Abstract
Brown spot is a devastating rice disease. Quantitative resistance has been observed in local varieties (e.g., ‘Tadukan’), but no economically useful resistant variety has been bred. Using quantitative trait locus (QTL) analysis of recombinant inbred lines (RILs) from ‘Tadukan’ (resistant) × ‘Hinohikari’ (susceptible), we previously found three QTLs (qBS2, qBS9, and qBS11) that conferred resistance in seedlings in a greenhouse. To confirm their effect, the parents and later generations of RILs were transplanted into paddy fields where brown spot severely occurred. Three new resistance QTLs (qBSfR1, qBSfR4, and qBSfR11) were detected on chromosomes 1, 4, and 11, respectively. The ‘Tadukan’ alleles at qBSfR1 and qBSfR11 and the ‘Hinohikari’ allele at qBSfR4 increased resistance. The major QTL qBSfR11 coincided with qBS11 from the previous study, whereas qBSfR1 and qBSfR4 were new but neither qBS2 nor qBS9 were detected. To verify the qBSfR1 and qBSfR11 ‘Tadukan’ resistance alleles, near-isogenic lines (NILs) with one or both QTLs in a susceptible background (‘Koshihikari’) were evaluated under field conditions. NILs with qBSfR11 acquired significant field resistance; those with qBSfR1 did not. This confirms the effectiveness of qBSfR11. Genetic markers flanking qBSfR11 will be powerful tools for marker-assisted selection to improve brown spot resistance.
Keywords: Bipolaris oryzae, brown spot, Oryza sativa L., QTL analysis, field resistance, rice
Introduction
Brown spot of rice, which is caused by the fungus Bipolaris oryzae (Breda de Haan) Shoemaker, is one of the most serious rice diseases worldwide. It can greatly decrease grain yield and quality, and was the main cause of the Bengal famine of 1943 (Padmanabhan 1973). The damage caused by brown spot generally becomes noticeable when the rice is produced in nutrient-deficient or otherwise unfavorable soil conditions (Katara et al. 2010). For example, yield losses due to brown spot ranged between 16 and 43% in a silicon-deficient Histosol (Datnoff et al. 1991).
In 2012, the area of Japan affected by the brown spot epidemic covered 146,636 ha, the fourth largest area after those of sheath blight (491,932 ha), leaf blast (251,241 ha), and neck blast (209,241 ha) (JPPA 2013). However, disease incidence has increased in some regions with silicon-deficient soils (NCRC 2010). One reason for the spread of brown spot is that the dose or application frequency of fungicide has decreased to permit low-input sustainable agriculture (NCRC 2010, Yamaguchi et al. 2007). The ear blight caused by brown spot has been frequently found in the Niigata region, where such agriculture has been performed over 90,000 ha using blast–resistant multi lines (Nagasawa 2009, Yamaguchi et al. 2007).
Although the application of fungicide and silicon soil amendments has reduced the severity of the disease, breeding varieties resistant to brown spot may be a more cost-effective alternative for disease control (Sato et al. 2008a). Though several studies have examined the genotypic variability of brown spot resistance in rice (Deren et al. 1994, Eruotor 1985, Misra 1985, Ohata and Kubo 1974, Yoshii and Matsumoto 1951), little information is available on the inheritance of this resistance (Katara et al. 2010, Sato et al. 2008a). Among the resistant varieties and lines, the indica cultivar ‘Tadukan’ shows a high level of partial resistance (Ohata and Kubo 1974, Yoshii and Matsumoto 1951). We previously identified three quantitative trait loci (QTLs) that contributed to brown spot resistance at the seedling stage using recombinant inbred lines (RILs) from a cross of ‘Tadukan’ with ‘Hinohikari’ (Sato et al. 2008a). The objectives of this study were to confirm the effects of these resistance QTLs under field conditions and to introduce them into a regionally adapted variety, such as ‘Koshihikari’. Based on the results of our field trials, we discuss the usefulness of marker-assisted selection (MAS) to improve brown spot field resistance in rice.
