Graphical abstract
Abbreviations: dTP, dual targeting peptide; mTP, mitochondrial targeting peptide; cTP, chloroplastic targeting peptide; aaRS, amino acyl-tRNA synthetase; ThrRS, threonyl tRNA synthetase; TOM, translocase of the outer mitochondrial membrane; TIM, translocase of the inner mitochondrial membrane; TOC, translocase of the outer envelope membrane of chloroplasts; TIC, translocase of the inner envelope membrane of chloroplasts; HSQC, heteronuclear single-quantum coherence
Keywords: Dual targeting, Chloroplasts and mitochondria, Protein import, NMR, Toc34 receptor
Highlights
-
•
The mechanism of dual targeting of proteins to mitochondria and chloroplasts is poorly understood.
-
•
The interaction between a dually targeted peptide and the chloroplastic receptor Toc34 was examined.
-
•
The interaction between AtThrRS-dTP(2–60) and AtToc34 involves residues throughout the entire targeting peptide sequence.
-
•
The interaction of AtThrRS-dTP(2–60) with AtToc34 is different to the interaction with AtTom20.
Abstract
Organellar proteins synthesized in the cytosol are usually selective for only one destination in a cell but some proteins are localized in more than one compartment, for example in both mitochondria and chloroplasts. The mechanism of dual targeting of proteins to mitochondria and chloroplasts is yet poorly understood. Previously, we observed that the dual targeting peptide of threonyl-tRNA synthetase in Arabidopsis thaliana (AtThrRS-dTP) interacts with the mitochondrial receptor AtTom20 mainly through its N-terminal part. Here we report on the interaction of AtThrRS-dTP with the chloroplastic receptor AtToc34, presenting for the first time the mode of interactions of a dual targeting peptide with both Tom20 and Toc34. By NMR spectroscopy we investigated changes in 15N HSQC spectra of AtThrRS-dTP as a function of AtToc34 concentration. Line broadening shows that the interaction with AtToc34 involves residues along the entire sequence, which is not the case for AtTom20. The N-terminal φχχφφ motif, which plays an important role in AtTom20 recognition, shows no specificity for AtToc34. These results are supported by import competition studies into both mitochondria and chloroplasts, in which the effect of peptides corresponding to different segments of AtThrRS-dTP on in vitro import of organelle specific proteins was examined. This demonstrates that the N-terminal A2-Y29 segment of AtThrRS-dTP is essential for import into both organelles, while the C-terminal L30-P60 part is important for chloroplastic import efficiency. In conclusion, we have demonstrated that the recognition of the dual targeting peptide of AtThr-tRNA synthetase is different for the mitochondrial and chloroplastic receptors.
1. Introduction
Correct transport of proteins to their respective organellar destinations is an essential process for biogenesis and cell function. The great majority of proteins are destined to one specific location in a cell and have specific targeting sequences, but some proteins, termed dual targeted proteins, despite the fact that they are encoded by one single gene can be targeted to two or multiple sub-cellular compartments. Among them, most are dually targeted to mitochondria and chloroplasts and to date over 100 identical proteins have been found in both mitochondria and chloroplasts by means of in vivo and in vitro studies [1–3]. The concept of dual targeting was first proposed in 1990 for a yeast leader peptide [4] and a few years later, glutathione reductase (GR) from Pisum sativum was identified to be a dual targeted protein to both mitochondria and chloroplasts [5,6]. An examination of proteins that are dually targeted revealed that they encode basic essential functions required in mitochondria and chloroplasts and are particularly enriched in the categories involving gene expression, such as DNA replication, transcription and protein synthesis. Furthermore, proteins associated with organellar quality control mechanisms have also been found within these categories [7,8]. Studies of the expression patterns of dual targeted proteins revealed that they are constitutively expressed across different tissues and across plant development [1]. There are many important perspectives on dual targeting concerning evolutionary pathways and the molecular mechanisms of dual targeting.
In general, nuclear encoded proteins destined to mitochondria are synthesized as precursor proteins with a N-terminal mitochondrial targeting peptide (mTP) that can be recognized by organellar receptors. In the same way, a chloroplastic targeting peptide (cTP) contains the necessary information to deliver a protein to chloroplasts [7]. For dual targeted proteins, resulting from the expression of one single gene, an ambiguous dual targeting peptide (dTP) at the N-terminus of the precursor protein, plays an important role. It has been demonstrated that dual targeted proteins use the same protein import machineries as organelle specific proteins and that the physicochemical properties of dTPs are intermediary in comparison to mTPs and cTPs. The largest difference among different targeting peptides occurs in the N-terminal part of the sequence. dTPs contain more serines as compared to mTPs, and more arginines as compared to cTPs in the N-terminal portion of the peptide [9,10]. Also, dTPs contain less negatively charged residues in comparison to both mTPs and dTPs. The ambiguous dTPs have been proposed to function by different mechanisms. Some dTPs have been suggested to harbour distinct organellar signals located in different domains within the sequence, such as, e.g., in GR [6] or Presequence Protease, PreP [11]. Other dTPs are recognized by mitochondrial and chloroplastic import machineries with the targeting information spread throughout the sequence [1–3].
In Arabidopsis thaliana, 18 aminoacyl-tRNA synthetases, aaRSs, are dually targeted to mitochondria and chloroplasts. Previously, the targeting determinants of the threonyl-tRNA synthetase dual targeting peptide, AtThrRS-dTP, were studied using several N- and C-terminal deletion variants and in vitro import into mitochondria and chloroplasts. These studies revealed that the shortest peptide that was capable of conferring dual targeting to mitochondria and chloroplasts was 60 amino acids long, AtThrRS-dTP(2–60) [12]. Moreover, AtThrRS-dTP(2–60) was shown to carry its dual targeting capacity without separate organelle-specific domains and CD spectra of the peptide revealed that it is mostly unstructured in aqueous environment, but has a propensity to form amphiphilic α-helix structure in membrane mimetic media [12]. In the present study we have focused on the determinants of the AtThrRS-dTP(2–60) for import into chloroplasts.
