Abstract
The ability to introduce different biophysical probes into defined positions in target proteins will provide powerful approaches for interrogating protein structure, function and dynamics. However, methods for site-specifically incorporating multiple distinct unnatural amino acids are hampered by their low efficiency. Here we provide a general solution to this challenge by developing an optimized orthogonal translation system that uses amber and evolved quadruplet-decoding transfer RNAs to encode numerous pairs of distinct unnatural amino acids into a single protein expressed in Escherichia coli with a substantial increase in efficiency over previous methods. We also provide a general strategy for labelling pairs of encoded unnatural amino acids with different probes via rapid and spontaneous reactions under physiological conditions. We demonstrate the utility of our approach by genetically directing the labelling of several pairs of sites in calmodulin with fluorophores and probing protein structure and dynamics by Förster resonance energy transfer.
Introduction
Genetically programming the efficient site-specific incorporation of multiple unnatural amino acids into proteins will facilitate a range of applications from Förster resonance energy transfer (FRET) studies1, 2 on protein conformational change and dynamics, to protein stapling3, 4 for improving therapeutic proteins, and ultimately, to the encoded cellular synthesis and evolution of unnatural polymers5.
Encoding multiple distinct unnatural amino acids in cells requires (1) mutually orthogonal aminoacyl-transfer RNA (tRNA) synthetase/tRNA pairs that recognize distinct amino acids and (2) blank codons that can be assigned to new amino acids6-8. We previously demonstrated that the Methanococcus janaschii tyrosyl-tRNA synthetase (MjTyrRS)/MjtRNA pair and its active site variants, and the pyrrolysyl-tRNA synthetase (PylRS)/tRNA pair from certain Methanosarcina species and their active site variants are mutually orthogonal in their aminoacylation specificity9 We created orthogonal ribosome-mRNA pairs in which the orthogonal ribosome is specifically directed to an orthogonal message10. These ribosomes, unlike natural ribosomes, are not responsible for the synthesis of the proteome, and can be altered to perform new functions9, 11, 12. We evolved an orthogonal ribosome (ribo-Q1)9 to decode quadruplet codons 13, 14 and amber codons, which are inefficiently decoded by natural ribosomes to provide a series of blank codons that can be used on orthogonal messages. By combining ribo-Q1 with an orthogonal message containing a quadruplet and amber codon, and AzPheRS*/tRNAUCCU (a derivative of MjTyrRS/tRNA) and PylRS/tRNACUA we were able to direct the incorporation of p-azidophenylalanine (4) and N6-[(2-propynyloxy)carbonyl]-L-lysine (2), and program the formation of a triazole crosslink9. Although ribo-Q1 provided a large increase in quadruplet-decoding efficiency, this increase was not exploited fully in the original system, in part because the MjAzPheRS*/tRNAUCCU pair has suboptimal efficiency and specificity in directing the incorporation of 4. Subsequent efforts to encode two distinct unnatural amino acids have used multiple stop codons to direct the incorporation of unnatural amino acids15-17. However, the efficiency of incorporation of amino acids in response to stop codons in natural translation is limited by competition with release factors11.
Here we evolve a series of Pyl tRNAs that provide optimal decoding of cognate quadruplet codons on ribo-Q1. We optimized orthogonal translation using the new tRNAs to provide a large increase in the efficiency of incorporating two distinct unnatural amino acids into a single polypeptide. We demonstrate the generality of our approach by encoding a matrix of unnatural amino acid pairs in proteins. Moreover, we demonstrate the first general route to the rapid and spontaneous labelling of proteins at genetically defined positions with two distinct probes under physiological conditions, and we demonstrate the utility of this approach for following conformational change in calmodulin (CaM) by FRET.
Results
Selection of quadruplet-decoding Pyl tRNA variants
PylRS, unlike MjTyrRS and its derivatives, does not recognize the anticodon stem loop of its cognate tRNA, Pyl tRNACUA18, 19. This may allow the anticodon stem loop of Pyl tRNA to be optimized for ribosomal decoding on ribo-Q1 without interfering with its aminoacylation by PylRS. We set out to select Pyl tRNA variants that are efficiently decoded on ribo-Q1. Previous work explored the selection of non-orthogonal tRNAs for decoding quadruplet codons on the natural ribosome20, and recently the selection of a tRNA, derived from Pyl tRNA, that decodes the AGGA codon on the natural ribosome has been reported21. We were interested in discovering (1) a number of extended anticodon/quadruplet codon pairs that are efficiently decoded on ribo-Q1 and (2) Pyl tRNA variants that are efficiently and specifically decoded on ribo-Q1.
We chose four quadruplet codons (AGGA, AGTA, TAGA and CTAG) as potential targets for decoding by evolved Pyl tRNAXXXX variants (in which the anticodon XXXX is complementary to a specific codon) on ribo-Q1. AGGA was chosen because it has previously been used for unnatural amino acid incorporation9. The other codons were chosen because the two overlapping triplets embedded within the corresponding quadruplet anticodon are not recognition elements for natural synthetases in the cell (Supplementary Fig. S1), and we hypothesized that this would facilitate the selection of Pyl tRNA variants that are not mis-aminoacylated by endogenous synthetases.
To select for Pyl tRNA variants that are able to efficiently decode the selected quadruplet codons on ribo-Q1 we first created saturation mutagenesis libraries, Pyl tRNA(N8)XXXX, in which the sequence of eight nucleotides in the anticodon stem loop was varied to all possible combinations (Fig. 1a and 1b). Nucleotides 30-33 and 37-40 were randomized for tRNA(N8)UCUA and 29-32 and 38-41 were randomized for Pyl tRNA(N8)UCCU, Pyl tRNA(N8)UACU, and Pyl tRNA(N8)CUAG (Fig. 1b). Each library contained at least 108 members, ensuring greater than 99.9% coverage.
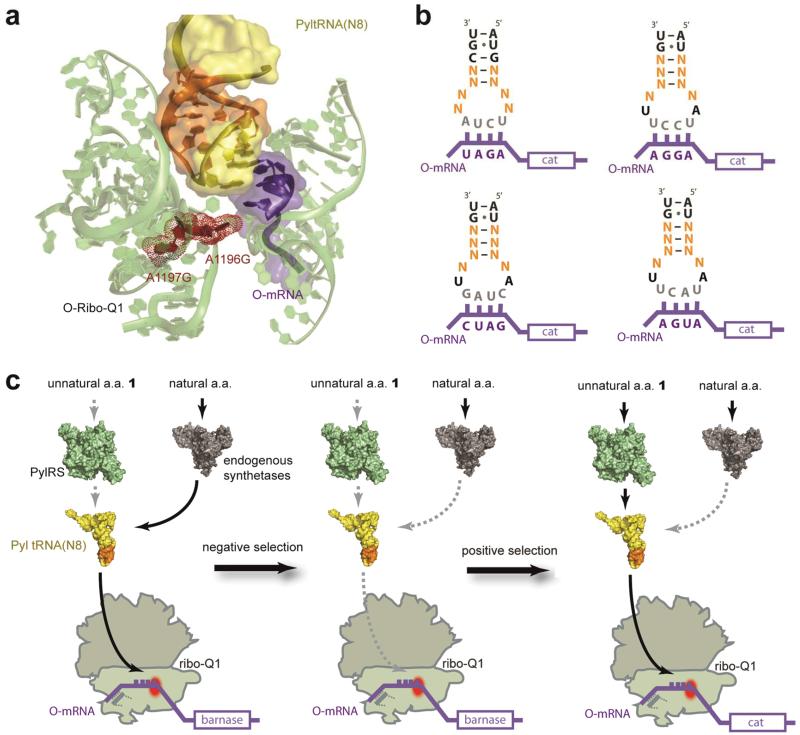
Figure 1. Optimizing Pyl tRNA(N8)XXXX for incorporating unnatural amino acids in response to quadruplet codons decoded by ribo-Q1.
