Abstract
Intracellular lipolysis is an important cellular process in key metabolic tissues, and while much is known about the enzymatic basis of lipolysis, our understanding of how these processes are organized and regulated within cells is incomplete. Lipolysis takes place on the surface of intracellular lipid droplets, which are now recognized as bona fide organelles, and a large number of proteins have been found to change their associations with lipid droplets in response to lipolytic stimulation. Intracellular lipolysis has critical spatial and temporal domains that can be investigated using high-resolution imaging of fixed and live cells. Here, we describe techniques for high-resolution imaging of native lipid droplet proteins, of dynamic trafficking and interaction of these proteins in model systems, and of intracellular fatty acid production using fluorescent reporters in live adipocytes.
Keywords: Adipocyte, Imaging, Trafficking, Fluorescence microscopy, Immunofluorescence
1. INTRODUCTION
Intracellular lipolysis is an important cellular process in key metabolic tissues, including adipose tissue, liver, and muscle. A considerable amount is known about the enzymatic basis for lipogenesis and lipolysis; however, our understanding of how these processes are organized and regulated within cells is incomplete. Until recently, cellular triglyceride was considered to be stored in droplets lacking biological structure or organization. Growing evidence, however, suggests that lipid droplets are specialized, heterogeneous organelles that perform distinct roles in lipid biosynthesis, in transport and mobilization (Brasaemle & Wolins, 2012; Murphy, Martin, & Parton, 2009; Wolins, Brasaemle, & Bickel, 2006), and more recently in cell signaling (Mottillo, Bloch, Leff, & Granneman, 2012; Zechner et al., 2012). A large number of proteins have been found to change their associations with lipid droplets in response to lipolytic stimulation (Brasaemle, Dolios, Shapiro, & Wang, 2004). How these proteins are coordinated to control lipid storage and utilization remains an important area of investigation.
The size and protein composition of lipid droplets vary among these key metabolic tissues (Bell et al., 2008; Granneman, Moore, Mottillo, & Zhu, 2009; Wolins et al., 2006) and likely reflect their specialized functions. A substantial body of evidence indicates that members of the perilipin family of proteins play a central role in coordinating the trafficking of lipolytic effectors at the surface of lipid droplets, where fatty acid mobilization takes place (Moore, Silver, Mottillo, Bernlohr, &Granneman, 2005). The expression pattern of certain perilipin family members varies among metabolic tissues, which likely reflects functional specialization (Granneman, Moore, Krishnamoorthy, & Rathod, 2009; Granneman, Moore, Mottillo, Zhu, & Zhou, 2011; Subramanian et al., 2004; Wang et al., 2011).
Intracellular lipolysis has critical spatial and temporal domains. For example, lipolysis takes place at the surface of lipid droplets, yet several critical effector proteins, like hormone-sensitive lipase (HSL) and adipose triglyceride lipase ATGL, are largely cytosolic (Sztalryd et al., 2003). Furthermore, unperturbed lipid droplets can vary in diameter from 200 to 300 nm in cardiac muscle to >50 µm in white adipocytes (Bosma et al., 2013; Granneman et al., 2007; Moore et al., 2005). In adipocytes, hormones and neurotransmitters stimulate lipolysis within seconds of application, and this activation involves substantial changes in trafficking and protein–protein interactions at the lipid droplet surface. The molecular details of these critical processes have not been fully developed; however, given the key role of lipolysis in metabolism, a fuller understanding of the mechanisms involved could lead to new therapies for cardiovascular and metabolic diseases. Advancing this goal requires the ability to perform high-resolution imaging of native lipid droplets and monitoring the dynamic trafficking and specific protein–protein interactions on physiologically relevant timescales. In the succeeding text, we describe the techniques for high-resolution imaging of native lipid droplet proteins, dynamic trafficking of these proteins in model systems, and imaging intracellular fatty acid production using fluorescent reporters in live cells.