Tom20 and Toc34 are the primary import receptors of the translocase of the outer membrane of mitochondria (TOM) and the translocase of the outer envelope of chloroplasts (TOC), respectively, in plants. They play key roles during protein import process as they can distinguish the information between mTPs and cTPs and thus allow the protein to be imported to the correct organelle [13–16]. Nonetheless, dTPs are recognized by both receptors, and the determinants for recognition are not yet understood. In a recent study, we investigated the interaction between AtThrRS-dTP(2–60) and AtTom20 using NMR spectroscopy [17]. Our results showed that the interaction between the AtThrRS-dTP(2–60) and the receptor fulfills the key characteristic for the interaction between Tom20 and a mTP: an amphiphilic helical region is involved, i.e., F9-V28. Moreover, this region contains an important mTP φχχφφ motif (L24RRFV28) where ϕ represents a hydrophobic/aromatic residue and χ represents any amino acid [18–20]. In addition to the similarities to Tom20–mTP interaction, a second segment in AtThrRS-dTP(2–60), L30-Q39, also takes part in AtTom20 recognition.
In contrast to interactions between mTPs and Tom20, which have been intensively studied, knowledge about Toc34–cTP interaction at a molecular level is yet limited. In Arabidopsis, two isoforms of this receptor have been identified, AtToc33 and AtToc34 [21]. The two isoforms show different expression profiles reflecting their different substrate specificities. AtToc33 is involved in import of photosynthetic proteins, whereas AtToc34 is involved in import of house-keeping proteins [16,22,23]. As AtThrRS belongs to the latter class of proteins, AtToc34 was chosen in this study for investigation of the interaction with AtThrRS-dTP(2–60). We have used a combination of NMR spectroscopy and biochemical studies to examine the interaction between AtThrRS-dTP(2–60) and AtToc34. In the biochemical studies, different segments of AtThrRS-dTP(2–60) were applied for in vitro import competition studies to mitochondria and chloroplasts.
Our studies revealed for the first time the molecular details on the interaction of a dTP with the chloroplastic receptor Toc34. Further, it exposed differences in the interaction of a dTP with Toc34 compared to the mitochondrial Tom20 receptor.
2. Results
2.1. Mapping the AtToc34 interaction sites in AtThrRS-dTP(2–60)
In order to understand the dual targeting ability of AtThrRS-dTP(2–60), the interaction between the peptide and AtToc34 was studied by NMR spectroscopy. To facilitate studies, a construct of AtToc34 (Toc34ΔTM252–6His) in which the transmembrane domain was deleted was used (hereafter referred to as AtToc34). The assignments of the spectrum have been obtained previously [17]. One may note that the chemical shifts of a few residues (e.g. Thr21 and Thr32) were sensitive to sample preparation. The assignments were, however, verified and the spectrum remained the same for the one batch of peptide. Changes in the 15N HSQC spectra of AtThrRS-dTP(2–60) peptide were monitored as a function of AtToc34 concentration, ranging from 0 to 1:1 AtToc34:AtThrRS-dTP(2–60) molar ratio. Spectra of the first few titration points (molar ratios of 0:1–0.5:1) are shown in Fig. 1. At a molar ratio of 0.5:1 most of the cross-peaks for AtThrRS-dTP(2–60) were not observed with these display parameters (Fig. 1D). The main effects on the cross-peaks were related to line broadening and intensity loss rather than large chemical shift changes (Fig. 1). The line broadening of the cross-peaks at moderate AtToc34 concentration indicates that binding to AtToc34 gives rise to much slower tumbling motion for AtThrRS-dTP(2–60) and that exchange processes between the on- and off-states most likely exist.
Fig. 1.
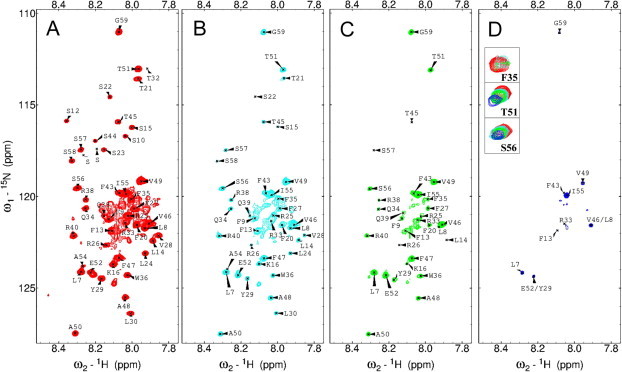
15N HSQC spectra from the titration of AtToc34 onto 80 μM 15N-ThrRS-dTP(2–60) recorded at 700 MHz 1H frequency, 298 K. (A) Assigned 15N HSQC spectrum of 80 μM ThrRS-dTP(2–60) in 50 mM sodium phosphate buffer at pH 6. (B–D) shows spectra recorded for mixtures of AtToc34 and ThrRS-dTP with molar ratios of 0.1:1 (B), 0.2:1 (C) 0.5:1 (D). All spectra were recorded under identical conditions and are shown with the same display parameters. Cross-peaks for F35, T51 and S56 are shown as insets in (D), red, cyan, green and blue correspond to 0, 0.1, 0.2 and 0.5 molar ratio of Toc34, respectively. Note that different contour levels were applied in the insets in order to be able to display peaks at higher Toc34 concentrations and that the crosspeak for F35 at 0.5:1 molar ratio has disappeared.