a. Nucleotides that are targeted for mutagenesis in the anticodon stem loop of Pyl tRNA(N8)XXXX are represented in orange on tRNA bound to the ribosome , and the rest of the tRNA is in yellow. rRNA in pale green with ribo-Q1 mutation sites in red, and mRNA in purple. The structural image is based on PDB ID 2J00, created with Pymol (www.pymol.org). b. The anticodon stem loop of Pyl tRNA(N8)XXXX. The nucleotides in orange are randomized in each library. Codon sequences and mRNAs are in purple, and anticodons in grey. c. Two-step selection procedure for identifying specific and efficient Pyl tRNA(N8)XXXX library members in orthogonal translation. A negative O-barnase selection followed by a positive O-cat selection. Negative selection in the absence of unnatural amino acids eliminates Pyl tRNA(N8)XXXX library members that are mis-aminoacylated with natural amino acids by endogenous aminoacyl synthetases. Subsequent positive selection enriches evolved Pyl tRNA(N8)XXXX library members that are aminoacylated with the added unnatural amino acid by PylRS and efficiently decoded at quadruplet codons by ribo-Q1. a.a, amino acid; O-mRNA, orthogonal mRNA; cat, chloramphenicol acetyl transferase.
We performed a two-step selection (Fig. 1c) to discover the desired tRNAs: a negative selection to remove Pyl tRNA(N8)XXXX library members that are substrates for endogenous synthetases (because all aminoacyl-tRNA synthetases, except for AlaRS, LeuRS and SerRS, recognize the anticodon stem loop of their cognate tRNA in E. coli22), and a positive selection to select for library members that are aminoacylated with an unnatural amino acid by PylRS and efficiently decoded on ribo-Q1 in response to a cognate quadruplet codon in the orthogonal mRNA.
To remove Pyl tRNA(N8)XXXX library members that are both substrates for endogenous synthetase and decoded on ribo-Q1 we created four negative selection reporter constructs (O-barnase3XXXX+45XXXX), each of which contain a barnase gene that is specifically translated by ribo-Q1 (Fig. 1c). Each of the four barnase genes contains two in-frame quadruplet codons (XXXX), where XXXX is the Watson-Crick complementary sequence to the specific anticodon of a tRNA(N8)XXXX. Cells contained ribo-Q1, O-barnase3XXXX+45XXXX, and PylRS/Pyl tRNA(N8)XXXX. If a Pyl tRNA(N8)XXXX library member is aminoacylated with a natural amino acid by an endogenous synthetase and can be efficiently decoded on O-barnase3XXXX+45XXXX by ribo-Q1, then full-length barnase will be produced, leading to cell death. Pyl tRNA(N8)XXXX library members that survive the selection are either not substrates for native synthetases and/or they are unable to decode their quadruplet codon on ribo-Q1.
To select for functional PylRS/Pyl tRNA(N8)XXXX pairs from the Pyl tRNA(N8)XXXX library members that survive the negative selection we created positive selection reporters that contains an orthogonal chloramphenicol acetyl transferase (cat) gene with an in-frame quadruplet codon (O-cat111XXXX)9, 20 (Fig. 1c). Cells contained ribo-Q1, O-cat111XXXX, and PylRS/Pyl tRNA(N8)XXXX. In the presence of 1, PylRS aminoacylated correctly transcribed, processed and folded Pyl tRNA(N8)XXXX library members with 1, and the quadruplet codon in cat is decoded by ribo-Q1. The two-step selection should lead to chloramphenicol resistance only if Pyl tRNA(N8)XXXX is efficiently and selectively aminoacylated with 1 by PylRS and is efficiently decoded in response to the cognate quadruplet codon by ribo-Q1. Importantly, unlike selections for quadruplet-decoding tRNAs on natural ribosomes, selection of quadruplet-decoding tRNAs directly on ribo-Q1 provides the possibility of selecting quadruplet-decoding tRNAs that are specific for the decoding centre of ribo-Q1, such tRNAs would further enhance the specificity of orthogonal translation.
We sequenced and phenotyped clones from each selection (Supplementary Fig. S2 and S3). On the basis of these initial experiments, the PylRS/evolved Pyl tRNAXXXX pairs giving the highest level of chloramphenicol resistance in cells containing ribo-Q1, O-cat111XXXX, in presence of 1 and lowest chloramphenicol resistance in the absence of 1 were characterized further.
The evolved Pyl tRNAUCUA contains a G–U pair in place of the parental G30–C40 pair, a G–C pair in place of the A31–U39 pair, and an A38U mutation. Although this library randomised eight positions, nucleotides at three positions (C32, U33 and A37), are identical to those found in the parent Pyl tRNACUA (Fig. 2a). The evolved Pyl tRNAUCCU, contains a C29–G41 pair in place of the parental G29–C41 pair. G30C and C40A replace the parental G30–C40 pair, and the evolved sequence contains an A31U mutation. C32, A38 and U39 are conserved between the parental Pyl tRNACUA and the evolved Pyl tRNAUCCU, these mutations are predicted to expand the anticodon loop and shorten the anticodon stem (Fig. 2a). In the evolved Pyl tRNAUACU, U29 and U41 replace the parental G29–C41 pair, a G31–C39 pair replaces the parental A31–U39 pair and A38 is mutated to U (Fig. 2a). In the evolved Pyl tRNACUAG, C31 and C39 replace the A31–U39 base pair. These mutations are predicted to expand the anticodon loop (Fig. 2a).
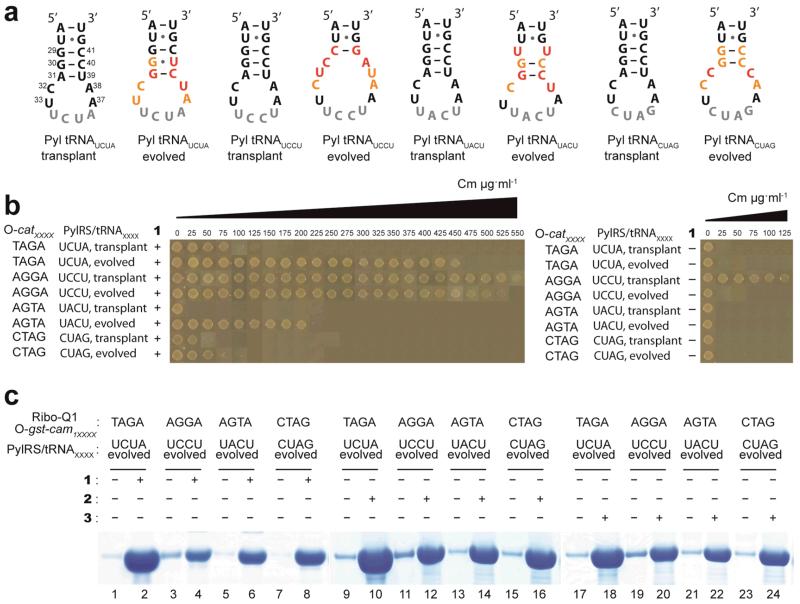
Figure 2. Evolved Pyl tRNA(N8)XXXX direct the efficient unnatural amino acids incorporation in response to quadruplet codons decoded by ribo-Q1.
a. The selected anticodon stem-loop sequences of evolved Pyl tRNAXXXX, and the corresponding transplant sequences. The anticodons are in gray, and the nucleotides mutated in the library are shown in colour. Positions where parental sequence is selected are in orange, and positions where new nucleotides are selected are in red. b, Evolved Pyl tRNAXXXX substantially enhance the incorporation of unnatural amino acids by ribo-Q1 in response to quadruplet codons when compared to the corresponding transplant Pyl tRNAXXXX. The unnatural amino acid-dependent decoding of quadruplet codons in the O-cat111XXXX was measured by survival on increasing concentrations of chloramphenicol (Cm). c. Diverse unnatural amino acids are efficiently incorporated in recombinant proteins in response to quadruplet codons using PylRS/Pyl tRNAXXXX with orthogonal translation. Full gels are given in Supplementary Fig. S4.