Materials and Methods
Fungal isolation and inoculation
The Iga-2 strain of Bipolaris oryzae was isolated from rice leaves collected in a research field at the Mie Prefecture Agricultural Research Institute (MPARI, Iga, Mie, Japan) using the technique described by Kihara and Kumagai (1994). The stock culture is maintained in MPARI. Inoculation, culture of the mycelia, and induction of conidiophore formation under irradiation with black light lamps were also based on the methods of Kihara and Kumagai (1994).
Plant materials
We used 110 RILs in the F8 generation derived from crosses between ‘Tadukan’ (resistant) and ‘Hinohikari’ (susceptible) in the present resistance testing and QTL mapping for field resistance to brown spot. The RILs were advanced from different F2 plants by means of single-seed descent; an intermediate generation (F5) had been previously used to identify three QTLs for brown spot resistance at the seedling stage (Sato et al. 2008a). On the basis of our initial field resistance and mapping results, we selected one RIL (THRIL50) that possessed putative QTLs for field resistance to brown spot, and crossed it with a susceptible variety, ‘Koshihikari’. One F1 plant was backcrossed three times with ‘Koshihikari’ to introduce the QTLs into the ‘Koshihikari’ background. We obtained 19 near-isogenic lines (NILs) in the BC3F5 generation, which were grown to evaluate their field resistance.
Field evaluation of brown spot resistance
In 2012 and 2013, brown spot resistance was evaluated for QTL mapping in a paddy field at MPARI, with two replications, following the procedure of Matsumoto et al. (2014). To initiate the disease in the field, spreader plants (cv. ‘Mienoyume’, susceptible) that had been inoculated with the Iga-2 strain were planted at a rate of three to five plants per hill, at a spacing of 30 × 30 cm among the plants. Then, each RIL and its parents were transplanted (11 hills, 30 × 15 cm) between the spreader rows. Disease scores were recorded 113 days after transplanting, using a scale from 0 to 9 (Supplemental Table 1). In 2013, the brown spot field resistance of the 19 NILs was examined using the same method.
DNA marker assays
Total DNA was extracted from the leaves using the CTAB method (Murray and Thompson 1980). To construct a linkage map, we used 169 polymorphic markers: 162 rice simple-sequence repeat (SSR) markers and 7 sequence-tagged site (STS) markers. SSR and STS analysis were carried out according to the protocol of Sato et al. (2008a). Linkage groups and the order of the markers were determined using version 3 of the MAPMAKER/EXP software (Lander et al. 1987). The resulting genetic linkage map was visualized by using a Microsoft Excel macro, MapDraw (Liu and Meng 2003). QTL analysis was performed using version 2.5 of Windows QTL cartographer (Wang et al. 2006) with the default composite interval mapping and control parameters, model 6 of the standard model, five control markers, a 10-cM window size, and the forward and backward regression model. We used genome-wide threshold values (α = 0.05) to detect putative QTLs on the basis of the results of 1000 permutations. To survey the genotypes of NILs that harbored putative brown spot resistance QTLs, single-nucleotide polymorphism (SNP) analysis was employed. A 384-plex set of SNP markers was selected from diverse accessions of cultivated Asian rice (Ebana et al. 2010). Genotyping was performed by using the GoldenGate BeadArray technology platform (Illumina Inc., San Diego, CA, USA). These SNPs were detected by using the Illumina Bead Station 500G system. All experimental procedures for the SNP typing followed the manufacturer’s instructions.