As can be seen in Fig. 1A and B, the peptide was very sensitive to the addition of AtToc34. At a AtToc34/AtThrRS-dTP(2–60) concentration ratio of 0.1:1, the intensities of most of the peaks were severely reduced. In order to examine the distribution of the effects along the sequence of the peptide, we calculated the intensities of cross-peaks of each residue in the spectra for the samples with AtToc34/AtThrRS-dTP(2–60) molar ratios of 0.1:1, 0.2:1 and 0.5:1 relative to the spectra for the sample containing exclusively AtThrRS-dTP(2–60) (Fig. 2A). The intensity loss observed for the cross-peaks was pronounced but not specific to any region in the sequence, which is different from previous results for the AtThrRS-dTP(2–60)–AtTom20 interaction [17]. Note that even the very C-terminal region, the region that was the least affected by AtTom20, was severely broadened out by the addition of AtToc34. Only 9 peaks had significant intensities remaining at a AtToc34/AtThrRS-dTP(2–60) molar ratio of 0.5:1 and these belong to L7, F13, R33, F43, V46/L8 (overlap), V49, Y29/E52 (overlap), I55, and G59 which are mostly hydrophobic residues and spread out all over the sequence. Contrary to the evident change in the cross-peak intensity, the initial chemical shift perturbation was minor at molar ratios below 0.5:1 (Fig. 2A and B). There were only 2 peaks with a chemical shift difference larger than cut-off values (ΔδHN > 0.01 and ΔδNH > 0.2), belonging to F35 and Q39 (Fig. 2B). Neither of them was among the ones that were previously observed to have weak structural propensities. Moreover, none of the residues from the φχχφφ motif, which is believed to be a AtTom20 recognition element [19], appears to be the most affected by addition of AtToc34. Taking the line-broadening and chemical shift effects of adding AtToc34 to AtThrRS-dTP(2–60) at the first few titration points together (Fig. 2C), we can summarize that the increase in line-width of peaks occurs all along the sequence with no specific region being more affected and that the chemical shift changes are in general minor. The main effect of addition of the receptor protein is line-broadening rather than chemical shift changes, even more so than previously observed with AtTom20, again indicating that the receptor interactions are characterized by dynamic events. The overall line-broadening effects were also much larger than what was previously found for the interaction with AtTom20.
Fig. 2.
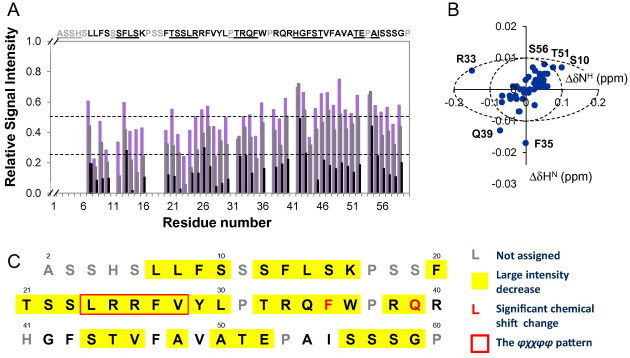
Effects on the ThrRS-dTP(2–60) 15N-HSQC spectrum upon addition of AtToc34. (A) The relative change in signal intensity of cross-peaks in 15N HSQC spectra upon titration. The bars indicate the relative change in signal intensity compared to intensities in a spectrum recorded for a 80 μM ThrRS-dTP(2–60) sample without AtToc34. Purple indicates a 1:0.1 molar ratio of ThrRS-dTP and AtToc34; Grey indicates a 1:0.2 molar ratio and black indicates a 1:0.5 molar ratio. (B) HN and NH chemical shift differences in the spectrum of 80 μM ThrRS-dTP(2–60) without AtToc34 and with 16 μM AtToc34 (AtToc34:ThrRS-dTP, 0.2:1). The shifts are reported as δwith–δwithout. The cut-off limits for significant chemical shift differences, chosen as ΔδHN > 0.01, and ΔδNH > 0.2 are shown. Residues with shift perturbations around this cut-off are indicated. (C) Summary of the effect of adding AtToc34 to ThrRS-dTP(2–60) at a molar ratio of 0.2:1. The positions with a significant change in chemical shifts by AtToc34 addition have letters in red. The φχχφφ motif, previously reported as a Tom20 recognition element, is indicated by the red frame. The positions that show a decrease in the signal intensity ratio of more than 0.5 are highlighted yellow. Letters in grey are prolines or unassigned residues.
Further, dramatic changes in the appearance of the spectra occurred as AtToc34/AtThrRS-dTP(2–60) molar ratio reached 1–1 (Fig. 3). Most of the peaks disappeared, and a subset of new, very weak peaks appeared with a larger apparent spectral dispersion in the 1H backbone amide region. This new spectrum is most likely from a combination of a bound state of the peptide, along with very weak peaks stemming from degradation products. The new weak peaks were not possible to assign, but at any rate these changes indicate that a structure in AtThrRS-dTP(2–60) is to some degree probably introduced by addition of the receptor AtToc34.
Fig. 3.
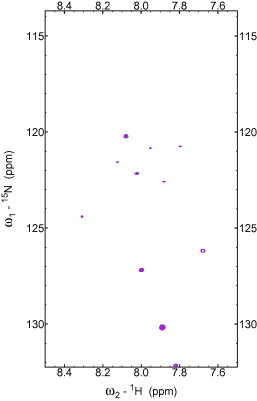
15N HSQC spectrum recorded for 80 μM 15N-ThrRS-dTP(2–60) at the presence of the same amount of AtToc34. The experimental condition and the display parameters are the same as that of the spectra shown in Fig. 1.