Enhanced unnatural amino acids incorporation at quadruplet codons
We next demonstrated the improved quadruplet decoding efficiency and specificity of the evolved Pyl tRNAXXXX variants, compared to the transplant Pyl tRNAXXXX, in which an extended quadruplet anticodon replaces CUA. Cells containing ribo-Q1, O-cat111XXXX, PylRS, 1, and either evolved Pyl tRNAXXXX or transplant Pyl tRNAXXXX were plated on increasing concentrations of chloramphenicol (Fig. 2b) to assess the quadruplet decoding efficiency of each tRNA.
Cells that contained the evolved Pyl tRNAUCUA were able to survive on chloramphenicol concentrations of up to 450 μg ml−1, which is substantially greater than the 75 μg ml−1 survival conferred by the transplant Pyl tRNAUCUA. This indicates that the evolved Pyl tRNAUCUA is more efficient at decoding the quadruplet codon TAGA than the transplant Pyl tRNAUCUA (Fig. 2b). Experiments in which the substrate for PylRS, 1, is omitted (Fig. 2b) demonstrate that the evolved Pyl tRNAUCUA is specifically aminoacylated by PylRS and not by endogenous aminoacyl-tRNA synthetases.
The transplant Pyl tRNAUCCU and evolved Pyl tRNAUCCU both conferred chloramphenicol resistance up to 500 μg ml−1 when cells were grown in the presence of 1. However, the transplant Pyl tRNAUCCU, unlike the evolved Pyl tRNAUCCU conferred significant chloramphenicol resistance in the absence of 1 (Fig. 2b). This indicated that although both the transplant Pyl tRNAUCCU and evolved Pyl tRNAUCCU can decode the AGGA codon, only the evolved Pyl tRNAUCCU is specifically recognized by PylRS and not by endogenous synthetases.
The transplant Pyl tRNAUACU and Pyl tRNACUAG conferred minimal chloramphenicol resistance; in contrast the evolved Pyl tRNAUACU and Pyl tRNACUAG were able to decode the quadruplet codons AGTA and CTAG and confer enhanced chloramphenicol resistance. Experiments in which 1 is omitted (Fig. 2b) demonstrate that the evolved Pyl tRNAUACA and Pyl tRNACUAG are specifically aminoacylated by PylRS and not by endogenous aminoacyl-tRNA synthetases.
We next demonstrated that the improved quadruplet-decoding efficiency of the evolved PylRS/Pyl tRNAXXXX pairs on ribo-Q1 translates into an increased production of recombinant protein bearing an unnatural amino acid. To provide a benchmark for the incorporation achieved using the evolved Pyl tRNAXXXX systems, we expressed and purified a fusion protein of glutathione-S-transferase and calmodulin (GST-CaM) site-specifically incorporating 4 from cells containing ribo-Q1, O-gst-cam1AGGA (a fusion gene between glutathione-S-transferase (gst) and cam in which the first codon of cam is replaced with an AGGA codon), (Supplementary Fig. S4), and MjAzPheRS*/tRNAUCCU (a synthetase/tRNA pair previously developed for incorporating 4 in response to AGGA codons)9. This led to approximately 13% full-length protein upon addition of 4, with approximately 7% full-length protein produced in the absence of 4 (Supplementary Fig. S4).
To compare the incorporation achieved with the evolved Pyl tRNAXXXX and the corresponding transplant Pyl tRNAXXXX, we expressed GST-CaM from cells containing ribo-Q1, O-gst-cam1XXXX (XXXX is complementary to the tRNA anticodon used), amino acid 1, and either transplant Pyl tRNAXXXX or evolved Pyl tRNAXXXX (Supplementary Fig. S4). The evolved Pyl tRNAUCUA, evolved Pyl tRNAUACU and evolved Pyl tRNACUAG substantially increased the efficiency of incorporation of 1 in response to TAGA, AGTA, CTAG codons respectively (Supplementary Fig. S4).
Both the evolved and the transplant Pyl tRNAUCCU led to comparable read-through of the AGGA codon in O-gst-cam1AGGA upon addition of 1 (Supplementary Fig. S4). However, the transplant Pyl tRNAUCCU was substantially mis-aminoacylated in the absence of 1 (Supplementary Fig. 4). This is consistent with the phenotypic data (Fig. 2b) and may demonstrate the advantage of the negative selection step in selecting synthetase/tRNA pairs for unnatural amino acid dependent incorporation at quadruplet codons.
To demonstrate that a range of unnatural amino acids can be incorporated in response to quadruplet codons using the evolved tRNAs, we compared the efficiency of incorporation using chemically distinct unnatural amino acids (Fig. 2c and Supplementary Fig. 4). The efficiencies of incorporating 1, 2 and 3 in response to quadruplet codons TAGA, AGGA, AGTA, and CTAG using their cognate evolved Pyl tRNAXXXX ranged from approximately 27% to 62%, and were all substantially greater than the unnatural amino acid incorporation efficiency using the MjAzPhe RS*/tRNAUCCU pair9 (Supplementary Fig. S4). Control experiments demonstrated that the unnatural amino acid incorporation efficiency was improved on providing multiple copies of PylRS (Supplementary Fig. S5), and was robust in different strains of E. coli (data not shown). The efficiency and specificity of incorporating 1 in response to the quadruplet codons using our evolved quadruplet-decoding systems is greater than the efficiency and specificity we observe when using PylRS/tRNA anticodon variants and wild-type ribosome to decode the ochre (UAA), opal (UGA), or AGGA codons15, 17,21 at the same position in GST-CaM (Supplementary Figs. S4 & S5)
Incorporating a matrix of unnatural amino acid pairs into proteins
Next, we optimized the incorporation of two distinct unnatural amino acids into proteins via ribo-Q1. Our starting system contains four plasmids and used ribo-Q1, MjAzPheRS*/tRNAUCCU, and PylRS/tRNACUA to read AGGA and amber codons on orthogonal messages (Fig. 3a and Supplementary Figs. S6 & S7).
