliferating and fail to undergo differentiation (Fig. 1), suggesting that 14-3-3 mediated Yap1 function is critical for maintain the keratinocyte proliferation. Although a stronger 14-3-3σ expression is evident in differentiating suprabasal cells of epidermis in vivo, phosphorylation of Yap1 is likely a key step to control Yap1 cytoplasmic retention by 14-3-3. Mutation of Yap1 Ser127 to alanine inhibits the interaction between Yap1 and 14-3-3 protein (Zhao et al., 2007). Both Lats1/2 and Mst1/2 have been traditionally considered as Yap1 upstream kinases, but are not required for epidermis proliferation and differentiation (Schlegelmilch et al., 2011), therefore the upstream kinase for Yap1 activation in skin is still unknown. Previous study showed that binding of the Yap1/14-3-3/α-catenin inhibited dephosphorylation of Yap1Ser127 by PP2A (Schlegelmilch et al., 2011). However, we detected only slightly reduction of Yap1Ser127 and no significant change of the total Yap1 protein level in high calcium induced Er/Er keratinocytes as compared with these in the WT keratinocytes at the same condition (see the Fig. 3b). Further studies are needed to determine whether phosphorylated Yap1 has any nuclear transcriptional coactivator activity.
Materials and Methods
Calculation of the number of BrdU-positive cells and their statistical analysis
In the Yap1 knockdown experiments, the number and percentage of BrdU-positive cells for 1000 randomly selected Yap1 shRNA lentivirus-transduced cells and for 1000 control lentivirus-transduced cells were determined in three independent experiments. All data are summarized as the mean ± standard deviation (SD). Student’s t test was performed to determine the significance of the differences between means. The differences were considered statistically significant if the p values were less than 0.05.
Lentiviral vector, viral production, and viral transduction
Expression vectors encoding the Yap1 shRNA and scrambled control shRNA were ordered from Open Biosystems, Inc. Lentivirus was produced by a four-plasmid (for 3rd-generation lentiviral vectors) transfection system as described by Tiscornia et al. (Tiscornia et al., 2006). Lentivirus was collected in keratinocyte serum-free medium and directly used to transduce primary keratinocytes overnight. Animals and other detailed methods are shown in the Supplementary Materials and Methods. All animal experiments were approved by Institutional Animal Care and Use Committee the Animal Care Committee at the University of Louisville (IACUC #12005)
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants EY019891 and EY021548 as well as a Research to Prevent Blindness Ernest & Elizabeth Althouse Special Scholar Award. Authors also thank Research to Prevent Blindness, Inc, New York, N.Y. for departmental support and Guirong Liu for tissue processing in the histology core of the Department of Ophthalmology and Visual Sciences.
Abbreviations
- Er
Repeated Epilation
- DMBA
7,12 dimethylbenzanthracene
- TPA
12-O-tetradecanoyl-phorbol-13-acetate
- E#
embryonic day #
- WT
wild type
- BrdU
5-bromo-2′-deoxyuridine
- Cy3
carbocyanine 3
- FITC
fluorescein isothiocyanate
- DAPI
diamidinophenylindole
- HE
hematoxylin-eosin
- CT
comparative threshold cycle
- SD
standard deviation
Footnotes
Conflict of Interest.
The authors state no conflict of interest.
References
- Basu S, Totty NF, Irwin MS, et al. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Molecular cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Benzinger A, Muster N, Koch HB, et al. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Molecular & cellular proteomics: MCP. 2005;4:785–95. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Current biology: CB. 2007;17:2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Hammond NL, Headon DJ, Dixon MJ. The cell cycle regulator protein 14-3-3sigma is essential for hair follicle integrity and epidermal homeostasis. The Journal of investigative dermatology. 2012;132:1543–53. doi: 10.1038/jid.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, et al. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat Genet. 2005;37:1210–2. doi: 10.1038/ng1652. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. The EMBO journal. 2000;19:6778–91. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, et al. Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15977–82. doi: 10.1073/pnas.0508310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Sambandam SA, Lu HJ, et al. 14-3-3sigma and p63 play opposing roles in epidermal tumorigenesis. Carcinogenesis. 2011;32:1782–8. doi: 10.1093/carcin/bgr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D, Hermeking H. Epigenetic silencing of 14-3-3sigma in cancer. Seminars in cancer biology. 2006;16:214–24. doi: 10.1016/j.semcancer.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Lu Q, Xin Y, Ye F, et al. 14-3-3sigma controls corneal epithelium homeostasis and wound healing. Investigative ophthalmology & visual science. 2011;52:2389–96. doi: 10.1167/iovs.09-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & development. 2007;21:886–97. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, et al. KRAS and YAP1 Converge to Regulate EMT and Tumor Survival. Cell. 2014;158:171–84. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YW, Hao Z, Hirao A, et al. 14-3-3sigma regulates B-cell homeostasis through stabilization of FOXO1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1555–60. doi: 10.1073/pnas.1017729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nature protocols. 2006;1:241–5. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- van Heusden GP. 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB life. 2005;57:623–9. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- Verdoodt B, Benzinger A, Popowicz GM, et al. Characterization of 14-3-3sigma dimerization determinants: requirement of homodimerization for inhibition of cell proliferation. Cell cycle. 2006;5:2920–6. doi: 10.4161/cc.5.24.3571. [DOI] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, et al. A structural basis for 14-3-3sigma functional specificity. The Journal of biological chemistry. 2005;280:18891–8. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- Xin Y, Lu Q, Li Q. 14-3-3sigma controls corneal epithelial cell proliferation and differentiation through the Notch signaling pathway. Biochemical and biophysical research communications. 2010;392:593–8. doi: 10.1016/j.bbrc.2010.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Lu Q, Li Q. 14-3-3sigma is required for club hair retention. The Journal of investigative dermatology. 2010;130:1934–6. doi: 10.1038/jid.2010.56. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2270–5. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, et al. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes & development. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–38. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.