Results
Phenotypic analysis of parental lines and their progeny
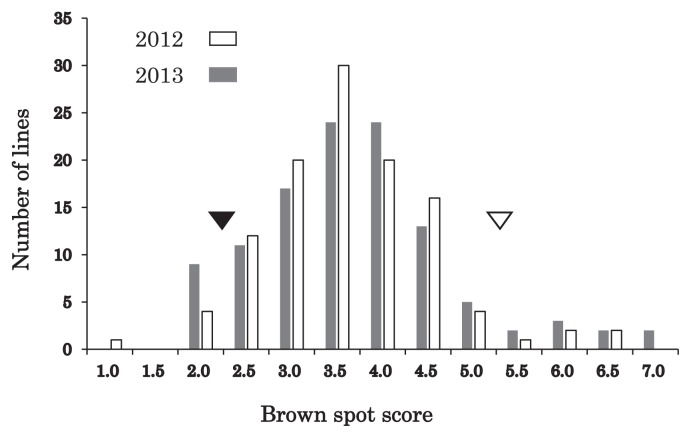
Distinct differences in brown spot field resistance were observed between the parental varieties, ‘Tadukan’ and ‘Hinohikari’: the mean disease scores in 2012 and 2013 were 2.2 ± 0.5 and 5.3 ± 0.8, respectively (Fig. 1, 2). The disease score ratings of the RIL populations in 2012 and 2013 were normally distributed, and a certain number of lines exhibited transgressive segregation in both directions, which indicated that the inheritance of field resistance was quantitative (Fig. 2).
Fig. 1.
Brown spot lesions observed in the Mie paddy field in 2012. The images were taken 109 days after transplanting. (A) Resistant variety ‘Tadukan’, (B) Susceptible variety ‘Hinohikari’.
Fig. 2.
Distribution of brown spot disease scores for 110 F8 RILs (Tadukan/Hinohikari) based on a 0 to 9 rating scale (Supplemental Table S1) in 2012 and 2013. The mean trait values for parents of the RILs, ‘Tadukan’ and ‘Hinohikari’, are indicated as black and white arrowheads, respectively.
QTLs for field resistance
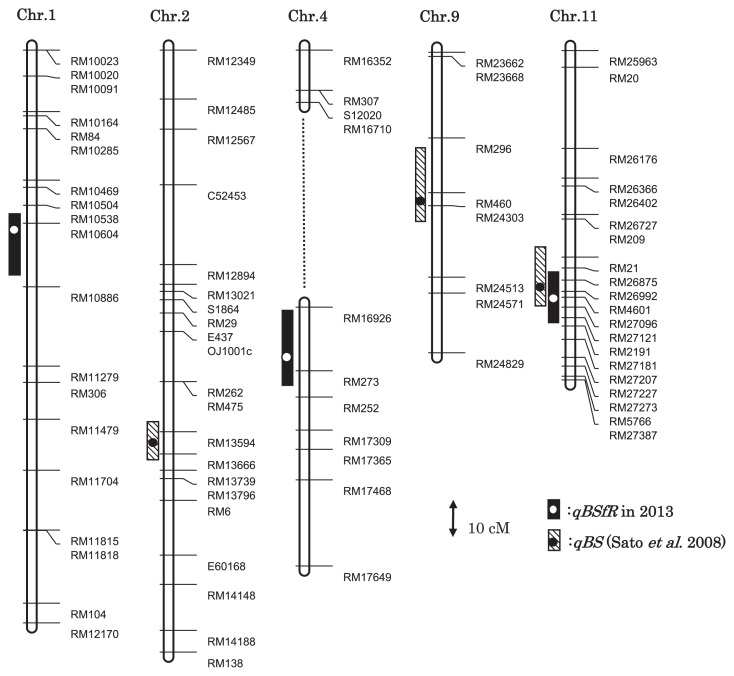
The 162 SSR and 7 STS polymorphic markers were used for map construction. This map covered a total genetic distance of 1286.4 cM and provided partial linkage groups for all chromosomes, except for small gaps in chromosomes 4, 6, and 8 (Data not shown). Three QTLs for field resistance (qBSfR1, qBSfR4, and qBSfR11) were identified based on the data from 2012 and 2013 (Table 1). These QTLs were located on chromosomes 1, 4, and 11. The ‘Tadukan’ alleles of qBSfR1 and qBSfR11 explained 10.4 to 12.3% and 17.9 to 19.2% of the total phenotypic variation, respectively; qBSfR4 from ‘Hinohikari’ accounted for 10.2 to 10.8% of the total variation. The position of the major QTL qBSfR11 coincided with that of the resistance allele qBS11 that was detected in ‘Tadukan’ at the seedling stage (Fig. 3; Sato et al. 2008a). However, the two other QTLs differed from the resistance alleles qBS2 and qBS9 reported by Sato et al. (2008a) (Fig. 3). Thus, in spite of the difference in growth stages analyzed in the two studies, a major resistance QTL from ‘Tadukan’ was detected in both experiments, whereas the relatively minor QTLs detected in the present study did not correspond to those in the previous study (Sato et al. 2008a).