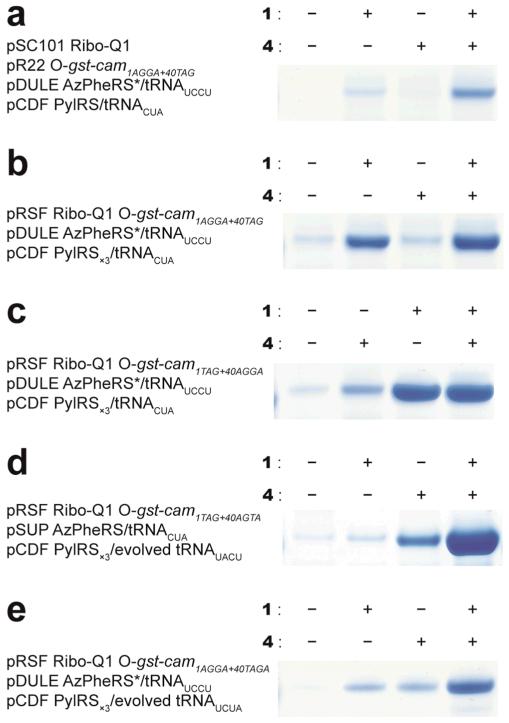
Figure 3. Efficient incorporation of multiple distinct unnatural amino acids into a single polypeptide.
a. Site-specific incorporation of 1 and 4. AGGA replaces the 1st codon and UAG replaces the 40th codon in the cam open reading frame of O-gst-cam to make O-gst-cam1AGGA+40TAG. Decoding of both the AGGA and TAG codons by ribo-Q1 produces full length Gst-CaM, and failure to decode these codons leads to premature termination of the polypeptide. b. and c. The site-specific incorporation efficiency of 1 and 4 is improved by reducing the number of plasmids. d. The newly evolved PylRS/tRNAUACU pair substantially increases the efficiency of double incorporation e. Incorporating two distinct unnatural amino acids using two distinct quadruplet codons. Full gels are given in Supplementary Fig. S7.
To improve the system, we first created a three-plasmid system using the same elements (Fig. 3b, c and Supplementary Fig S6, & S7). This increased the fraction of full-length protein produced, but also resulted in a high level of unnatural amino acid independent read-through of the AGGA codon. To increase the fidelity and efficiency of the system for incorporating two distinct unnatural amino acids we used the evolved Pyl tRNAUACU, developed in this work, in combination with MjAzPheRS/tRNACUA23. This led to a further substantial increase in the efficiency and specificity of the system, and double incorporation efficiencies in excess of 20% (Figure 3d, and Supplementary Fig. S6, & S7). The efficiency of double incorporation using evolved Pyl tRNAUCCU or evolved Pyl tRNACUAG was comparable to the efficiency in experiments using evolved Pyl tRNAUACU (Supplementary Fig. S7). By combining the MjAzPheRS*/tRNAUCCU pair and the PylRS/tRNAUACU pair with ribo-Q1, we directed the cellular incorporation of multiple distinct unnatural amino acids in response to multiple distinct quadruplet codons (Fig. 3e and Supplementary Fig. S7).
To demonstrate the generality of the approach for incorporating chemically diverse pairs of unnatural amino acids into proteins, we incorporated every combination of three distinct unnatural PylRS substrates and four distinct substrates for MjTyrRS variants (Figure 4 and Supplementary Fig. S8) using the orthogonal translation system described in Fig. 3d. The resulting matrix of 12 distinct pairs of unnatural amino acids, including unique chemical handles (azides23, alkenes24, alkynes25, 26, tetrazines27) and a photocrosslinker (benzophenone28) were efficiently and specifically incorporated into GST-CaM, with full length protein yields from 1.5-2 mg L−1 of culture (for comparison the yield of wild-type GST-CaM was approximately 4 mg L−1 of culture). There is some read-through of the codons used to direct unnatural amino acid incorporation in the absence of unnatural amino acid, as previously described9, 11. However, electrospray ionization mass spectrometry (ESI-MS) and matrix-assisted laser desorption mass spectrometry (MALDI-MS) (Supplementary Fig. S8) conclusively demonstrated the genetically programmed incorporation of both added unnatural amino acids, without measureable mis-incorporation of natural amino acids.
Figure 4. Efficient incorporation of a matrix of pairs of unnatural amino acids, including photocrosslinkers and chemical handles (azides, alkenes, alkynes, tetrazines) demonstrates generality.
Cells contained O-gst-cam1TAG+40AGTA and ribo-Q1 expressed from an RSF plasmid, the pSUP MjAzPheRS/tRNACUA plasmid (or a variant specific for the relevant substrate) and the pCDF PylRS/evolved tRNAUACU plasmid. All combinations of PylRS substrates (1-3) and MjTyrRS active site variant substrates (4-7) were incorporated in 3×4 matrix. We further confirmed the incorporation of distinct unnatural amino acids by ESI and MALDI mass spectrometry (Supplementary Fig. S8). Full gels are given in Supplementary Fig. S8. We observed an additional peak for protein samples with 4 corresponding to the reduction of the azide to an amine.
We compared the efficiency of incorporating 1 and 4 in response to two stop codons on the natural ribosome15, 17 and to (using an MjTyrRS variant specific for 4 (ref. 23) PylRS/tRNAUUA and PylRS/tRNAUCA to direct the incorporation of 1 in response the ochre or opal codon) to our system. Double incorporation in response to two stop codons was not detectable in GST-CaM (Supplementary Fig. S7), consistent with the low efficiency with which both PylRS/tRNAUUA and PylRS/tRNAUCA decode their cognate stop codons (Supplementary Fig. S5). Comparison of the experiments using two stop codons with the experiments using our optimized system with amber and quadruplet codons decoded on the evolved orthogonal ribosome (Supplementary Fig. S7) reveals the advantages of the approach reported herein in GST-CaM.
Programming mutually orthogonal chemical handles into proteins
The ability to program multiple unnatural amino acids into proteins raises the possibility of programming pairs of mutually orthogonal functional groups into proteins that can be specifically and quantitatively labelled with a range of probes under conditions that maintain the native fold of proteins (physiological pH, temperature and pressure). This would facilitate a range of applications, including FRET to study protein structure, conformation and dynamics1. Efforts to double-label proteins at genetically encoded unnatural amino acids have relied on encoding ketones to react with alpha-effect nucleophiles, and encoding azides to react with strained alkynes16, 17. Unfortunately, ketone labelling is very slow (with rate constants (r) of approximately 10−4 M−1s−1) and commonly requires low pH, which makes it incompatible with the folding and stability of many proteins. Moreover genetically encoded azides are succeptible to in vivo reduction (Figure 4)29, which makes their quantitative labelling problematic.
Inverse electron-demand Diels-Alder reactions allow the spontaneous and rapid coupling of a number of strained alkenes or alkynes and tetrazines30, 31, and provide a promising route to rapid and quantitative labelling of proteins 27, 32-36. The rates of these reactions depend on the tetrazine substituents and the strain/sterics of the alkene or alkyne37, 38. We reasoned that it may be possible to tune the reactivity of tetrazines and strained alkenes in encoded amino acids such that they do not react with each other in the protein, but can be specifically and rapidly labelled with added probes.
