Abstract
Review on SERPINB3, with data on DNA/RNA, on the protein encoded and where the gene is implicated.
Identity
Other names
HsT1196, SCC, SCCA-1, SCCA-PD, SCCA1, T4-A.
HGNC (Hugo)
SERPINB3.
Location
18q21.33.
Local order
According to Entrez-Gene, SERPINB3 maps to NC_000018.10 in the region between 63655197 and 63661963, complement and span 6767 bases. Flanked by SERPINB4 and SERPINB11.
Note
SERPINB3, also known as SCCA1, encodes members of the serpins family. The serpins are a family of protease inhibitors originally grouped together as serine protease inhibitors, most of which are secreted [1]. The clade B serpins comprise a number of proteins including SERPINB3. In the early 1990’s it was recognized as circulating “squamous cell carcinoma antigen” (SCCA1) that was present in a substantial fraction of sera from patients bearing squamous cell cancers of the cervix [2]. Later on it was found to be associated with other types of cancer of epithelial or endodermal origins, including lung cancer, head and neck cancer, and hepatocellular carcinoma [3,4].
DNA/RNA
Description
According to the NCBI map viewer, the gene is located on chromosome 18q21.3 (NCBI Reference Sequence: NC_000018.10) and encompasses 6767 bp (Fig. 1).
Figure 1.
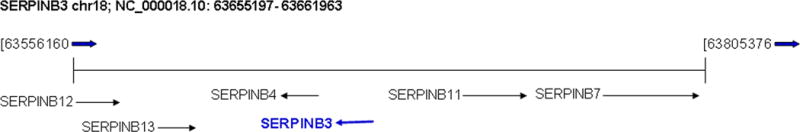
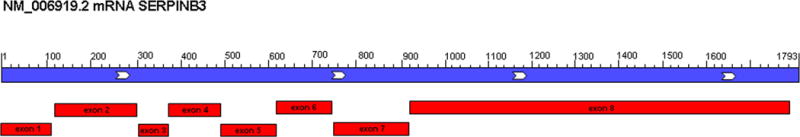
A. SERPINB3 maps in chromosome 18q21.3 (NC_000018.10) in the region between 63655197 and 63661963. Local order and flanked genes are reported. B. Map of a SERPINB3 transcript mRNA (NM_006919.2) showing its organization in 8 exons
Transcription
The SERPINB3 gene comprises eight exons and seven introns which commonly encoded a 1,793 kb mRNA. The ATG start is located in exon 2 with the stop codon in exon 8.
Transcription control is regulated by STAT3. STAT3 occupies the promoter of SERPINB3/B4 and siRNA removal of SERPINB3/B4 mRNA caused cell death in HN13 head and neck cancer cells. Thus persistently activated STAT3 is a required part of the continuous activation of SERPINB3/B4 genes, which protects tumor cells from dying [5].
Moreover recent mechanistic experiments and ChIP assays reveal that SERPINB3 increased expression in response to hypoxic conditions is specifically mediated by the binding of HIF-2α to the SERPINB3 promoter [6].
Pseudogene
No known pseudogene.
Protein
Description
SERPINB3 encodes a 390 amino acid 44,56 Kda protein, which shows sequence homology to the ovalbumin family of serine protease inhibitors (Ov-serpin) [7], a subfamily of the large serpin superfamily. Serpins have a highly ordered tertiary structure defined by the crystal structure of the prototype molecule α1-antitrypsin, consisting of nine α-helices and three β-sheets arranged in a stressed configuration with the reactive center, which has the unusual feature of being the most variable region, located in an exposed loop [8]. The mechanism of protease inhibition by serpins involves a profound change in conformation, initiated by interaction of the protease with the reactive site loop of the serpin (RSL) (amino acids 340–368). RSL consists of a loop projecting from the body of the protein, comprising a hinge region and a variable RSL [9]. Biochemical analysis of recombinant SERPINB3 shows that it is a potent cross-class inhibitor of papain-like cysteine proteinases such as cathepsin L, cathepsin S and cathepsin K [10].
An isoform produced by alternative splicing has been reported. The sequence of this isoform differs from the canonical sequence for 205–256 amino acid missing (Fig. 2).
Figure 2. SERPINB3 protein mature chain.
