Abstract
Because grain shape is an important component of rice grain yield, the discovery of genes related to rice grain shape has attracted much attention of rice breeding programs. In recent years, some of these genes have been cloned and studied. They have been found not only regulate grain shape by changing the shape of the spikelet hull, but also regulate endosperm development through control of cell division using different molecular mechanisms. In this paper, we review the recent research on genes related to rice grain shape and their possible regulatory mechanisms.
Keywords: regulatory mechanisms, rice, grain shape
Introduction
Rice grain shape is a determinant of grain weight and an important component of grain yield, along with the number of panicles per plant and the number of grains per panicle. It is composed of three elements: grain length, width and thickness (Xing et al. 2002). As rice is a major cereal crop worldwide, increasing its yield is considered an overriding objective of rice breeding programs. Benefiting from the use of semi-dwarf genes and hybrid rice technologies, rice yields have been elevated to new levels (Xing and Zhang 2010). However, with increasing consumption due to the growing global population, it has been projected that global food production must increase 70% by 2050. This is made even more challenging by the decreasing availability of arable land. Not surprisingly, food insecurity has increased in recent years (Godfray et al. 2010). Thus, research focused on improving grain yields is necessary to solve these problems.
Rice yield is dependent on several factors, including number of plants per unit area, number of grains per panicle and grain weight, which is largely determined by grain size (Ikeda et al. 2013, Xing et al. 2002). Grain size directly affects to rice yield and is an important determinant of rice quality (Tan et al. 2000). Elucidating the genetic mechanisms affecting grain shape has great significance to breed high-yielding rice varieties. In recent years, much research has been devoted to the study of the identification, localization, cloning and functional analysis of genes involved grain shape and great progress has been made (Miura et al. 2011).
The development of molecular marker techniques, genome mapping and quantitative trait locus (QTL) analysis has helped advance the study of grain shape and weight. High-density genetic linkage maps have been constructed using restriction fragment length polymorphism (RFLP) and simple sequence repeat markers (SSR) methodologies. Over 400 QTLs related to grain shape and weight have been mapped to chromosomes, and over 160,100 and 95 QTLs have been associated with 1000-grain weight (KGW), grain length and grain width, respectively (Huang et al. 2013). However, to date, only a few genes have been fine-mapped or identified. Seven genes associated with grain shape have been isolated using map-based cloning strategies: GRAIN WIDTH 2,5 and 8 (GW2, Song et al. 2007; GW5, Shomura et al. 2008, Wan et al. 2008, Weng et al. 2008; GW8, Wang et al. 2012); GRAIN SIZE 3 (GS3, Fan et al. 2006); GRAIN SIZE ON CHROMOSOME 5 (GS5, Li et al. 2011); GRAIN LENGTH 3 (qGL3, Qi et al. 2012, Zhang et al. 2012); and THOUSAND-GRAIN WEIGHT 6 (TGW6, Ishimaru et al. 2013). The isolation of these genes has enhanced our knowledge of the molecular regulatory mechanisms responsible for grain size and offers potential for future high-yield rice breeding (Song et al. 2008).
In this article, we review possible genetic regulatory mechanisms involved in determining rice grain shape, including heterotrimeric GTP-binding protein (G-protein) signaling pathways, protein phosphatase regulatory pathways, ubiquitin (Ub)/26S proteasome pathways, gene expression levels and Phytohormones.
Heterotrimeric G-protein signaling and grain shape
GS3 is a negative regulator of grain size, with major effects on grain length and weight, and minor effects on grain width and thickness (Fan et al. 2006). The complete GS3 protein contains a plant-specific organ size regulation (OSR) domain in the N terminus, a transmembrane domain, and both a tumor necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family cysteine-rich domain, and a von Willebrand factor type C (VWFC) in the C terminus (Mao et al. 2010). The OSR domain is the key negative regulator of grain length, whereas the two C-terminal domains have an inhibitory effect on OSR function (Mao et al. 2010).
Three alleles in the GS3 locus have been identified in different rice varieties. Rice varieties such as Oryza sativa L. ‘Zhenshan97’ which have a normal grain length, carry a GS3 allele that encodes a complete transmembrane protein and regulates cell number in the upper epidermis of the glume (Takano-Kai et al. 2013). Rice varieties such as Oryza sativa L. ‘Minghui63’, which have long grains, contain a GS3 allele characterized by a C-to-A substitution at 165bp that causes premature termination of the predicted protein, resulting in a complete loss of the functional protein domain. This allele is common in rice with long grains. The third GS3 allele also encodes a truncated protein, this one having only the OSR and transmembrane domains. It exists in varieties such as Oryza sativa L. ‘Chuan7’, which has very short grains (Fan et al. 2006, 2009, Mao et al. 2010).