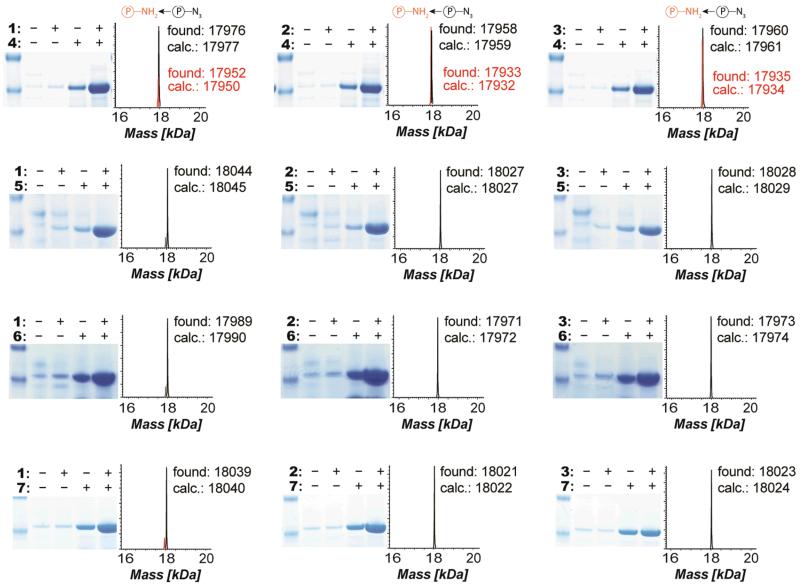
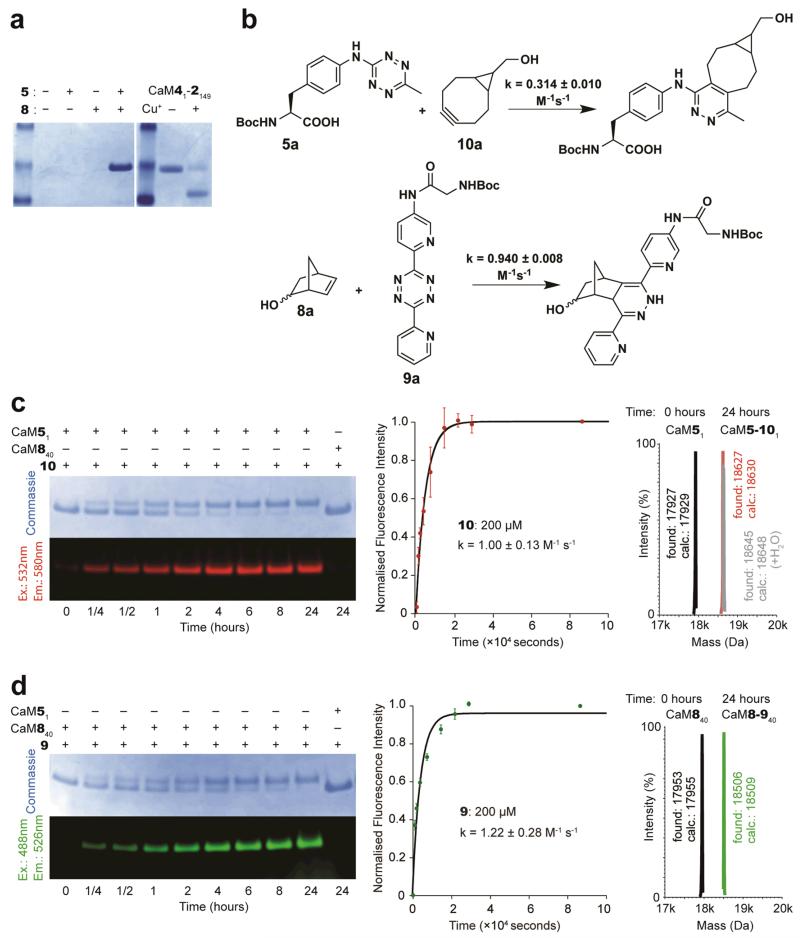
As a model for reaction between 5 and 8, we investigated the rate of reaction between 5-norbornene-2-ol (8a) and the t-butyloxycarbonyl (Boc) protected amino acid containing a tetrazine (5a) that is deactivated in inverse electron-demand Diels-Alder reactions by virtue of electron donation from the aniline moiety (Supplementary Fig. S9). The reaction is very slow, and we estimate that the rate constant for this reaction is approximately 10−5 M−1s−1 (Supplementary Fig. S9). To demonstrate that we can genetically encode 5 and 8 in a single polypeptide, we grew cells containing O-gst-cam1TAG+149AGTA, ribo-Q1, MjTetPheRS (an MjTyrRS variant specific for 5)/tRNACUA, NorKRS (a PylRS variant39 for the incorporation of 8)/evolved Pyl tRNAUACU in the presence of 5 and 8. The production of full-length protein (CaM51-8149) was dependent on the addition of both 5 and 8 (Fig. 5a).
Figure 5. 5 and 8 do not react with each other in a protein, but can be efficiently labelled with 10 and 9.
a. 5 and 8 do not react when placed in proximity within a protein. CaM51-8149 was purified from cells bearing pRSF ribo-Q1 O-gst-cam1TAG+149AGTA, pSUP MjTetPheRS/tRNACUA, pCDF NorKRS×3/evolved tRNAUACU. CaM41-2149 undergoes a Cu(I) catalyzed click reaction to cyclize the protein (right gel panel). CaM51-8149 does not cyclize, as judged by mobility shift (compare left and right panels) and ESI-MS (Supplementary Fig. S10). b. Rate constants for the indicated reactions. c. CaM51 and CaM840 were incubated with 100 molar equivalents of 10 at 25°C. Only CaM51 was labelled with 10, yielding CaM5-101. Labelling was visualized using a Typhoon Imager and the resulting time-dependent fluorescence was used to calculate the on-protein labelling rate constant. All measurements were repeated twice and the error bars represent the standard deviation. ESI-MS confirmed that protein labelling is quantitative. d. CaM51 and CaM840 were incubated with 100 molar equivalents of 9 at 25°C. Only CaM840 was labelled with 9, which yielded CaM8-940. The labelling reaction was analysed as described in c. ESI-MS confirmed that labelling was quantitative.
CaM51-8149 migrated as the linear protein in SDS-PAGE (Fig. 5a) suggesting that 5 and 8 do not react, even when they are placed in proximity by the protein structure. The incorporation of 5 and 8 in the protein without cyclization was further confirmed by ESI-MS (Supplementary Fig. S10). These results demonstrate that 5 and 8 do not react even when placed at positions in a protein that facilitate the proximity-accelerated Cu (I) catalysed azide-alkyne cycloaddition between encoded amino acids (Fig. 5a), which is consistent with the rate of reaction between 5 and 8 being orders of magnitude slower than rates reported for Cu(I)-catalysed azide-alkyne cycloadditions40.
To demonstrate that amino acid 5a can be conjugated efficiently to bicyclononyne probes, we investigated the rate of the reaction between 5a and 10a (Fig. 5b). This reaction proceeded with a rate constant of 0.314 ± 0.010 M−1s−1 in MeOH/H2O (55/45) at room temperature (Fig. 5b and Supplementary Fig. S9). Previously we have demonstrated that the rate constant for the reaction of 8a with the activated tetrazine 9a (Fig. 5b) is 0.94 ± 0.008 M−1s−1 in MeOH/H2O (5/95) at room temperature (for comparison this rate constant is 0.47 ± 0.070 M−1s−1 in MeOH/H2O (55/45))33. These experiments demonstrate that the tetrazine and norbornene functionalities embedded in 5 and 8 can be labelled with strained alkynes and appropriately activated tetrazines, respectively.
We next synthesized BODIPY-TMR-X bicyclononyne conjugate 10 and demonstrated that it can be used to label 5- bearing CaM. The on-protein rate constant for this reaction is 1.00 ± 0.13 M−1s−1. ESI-MS demonstrates the quantitative single labelling of the protein (Fig. 5c). Similarly, we synthesized BODIPY-FL tetrazine conjugate 9 and demonstrated that it can be used to rapidly label 8- bearing CaM. The on-protein rate constant for this reaction is 1.22 ± 0.28 M−1s−1. ESI-MS demonstrated the quantitative single labelling of the protein (Fig. 5d). Control experiments demonstrate that bearing 8-CaM is not labelled with 10, and bearing 5-CaM is not labelled with 9. These experiments demonstrate that the rate constants measured for small-molecule labelling translate into efficient protein labelling and that the labelling reagents are specific for their targeted unnatural amino acid in proteins. The rate constants for the reaction between 5 and 10, and between 8 and 9 are two to four orders of magnitude faster than ketone labelling reactions, label metabolically stable unnatural amino acids at physiological pH, do not use transition-metal catalysis, and generate nitrogen gas as the only by-product, making them compatible with most proteins.