2. METHODS TO IMAGE AND COLOCALIZE ENDOGENOUS LIPOLYTIC EFFECTORS IN ADIPOCYTES
2.1. Materials
2.1.1 Fat cell isolation from adipose tissue, fat cells floated to Matrigel, and microdissected muscle fibers
Collagenase type I (Worthington Biochemical, Cat. # CLS-1)
Bovine serum albumin (BSA) Fraction V, fatty acid-free (Roche, Cat. # 03117057001)
(−)-N6-(2-phenylisopropyl)adenosine (PIA; Sigma-Aldrich, Cat. # P4532)
Matrigel (BD Biosciences, Cat. # 354234)
Isolation of mature adipocytes from mouse adipose tissue is carried out according to the method of Rodbell (1964). Briefly, perigonadal fat pads from two 3-month-old C57/Bl6 mice are removed and finely minced with scissors. The tissue is then digested with 1.5 mg/ml collagenase in isolation buffer (DMEM/10 mM HEPES, pH 7.4/1% BSA/100 nM PIA) for 30 min at 37 °C with vigorous shaking. The addition of PIA suppresses lipolysis and improves adipocyte yield (Viswanadha & Londos, 2006). Adipocytes are separated from stromal cells by gentle centrifugation, and the floating adipocytes are then washed three times with the isolation buffer. For imaging purposes, it is best to embed the cells in Matrigel. Isolated cells are resuspended in 1 ml ice-cold Matrigel and plated on 25 mm glass coverslips in a six-well culture plate and allowed to solidify at 37 °C for 10 min. The cells are then incubated overnight in a 1:1 mixture of isolation buffer and growth medium (DMEM/10%FCS/1% penicillin and streptomycin) at 37 °C under 5% CO2 atmosphere. Alternatively, a single 50 µl drop of adipocytes, isolated as described earlier, is applied to the center of a Matrigel-coated coverslip. The coverslip is then inverted to create a hanging drop that allows floating fat cells to contact the surface of the Matrigel. Cells can then be fixed to the coverslips with 4% paraformaldehyde and processed for immunofluorescence, as detailed in the succeeding text.
2.1.2 Cell culture of differentiated adipocytes
Three-dimensional cultures of 3T3-L1 adipocytes (American Type Culture Collection (ATCC), Cat. # CL-173) are prepared as follows. 3T3-L1 pre-adipocytes are differentiated in 10 cm plates for 5 days as described (Zebisch, Voigt, Wabitsch, & Brandsch, 2012). Adipocytes are trypsinized and resuspended in 1 ml of ice-cold Matrigel. Forty microliters of the cell suspension are placed on 12 mm glass coverslips in a 24-well culture plate and allowed to solidify at 37 °C for 10 min. Growth medium is added and the cells are maintained in this medium for two more weeks. At the end of this period, over 75% of the cells will develop 2–3 major lipid droplets with diameters >20 µm.
2.1.3 Lipolytic activation
Typical experiments examine the effects of agents that activate lipolysis via stimulation of the protein kinase A (PKA) signaling pathway. A variety of agents are effective in this regard, including the nonselective beta adrenergic agonist isoproterenol (100 nM; Sigma-Aldrich, Cat. # I2760) or the beta3-selective agonist CL 316,243 (100 nM; Sigma-Aldrich, Cat. # C5976). Strong activation of lipolysis can be achieved by activating adenylyl cyclase and inhibiting phosphodiesterases with a mixture of forskolin (10 µM; Sigma-Aldrich, Cat. # F6886) and 3-isobutyl-1-methylxanthine (IBMX, 1 mM; Sigma-Aldrich, Cat. # I5879), respectively. In a typical experiment, 3T3-L1 adipocytes or isolated mature adipocytes are rinsed with PBS and pre-incubated with DMEM/20 mM HEPES, pH 7.4/3 nM PIA for 1 h at 37 °C. A 10× concentrated stock of lipolytic-stimulating reagents is added to one set of wells to achieve appropriate final concentrations of activators. The other set of wells (unstimulated control) receives additional PIA at a final concentration of 200 nM, which serves to reduce PKA activation and inhibit lipolysis. Cells are incubated at 37 °C in a water bath for 5–30 min and then fixed with 4% paraformaldehyde for 20 min and processed for indirect immunofluorescence microscopy.
2.2. Methods
2.2.1 Fluorescence and immunofluorescence labeling
2.2.1.1 Materials
Antibodies to lipolytic effector proteins can be purchased from (or produced by) a variety of commercial sources: anti-phospho-HSL (Cell Signaling Technology, Cat. # 4139), guinea pig anti-PLIN2 (Fitzgerald Industries International), goat anti-PLIN1 (Everest Biotech, EB07728), rabbit anti-ATGL (Cell Signaling Technology, Cat. # 2439), and rabbit anti-phospho-(Ser/Thr) PKA substrate (Cell Signaling Technology, Cat. # 9621). Our lab has found it cost-effective to contract the production of affinity-purified polyclonal antibodies from commercial sources (Proteintech, Chicago, IL). HCS LipidTOX Deep Red (Cat. # H34477), Bodipy FL C16 (Cat. # D3821), and MitoTracker Red CMXRos (Cat. # M-7512) are purchased from Invitrogen.