GS3 is a homolog of AGG3, the G protein γ-subunit of Arabidopsis, and the OSR domain is a γ-like domain (Li et al. 2012). Heterotrimeric G-proteins are comprised of Gα, Gβ and Gγ subunits and act as important molecular swithes in multiple growth and development pathways in eukaryotic cells. G-proteins exist in three conformational states (depending on whether they are bound to GDP, GTP or no nucleotide) that determine G-protein activity. Mammals have many genes encoding G-protein subunits (Offermanns 2000), whereas plants seem to harbor relatively few such genes. In rice, one α subunit gene (RGA1), one β subunit gene (RGB1) and five γ subunit genes (RGG1, RGG2, GS3, DEP1 and OSGGC2) have been identified (Ishikawa et al. 1995, 1996, Kato et al. 2004, Li et al. 2012). In contrast with Arabidopsis AGG3, which positively regulates seed and organ size, rice GS3 negatively regulates grain length and weight. As one of the five Gγ genes in rice, it is possible that negative regulation by GS3 involves interaction of GS3 with Gβ to form a Gβγ dimer (Smrcka 2008).
Protein Phosphatases and grain shape
qGL3/GL3.1 is a major QTL for rice grain length and weight, and a minor QTL for grain width and thickness, and encodes a Ser/Thr phosphatase of the protein phosphatase kelch-like (PPKL) family (Qi et al. 2012, Zhang et al. 2012). PPKL is a newly identified phosphatase family characterized by kelch-like domain repeats in the N terminus (Kutuzov and Andreeva 2002). A comparison of the coding region of OsPPKL1 from Oryza sativa L. ‘N411’ and ‘WY3’, which have larger grains, with Oryza sativa L. ‘93-11’ and ‘Fengaizhan-1’ (FAZ1), which have smaller grains, revealed four point mutations. Compared to 93-11 and FAZ1, a nucleotide transversion in the 10th exon and a nucleotide transition in the 11th exon leads to amino acid residue changes from Asp to Glu and His to Tyr respectively, in N411 and WY3. The mutation in the 10th exon is necessary for producing longer grains and acts by accelerating cell division (Qi et al. 2012). This mutation site has been detected in only a few rice varieties, such as Oryza sativa L. ‘N411’, ‘WY3’, ‘DT108’, ‘Jizi1560’ and ‘Jizi1581’ (Qi et al. 2012, Zhang et al. 2012). A transformation study of different qGL3/GL3.1 domains indicated that the Kelch domain is responsible for the negative regulatory function of OsPPKL193-11 (Zhang et al. 2012).
Ser/Thr phosphatases catalyze dephosphorylation of Ser and Thr residues, and act in opposition to Ser/Thr kinases, which catalyze the transfer of γ phosphates from ATP or GTP to Ser or Thr. Protein phosphorylation is an important posttranslational modification that occurs in almost a third of all proteins and is critical for transmission of extracellular signals into cells (Zolnierowicz and Bollen 2000). It is involved in multiple regulatory processes, such as protein activation, assembly and disassembly of protein complexes, protein degradation and subcellular localization of proteins (Olsen et al. 2006). The onset of mitosis requires phosphorylation of many proteins.
Cyclin-T1;3 is a substrate of GL3.1 in the grain length regulation pathway. The GL3.1 small grain allele has strong Cyclin-T1;3 dephosphorylation activity that results in almost complete dephosphorylation of Cyclin-T1;3 substrate and allows for a normal level of mitosis to produce ordinary grain. In comparison, the GL3.1 large grain allele has weaker Cyclin-T1;3 dephosphorylation activity, accelerates transformation from the G1 phase of mitosis to G2, and promotes formation of a longer spikelet hull. Thus, in some large-grain rice varieties, weaker GL3.1 dephosphorylation activity may result in accumulation of phosphorylated substrates that promote DNA synthesis and accelerate cell division, thereby producing longer grains (Qi et al. 2012).