Site-specific dual labelling of proteins under physiological conditions
FRET probes have been introduced via in vitro translation2, which allows access to small amounts of protein labelled with particular fluorophores. Native chemical ligation has been used to introduce FRET probes into proteins 41, and this has been combined with unnatural amino acid mutagenesis to introduce a limited range of short-wavelength probes42. These approaches generally limit the introduction of at least one of the probes to the termini of proteins.
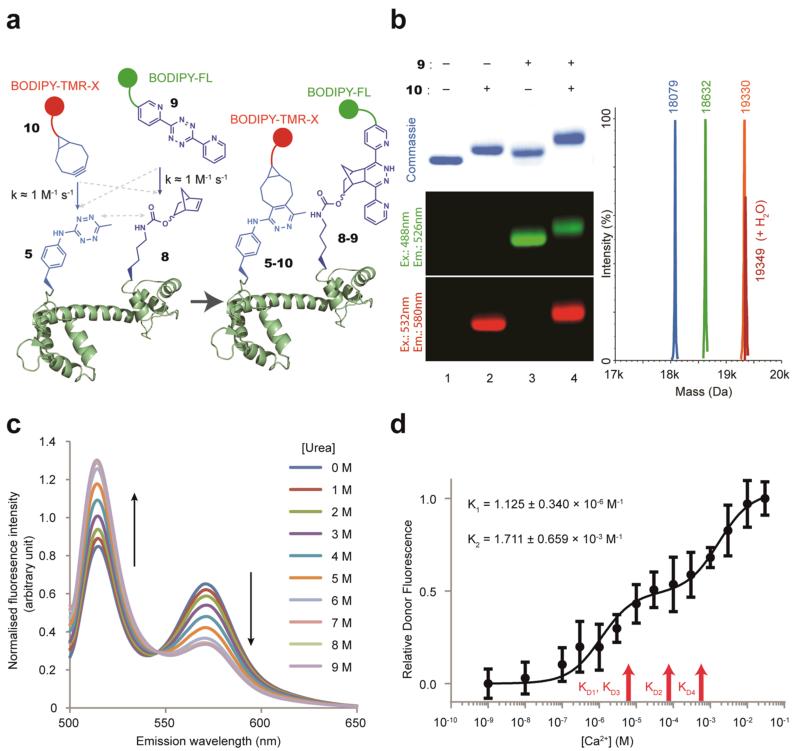
We demonstrated that the reactions we have characterized can be used to rapidly and spontaneously label proteins with two distinct fluorophores for FRET studies (Fig. 6). We chose BODIPY-FL (donor) and BODIPY-TMR-X (acceptor) as FRET probes because of the significant overlap between their emission and excitation spectra. We labelled 8 at position 149 in CaM51-8149 with a BODIPY-FL activated tetrazine conjugate 9, and labelled 5 at position 1 in CaM51-8149 with a BODIPY-TMR-X bicyclononyne conjugate 10, to create a doubly labelled protein (CaM5-101-8-9149). The labelling with each fluorophore was quantitative as judged by SDS-PAGE, fluorescence gel imaging and ESI-MS (Supplementary Fig. S10).
Figure 6. Site-specific double-labelling of CaM with a FRET pair to follow changes in protein conformation.
a. Strategy for protein double-labelling via inverse electron-demand Diels-Alder reactions. b. Quantitative and site-specific double labelling of CaM51-840. Purified CaM51-840 (Lane 1) was labelled with 100 equivalents (200μM) of 10, which yielded CaM5-101-840 (Lane 2) or 100 equivalents 9, which yielded CaM51-8-940 (Lane 3). CaM51-8-940 was labelled with 100 equivalents of 10, which yielded CaM5-101-8-940 (Lane 4). Labelling was visualised by fluorescence imaging and led to a mobility shift. All labelling reactions were quantitative, as confirmed by ESI-MS. CaM51-840 (blue peak; calculated Mass=18081 Da, observed Mass=18079 Da), CaM51-8-940 (green peak; calculated Mass=18635, observed Mass=18632), CaM5-101-8-940 (orange peak; calculated Mass=19336, observed Mass=19330). c. Fluorescence spectra of CaM5-101-8-9149 (following donor excitation at 485 nm) in the presence of increasing concentrations of urea. d. The relative donor-fluorescence intensity from doubly labelled CaM5-101-8-940 as a function of Ca2+ concentration. All measurements were repeated at least six times and the error bars represent the standard deviation. K1, K2 are for the observed transitions. R2= 0.9005. KD1, KD2, KD3 and KD4 are the reported KD (dissociation constants) values for sequential Ca2+ binding 51.
The fluorescence emission spectra of CaM5-101-8-9149 shows two distinct peaks when the sample is excited at 485 nm: one peak corresponds to emission from BODIPY-FL (donor, 515 nm), and the other peak corresponds to emission from BODIPY-TMR-X (acceptor, 570 nm) (Supplementary Fig. S10). In contrast, the fluorescence spectra of singly labelled protein with either BODIPY-FL (donor) at position 149 (CaM51-8-9149) or BODIPY-TMR-X (acceptor) at position 1 (CaM5-101-8149) shows minimal emission at 570 nm when the samples are excited at 485 nm (Supplementary Fig. S10). These data are consistent with FRET between the donor and acceptor fluorophores in CaM5-101-8-9149.
Using FRET to follow protein conformational change
The observed FRET signal for CaM5-101-8-9149 is sensitive to increasing concentrations of urea (Fig. 6c and Supplementary Fig. S10); the donor fluorescence signal increases and the acceptor fluorescence signal decreases as the urea concentration is increased. The observed decrease in FRET is consistent with the fluorophores moving further apart in the denatured state and this experiment demonstrates that protein conformational changes can be followed with our protein-labelling strategy.
CaM senses calcium in the cell and modifies its interactions with protein-binding partners43. CaM mediates many important processes, including short- and long-term memory, and the immune response44. CaM contains four EF-hand motifs, each of which binds one calcium ion45, 46. There are two EF-hand motifs in the N-terminal domain and two EF- hand motifs in the C-terminal domain45, 46. The two EF-hands within each domain bind calcium ions cooperatively, although the N- and C-terminal domains bind calcium independently47.
Structures of CaM have been solved in the presence of four calcium ions and in the absence of calcium ions46, 48, 49. Two distinct structures have been reported in the presence of four calcium ions45, 46, and single molecule solution measurements demonstrate that the protein is highly dynamic50. Moreover, much less is known about the structure of CaM with one, two or three calcium ions bound, though intermediate states of calcium binding are physiologically relevant in modulating the selectivity of CaM’s interactions with binding partners and signal propagation as a function of cellular calcium.
Although cellular calcium sensors based on FRET between fluorescent protein fusions, such as Chameleon, take advantage of global structural transitions in CaM that result from binding four calcium ions and movements of whole domains with respect to each other, it has been challenging to observe directly the local conformational change within domains as a function of calcium binding at individual sites because it is challenging to quantitatively install pairs of small molecule fluorophores at defined sites in the protein. These local changes are important because they provide the molecular basis for understanding cooperative binding of calcium to CaM and because of their biological significance.
To investigate local conformational changes in the N-terminal domain of CaM as a function of calcium concentration, we labelled CaM51-840 (Fig. 6b & Supplementary Fig. S11) with 9 and 10. Quantitative labelling of CaM was confirmed by fluorescence gel imaging and ESI-MS (Fig. 6b & Supplementary Fig. S11). The fluorescence spectra of the doubly labelled protein (CaM5-101-8-940) and singly labelled proteins (CaM5-101-840 and CaM51-8-940) are consistent with FRET between the donor and acceptor fluorophores in CaM5-101-8-940 (Supplementary Fig. S11).