2.2.1.2 Fluorescence staining procedures
To preserve lipid droplet structures, cells are typically fixed in 4% paraformaldehyde prepared in PBS for 15 min at room temperature before immunocytochemistry. It has been reported that other common fixation procedures, such as cold methanol or acetone, may induce lipid droplet fusion and thus should be avoided (DiDonato & Brasaemle, 2003). For colocalization experiments, it is best to use primary antisera raised in different species. This, however, is not always possible. To examine colocalization of two proteins using antibodies raised in the same species, we use the procedure of Negoescu et al. (1994). The key to this approach is the application of saturating levels of a monovalent secondary antibody when detecting the first antigen. Fixed cells are permeabilized with PBS containing 5% normal goat serum and 0.03% saponin (permeabilization buffer) for 1 h. Cells are then incubated for 1 h with primary antibodies against the first antigen. Cells are washed four times with PBS over a 40 min period and then incubated for 1–3 h with 3 µg/ml Cy3-conjugated goat anti-rabbit Fab fragment (Jackson Immunoresearch, Cat. # 111–167). Coverslips are washed as before and then incubated for 1 h with primary antibodies against the second antigen. The coverslips are again washed and incubated for 45 min with 2 µg/ml Alexa Fluor 488-conjugated goat anti-rabbit (Fab)2 antibodies (Invitrogen, Cat. # A11070). Washed slides are postfixed with 1% paraformaldehyde for 15 min. Affinity-purified primary antibodies are typically applied at a concentration of 1–3 µg/ml. All antibodies are diluted in permeabilization buffer, and incubations are at room temperature. Depending on the antigens, it may be necessary to adjust the antibody concentrations and time of incubation to achieve optimal results. For each pair of antibodies used, control experiments must be performed in which one of the primary antibodies is omitted in order to assure that the observed signals are not due to cross-reactivity. Omission of a primary antibody should completely eliminate fluorescent signals in its corresponding channel.
It is often desirable to counterstain the coverslips with organelle markers. For lipid droplets, we use either 0.25 µg/ml BODIPY FL C16 or a 1:200 fresh dilution of HCS LipidTOX Deep Red neutral lipid stain. These stains may be included in the last secondary antibody incubation during the immunofluorescence procedure. Although Nile Red can also be used to label lipid droplets, its use as a counterstain is limited due to the broad spectra that overlap with other fluorophores. If Nile Red is to be used, it must be applied after other fluorescent images have been acquired. Live cells may also be counterstained with 100–500 nM of freshly diluted MitoTracker CMXRos.
2.2.2 Microscopy and image analysis
2.2.2.1 Microscopes and lenses
A variety of high-quality confocal microscopes are commercially available for static imaging of lipolytic effectors. The exact choice depends on specific applications. Spinning disc confocal microscopes offer high-speed imaging that minimizes photobleaching and phototoxicity and are suitable for imaging protein translocation and changes in protein–protein interactions in live cells (see Section 3 in the succeeding text). A laser-scanning confocal microscope is useful for high-quality static imaging of fixed samples and dynamic imaging in live cells where precise laser control is needed, for example, in fluorescence recovery after photobleaching (FRAP) and fluorescence activation studies. In the majority of our studies, images are acquired with the Olympus IX81 microscope equipped with automated filter controls and a spinning disc confocal unit. The use of efficient high-numerical-aperture (NA) lenses is recommended.
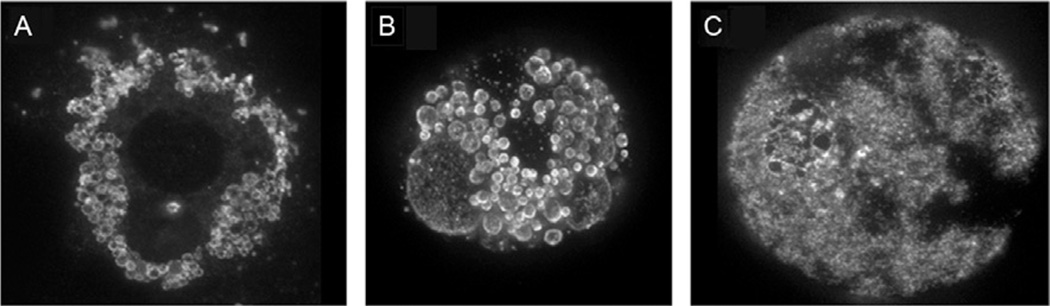
A number of special considerations should be taken into account when imaging lipid droplet proteins. Due to the thickness of the droplets, it is important to use confocal imaging mode with multiple z sections. Lipid droplet proteins reside on the droplet surface, so optical sections that capture the surface provide the best information regarding the distribution of proteins. For freshly isolated adipocytes, excellent images can be obtained by stacking a series of 20–30 individual confocal slices captured at 0.4 µm intervals using a 60 × 1.2 NA water objective. The approach avoids imaging the fluorescence at the droplet equator, where fluorescence intensity is greatest but spatial resolution is poor. It may not be feasible to perform surface imaging of droplets smaller than 1–2 µm in diameter. Under these conditions, the most intense fluorescence signal is obtained at the droplet equator, and the resulting signals are nearly perfectly round rims of fluorescence. An example of images captured with the earlier considerations is shown in Fig. 15.1. Figure 15.1A illustrates a 2D culture of 3T3-L1 adipocytes grown on glass coverslip and stained with anti-PLIN1. Most of the droplets are small and the fluorescent staining appears as perfectly round rims. When grown in 3D Matrigel (Fig. 15.1B), 3T3-L1 adipocytes develop two to three large central droplets surrounded by numerous smaller droplets. PLIN1 is preferentially localized to the smaller droplets. In isolated adipocytes (Fig. 15.1C), PLIN1 is not uniformly distributed on the surface of the central droplet and instead shows a patchy distribution.