Ubiquitin/26S proteasome pathway and grain shape
GW2 has been identified as a major QTL for grain width and weight. It encodes a RING-type domain with E3 ubiquitin ligase activity. The absence of one nucleotide in the 4th exon of GW2 in Oryza sativa L. WY3 results in the premature production of the protein and production of large grains. GW2 plays a negative role in controlling cell division through the ubiquitin-proteasome proteolytic pathway, and this function relies on the RING domain in the N terminus. The truncated GW2 protein found in WY3 contains the complete E3 functional region, but cannot negatively regulate grain width because of the absence of the substrate-binding region. Morphological observations of cells in a large-grain GW2 mutant indicate that an increase in cell numbers within spikelet hulls and accelerated filling are responsible for the observed increases in grain width, weight and yield (Song et al. 2007).
qSW5/GW5 is another major gene controlling grain width and weight, and is located in chromosome 5 (Shomura et al. 2008, Wan et al. 2008, Weng et al. 2008). Polymorphism studies in different rice varieties have shown that the deletion of 1212 base within qSW5/GW5 plays an important role in changing grain shape and increasing grain weight by controlling cell number in the outer glume of the rice flower (Shomura et al. 2008). qSW5/GW5 encodes a nucleoprotein containing 144 amino acid residues with an arginine-rich domain. A yeast two-hybrid experiment indicated that qSW5/GW5 interacts with polyubiquitin, suggesting that qSW5/GW5 activity in the ubiquitin-proteasome pathway regulates cell division during seed development (Weng et al. 2008).
The ubiquitin/26S proteasome system is the most important proteasome proteolytic pathway in eukaryotic organisms. This pathway participates widely in hormone signaling, regulation of chromatin structure and transcription, responses to abiotic and biotic environmental challenges, including pathogens (Vierstra RD 2009). In particular, it plays an important part in seed size determination in plants (Du et al. 2014). Ubiquitin is a highly conserved protein with 76 amino acid residues. Ubiquitination is mediated by three enzymes: ubiquitin-activating enzyme E1, which is responsible for activation of free ubiquitins; ubiquitin-conjugating enzyme E2, which is responsible for combining the ubiquitins activated by E1; and ubiquitin-protein ligase E3, which identifies targets and mediates ubiquitin transfer (Adam et al. 2009). In plants, E3 ubiquitin ligases can be divided into four types, each containing different domains, Homology to E6AP C Terminus (HECT), Real Interesting New Gene/U-Box (RING/U-Box), a complex of Skp1, CDC53, and F-box protein (SCF) and anaphase-promoting complex (APC), based on subunit composition and mechanism of action (Smalle and Vierstra 2004). The first two types are single polypeptides and the last two types are protein complexes with several subunits. There are at least 1,300 E3 ubiquitin-protein ligase genes in the rice genome (Du et al. 2009), including 425 genes encoding for RING finger proteins (Lim et al. 2010). The RING domain contains four pairs of zinc ligands formed by Cys and His, which can bind zinc ions and form C3H2C3 (RING-H2) or C3H1C4 (RING-HC) structures with the same conserved His at metal ligand position 4, and either a Cys or His residue at metal ligand position 5 (Zheng et al. 2000). The RING-type E3 ubiquitin ligase encoded by GW2 is different from any of the known structures. It is characterized by a Cys residue at metal ligand position 5 and a His residue at metal ligand position 6 (C5H1C2). This structure was also identified in maize and wheat, suggesting that GW2 represents a conserved RING domain in plants (Song et al. 2007).
Regulation of the plant cell cycle is precise and complicated. It involves the participation of many proteins, the most important being cyclin-dependent kinases (CDKs), CDK-activating kinases and CDK inhibitors. The functions of these regulatory factors are controlled by the 26S proteasome and specific E3 ubiquitin ligases (Smalle and Vierstra 2004). The E3 ubiquitin ligases that participate in cell cycle regulation are mainly APC and SCF types. SCF-type E3 ubiquitin ligases are responsible for interactions between ubiquitins and cyclins of G1/S and certain CDK inhibitors, and participate in the ubiquitination proteasome as regulatory factors in G1/S (Huang et al. 2005, Koepp et al. 2001, Wei et al. 2005, Welcker et al. 2004). APC-type E3 ubiquitin ligases are responsible for coupling ubiquitins with cyclins in M phase and to other factors necessary for mitosis. They are also involved in separation of sister chromatids, depolymerization of spindle fibers, cytokinesis and reentry into G1 phase (Ban et al. 2007, Lukas et al. 1999, Wei et al. 2004).