We observe two discrete transitions of comparable magnitude in the FRET spectra of CaM5-101-8-940 as a function of calcium concentration (Fig. 6d, and Supplementary Fig. S11). The first transition (K1= 1.125 ± 0.34 ×10−6 M) is close to the binding constants reported for the first three calcium ions binding to CaM in recent isothermal titration calorimetry (ITC) measurements51 (KD1=3.69 ± 0.53 X 10−6 M, KD2= 7.09 ± 1.206 ×10−5, KD3= 5.21 ± 0.841 ×10−6) and within the range reported in the literature52-54. As calcium binding at each EF-hand motif within the N- or C-terminal domain of CaM is cooperative, but binding of calcium to the distinct domains of CaM is thermodynamically independent47, it seems probable that the first transition we observe with probes in the N-terminal domain results from the binding of the third calcium ion to CaM: the first binding event in the N-terminal domain. However, we cannot rule out contributions to the FRET change we observe from conformational changes in the N-terminal domain that result from calcium binding to the C-terminal EF hand motifs of CaM (KD1 and KD2). The first conformational change of the N-terminal domain may set up the protein to bind the fourth calcium ion.
The second transition (K2 = 1.711 ± 0.659 × 10−3 M) is close to binding constants reported recently for binding the fourth calcium ion to CaM (ITC study, KD4 = 0.44 ± 0.0465 ×10−3 M), and the range of literature values for KD452-54, which suggests that this conformational change may result from binding the fourth and final calcium ion51.
Our data report on local conformational changes within a domain as a result of calcium-binding events at distinct sites, and allow us to observe discrete structural transitions as a result of sequential ligand-binding events at distinct sites. The FRET changes we observe are reproducibly small (Supplementary Fig. S11), consistent with one of the two X-ray structures of four calcium ions bound to CaM45, 46. However, FRET measurements depend on both probe separation and assumptions about probe orientation, our data do not distinguish unambiguously between the divergent structures of CaM bound to four calcium ions, which single-molecule studies demonstrate from part of an ensemble of structures in solution50.
Discussion
We evolved a series of quadruplet decoding Pyl tRNAXXXX that efficiently and specifically read their cognate quadruplet codons on messages decoded by ribo-Q1. In combination with PylRS or its evolved variants, these tRNAs direct the efficient and specific incorporation of diverse unnatural amino acids into proteins. We created an optimized orthogonal translation system that takes advantage of the new quadruplet-decoding tRNAs and directs the efficient incorporation of a matrix of pairs of unnatural amino acids in E. coli. By carefully characterizing the kinetics of inverse electron-demand Diels-Alder reactions we designed a strategy for encoding a tetrazine and a norbornene that do not react with each other in proteins, but can be labelled rapidly with complementary bicyclononynes and electron-deficient tetrazines. This provides the first modular route to install diverse pairs of probes into proteins at essentially any genetically controlled pair of sites in proteins at physiological temperature, pressure and pH. The reactions proceed spontaneously without generating toxic side products or using toxic catalysts. We demonstrate the utility of our strategy by site-specifically labelling CaM with FRET probes and reveal that this allows local conformational changes within a domain that result from sequential ligand-binding events at distinct sites to be observed. The modular approach we have developed will find utility in labelling proteins with a range of fluorophores, both for ensemble and single-molecule FRET experiments and for other biophysical measurements. Future work will focus on further extending and applying the approaches we have developed to address outstanding challenges in defining the molecular changes that underpin ligand binding, cooperativity, allostery and catalysis.
Methods
Protocols for chemical synthesis of amino acids, and fluorescent labelling probes as well as protocols for cloning, protein expression, protein labelling, mass spectrometry, determination of rate constants, and fluorescence measurements are provided in the Supplementary material along with detailed descriptions of all experimental procedures. The PylRS/tRNA pairs are derived from M.barkeri unless otherwise stated.
Selection of Pyl tRNA(N8)XXXX for enhanced quadruplet decoding in the ribo-Q1 orthogonal mRNA system
We transformed each PyltRNA(N8)XXXX library into GeneHogs E. coli cells bearing the matching O-barnase3XXXX+45XXXX plasmid and RSF ribo-Q1 plasmid. The transformed cells were recovered in 1 ml SOB medium containing 2% glucose for 1 hour at 37°C. The transformation was used to inoculate 200 ml of LB-GKST (LB media with 2% glucose, 12.5 μg ml−1 kanamycin, 18.8 μg ml−1 spectinomycin, and 6.25 μg ml−1 tetracycline) and incubated overnight (37°C, 250 rpm, 16 h). The overnight culture (2ml) was pelleted by centrifugation (3,000g) and washed with LB to remove glucose, prior to diluting to OD600=0.1 in LB-KST (LB media with 12.5 μg ml−1 kanamycin, 18.8 μg ml−1 spectinomycin, and 6.25 μg ml−1 tetracycline), and incubated (37°C, 250 rpm, 2-3 h) until OD600 reached 0.5. Isopropyl β-D-1thiogalactopyranoside (IPTG (1 mM final concentration)) was added to induce the expression of ribo-Q1 ribosomal RNA (rRNA) at OD600≈0.5. The culture was incubated (37°C, 250 rpm) until the OD600 reached approximately 2. Aliquots (500 μl) of the culture were serial diluted and plated on LB-KST agar plate supplemented with 1 mM IPTG. The plates were incubated at 37°C for 16 hours. Surviving colonies were scraped from the plate and plasmid DNA was extracted by Qiagen Miniprep Kit to make the post-negative selection pool.
The negative selection was followed by a positive selection. The post negative selection Pyl tRNA(N8)XXXX pools were cleansed of non pCDF PylRS/tRNAXXXX plasmids by digestion with SacII to remove the O-barnase3XXXX+45XXXX and pRSF ribo-Q1 plasmid. The cleansed post-negative selection Pyl tRNA(N8)XXXX pools were electroporated into GeneHog cells bearing the matching O-cat111XXXX and pRSF ribo-Q1 plasmids. The transformed cells were recovered in 1 ml SOB medium containing 2% glucose for one hour at 37°C. The transformation was used to inoculate 200 ml of LB-GKST and incubated overnight (37°C, 250 rpm, 16 h). The overnight culture (2 ml) was pelleted by centrifugation (3,000g) and washed with LB to remove glucose, prior to diluting to OD600=0.1 in LB-KST, and incubated (37°C, 250 rpm, 2-3 h) until OD600 reached 0.5. IPTG (1 mM final concentration) and 1 (8 mM final concentration) were added to induce the expression of ribo-Q1 rRNA and facilitate aminoacylation of Pyl tRNA(N8)XXXX library members by PylRS at OD600≈0.5. The culture was incubated (37°C, 250 rpm) until the OD600 reached approximately 2. Aliquots (500 μl) of the culture were serial diluted and plated on LB-KST agar plates supplemented with 1mM IPTG, 8mM 1, and chloramphenicol of different concentrations (100 μg ml−1, 150 μg ml−1, 200 μg ml−1). The plates were incubated at 37°C for 40 hours.
To separate the selected pCDF PylRS/tRNA(N8)XXXX plasmid from the O-cat111XXXX and pRSF ribo-Q1 plasmids, total plasmid DNA from selected clones was purified, digested with SacII, transformed into DH10B cells and plated on LB agar supplemented with spectinomycin (75 μg ml−1). Individual colonies from the spectinomycin plates were incubated overnight in LB media supplemented with spectinomycin (75 μg ml−1) and replica plated onto three sets of LB agar plates supplemented with spectinomycin (75 μg ml−1), or tetracycline (25 μg ml−1), or kanamycin (50 μg ml−1). After 16 h, colonies were observed only on spectinomycin plates but not on tetracycline or kanamycin plates. The DNA from separated pCDF PylRS/tRNA(N8)XXXX clones was purified, confirmed by restriction digestion and DNA sequencing.