In summary, we observed that adding AtToc34 mainly induced line-broadening and intensity loss for a large portion of the targeting sequence, indicating, in contrast to what has previously been observed for the interaction with AtTom20, that most of the sequence is involved in the interaction.
2.2. Import capacities of different segments of AtThrRS-dTP
To confirm specific interaction of the dual targeting peptide of AtThrRS with AtToc34, we have added AtToc34, to the isolated chloroplasts prior to the import of precursor protein containing 100 N-terminal amino acids of the AtThrRS precursor fused to a passenger protein, red fluorescent protein, RFP, AtThrRS(1–100)-RFP. As shown in Fig. 4, AtToc34 inhibited import of AtThrRS(1–100)-RFP (lanes 4–5), revealing specific binding of Toc34 to the dual targeting peptide of AtThrRS. As controls, we show that the AtThrRS(1–100)-RFP precursor was effectively imported into chloroplasts without AtToc34 pre-incubation (lanes 2–3) or with addition of BSA, (lanes 6–7).
Fig. 4.
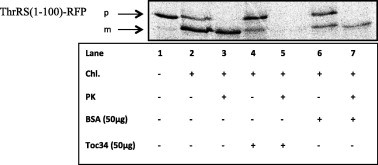
Effect of Toc34 on in vitro import of AtThrRS(1–100)-RFP into chloroplasts. Lane 1, in vitro transcription/translated product only (10% of input). Lane 2, in vitro transcription/translated product incubated with chloroplasts. Lane 3, the same as lane 2, but treated with Proteinase K (PK) after the import reaction, for detection of protected translation product inside the organelle. Lanes 4 and 5 are as lane 2 and 3 but after addition of 50 μg of the purified Toc34 to the chloroplasts. Lanes 6 and 7 are as lanes 4 and 5 but after addition of 50 μg of BSA instead of Toc34.
Further, we studied the interaction of different parts of the AtThrRS-dTP peptide with organellar receptors, Tom20 and Toc34. Peptides derived from different segments of the AtThrRS-dTP were synthesized and their effects were tested on mitochondrial and chloroplastic in vitro import of organelle-specific proteins that use Tom20 and Toc34 during import. Peptides corresponding to the N- and C-terminal segments of AtThrRS-dTP were selected on the basis of the previously established structural properties of AtThrRS-dTP(2–60) and its interactions with AtTom20 [12,17] and AtToc34 (present study). As mentioned previously, AtThrRS-dTP(2–60) has the propensity to form an amphiphilic α-helical structure in the F9-V28 region and it contains a previously identified mTP φχχφφ motif (ϕ represents a hydrophobic/aromatic residue and χ represents any amino acid). Therefore, the following peptides were used for the import competition studies: AtThrRS-dTP(1–29), AtThrRS-dTP(1–19), AtThrRS-dTP(20–29) and AtThrRS-dTP(30–60) (Fig. 5A). We pre-incubated the isolated organelles with the AtThrRS-dTP(2–60) derived peptides prior to import of two organelle-specific precursors, either the mitochondrial precursor of the F1β subunit of the ATP synthase, pF1β (Fig. 5B) or the chloroplastic precursor of the small subunit of Rubisco, pSSU (Fig. 5C). The import into both organelles was strongly inhibited (73% for mitochondria and 88% for chloroplasts) by the N-terminal amphiphilic helical AtThrRS-dTP(1–29) peptide (lanes 4 and 5). The truncated versions of the AtThrRS-dTP(1–29) peptide, AtThrRS-dTP(1–19) and AtThrRS-dTP(20–29), exhibited only a weak inhibitory effect (⩽22%) on both organelles (lanes 6–9). Interestingly, the C-terminal unstructured AtThrRS-dTP(30–60) part of the dual targeting peptide revealed very strong inhibition (90%) of the chloroplast import and affected the mitochondrial import to much lower extent (25%) (lanes 10 and 11).
Fig. 5.
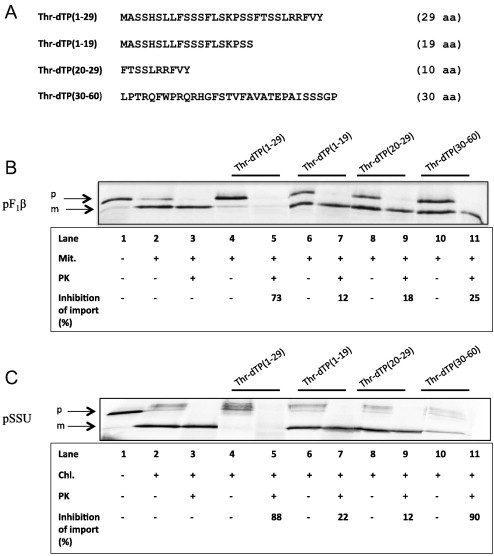
Effect of AtThrRS-dTP(1–60) peptide fragments on in vitro import of the pF1β into mitochondria and pSSU into chloroplasts. Organelles were preincubated with 20 μg of each peptide fragment prior to the incubation with the precursors. (A) Sequences of peptide fragments derived from AtThrRS-dTP(1–60). (B) Lane 1, in vitro transcription/translated product only (10% of input). Lane 2, in vitro transcription/translated product incubated with mitochondria. Lane 3, the same as lane 2, but treated with PK after the import reaction, for detection of protected translation product inside the organelle. Lanes 4–5, 6–7, 8–9 and 10–11 are the same as lanes 2–3, but mitochondria were pre-incubated with 20 μg of each peptide fragment prior to import. (C) The same as in (B) but with chloroplasts instead of mitochondria in the reaction mixtures.