Figure 15.1.
Confocal images showing distribution of perilipin 1 in (A) 2D culture of 3T3-L1 adipocytes grown on glass coverslips, (B) 3D culture of 3T3-L1 adipocytes in Matrigel, and (C) mature adipocytes isolated from perigonadal fat pads of mice. Shown are stacked images of 10–30 confocal z-slices. See text for details.
2.2.2.2 Image analysis
Quantitation of fluorescence intensities and colocalization of fluorescence is determined by line scan analysis of merged confocal images using IPlabs software (Scanalytics, BD Biosciences), with the analyst blind to the labeling conditions. For colocalization studies, it is important to avoid displacement artifacts resulting from inadequate color correction of the objective or poorly aligned filters. This can often be recognized by a loss of pixel registration and the uniform displacement of color between fluorescence signals during color merging. Many of the available software packages can be used to correct displacement artifacts. Multicolor fluorescent latex microspheres added at a concentration of several particles per view field to the specimen provide a convenient reference point to ensure proper pixel registration. Pearson’s linear regression analyses are performed for each scan using the GraphPad Prism 5 software (GraphPad Software), and coefficients of determination (r2) are averaged for each cell, with n = number of cells examined. Lipid droplet size is determined by measuring the cross-sectional length of respective immunofluorescence signals.
3. IMAGING PROTEIN TRAFFICKING AND PROTEIN–PROTEIN INTERACTIONS IN LIVE MODEL CELLS
3.1. Materials
3.1.1 Fluorescent fusion proteins
The panel of fluorescent fusion proteins that we have developed for imaging lipolytic trafficking utilizes fusions with enhanced yellow fluorescent protein (EYFP), enhanced cyan fluorescent protein (ECFP), and mCherry. The fluorescence emissions of each of these are readily resolved using standard filter sets, and the ECFP and EYFP can be used as fluorescence resonance energy transfer (FRET) donor and acceptors for monitoring close molecular interactions (Day & Davidson, 2012). Because tagging with fluorescent proteins can influence targeting, it is important to confirm correct targeting by comparing subcellular distribution to that of endogenous protein (e.g., in 3T3-L1 adipocytes). This also allows for assessing magnitude of overexpression. Fluorescent fusion proteins used in our lab are based on the Clontech ECFP/EYFP C1 and N1 vectors that allow placing the fluorescent protein on the N- or C-terminus of the targeted protein, respectively. We have found that the following fusions work well in a variety of cell types: PLIN1–EYFP, PLIN1–ECFP, and PLIN1–mCherry; ECFP–HSL, EYFP–HSL, and mCherry–HSL; ECFP–ABHD5, EYFP–ABHD5, mCherry–ABHD5, and ABHD5–mCherry; ECFP–ATGL, EYFP–ATGL, mCherry–ATGL, and ATGL-EYFP.
3.1.2 Bimolecular fluorescence complementation constructs
Bimolecular fluorescence complementation is a protein complementation methodology that assesses protein–protein interactions in live cells (Hu, Chinenov, & Kerppola, 2002; Magliery et al., 2005). The basic methodology involves fusing N-terminal (Yn = amino acid 1–158) and C-terminal (Yc = amino acids 155–239) fragments of EYFP (or Venus) to proteins of interest. Close interaction of test proteins allows spontaneous folding of the reporter fragments into an active protein. We found that the following protein pairs produce complemented fluorescence: Yn-ABHD5 + PLIN1-Yc; Yn-ABHD5 + PLIN5-Yc; Yn-ABHD5 + ATGL-Yc; Yn-ABHD5 + S47A ATGL-Yc; Yn-HSL + PLIN1-Yc; PLIN1-Yn + PLIN1-Yc.