Although many simple RING-type E3 ubiquitin ligases were identified through genome-wide studies of rice, the regulatory mechanisms of these genes have not been well studied, and information was limited to only some of these genes participating in disease resistance and response to abiotic stressors (Koiwai et al. 2007). GW2 was first discovered as a simple RING-type E3 ubiquitin ligase that controlled rice development through regulation of the cell cycle. The substrates and regulatory mechanisms of E3 are still pooly understood, and need to be studied further.
Gene expression levels and grain shape
GS5 is a major QTL for grain width, weight and filling that was identified on chromosome 5 using a double haploid population derived from a cross between Zhenshan 97 (ZS97, wide grain) and Oryza sativa L. ‘H94’ (narrow grain) (Li et al. 2011). GS5 encodes a putative serine carboxypeptidase belonging to the peptidase S10 family. In as comparison of cross-sections from the central part of the palea/lemma of the spikelet of near-isogenic lines (NILs), it was found that the inner parenchyma cell layer of NIL (ZS97) contains a greater number of cells than that of NIL (H94) (Li et al. 2011). Some important cell cycle-related genes involved in the G1/S phase are upregulated when GS5 is overexpressed, and downregulated in loss-of-function GS5 mutants. These facts indicate that GS5 acts as a positive regulatory factor in the cell cycle and that high GS5 expression can increase the cell number by regulating mitosis, thereby controlling grain size. An analysis of the open reading frames (ORFs) and promoter regions has shown that a sequence polymorphism in the promoter region, not in the ORF region, is responsible for the effect of GS5 on grain width (Li et al. 2011).
GW8 was is a major QTL found on chromosome 8 that aids in controlling grain width and yield. This QTL is located in a 7.5-kb region of the genome that contains the first exon of LOC_Os08g41940 and the promoter region. The candidate gene OsSPL16 encodes SQUAMOSA promoter-binding protein-like 16 which belongs to the SQUAMOSA promoter binding proteins (SBP) family of transcriptional factors. Although six polymorphisms were detected in the DNA sequence of Oryza sativa L. ‘Basmati385’ (slender grain) and Oryza sativa L. ‘HJX74’ (wide grain), none influenced grain width. The polymorphism that contributes greatly to the slender grain trait of Basmati385 is a 10-bp deletion in the OsSPL16 promoter. When the grain shape-related genes GW2, GS3 and GS5 were expressed at similar levels, the cell cycle related genes in G1/S phase showed higher expression level in NIL-GW8 than in NIL-gw8 with no expression differences detected in G2/M phase. These data, combined with the morphological observations, indicate that GW8 regulates the cell cycle, promoting latitudinal growth by increasing cell proliferation and inhibiting longitudinal growth by repressing cell elongation (Wang et al. 2012).
It is interesting to note that functional mutations in the two positive regulators of grain shape (GW8 and GS5) are in the promoter region, whereas functional mutations in the negative regulators (GW2, GS3, qGL3/GL3.1 and GW5/qSW5) are in the ORF. Moreover, GW8 and GS5 seemingly regulate cell division and grain shape by changing expression of genes involved in the G1/S phase. The genes upregulated by higher expression of GS5 were also increased in NIL-GW8 (Li et al 2011, Wang et al. 2012, Table 1), suggesting that the two genes may regulate grain shape using similar molecular mechanisms that regulate the cell cycle.
Table 1.
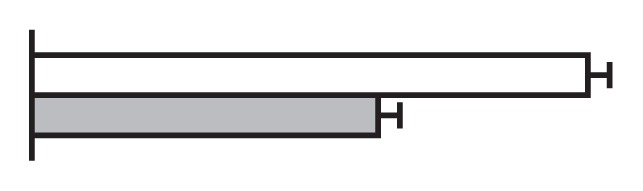
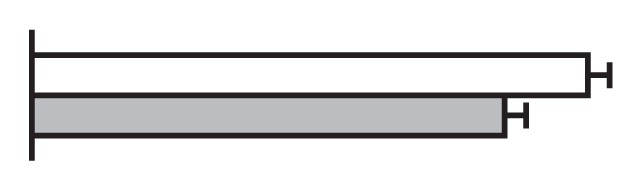
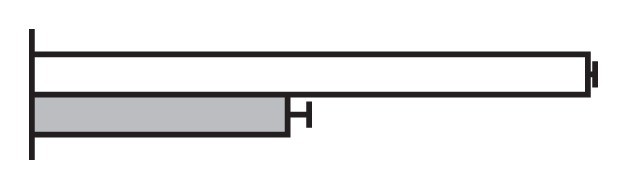
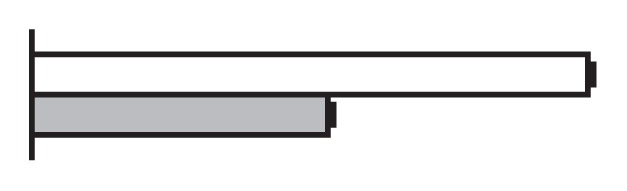
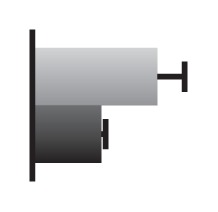
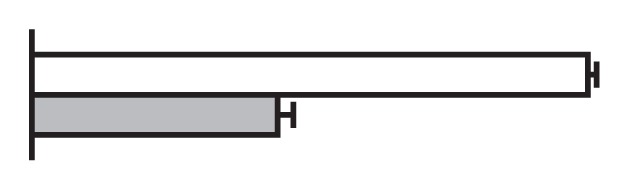
Regulation by GS5 and GW8 of the expression of genes involved in the cell cycle (Li et al. 2011, Wang et al. 2012)
The values were expressed relative to the level of transcript in NIL-GW8 set to be one. White bars, NIL-GW8; gray bars, NIL-gw8.