Additionally, we performed CD spectroscopy measurements to analyze secondary structures of the AtThrRS-dTP(2–60) peptide fragments (Fig. 6). We do indeed see that AtThrRS-dTP(1–29) possesses high helical propensity, whereas AtThrRS-dTP(30–60) and the truncated peptides AtThrRS-dTP(1–19) and AtThrRS-dTP(20–29) do not. Taken together, our results imply that the mitochondrial receptor Tom20 interacts mostly with the N-terminal portion of the AtThrRS-dTP(2–60) peptide, whereas the chloroplastic receptor Toc34 interacts with both the N-and C-terminal segments of the dual targeting peptide. The interaction of Toc34 with the C-terminal part of AtThrRS-dTP(2–60) shows that a helical structure is not required.
Fig. 6.
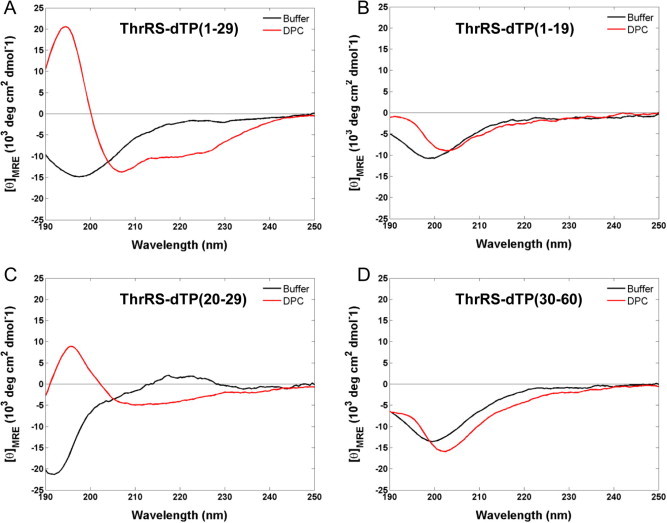
CD spectra recorded for ThrRS-dTP peptide fragments: (A) ThrRS-dTP(1–29), (B) ThrRS-dTP(1–19), (C) ThrRS-dTP(20–29) and (D) ThrRS-dTP(30–60). Spectra were recorded for peptides with a concentration of 125 μM in 50 mM phosphate buffer (black), and 100 mM DPC (red) at 25 °C.
3. Discussion
Due to the inherent similarity of amino acid composition of mitochondrial targeting peptides (mTPs) and chloroplastic targeting peptides (cTPs), it is difficult to distinguish the specific components in a dual targeting peptide (dTP) for either mitochondria or chloroplasts. Sequence analyses have shown that most differences between mTPs and cTPs are located in the N-terminal portion of TPs, where arginines are overrepresented in mTPs, and that serine and proline residues are more abundant in cTPs [9]. The importance of arginine in mTPs for mitochondria recognition is still a debate and these residues seem to be mostly involved in transport through the TOM and TIM complexes [24,25]. The selectivity of cTPs for chloroplastic import has not been abolished by removal of serines [26], and the role of proline residues is most probably to keep the targeting peptide in an unstructured flexible conformation. Both mTPs and cTPs are intrinsically disordered in aqueous environment. Nevertheless, an α-helix in the N-terminal region has been observed for most of mTPs in membrane mimetics, e.g., in the alternative oxidase (AOX) targeting peptide [27] and the F1β mTP [28]. Moreover, the helical structure of mTPs is amphiphilic, which appears to be very important for recognition by the Tom20 receptor. The hydrophobic side of the amphiphilic helix has been reported to constitute the Tom20 binding sites [17,28]. In contrast to mTPs, cTPs rarely form helical structures although exceptions do exist (one of them is exemplified by the transit peptide in Rubisco activase enzyme) [29]. In previous work, the dual targeting peptide of AtThr-tRNA synthetase was shown to be unstructured in buffer but had a propensity to form an amphiphilic helix in the N-terminal part in membrane mimetic media [17]. Moreover, in a recent study, it has been suggested that the enrichment of arginines in mTPs may function as an “avoidance signal” for chloroplast import [30].
We now turn to a comparison of the binding of AtThrRS-dTP(2–60) to the two receptors. A summary of the effects of the two receptors on the line-broadening and chemical shift differences observed for AtThrRS-dTP(2–60) is shown (Fig. 7). A φχχφφ motif has previously been identified by NMR studies for a group of mTPs upon their interaction with Tom20 and was shown to be important for the interaction [19]. In this motif ϕ represents a hydrophobic/aromatic residue and χ represents any amino acid residue, but usually one with a long aliphatic side-chain. In the present study we find that the φχχφφ motif is not as important for AtToc34 recognition as it is for AtTom20 recognition. On the contrary, it appears that there are no particular residues or regions in the sequence of AtThrRS-dTP(2–60) that are specifically affected by the AtToc34 interaction (Fig. 7). The significant line-broadening together with the corresponding drop in cross-peak intensity observed for most peaks indicate that most of the AtThrRS-dTP(2–60) sequence participates directly in the interaction, or at least that the interaction affects the whole sequence equally.
Fig. 7.