Table 1.
Putative QTLs for field resistance to brown spot of rice detected by using RILs (Tadukan/Hinohikari)
| Years | QTL | Chromosome | Peak position (cM) | Nearest marker | LOD score | Varience explained of total (%) | Additive effect a |
|---|---|---|---|---|---|---|---|
| 2012 | qBSfR 1 | 1 | 40.7 | RM10604 | 4.8 | 10.4 | −0.3 |
| qBSfR 4 | 4 | 91.4 | RM273 | 4.3 | 10.2 | 0.3 | |
| qBSfR 11 | 11 | 52.4 | RM26992 | 8.0 | 17.9 | −0.4 | |
|
| |||||||
| 2013 | qBSfR 1 | 1 | 44.7 | RM10604 | 4.9 | 12.3 | −0.4 |
| qBSfR 4 | 4 | 92.4 | RM273 | 4.8 | 10.8 | 0.3 | |
| qBSfR 11 | 11 | 56.6 | RM27096 | 8.6 | 19.2 | −0.5 | |
Negative values mean that the ‘Tadukan’ allele decreases the disease score (i.e. improves field resistace).
Fig. 3.
Positions of the QTLs for brown spot resistance in the rice chromosomes. The dotted line shows a gap in chromosome 4. Circles and bars represent the LOD peaks of the QTLs and their 1.5-LOD support intervals (Dupuis and Siegmund 1999), respectively.
Verification of QTLs for field resistance
To verify the effects of the ‘Tadukan’ resistance QTLs, 19 NILs from the BC3F5 generation, which were derived from the same BC3F1 plant, were genotyped using SNP markers and their disease phenotypes were evaluated in a field test (Table 2). ‘Koshihikari’, the susceptible recurrent parent of the NILs, is a parent of ‘Hinohikari’ (‘Aichi 40’/’Koshihikari’); there were no markers around the qBSfR4 region that were polymorphic between ‘Koshihikari’ and ‘Hinohikari’ (data not shown). Since all of the 19 lines possessed the same allele as ‘Koshihikari’ at the qBSfR4 locus, we have not further explored the effectiveness of qBSfR4 itself.
Table 2.
Genotypes of 13 SNP markers and disease scores for brown spot of rice in the BC3F5 lines (Koshihikari*3//Tadukan/Hinohikari)
| Lines | Genotype of SNP Marker in the BC3F5 linesa | Disease score c | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| qBSfR 1 region | qBSfR 4 region | qBSfR 11 region | ||||||||||||
|
|
|
|
||||||||||||
| AE01003285 (7.8)b | P0634_1 (9.5)b | ad01004243 (10.0)b | ah01000747 (10.7)b | P0709_2 (19.7)b | ad04008446 (22.2)b | ad04008616 (23.5)b | ad04009558 (25.8)b | AE11002117 (17.9)b | ah11000824 (21.0)b | P0511 (22.9)b | AE11005627 (25.3)b | ah11000941 (25.6)b | ||
| Tadukan | A | A | A | A | A | A | A | A | A | A | A | A | A | 2.5 ± 0.7 ** |
| Koshihikari | B | B | B | B | B | B | B | B | B | B | B | B | B | 6.0 ± 0.0 |
|
| ||||||||||||||
| R307-240-4 | A | A | A | A | B | B | B | B | B | B | B | B | B | 5.0 ± 0.0 |
| R307-147-2 | A | A | A | A | B | B | B | B | B | B | B | B | B | 5.5 ± 0.7 |
|
| ||||||||||||||
| R307-89-9 | B | B | B | B | B | B | B | B | B | A | A | B | B | 5.5 ± 0.7 |
| R82-1 | B | B | B | B | B | B | B | B | B | A | A | A | A | 4.0 ± 0.0 * |
| R82-2 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R82-3 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R82-4 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R92-2 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R92-3 | B | B | B | B | B | B | B | B | B | A | A | A | A | 4.0 ± 0.0 * |
| R103-1 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R103-2 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.0 ± 0.0 ** |