The values were expressed relative to the level of transcript in negative segregants set to be one. gray bars, GS5 overexpressor; Black bars, negative segregants.
Phytohormones and grain shape
TGW6 is a newly cloned gene from kasalath, a small-grain indica variety, and affects grain length and weight (Ishimaru et al. 2013). It encodes a protein with indole-3-acetic acid (IAA)-glucose hydrolase activity. The tgw6 allele of Oryza sativa L. ‘Nipponbare’ can affect the timing of the transition from the syncytial to the cellular phase by controlling the supply of IAA, and also limits cell number and grain length. A single nucleotide deletion at 313bp in the TGW6 allele derived from kasalath is a functional single nucleotide polymorphism that produces truncated protein and results in the loss of the negative regulation of grain shape. NIL (TGW6) not only contains more endosperm cell layers than Nipponbare, based on the identical cell length, but also has greater dry weight throughout grain development (Ishimaru et al. 2013). During rice seed development, the primary endosperm nucleus undergoes a series of mitosis without cytokinesis to produce coenocytes. After several rapid rounds of this nuclear proliferation, mitosis in the coenocytes halts and cellularization (a special kind of cytokinesis that form single cells) occurs. The duration of the syncytial phase and timing of cellularization are very influential factors in determining grain size and weight (Mizutani et al. 2010). The IAA-glucose hydrolase encoded by TGW6 in Nipponbare hydrolyzes IAA-glucose into IAA and glucose. The IAA content in NIL (TGW6) is only 16.6% of that in Nipponbare, and the expression of many auxin-responsive genes is lower. TGW6 from Nipponbare controls cell division to limit cell number and grain length through these IAA effects, and at the same time, suppresses expression of genes related to starch synthesis (Ishimaru et al. 2013). In contrast with the above-mentioned genes (GW2, GW5/qSW5, GS3, qGL3/GL3.1, GS5 and GW8), all of which regulate endosperm indirectly by controlling the size of spikelet hulls, TGW6 regulates endosperm cell number directly, and effects starch synthesis and grain weight (Ishimaru et al. 2013).
Summary of regulatory mechanisms of grain shape
Despite many advances in the identification and molecular characterization of genes that affect grain shape, many aspects of the genetic and molecular mechanisms underlying their action remain unclear.
As one of the five γ-subunits in rice, GS3 controls transmembrane signal transmission, which regulates grain shape by changing its own structure. An expression study of GS3 indicates that the regulatory function is realized through control of ovule development (Takano-Kai et al. 2009). However, determining where and how regulation occurs requires additional morphological and molecular research. In addition, there are differences in growth and morphological phenotypes between Arabidopsis and rice G-protein mutants (for instance, GS3 in rice and AGG3 in Arabidopsis), and some proteins involved in heterotrimeric G-protein signaling pathways in Arabidopsis do not exist in rice. These findings suggest that cereals may use G-protein signaling mechanisms that are distinct from those in other flowering plants (Urano et al. 2013). More experiments are needed to identify and characterize the constituents and functions of G-protein that regulate grain shape.
qGL3/GL3.1 encodes a Ser/Thr phosphatase that dephosphorylates proteins. The CDKs, key regulators of the cell cycle, are a group of Ser/Thr phosphatases that act with cyclin to regulate the cell cycle. Thus, phosphorylation and dephosphorylation of substrates are important in the cell cycle and are necessary for G1-S and G2-M phase changes in plant cells (Veylder et al. 2003). qGL3/GL3.1 dephosphorylates its substrate, cyclin-T1;3, which regulates grain length by affecting the cell division rate in the spikelet hull.