3.2. Methods
3.2.1 Considerations
As mentioned earlier, major considerations concern faithful targeting of fusion proteins and analysis in an appropriate cellular background. Fluorescent reporters can be used as tracers to monitor protein trafficking when expressed at relatively low levels. Experiments that involve assessment of protein–protein interactions require the use of cells lacking endogenous expression, since endogenous proteins will compete and reduce signals. For this purpose, one can use 3T3-L1 preadipocytes. The imaging of lipid droplets in transfected preadipocytes is greatly facilitated by supplementing the media with 400 µM oleic acid that has been complexed to BSA. When using transfected cells, it is important to keep expression levels as low as possible to avoid mistargeting. For example, PLIN1 accumulates on the nuclear envelope and ER when moderately overexpressed or when the triglyceride supply is limiting. Lastly, fluorescence imaging of lipid droplet protein trafficking can be complicated by lipase activity of transfected proteins. For example, it is very difficult for cells coexpressing ABHD5 and ATGL to accumulate lipid droplets. Under these conditions, it is useful to use lipase dead (e.g., ATGL S47A) mutants.
3.2.2 Transfection
Two days before imaging, cells are trypsinized and plated into six-well cluster plates containing 25 mm glass coverslips (# 1.5). Cells are allowed to attach overnight and are transfected the following day. Efficient transfection can be achieved using any of several commercially available transfection reagents. For 3T3-L1 cells, the use of Lipofectamine LTX with Plus reagent (Invitrogen, Cat. # 15338-100; following the manufacturer’s instructions) yields 10–40% transfection efficiency. For preadipocytes, 400 µM oleic acid/0.5% BSA is added immediately after cells are returned to full media following transfection.
3.2.3 Imaging
Cells are imaged 1–2 days following transfection. A single coverslip is removed from the cluster plate using fine forceps and placed in a recording chamber, the design of which may vary depending on coverslip configuration. For 25 mm coverslips, we use an Attofluor cell chamber (Invitrogen, Cat. # A-7816), which allows imaging in a static bath. Krebs–Henseleit buffer containing 25 mM HEPES and 1% BSA (Sigma-Aldrich, Cat. # K4002) is gently added immediately after securing the coverslip into the chamber. Cells are typically imaged at room temperature in a normal atmosphere for up to 150 min.
3.2.3.1 Colocalization of fluorescent proteins
Routine imaging is performed using an Olympus IX81 equipped with a spinning disc confocal unit and 40× (0.9 NA) or 60× (1.2 NA) apochromatic objectives. Static colocalization of fluorescent proteins can be performed as detailed earlier. For typical trafficking experiments using lipolysis activators, images are captured at a rate of 1–2 frames per minute in confocal mode. Excitation and camera gain are set so that each exposure is less than 1 s and is usually on the order of 100–200 ms. It is important to match the capture frequency to the phenomenon under study: too slow will miss important events, whereas too fast may unnecessarily bleach the sample. A typical experimental design is to capture three or more control images, then add lipolysis activators or vehicle, and continue imaging for 10–30 min. Quantification of protein trafficking and colocalization is performed as follows. Lipid droplets are usually found as clusters in the perinuclear region. First, images are merged from the channels of interest, for example, PLIN1–EYFP and ECFP–HSL. The largest clusters of lipid droplets (detected in the PLIN1–EYFP channel) are outlined, and fluorescence intensity values of individual pixels in the ECFP and EYFP channels are indexed and imported into GraphPad Prism 5. Linear regression analyses are performed on indexed intensity values of the same droplet region for each captured frame. The extent of colocalization is evaluated pixel by pixel using Pearson’s linear regression. Since PLIN1–EYFP remains bound to the LD surface, the magnitude of translocation to droplets by soluble proteins (e.g., HSL or ATGL) can be assessed by an increase in the slope of the linear regression, that is, the amount of soluble protein bound per unit PLIN1–EYFP. The actual slope values under control conditions are somewhat arbitrary, since fluorescence intensities vary according to capture parameters and amounts of reporters present in any given cell. Therefore, to normalize across cells and experiments, the effect of stimulation is calculated as a ratio of stimulated slope to the control slope.
3.2.3.2 Fluorescence resonance energy transfer
The experimental setup for FRET is identical to the translocation analysis just described. FRET is performed using the three filter method (Gordon, Berry, Liang, Levine, & Herman, 1998) and net FRET calculated using the FRET extension of the IPlabs software. Calculation of FRET signals involves determining FRET constants, which quantifies the fluorescence bleed-through between donor (ECFP) and acceptor (EYFP) channels. FRET constants are determined for each experimental day using 3T3-L1 cells transfected with single-fluorescent constructs. For a given experimental day, all acquisition parameters (exposure, camera gain, neutral density filters, etc.) are equivalent for channels and all cell records. To quantify the net FRET signals, the subcellular domain containing PLIN–EYFP lipid droplets is defined for each frame using the autosegmentation tool of IPlabs, and the calculated net FRET values of those pixels are summed for each cell in every frame. Values for each experiment are normalized to the maximal FRET values obtained for each cell. The adequacy of the FRET constants in eliminating bleed-through can be verified with independent singly transfected cells. In addition, FRET signals should only be observed at droplets containing ECFP, and photobleaching of the EYFP acceptor should eliminate the calculated FRET signals.