| R103-3 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R103-4 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.0 ± 0.0 ** |
| R138-2 | B | B | B | B | B | B | B | B | B | A | A | A | A | 3.0 ± 0.0 ** |
|
| ||||||||||||||
| R225-1 | A | A | A | A | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R225-2 | A | A | A | A | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R225-3 | A | A | A | A | B | B | B | B | B | A | A | A | A | 4.0 ± 0.0 * |
| R225-4 | A | A | A | A | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
| R306-1 | A | A | A | A | B | B | B | B | B | A | A | A | A | 3.5 ± 0.7 ** |
Genotypes of the SNP markers are represented by A (gray shading) for homozygous ‘Tadukan’ and B (white) for homozygotes ‘Koshihikari’.
Numbers in parentheses beside the SNP markers indicate their physical map position (Mbp) in the chromosomes, according to the number of International Rice Genome Sequence Project (IRGSP) ver.1.0.
Disease scores for brown spot are showed as Mean ± SD.
indicate a significant difference from ‘Koshihikari’ (Dunnet’s test, P < 0.05 and P < 0.01, respectively).
The mean disease scores of ‘Tadukan’ and ‘Koshihikari’ were 2.5 and 6.0, respectively. Two lines that possessed the ‘Tadukan’ allele of qBSfR1 showed slightly lower disease scores (i.e., slightly better resistance) than ‘Koshihikari’, with values of 5.0 for R307-240-4 and 5.5 for R307-147-2. However, the difference was not significant for either line. Among the 12 lines that possessed the major resistance QTL from ‘Koshihikari’ (qBSfR11), 11 lines had significantly lower disease scores than ‘Koshihikari’ (3.0 to 4.0) and were considered resistant; R307-89-9, with a disease score of 5.5, was considered to be as susceptible as ‘Koshihikari’. From this comparison of the phenotypes and graphical genotypes, it seems likely that qBSfR11 lies around the SNP marker interval; AE11005627-AE11000941. Although 5 lines (R225-1, -2, -3, and -4, and R306-1) harbored both of the ‘Tadukan’ resistance QTLs (qBSfR1 and qBSfR11), they showed disease scores (3.5 to 4.0) that were not significantly different from the 11 lines that possessed only qBSfR11 from ‘Tadukan’ (3.0 to 4.0), so no practical gene pyramiding effect was observed in our experiment.
Discussion
Many agriculturally important rice varieties are susceptible to brown spot, and show yield losses of 16 to 43% in inoculated plots (Datnoff et al. 1991, Yamaguchi et al. 1992). No major genes conferring immunity to brown spot have been identified, possibly because of the absence of physiological races of the fungus (Eruotor 1985, Sreegharan and Menon 1974). However, a few resistant varieties, such as ‘Tadukan’, offer sufficiently high quantitative resistance that they will be agriculturally useful (Eruotor 1985, Ohata and Kubo 1974, Yoshii and Matsumoto 1951). Despite their usefulness, genetic information about their resistance has been limited until our previous and present studies. Previously, we identified QTLs for resistance at the seedling stage in a greenhouse study by using an earlier generation of the ‘Tadukan’ and ‘Hinohikari’ RILs than those that were used in the present study (Sato et al. 2008a). In the present study, we aimed to confirm the location and effect of these previously reported resistance QTLs and identify additional QTLs for resistance under field conditions.