Supplementary Material
Acknowledgements
Medical Research Council (U105181009, UD99999908). We thank Sew-Peak Chew (MRC-LMB Mass Spectrometry) for obtaining MALDI data.
References
- 1.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 2.Kajihara D, et al. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 3.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, Roller PP. Cyclization strategies in peptide derived drug design. Curr Top Med Chem. 2002;2:325–341. doi: 10.2174/1568026023394209. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew Chem Int Ed Engl. 2012;51:2288–2297. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- 6.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nature Publishing Group. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 7.Chin JW. Molecular biology. Reprogramming the genetic code. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- 8.Chin JW. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu Rev Biochem. 2014;83 doi: 10.1146/annurev-biochem-060713-035737. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 9.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 10.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 11.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nature biotechnology. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 12.An W, Chin JW. Synthesis of orthogonal transcription-translation networks. P Natl Acad Sci USA. 2009;106:8477–8482. doi: 10.1073/pnas.0900267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkins JF, Bjork GR. A Gripping Tale of Ribosomal Frameshifting: Extragenic Suppressors of Frameshift Mutations Spotlight P-Site Realignment. Microbiol Mol Biol R. 2009;73:178. doi: 10.1128/MMBR.00010-08. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl G, McCarty GP, Farabaugh PJ. Ribosome structure: revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan W, et al. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 16.Wu B, Wang Z, Huang Y, Liu WR. Catalyst-free and site-specific one pot dual-labeling of a protein directed by two genetically incorporated noncanonical amino acids. Chembiochem : a European journal of chemical biology. 2012;13:1405–1408. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry. 2013 doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci U S A. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R, Krzycki JA. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J Biol Chem. 2012;287:32738–32746. doi: 10.1074/jbc.M112.396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magliery TJ, Anderson JC, Schultz PG. Expanding the genetic code: Selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu W, Schultz PG, Guo J. An Expanded Genetic Code in Mammalian Cells with a Functional Quadruplet Codon. ACS Chem Biol. 2013 doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Chin JW, et al. Addition of p-Azido- l-phenylalanine to the Genetic Code of Escherichiacoli. Journal of the American Chemical Society. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 24.Yanagisawa T, et al. Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode Nε-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification. Chemistry & Biology. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen DP, et al. Genetic Encoding and Labeling of Aliphatic Azides and Alkynes in Recombinant Proteins viaa Pyrrolysyl-tRNA Synthetase/tRNA CUAPair and Click Chemistry. Journal of the American Chemical Society. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 26.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorganic & Medicinal Chemistry Letters. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 27.Seitchik JL, et al. Genetically Encoded Tetrazine Amino Acid Directs Rapid Site-Specific in VivoBioorthogonal Ligation with trans-Cyclooctenes. Journal of the American Chemical Society. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proceedings of the National Academy of Sciences. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasmal PK, et al. Catalytic azide reduction in biological environments. ChemBioChem. 2012;13:1116–1120. doi: 10.1002/cbic.201100719. [DOI] [PubMed] [Google Scholar]
- 30.Blackman ML, Royzen M, Fox JM. Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels–Alder Reactivity. Journal of the American Chemical Society. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devaraj NK, Weissleder R. Biomedical Applications of Tetrazine Cycloadditions. Acc Chem Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014 doi: 10.1021/cr400355w. in press. [DOI] [PubMed] [Google Scholar]
- 33.Lang K, et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang K, et al. Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels–Alder Reactions. Journal of the American Chemical Society. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plass T, et al. Amino Acids for Diels-Alder Reactions in Living Cells. Angewandte Chemie International Edition. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 36.Borrmann A, et al. Genetic Encoding of a Bicyclo[6.1.0]nonyne-Charged Amino Acid Enables Fast Cellular Protein Imaging by Metal-Free Ligation. ChemBioChem. 2012;13:2094–2099. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]
- 37.Balcar J, Chrisam G, Huber FX, Sauer J. Reaktivitaet von Stickstoff-Heterocyclen gegenueber Cyclooctin als Dienophil. Tetrahedron Letters. 1983;24:1481–1484. [Google Scholar]
- 38.Thalhammer F, Wallfahrer U, Sauer J. Reaktivitaet einfacher offenkettiger und cyclischer Dienophile bei Diels-Alder Reaktionen mit inversem Elektronenbedarf. Tetrahedron Letters. 1990;31:6851–6854. [Google Scholar]
- 39.Kaya E, et al. A Genetically Encoded Norbornene Amino Acid for the Mild and Selective Modification of Proteins in a Copper-Free Click Reaction. Angewandte Chemie International Edition. 2012;51:4466–4469. doi: 10.1002/anie.201109252. [DOI] [PubMed] [Google Scholar]
- 40.Lang K, Chin JW. Bioorthogonal reactions for labeling proteins. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 41.Hellstrand E, et al. Forster resonance energy transfer studies of calmodulin produced by native protein ligation reveal inter-domain electrostatic repulsion. FEBS J. 2013;280:2675–2687. doi: 10.1111/febs.12269. [DOI] [PubMed] [Google Scholar]
- 42.Wissner RF, Batjargal S, Fadzen CM, Petersson EJ. Labeling proteins with fluorophore/thioamide Forster resonant energy transfer pairs by combining unnatural amino acid mutagenesis and native chemical ligation. J Am Chem Soc. 2013;135:6529–6540. doi: 10.1021/ja4005943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 44.Carafoli E. Calcium signaling: a tale for all seasons. Proc Natl Acad Sci U S A. 2002;99:1115–1122. doi: 10.1073/pnas.032427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor DA, Sack JS, Maune JF, Beckingham K, Quiocho FA. Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution. J Biol Chem. 1991;266:21375–21380. doi: 10.2210/pdb4cln/pdb. [DOI] [PubMed] [Google Scholar]
- 46.Fallon JL, Quiocho FA. A closed compact structure of native Ca(2+)-calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Linse S, Helmersson A, Forsen S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 48.Babu YS, et al. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 49.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nat Struct Biol. 2001;8:990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 50.Johnson CK. Calmodulin, conformational states, and calcium signaling. A single-molecule perspective. Biochemistry. 2006;45:14233–14246. doi: 10.1021/bi061058e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu G, Gao Z, Dong A, Yu S. Calcium-induced changes in calmodulin structural dynamics and thermodynamics. Int J Biol Macromol. 2012;50:1011–1017. doi: 10.1016/j.ijbiomac.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 52.Porumb T. Determination of calcium-binding constants by flow dialysis. Anal Biochem. 1994;220:227–237. doi: 10.1006/abio.1994.1332. [DOI] [PubMed] [Google Scholar]
- 53.Haiech J, Klee CB, Demaille JG. Effects of cations on affinity of calmodulin for calcium: ordered binding of calcium ions allows the specific activation of calmodulin-stimulated enzymes. Biochemistry. 1981;20:3890–3897. doi: 10.1021/bi00516a035. [DOI] [PubMed] [Google Scholar]
- 54.Stefan MI, Edelstein SJ, Le Novere N. An allosteric model of calmodulin explains differential activation of PP2B and CaMKII. Proc Natl Acad Sci U S A. 2008;105:10768–10773. doi: 10.1073/pnas.0804672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.