FRET is fundamentally a method for measuring the proximity of fluorescent molecules, and it is possible to observe FRET signals owing to molecular crowding at the droplet surface. It is therefore important to keep expression levels well below the maximum that can be targeted to droplets and to demonstrate that FRET occurs across a range of expression and is not affected by coexpression of untagged, control proteins that are also targeted to droplets.
3.2.3.3 Fluorescence recovery after photobleaching
Lipolytic proteins have different affinities for the lipid droplet surface, and the amount of time that a given protein resides on the lipid droplet surface can vary according to physiological conditions. FRAP is a technique for determining the temporal association of proteins in specific subcellular compartments, like the lipid droplet surface. The basic experimental design is to use laser excitation to bleach a specific patch of lipid droplets and then to record the recovery of fluorescence over time as a measure of dynamic exchange on the droplet surface. FRAP experiments are performed using a Leica TCS SP5 laser-scanning microscope with a 63 × (1.4 NA) oil objective. FRAP is performed using the Leica FRAP wizard module. Briefly, a region of interest (ROI) on a patch of lipid droplets is delineated and bleached by 1–2 scans (2.6 s/scan) with full laser power from the 405, 458, and 496 nm lasers. Fluorescence recovery for proteins with ECFP tags is then monitored for up to 15 min by 5% 405 nm laser excitation and 454–493 nm emission. For each FRAP experiment, fluorescence intensities are monitored in the bleached ROI and an unbleached ROI on droplets in the same focal plane. Monitoring beached and unbleached areas allows for one to correct for bleaching that may occur during the recovery/image acquisition phase of the experiment. Average fluorescence intensity of the bleached ROI is compared to prebleach level (with 100% bleach and 0% recovery = first postbleach scan), with correction for bleaching during acquisition. Rate and extent of recovery are estimated by fitting recovery values to a single exponential association function by nonlinear regression.
3.2.3.4 Bimolecular fluorescence complementation
3T3-L1 preadipocytes are grown on 18 mm coverslips and transfected with 500 ng each of the complementary Yn and Yc fusion constructs, along with 100 ng of an ECFP tracer to identify transfected cells. For qualitative assessment of protein–protein interactions, 3T3-L1 preadipocytes are cultured for 24 h following transfection at 32 °C in growth media supplemented with oleic acid and then fixed in PBS containing 1% paraformaldehyde. Reduced temperature facilitates fluorophore maturation (Hu et al., 2002). Transfected cells are identified by ECFP fluorescence and cells scored as to the presence or absence of EYFP fluorescence. The rate of false positives arising from cellular autofluorescence (i.e., background) is usually <2%.