By using the newly constructed partial linkage map, we were able to identify three QTLs (qBSfR1, qBSfR4, and qBSfR11) for field resistance in the RILs (Fig. 3, Table 1). The ‘Tadukan’ alleles at the qBSfR1 and qBSfR11 loci significantly decreased the disease score (Table 1). We previously reported that three QTLs (qBS2, qBS9, and qBS11) contributed to resistance at the seedling stage and that ‘Tadukan’ provided the resistance alleles for qBS9 and qBS11 (Sato et al. 2008a). Of these QTLs, only the major QTL on chromosome 11 was stably found in both the greenhouse and field studies, whereas the minor loci were detected in only one of the two environments. This discrepancy could be due to differences in the growth stage or growth conditions. Similarly, the indica rice variety ‘Kasalath’ showed moderate resistance to brown spot at the seedling stage in a greenhouse, and genetic analysis revealed a gene on chromosome 9 that conferred moderate resistance (Sato et al. 2008b). However, the resistance during early vegetative stages was not found in an infected field (Matsumoto et al. 2014). Thus, to confirm the effect of resistance QTLs in a rice breeding program, it would be necessary to conduct field trials to confirm greenhouse results.
We created 19 NILs that contained the two resistance QTLs (qBSfR1 and qBSfR11) from ‘Tadukan’ in the susceptible variety ‘Koshihikari’, and evaluated their phenotypes in a field test (Table 2). Since the 11 NILs that possessed the qBSfR11 allele region from ‘Tadukan’ showed significantly higher field resistance than the parent (Table 2), this QTL appears likely to be agronomically useful. Based on the genotype of the “susceptible” line that harbors the ‘Tadukan’ qBSfR11 region, R307-89-9, it is likely that qBSfR11 lies around the SNP marker interval; AE11005627–AE11000941 (Table 2). In future work, this genetic information will facilitate the precise mapping and cloning of one or more resistance genes in the region. On the other hand, as none of the NILs that possessed the ‘Tadukan’ qBSfR1 allele showed significantly improved disease scores (Table 2), qBSfR1 is likely to be a false field resistance locus.
Katara et al. (2010) recently identified 10 QTLs for field resistance in double-haploid lines derived from CT9993/IR62266, and 3 of these QTLs (on chromosomes 2, 9, and 11) were located in similar marker intervals to those of the QTLs in our previous and present studies (qBS2, qBS9, qBS11, and qBSfR11). In addition, qBSfR4 from the present study was near the BSq4.1v locus mapped by Katara et al. (2010). This work in India confirms that QTL analysis for brown spot resistance can provide an effective framework for MAS in a rice breeding program. The QTL qBSfR11 whose existence and location we confirmed in the present study could be useful in MAS-supported breeding to enhance resistance to brown spot disease. Now we promote strongly the MAS in our breeding programs to breed the practical varieties provided with resistance in near future.
Acknowledgments
We thank the field managers (Mie Prefecture Agricultural Research Institute) for their support during our study. We also thank Mrs. T. Inoue for her technical assistance in the SSR and STS analysis. We are grateful to Dr. T. Ando and Dr. S. Fukuoka (National Institute of Agrobiological Sciences, Japan) for their support during the SNP analysis. This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Project for “Development of mitigation and adaptation techniques to global warming in the sectors of agriculture, forestry, and fisheries”, Rice #2006 and #3003).