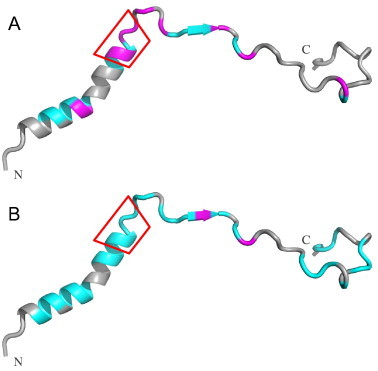
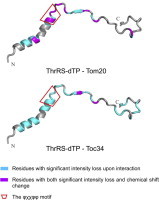
Comparison of the distribution of affected cross-peak intensities and chemical shifts of the ThrRS-dTP(2–60) in 15N-HSQC spectrum upon addition of AtTom20 (A) and AtToc34 (B). The structure of the dTP was constructed by using PyMOL and was based on secondary structural propensity information from previous work [17]. Residues with only significant intensity loss (⩾50%) after addition of receptors are marked in cyan and residues with both large intensity loss and chemical shift differences (ΔδHN ⩾ 0.01 and ΔδNH > 0.1 are marked in magenta. The φχχφφ motif is indicated by a red box.
These results are additionally supported by biochemical studies, in which organelle-specific in vitro import was studied. Different peptides derived from the AtThrRS-dTP(2–60) sequence were pre-incubated with mitochondria and chloroplasts prior to import reactions. In agreement with the NMR results, these data revealed that the mitochondrial import was mostly inhibited by the N-terminal helical segment of the AtThrRS-dTP(2–60), whereas the import into chloroplasts was equally affected by both the N- and C-terminal peptides. CD spectroscopy measurements of the peptide fragments showed that the N-terminal 29 amino acid residues of the dTP have helical propensity, whereas the C-terminal 30 amino acid residues do not (cf. Fig. 6). The two small peptide fragments corresponding to the N- and C-terminal part of AThrRS-dTP(1–29), which break the presumed amphiphilic α-helical region F9-V28, show no or very low helical propensity. As the peptide fragments used in the in vitro import competition studies possess structures that previously have been described as determinants for receptor recognition, it gives strong evidence that the inhibition of import occurs at the receptor level [27,28,31,32].
Previous studies have shown that cTPs with reversed sequences but with the same composition can be recognized by the TOC machinery, based on their physico-chemical properties [31,33]. cTPs are on average longer and also more flexible than mTPs, which might be related to the fact that there are several receptors on the chloroplast outer envelope membrane that cooperatively can bind to the cTPs [34,35]. Furthermore, cTPs can bind to a so called “guidance-complex” consisting of 14-3-3 proteins and Hsp70 as well as to Hsp90 in the cytosol which enhances the import kinetics [36,37]. This might require less specific interaction with each of the receptors but would constitute a very high fidelity. In the present study, we observe that the dual targeting peptide of ThrRS reveals a more specific interaction to Tom20 involving the φχχφφ in the N-terminal region, whereas the interaction to Toc34 involves the whole peptide sequence. Thus, it is therefore likely that the accumulated physico-chemical properties of the AtThrRS-dTP sequence play a role in the AtToc34/AtThrRs-dTP interaction. As the mitochondrial surface area is roughly 10 times less than the chloroplastic surface area, the dTPs should be required to bind more specifically to Tom20 (via the φχχφφ motif) in order to “compete” with chloroplasts for the same precursor protein.
In summary, our work has for the first time presented the interaction of a dual targeting peptide with the Toc34 receptor on the molecular level. It also enabled a comparison of the interaction of an ambiguous targeting peptide directing the protein to both organelles with the mitochondrial Tom20 and chloroplastic Toc34 receptors and clearly revealed different modes of action with the two receptors.
4. Materials and methods
4.1. Sample preparation
Cloning of the construct of ThrRS-dTP(1–60) and purification of ThrRS-dTP(2–60) were done as described previously by Berglund et al. [12]. In short, GST-ThrRS-dTP(1–60) was overexpressed as a fusion protein in Escherichia coli BL21-DE3 cells, forming inclusion bodies. The inclusion bodies containing the insoluble protein were purified and ThrRS-dTP(2–60) was obtained by CNBr cleavage, which cuts peptide bonds after the methionines in ThrRS-dTP(1–60), as described earlier [38]. The ThrRS-dTP(2–60) was purified using cation exchange chromatography. 15N-labelled ThrRS-dTP(2–60) were overexpressed in a minimal culture medium with 15N-enriched NH4Cl (1 g/L) and glucose (2 g/L). Dialyzed peptide was lyophilized and dissolved in a solution containing 50 mM sodium phosphate buffer (pH 6.0) and 10% (v/v) D2O. The final concentration of the peptide was about 80 μM.
E. coli BL21-DE3 cells harbouring the plasmid pET21d-Toc34ΔTM252–6His, with the transmembrane domain removed, were grown at 37 °C in Luria-Bertani medium, to a density of A600 between 0.6 and 0.8. Expression of AtToc34ΔTM252–6His was achieved by induction with Isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM overnight, at 12 °C. AtToc34ΔTM252 was purified by Ni-NTA column (GE Healthcare; www.gehealthcare.com). The purified protein was dialyzed against 20 mM sodium phosphate buffer (pH 6.0), lyophilized and dissolved in H2O to a concentration of 1 mM in 100 mM sodium phosphate buffer (pH 6.0).
4.2. NMR spectroscopy
All NMR experiments were carried out at 25 °C on a 700 MHz Bruker Avance spectrometer, equipped with cryogenically cooled probe. The data were processed and analyzed with TopSpin 3.0, NMRpipe and Sparky 3 [39]. For Toc34 interaction studies, AtToc34 was titrated into a ThrRS-dTP(2–60) sample. The titration was started by recording a 2D 15N HSQC spectrum of a sample of 400 μl 80 μM 15N-labeled ThrRS-dTP(2–60) in 50 mM phosphate buffer (pH 6.0). Later, 4.6 μl, 4.6 μl, 13.8 μl and 23 μl of 0.7 mM, unlabeled AtToc34 was added to the ThrRS-dTP(2–60) sample in steps, giving molar ratios of AtToc34:ThrRS-dTP(2–60) of 0.1:1, 0.2:1, 0.5:1 and 1:1. The overall volume change using this procedure was less than 10%. To monitor possible effects of small pH changes induced by the addition of the solution containing AtToc34, additional experiments on another 15N-labeled ThrRS-dTP(2–60) sample were made at both pH 6.0 and pH 6.5. The results show that no apparent influence on the ThrRS-dTP(2–60) are observed due to small changes in pH.