Acting as an E3 ubiquitin ligase, GW2 functions in ubiquitin/26S proteasomal pathway and negatively regulates the cell division in spikelet hulls. The protein encoded by GW5/qSW5 physically interacts with polyubiquitin, affecting cell division to regulate cell number in the outer glume and thus determine grain width. This indicates that GW5/qSW5 and GW2 may share a similar mechanism in the ubiquitin-proteasome pathway for grain width regulation. The two kinds of ubiquitin ligase complexes, SCF and APC, are involved in cell cycle regulation. GW2 is regarded as a new type of simple ubiquitin ligase that also aids in cell cycle regulation. However, the substrates of GW2 and GW5/qSW5 and their downstream effects have yet to be elucidated.
Unlike other genes related to rice grain shape, the IAA-glucose hydrolase encoded by TGW6 regulates cell division by controlling the supply of IAA, affecting cell number in the endosperm and thus grain length, instead of affecting the length of the spikelet hulls. IAA plays a key role in cell division and elongation (Leyser 2002). It can induce the expression of cyclin-dependent kinase CDKA1;1 (Zhang et al. 1996) and CYCA2;2, and is also upregulated by IAA during lateral root development in alfalfa (Roudier et al. 2003). In the root tip of maize, altering the distribution of auxin can activate the root apical meristem quiescent center (QC), a group of mitotically inactive cells, stimulating cells to transition rapidly from G1 to M phase (Jiang et al. 2003). In rice, TGW6 regulates the cell cycle by affecting the timing of the transition from the syncytial to the cellular phase. Although this could modify the expression of many important cell-cycle-related genes, the specific target genes and underlying mechanisms require further study.
The expression levels GS5 and GW8 positively regulate grain shape. High expression levels of GS5 and GW8 induce expression of important genes in the G1/S phase, accelerate cell division and change grain shape. Some genes regulated by GS5 in the G1/S phase were also influenced by GW8. Thus, these two genes may regulate grain shape through similar molecular regulatory pathway.
GW2, GS3, qGL3, GW5/qSW5, GS5, GW8 and TGW6 regulate the cell cycle by promoting or suppressing cell division, thus affecting cell numbers in rice grains, in the latitudinal or longitudinal directions. Additionally, many other genes regulating rice grain shape have been found by mutant analyses. Studies of rice dwarf mutants led to the identification of D1, which encodes a Gα subunit (Ashikari et al. 1999, Fujisawa et al. 1999); D2 and D11, which encode novel cytochrome P450 CYP90D2 and CYP724B1, respectively (Hong et al. 2003, Tanabe et al. 2005); and D61/OsBRI1, which encodes a putative Brassinosteroid (BR) receptor kinase (Yamamuro et al. 2000). Screening of small and round rice seed mutants led to identification of SRS1, which encodes a novel protein (Abe et al. 2010); SRS3, which encodes a kinesin 13 protein (Kitagawa et al. 2010); and SRS5 (Segami et al. 2012), which encodes an α-tubulin protein. Investigation of an activation-tagged rice line led to identification of SHORT GRAIN 1 (SG1), which encodes a protein of unknown function (Nakagawa et al. 2012). Some of these genes are related to both G-protein and BR signaling pathways.
In the complex signal transduction pathways of plants, interactions among different pathways are possible. Thus, as research progresses, interactions will likely be discovered among the different pathways regulating grain shape. Improving our knowledge of the genes involved in determining grain shape will not only promote progress in rice breeding programs, but will enhance the our general understanding of plant cell cycle regulation.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (No. 31271678), China Postdoctoral Science Foundation (No: 2013M531297) and Jiangsu Agricultural Scientific Self-Innovation Fund (Grant No. CX[11]4023).