4. IMAGING INTRACELLULAR FATTY ACID PRODUCTION
4.1. Materials
4.1.1 Intracellular fatty acid bipartite sensor constructs
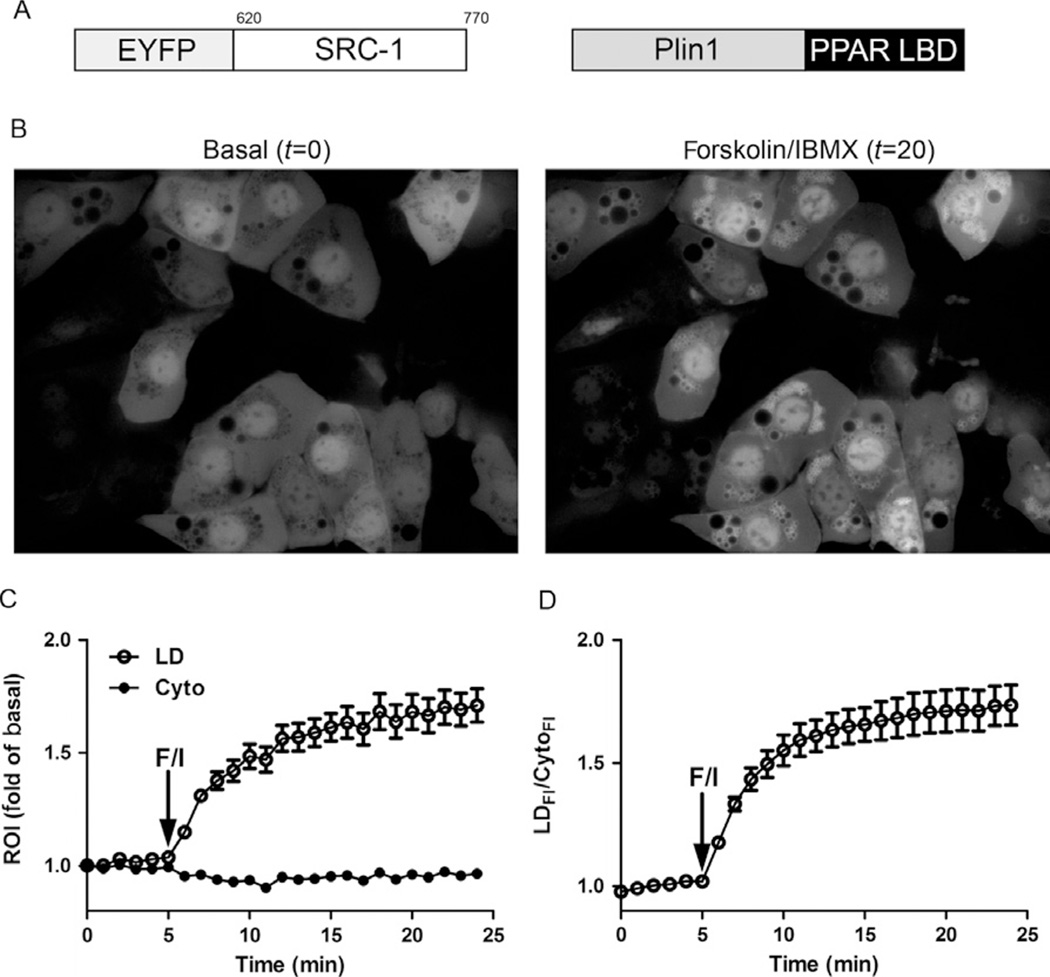
Intracellular production of fatty acids can be monitored using a bimolecular system (Mottillo et al., 2012) (Fig. 15.2A) consisting of the ligand-binding domain (LBD) of mouse PPAR-α fused to the C-terminus of full-length PLIN1 and the nuclear receptor binding domain of steroid receptor coactivator 1 (SRC-1) fused to the C-terminus of EYFP (Clontech). In this system, the binding of fatty acids to the PPAR-α LBD recruits EYFP to lipid droplets via ligand-dependent interactions of the LxxLL motifs in SRC-1 (Fig. 15.2A). The “sensor” LBD component can be additionally targeted to other intracellular compartments such as ER (McFie, Banman, Kary, & Stone, 2011) and mitochondria (Means et al., 2011).
Figure 15.2.
Fluorescence sensor for imaging intracellular fatty acid production. (A) A schematic of the constructs used for the fluorescence fatty acid sensor (amino acids for SRC1 are shown). PLIN1, perilipin-1; LBD, ligand-binding domain. (B) EYFP fluorescence images of the lipid droplet fatty acid sensor stably expressed in BA cells. Basal image capture at t = 0 and at t = 20 min, 15 min after addition of forskolin/IMBX (F/I). (C) Quantification of the region of interest (ROI) from lipid droplet (LD) or cytoplasm (Cyto) and expressed as a fold of basal. (D) Data were further expressed as a ratio of the fluorescent intensity from the LD (LDFI) over the fluorescence intensity from the cytoplasm (CytoFI). (For color version of this figure, the reader is referred to the online version of this chapter.)
4.2. Methods
4.2.1 Cell culture and transfections
Immortalized brown adipocytes are grown and differentiated as previously described (Uldry et al., 2006). Differentiated brown adipocytes (4–5 days postinduction) are transfected with Lipofectamine LTX with Plus reagent. Twenty-five millimeter coverslips are placed in wells of a six-well (35 mm) cluster plate. A transfection mixture is prepared containing (for each well) 0.75 µg of PPAR–PLIN1, 0.75 µg of EYFP-SRC1, 1.5 µl of Plus reagent, and 4 µl of LTX (Invitrogen) in 1 ml of growth media. Differentiated brown adipocyte cultures are trypsinized and resuspended at 12 million cells/ml of full media. The cells (200 µl) are applied to individual wells containing transfection mixture and incubated overnight. The media is changed the following day and cultures are imaged 1–2 days posttransfection.