4.3. In vitro import
The precursors: pF1β (Nicotiana plumbaginifolia), pSSU (Spinacia oleracea) and AtThrRS(1–100)-RFP were expressed in a coupled transcription/translation system in the presence of [35S]-methionine according to the manufacturer’s instructions (Promega). AtThrRS-dTP(1–60) derived peptide fragments were synthesized by GenicBio (http://www.genicbio.com). Mitochondria and chloroplasts were isolated as described in [40] and [41], respectively. In vitro import reactions were performed according to [42] and [41]. In order to study the effect of AtToc34 on in vitro import of AtThrRS(1–100)-RFP, purified AtToc34ΔTM252 and BSA (25, 50 and 75 μg) were added to the import reactions. In order to study the effect of AtThrRS-dTP(1–60) peptide fragments on the in vitro import of pF1β, and pSSU, isolated mitochondria and chloroplasts were pre-incubated with 20 μg of each peptide for 10 min on ice, prior to addition of the precursor. Import efficiency was calculated as a ratio between mature protein after PK treatment/input protein. To assess inhibition of import by peptide fragments, import efficiency of each reaction with peptide fragments was divided by the import efficiency of the reaction without peptides. Each experiment was repeated three times, gels were quantified and mean values were taken for calculation of inhibition of import.
4.4. CD spectroscopy
CD measurements were performed for the peptide fragments used in the in vitro import assay using Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK) at 25 °C in the range of 190–250 nm. The time per points was 2 s, with 1 nm bandwidth and 0.5 nm step resolution. A quartz cuvette of 0.05 mm path length was used. 10–15 spectra of each of the four ThrRS-dTP fragments in 50 mM phosphate buffer (pH 6.0) or 100 mM DPC (pH 6.0) were recorded and averaged. The concentration of the peptide was about 125 μM. A background spectrum of the solvent was subtracted from the peptide spectrum.
Acknowledgements
This work was supported by grants from the Swedish Research Council (to L.M. and E.G.) and by a grant from the Carl Trygger Foundation. Support from the Knut and Alice Wallenberg Foundation for the NMR spectrometer is gratefully acknowledged. We are grateful to Prof. Jürgen Soll for the generous gift of the Toc34-deltaTM clone. W.Y., E.S., E.G. and L.M. conceived and designed the project. W.Y. and E.S. acquired and analyzed the data. W.Y., E.S., E.G. and L.M. interpreted the data and wrote the paper.
References
- 1.Carrie C., Giraud E., Whelan J. Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- 2.Carrie C., Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta. 2013;1833:253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 3.Silva-Filho M.C. One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr. Opin. Plant Biol. 2003;6:589–595. doi: 10.1016/j.pbi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Huang J., Hack E., Thornburg R.W., Myers A.M. A yeast mitochondrial leader peptide functions in vivo as a dual targeting signal for both chloroplasts and mitochondria. Plant Cell. 1990;2:1249–1260. doi: 10.1105/tpc.2.12.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creissen G., Reynolds H., Xue Y., Mullineaux P. Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J. 1995;8:167–175. doi: 10.1046/j.1365-313x.1995.08020167.x. [DOI] [PubMed] [Google Scholar]
- 6.Rudhe C., Clifton R., Whelan J., Glaser E. N-terminal domain of the dual-targeted pea glutathione reductase signal peptide controls organellar targeting efficiency. J. Mol. Biol. 2002;324:577–585. doi: 10.1016/s0022-2836(02)01133-6. [DOI] [PubMed] [Google Scholar]
- 7.Glaser E., Soll J. Targeting signals and import machinery of plastids and plant mitochondria. In: Daniell H., Chase C., editors. Molecular Biology and Biotechnology of Plant Organelles Chloroplasts and Mitochondria. Springer; 2004. pp. 385–417. [Google Scholar]
- 8.Christensen A.C., Lyznik A., Mohammed S., Elowsky C.G., Elo A., Yule R., Mackenzie S.A. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell. 2005;17:2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhushan S., Kuhn C., Berglund A., Roth C., Glaser E. The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett. 2006;580:3966–3972. doi: 10.1016/j.febslet.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X.P., Glaser E. Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 2002;7:14–21. doi: 10.1016/s1360-1385(01)02180-x. [DOI] [PubMed] [Google Scholar]
- 11.Bhushan S., Lefebvre B., Ståhl A., Wright S.J., Bruce B.D., Boutry M., Glaser E. Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 2003;4:1073–1077. doi: 10.1038/sj.embor.7400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berglund A., Spånning E., Biverståhl H., Maddalo G., Tellgren-Roth C., Mäler L., Glaser E. Dual targeting to mitochondria and chloroplasts: characterization of Thr–tRNA synthetase targeting peptide. Mol. Plant. 2009;2:1298–1309. doi: 10.1093/mp/ssp048. [DOI] [PubMed] [Google Scholar]
- 13.Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R., Hulett J.M., Lithgow T., Whelan J. Protein import into mitochondria: origins and functions today. Mol. Membr. Biol. 2005;22:87–100. doi: 10.1080/09687860500041247. [DOI] [PubMed] [Google Scholar]