Literature Cited
- Abe, Y., Mieda, K., Ando, T., Kono, I., Yano, M., Kitano, H. and Iwasaki, Y. (2010) The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 85: 327–339. [DOI] [PubMed] [Google Scholar]
- Adam, C., Ewan, R., Joelle, M., GudiPati, V. and Ari, S. (2009) E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 60: 1123–1132. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Wu, J., Yano, M., Sasaki, T. and Yoshimura, A. (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96: 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, K.H., Torres, J.Z., Miller, J.J., Mikhailov, A., Nachury, M.V., Tung, J.J., Rieder, C.L. and Jackson, P.K. (2007) The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev. Cell 13: 29–42. [DOI] [PubMed] [Google Scholar]
- Du, L., Li, N., Chen, L., Xu, Y., Li, Y., Zhang, Y., Li, C. and Li, Y. (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26: 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z., Zhou, X., Li, L. and Su, Z. (2009) plantsUPS: a database of plants’ Ubiquitin Proteasome System. BMC Genomics 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X. and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fan, C., Yu, S., Wang, C. and Xing, Y. (2009) A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118: 465–472. [DOI] [PubMed] [Google Scholar]
- Fujisawa, Y., Kato, T., Ohki, S., Ishikawa, A., Kitano, H., Sasaki, T., Asahi, T. and Iwasaki, Y. (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 96: 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray, H.C., Beddington, J.R., Crute, I.R., Haddad, L., Lawrence, D., Muir, J.F., Pretty, J., Robinson, S., Thomas, S.M. and Toulmin, C. (2010) Food security: the challenge of feeding 9 billion people. Science 327: 812–818. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H. and Matsuoka, M. (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Regan, K.M., Wang, F., Wang, D., Smith, D.I., van Deursen, J.M. and Tindall, D.J. (2005) Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 102: 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R., Jiang, L., Zheng, J., Wang, T., Wang, H., Huang, Y. and Hong, Z. (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18: 218–226. [DOI] [PubMed] [Google Scholar]
- Ikeda, M., Miura, K., Aya, K., Kitano, H. and Matsuoka, M. (2013) Genes offering the potential for designing yield-related traits in rice. Curr. Opin. Plant Biol. 16: 213–220. [DOI] [PubMed] [Google Scholar]
- Ishikawa, A., Tsubouchi, H., Iwasaki, Y. and Asahi, T. (1995) Molecular cloning and characterization of a cDNA for the alpha subunit of a G protein from rice. Plant Cell Physiol. 36: 353–359. [DOI] [PubMed] [Google Scholar]
- Ishikawa, A., Iwasaki, Y. and Asahi, T. (1996) Molecular cloning and characterization of a cDNA for the b subunit of a G protein from rice. Plant Cell Physiol. 37: 223–228. [DOI] [PubMed] [Google Scholar]
- Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., Kashiwagi, T., Ujiie, K., Shimizu, B., Onishi, A.et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45: 707–711. [DOI] [PubMed] [Google Scholar]
- Jiang, K., Meng, Y.L. and Feldman, L.J. (2003) Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development 130: 1429–1438. [DOI] [PubMed] [Google Scholar]
- Kato, C., Mizutani, T., Tamaki, H., Kumagai, H., Kamiya, T., Hirobe, A., Fujisawa, Y., Kato, H. and Iwasaki, Y. (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 38: 320–331. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K., Kurinami, S., Oki, K., Abe, Y., Ando, T., Kono, I., Yano, M., Kitano, H. and Iwasaki, Y. (2010) A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol. 51: 1315–1329. [DOI] [PubMed] [Google Scholar]
- Koepp, D.M., Schaefer, L.K., Ye, X., Keyomarsi, K., Chu, C., Harper, J.W. and Elledge, S.J. (2001) Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294: 173–177. [DOI] [PubMed] [Google Scholar]
- Koiwai, H., Tagiri, A., Katoh, S., Katoh, E., Ichikawa, H., Minami, E. and Nishizawa, Y. (2007) RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J. 51: 92–104. [DOI] [PubMed] [Google Scholar]
- Kutuzov, M.A. and Andreeva, A.V. (2002) Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell. Signal. 14: 745–750. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2002) Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53: 377–398. [DOI] [PubMed] [Google Scholar]
- Li, S., Liu, W., Zhang, X., Liu, Y., Li, N. and Li, Y. (2012) Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signal. Behav. 7: 1357–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., Shao, D., Xu, C., Li, X., Xiao, J.et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Lim, S.D., Yim, W.C., Moon, J.C., Kim, D.S., Lee, B.M. and Jang, C.S. (2010) A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes. Plant Mol. Biol. 72: 369–380. [DOI] [PubMed] [Google Scholar]
- Lukas, C., Sørensen, C.S., Kramer, E., Santoni-Rugiu, E., Lindeneg, C., Peters, J.M., Bartek, J. and Lukas, J. (1999) Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature 401: 815–818. [DOI] [PubMed] [Google Scholar]
- Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., Li, X. and Zhang, Q. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 107: 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Ashikari, M. and Matsuoka, M. (2011) The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci. 16: 319–326. [DOI] [PubMed] [Google Scholar]
- Mizutani, M., Naganuma, T., Tsutsumi, K. and Saitoh, Y. (2010) The syncytium-specific expression of the Orysa; KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J. Exp. Bot. 61: 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, H., Tanaka, A., Tanabata, T., Ohtake, M., Fujioka, S., Nakamura, H., Ichikawa, H. and Mori, M. (2012) Short grain1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns, S. (2000) Mammalian G-protein function in vivo: new insights through altered gene expression. Rev. Physiol. Biochem. Pharmacol. 140: 63–133. [DOI] [PubMed] [Google Scholar]
- Olsen, J.V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P. and Mann, M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648. [DOI] [PubMed] [Google Scholar]
- Qi, P., Lin, Y.S., Song, X.J., Shen, J.B., Huang, W., Shan, J.X., Zhu, M.Z., Jiang, L., Gao, J.P. and Lin, H.X. (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22: 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier, F., Fedorova, E., Lebris, M., Lecomte, P., Gyorgyey, J., Vaubert, D., Horvath, G., Abad, P., Kondorosi, A. and Kondorosi, E. (2003) The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 131: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami, S., Kono, I., Ando, T., Yano, M., Kitano, H., Miura, K. and Iwasaki, Y. (2012) Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S. and Yano, M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Smalle, J. and Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Smrcka, A.V. (2008) G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell. Mol. Life Sci. 65: 2191–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Song, X.J. and Ashikari, M. (2008) Toward an optimum return from crop plants. Rice 1: 135–143. [Google Scholar]
- Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., Kanamori, H., Padhukasahasram, B., Bustamante, C., Yoshimura, A., Doi, K.et al. (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai, N., Jiang, H., Powell, A., McCouch, S., Takamure, I., Furuya, N., Doi, K. and Yoshimura, A. (2013) Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 63: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y.F., Xing, Y.Z., Li, J.X., Yu, S.B., Xu, C.G. and Zhang, Q.F. (2000) Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 101: 823–829. [DOI] [PubMed] [Google Scholar]
- Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., Yoshimura, A., Kitano, H., Matsuoka, M., Fujisawa, Y.et al. (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano, D., Chen, J.G., Botella, J.R. and Jones, A.M. (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veylder, L.D., Joubès, J. and Inzé, D. (2003) Plant cell cycle transitions. Curr. Opin. Plant Biol. 6: 536–543. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wan, X., Weng, J., Zhai, H., Wang, J., Lei, C., Liu, X., Guo, T., Jiang, L., Su, N. and Wan, J. (2008) Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in arecombination hotspot region on chromosome 5. Genetics 179: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., Zeng, R., Zhu, H., Dong, G., Qian, Q.et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44: 950–954. [DOI] [PubMed] [Google Scholar]
- Wei, W., Ayad, N.G., Wan, Y., Zhang, G.J., Kirschner, M.W. and Kaelin, W.G.Jr. (2004) Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428: 194–198. [DOI] [PubMed] [Google Scholar]
- Wei, W., Jin, J., Schlisio, S., Harper, J.W. and Kaelin, W.G.Jr. (2005) The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8: 25–33. [DOI] [PubMed] [Google Scholar]
- Welcker, M., Orian, A., Jin, J., Grim, J.A., Harper, J.W., Eisenman, R.N. and Clurman, B.E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101: 9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., Lei, C., Zhang, X., Cheng, Z., Guo, X.et al. (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Tan, Y., Hua, J., Sun, X., Xu, C. and Zhang, Q. (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 105: 248–257. [DOI] [PubMed] [Google Scholar]
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61: 421–442. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H. and Matsuoka, M. (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K., Letham, D.S. and John, P.C. (1996) Cytokinin controls the cell cycle at mitosis by stimulating the tyrosine dephosphorylation and activation of p34cdc2-like H1 histone kinase. Planta 200: 2–12. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., Wu, Y., Tang, H., Qian, Q., Li, J.et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109: 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., Wang, P., Jeffrey, P.D. and Pavletich, N.P. (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539. [DOI] [PubMed] [Google Scholar]
- Zolnierowicz, S. and Bollen, M. (2000) Protein phosphorylation and protein phosphatases De Panne, Belgium, September 19–24, 1999 EMBO J. 19: 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]