4.2.2 Imaging and analysis
Images are acquired as described earlier for EYFP fluorescence using a 40× (0.9 NA) or 60× (1.2 NA) water immersion lens. The EYFP reporter is imaged every 30–60 s over a 30 min time course. Typically, basal fluorescence is recorded for the first 5 min, followed by stimulation of lipolysis with forskolin/IBMX over 20 min, after which synthetic PPAR-α ligand (Wy 14,643, 100 µM; Enzo Life Sciences, Cat. # BML-GR200-0050) is added for 5 min to determine maximal response. Examples of images from brown adipocytes that express the lipid droplet reporter are shown before (t = 0 min; Fig. 15.2B) and after stimulation with forskolin/IBMX (t = 20 min; Fig. 15.2B). The ROI (i.e., lipid droplets or cytoplasm) from two to three frames for each treatment (i.e., basal, forskolin/IBMX, or ligand) is quantified using an appropriate program such as ImageJ. Data are initially normalized by subtracting basal fluorescence or by expressing as a fold of basal fluorescence (Fig. 15.2C). Depending on the experiment, data can be also normalized to the maximal effect of synthetic PPAR ligand to probe effects such as inhibition of lipolysis (Mottillo et al., 2012). Additionally, data can be expressed as the fluorescence intensity of the LD (LDFI) relative to the intensity of the cytoplasm (CytoFI), which controls for nonspecific changes in fluorescence intensity due to minor changes in excitation or focal plane (Fig. 15.2D).
REFERENCES
- Bell M, Wang H, Chen H, McLenithan JC, Gong DW, Yang RZ, et al. Consequences of lipid droplet coat protein downregulation in liver cells: Abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57(8):2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma M, Sparks LM, Hooiveld GJ, Jorgensen JA, Houten SM, Schrauwen P, et al. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. Biochimica et Biophysica Acta. 2013;1831(4):844–852. doi: 10.1016/j.bbalip.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. The Journal of Biological Chemistry. 2004;279(45):46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. Packaging of fat: An evolving model of lipid droplet assembly and expansion. The Journal of Biological Chemistry. 2012;287(4):2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, Davidson MW. Fluorescent proteins for FRET microscopy: Monitoring protein interactions in living cells. Bioessays. 2012;34(5):341–350. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. The Journal of Histochemistry and Cytochemistry. 2003;51(6):773–780. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophysical Journal. 1998;74(5):2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. The Journal of Biological Chemistry. 2007;282(8):5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) The Journal of Biological Chemistry. 2009;284(50):34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. The Journal of Biological Chemistry. 2009;284(5):3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. The Journal of Biological Chemistry. 2011;286(7):5126–5135. doi: 10.1074/jbc.M110.180711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Molecular Cell. 2002;9(4):789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: Scope and mechanism. Journal of the American Chemical Society. 2005;127(1):146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- McFie PJ, Banman SL, Kary S, Stone SJ. Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. The Journal of Biological Chemistry. 2011;286(32):28235–28246. doi: 10.1074/jbc.M111.256008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(48):E1227–E1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HP, Silver RB, Mottillo EP, Bernlohr DA, Granneman JG. Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase. The Journal of Biological Chemistry. 2005;280(52):43109–43120. doi: 10.1074/jbc.M506336200. [DOI] [PubMed] [Google Scholar]
- Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) α and δ in brown adipocytes to match fatty acid oxidation with supply. Journal of Biological Chemistry. 2012;287(30):25038–25048. doi: 10.1074/jbc.M112.374041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochimica et Biophysica Acta. 2009;1791(6):441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Negoescu A, Labat-Moleur F, Lorimier P, Lamarcq L, Guillermet C, Chambaz E, et al. F(ab) secondary antibodies: A general method for double immunolabeling with primary antisera from the same species. Efficiency control by chemiluminescence. The Journal of Histochemistry and Cytochemistry. 1994;42(3):433–437. doi: 10.1177/42.3.7508473. [DOI] [PubMed] [Google Scholar]
- Rodbell M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. The Journal of Biological Chemistry. 1964;239:375–380. [PubMed] [Google Scholar]
- Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. The Journal of Biological Chemistry. 2004;279(40):42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, et al. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. The Journal of Cell Biology. 2003;161(6):1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Viswanadha S, Londos C. Optimized conditions for measuring lipolysis in murine primary adipocytes. Journal of Lipid Research. 2006;47(8):1859–1864. doi: 10.1194/jlr.D600005-JLR200. [DOI] [PubMed] [Google Scholar]
- Wang H, Bell M, Sreenevasan U, Hu H, Liu J, Dalen K, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. The Journal of Biological Chemistry. 2011;286(18):15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Letters. 2006;580(23):5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Zebisch K, Voigt V, Wabitsch M, Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Analytical Biochemistry. 2012;425(1):88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, et al. FAT SIGNALS—Lipases and lipolysis in lipid metabolism and signaling. Cell Metabolism. 2012;15(3):279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]