Literature Cited
- Datnoff, L.E., Raid, R.N., Snyder, G.H. and Jones, D.B. (1991) Effect of calcium silicate on blast and brown spot intensities of rice. Plant Dis. 75: 729–732. [Google Scholar]
- Deren, C.W., Datnoff, L.E., Snyder, G.H. and Martin, F.G. (1994) Silicon concentration, disease response and yield components of rice genotypes grown on flooded organic Histosol. Crop Sci. 34: 733–737. [Google Scholar]
- Dupuis, J. and Siegmund, D. (1999) Statistical methods for mapping quantitative trait lici from a dense set of markers. Genetics 151: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebana, K., Yonemaru, J., Fukuoka, S., Iwata, H., Kanamori, H., Namiki, N., Nagasaki, H. and Yano, M. (2010) Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza sativa L.). Breed. Sci. 60: 390–397. [Google Scholar]
- Eruotor, P.G. (1985) Varietal reaction of rice to isolates of Cochliobolus miyabeanus. Indian Phytopath. 39: 62–64. [Google Scholar]
- JPPA (2013) Epidemic and controlling areas in 2012. In: Japan Plant Protection Association (eds.) Catalogue of agricultural chemicals, Tokyo, pp. 572–574. [Google Scholar]
- Katara, J.L., Sonah, H., Deshmukh, R.K., Chaurasia, R. and Kotasthane, A.S. (2010) Molecular analysis of QTLs associated with resistance to brown spot in rice (Oryza sativa L.). Indian J. Genet. 70: 17–21. [Google Scholar]
- Kihara, J. and Kumagai, T. (1994) Ecotypes of the fungus Bipolaris oryzae with various responses of the mycochrome system. Physiol. Plant. 92: 689–695. [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E. and Etoh, T. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Liu, R.H. and Meng, J.L. (2003) MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. Heraditas 25: 317–321. [PubMed] [Google Scholar]
- Matsumoto, K., Ota, C., Yamakawa, T. and Sato, H. (2014) A new method for evaluating field resistance to brown spot in rice and selection of standard rice varieties for evaluating the resistance. Breed. Res. 16 (Suppl. 1): 109. [Google Scholar]
- Misra, A.K. (1985) Effect of intercepting populations of resistant cultivars on reducing brown spot disease build up in a susceptible rice cultivar. Indian Phytopath. 38: 66–69. [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, Y. (2009) A core technology of low-input sustainable agriculture, Koshihikari BL. In: Kubota co. (eds.) U, dreaming of agriculture, Hyogo, West Japan ver. 20. pp. 3–4. [Google Scholar]
- NCRC (2010) Silicon amendments stabilized the effect of fungicide to brown spot disease in rice. Niigata prefecture agriculture forest and fisheries research results information. http://www.ari.pref.niigata.jp/nourinsui/seika10/index.html.
- Ohata, K. and Kubo, C. (1974) Studies on the mechanism of disease resistance of rice varieties to Cochliobolus miyabeanus. Bull. Shikoku Agric. Exp. Stn. 28: 17–57. [Google Scholar]
- Padmanabhan, S.Y. (1973) The great Bengal famine. Annu. Rev. Phytopathol. 11: 11–26. [Google Scholar]
- Sato, H., Ando, I., Hirabayashi, H., Takeuchi, Y., Arase, S., Kihara, J., Kato, H., Imbe, T. and Nemoto, H. (2008a) QTL analysis of brown spot resistance in rice (Oryza sativa L.). Breed. Sci. 58: 93–96. [Google Scholar]
- Sato, H., Ando, I., Takeuchi, Y., Hirabayashi, H., Arase, S., Kihara, J. and Nemoto, H. (2008b) Genetic analysis of brown spot resistance in rice using the 39 chromosome segment substitution lines (CSSLs) derived from crossing Koshihikari and Kasalath. Breed. Res. 10 (Suppl. 1): 274. [Google Scholar]
- Sreegharan, A. and Menon, M.R. (1974) Studies on the isolates of Helminthosporium oryzae. Indian Phytopath. 27: 131–133. [Google Scholar]
- Wang, S., Basten, C.J. and Zeng, Z.-B. (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Cited 23 Oct 2006. [Google Scholar]
- Yamaguchi, J., Matsuzaki, M. and Inada, M. (1992) An epiphytoic of brown spot at seedling in Saga prefecture in 1991. Kyushu Agric. Res. 54: 81. [Google Scholar]
- Yamaguchi, Y., Nakano, K. and Saito, R. (2007) An outbreak of rice brown spot at Niigata prefecture Kaetsu district in 2005 and 2006. The Association for Plant Protection of Hokuriku 59: 4. [Google Scholar]
- Yoshii, H. and Matsumoto, M. (1951) Studies on the resistance to Helminthosporiose of the rice varieties introduced to Japan (1). Bull. Matsuyama Agric. College 6: 25–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.