- 15.Benz J.P., Soll J., Bölter B. Protein transport in organelles: the composition, function and regulation of the Tic complex in chloroplast protein import. FEBS J. 2009;276:1166–1176. doi: 10.1111/j.1742-4658.2009.06874.x. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- 17.Ye W., Spånning E., Unnerståle S., Gotthold D., Glaser E., Mäler L. NMR investigations of the dual targeting peptide of Thr-tRNA synthetase and its interaction with the mitochondrial Tom20 receptor in Arabidopsis thaliana. FEBS J. 2012;279:3738–3748. doi: 10.1111/j.1742-4658.2012.08735.x. [DOI] [PubMed] [Google Scholar]
- 18.Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 19.Muto T., Obita T., Abe Y., Shodai T., Endo T., Kohda D. NMR identification of the Tom20 binding segment in mitochondrial presequences. J. Mol. Biol. 2001;306:137–143. doi: 10.1006/jmbi.2000.4397. [DOI] [PubMed] [Google Scholar]
- 20.Perry A.J., Hulett J.M., Likic V.A., Lithgow T., Gooley P.R. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 2006;16:221–229. doi: 10.1016/j.cub.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 21.Jelic M., Soll J., Schleiff E. Two Toc34 homologues with different properties. Biochemistry. 2003;42:5906–5916. doi: 10.1021/bi034001q. [DOI] [PubMed] [Google Scholar]
- 22.Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell. 2003;15:1859–1871. doi: 10.1105/tpc.012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt O., Pfanner N., Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 25.Murcha M.W., Kmiec B., Kubiszewski-Jakubiak S., Teixeira P.F., Glaser E., Whelan J. Protein import into plant mitochondria: signals, machinery, processing, and regulation. J. Exp. Bot. 2014;65:6301–6335. doi: 10.1093/jxb/eru399. [DOI] [PubMed] [Google Scholar]
- 26.Nakrieko K., Mould R.M., Smith A.G. Fidelity of targeting to chloroplasts is not affected by removal of the phosphorylation site from the transit peptide. Eur. J. Biochem. 2004;271:509–516. doi: 10.1046/j.1432-1033.2003.03950.x. [DOI] [PubMed] [Google Scholar]
- 27.Rimmer K.A., Foo J.H., Ng A., Petrie E.J., Shilling P.J., Perry A.J., Mertens H.D.T., Lithgow T., Mulhern T.D., Gooley P.R. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J. Mol. Biol. 2011;405:804–818. doi: 10.1016/j.jmb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Moberg P., Nilsson S., Stahl A., Eriksson A., Glaser E., Mäler L. NMR solution structure of the mitochondrial F1beta presequence from Nicotiana plumbaginifolia. J. Mol. Biol. 2004;336:1129–1140. doi: 10.1016/j.jmb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Bruce B.D. The role of lipids in plastid protein transport. Plant Mol. Biol. 1998;38:223–246. [PubMed] [Google Scholar]
- 30.Ge C., Spanning E., Glaser E., Wieslander A. Import determinants of organelle-specific and dual targeting peptides of mitochondria and chloroplasts in Arabidopsis thaliana. Mol. Plant. 2014;7:121–136. doi: 10.1093/mp/sst148. [DOI] [PubMed] [Google Scholar]
- 31.Bruce B.D. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta. 2001;1541:2–21. doi: 10.1016/s0167-4889(01)00149-5. [DOI] [PubMed] [Google Scholar]
- 32.von Heijne G., Nishikawa K. Chloroplast transit peptides. The perfect random coil? FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- 33.Chotewutmontri P., Reddick L.E., McWilliams D.R., Campbell I.M., Bruce B.D. Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell. 2012;24:3040–3059. doi: 10.1105/tpc.112.098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommer M., Rudolf M., Tillmann B., Tripp J., Sommer M.S., Schleiff E. Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ. 2013;36:970–983. doi: 10.1111/pce.12030. [DOI] [PubMed] [Google Scholar]
- 35.Aronsson H., Jarvis P. Dimerization of TOC receptor GTPases and its implementation for the control of protein import into chloroplasts. Biochem. J. 2011;436:e1–e2. doi: 10.1042/BJ20110659. [DOI] [PubMed] [Google Scholar]
- 36.May T., Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fellerer C., Schweiger R., Schongruber K., Soll J., Schwenkert S. Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant. 2011;4:1133–1145. doi: 10.1093/mp/ssr037. [DOI] [PubMed] [Google Scholar]
- 38.Pavlov P., Moberg P., Zhang X., Glaser E. Chemical cleavage of the overexpressed mitochondrial F1beta precursor with CNBr: a new strategy to construct an import-competent preprotein. Biochem. J. 1999;341:95–103. [PMC free article] [PubMed] [Google Scholar]
- 39.Goddard T., Kneller D. University of California; San Francisco: 2004. SPARKY 3. [Google Scholar]
- 40.Hamasur B., Birgersson U., Eriksson A., Glaser E. Large-scale purification procedure of spinach leaf mitochondria—isolation and immunological studies of the F1–ATPase. Physiol. Plant. 1990;78:367–373. [Google Scholar]
- 41.Bruce B.D., Perry S., Froehlich J., Keegstra K. Plant Molecular Biology Manual. Springer; 1994. In vitro import of proteins into chloroplasts; pp. 497–511. [Google Scholar]
- 42.Whelan J., Knorpp C., Glaser E. Sorting of precursor proteins between isolated spinach leaf mitochondria and chloroplasts. Plant Mol. Biol. 1990;14:977–982. doi: 10.1007/BF00019394. [DOI] [PubMed] [Google Scholar]