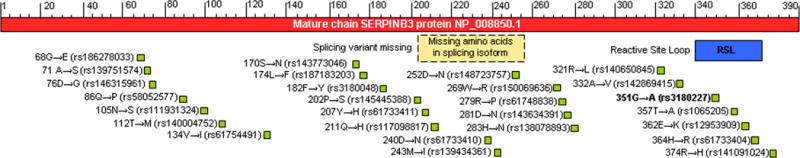
Site of Reactive Center Loop (RSL) (blue), and described variants (green) are indicated. Potential site of splicing variant missing amino acids 205–256 are also reported (yellow).
Expression
SERPINB3 is expressed in the spinous and granular layers of normal squamous epithelium, in several organs including: epithelium of the tongue, esophagus, tonsil, cervix uterine, vagina, Hassal’s corpuscles of the thymus and some areas of the skin. SERPINB3 was also detected in saliva, respiratory secretions and amniotic fluid samples from healthy individuals [11,12]. Moreover, SERPINB3 was recently reported to be expressed on CD27+ B lymphocytes [13].
In particular, immunohistochemistry analysis revealed positive staining in sweat glands in the dermis of the skin, endothelial cells of the veins and arteries walls in the intestine.
Within the normal liver, SERPINB3 protein expression was seen in portal interlobular ducts, in the walls (myocytes of the media) of the large and medium sized hepatic arteries and sometimes in the endothelial cells of the portal veins.
Normal hepatocytes, sinusoidal cells and Kupffer cells do not exhibit any reactivity, except some hepatocytes in the limiting plate that can show focal faint positivity [14]. HepCAM positive liver stem cells from both foetal and adult livers also express SERPINB3 [15].
Initially, SERPINB3 was discovered as a serological marker for advanced squamous cell tumors in the cervix [16], and was later found to be associated with other types of cancer with epithelial or endodermal origins. Moreover, elevated expression of SERPINB3 is associated with high-grade breast carcinoma and correlates with estrogen receptor/progesterone receptor double negative tumors as well as with a poor prognosis for breast cancer patients [17].
Localization
SERPINB3 may be found in cytoplasmic and pericellular locations [16]. Moreover an additional surface localization for this serpin has been reported [18], [13]. Although it was initially reported that SERPINB3 is a cytosolic protein, its nuclear localization has been also described recently, expanding the potential range of physiological functions of this molecule. Under certain conditions, such as following exposure to UV irradiation, SERPINB3 is translocated into the nucleus. Although SERPINB3 does not possess a nuclear localization signal, it binds with c-Jun NH2-terminal kinase-1 (JNK1), and upon JNK1 activation SERPINB3 enters the nucleus [19].
In addition, other studies have shown that SERPINB3 may be secreted in serum and can predict HCC development in patients with cirrhosis [20].
Function
SERPINB3 is physiologically involved in the regulation of differentiation in normal squamous epithelium and is overexpressed in neoplastic tissue of epithelial origin, where it might be involved in the apoptotic pathway as a protease inhibitor [21].
Regarding their role in normal epithelia, it has been suggested that SCCA isoforms may protect from bacterial, viral cystein proteases [21], and mast cell chymase [22]. As a protease inhibitor, SERPINB3 is able to inhibit cysteine proteases (cathepsins L, S, K and papain) [10], and in cancer cells it confers resistance to drug-induced apoptosis by inhibiting lysosomal cathepsin proteases [23]. However, under a variety of stress conditions SERPINB3 displays an anti-apoptotic function unrelated to its proteinase inhibition activity [24, 25]. Indeed, SERPINB3 protects cells from exposure to radiation through an inhibitory effect either on the MAP family of c-Jun terminal kinase (JNK) [19] and the interaction between SERPINB3 and JNK1 was also supported by the crystallographic study [26]; or with a decreased phosphorylation of the proapoptotic p38 mitogen-activated protein kinase on p38 [27]. In epithelial ovarian cancer cells exposed to cisplatin, SERPINB3 expression is associated with drug resistance and poor progression-free survival [28,29], whereas it inhibits the release of mitochondrial cytochrome c in squamous cell carcinoma after treatment with TNF-α [30], or with DNA alkylating agents [28].
Moreover, SERPINB3 expression is associated with poor survival in patients with breast cancer treated with anthracycline-based neoadjuvant chemotherapy [31] and in patients with epithelial ovarian cancer a high SERPINB3 expression is a prognostic factor for platinum resistance and shorter progression-free survival [32].
In addition, recent results, reported that SERPINB3 protects from oxidative damage by chemotherapeutics through inhibition of mitochondrial respiratory complex I [25].
Experiments carried out with serum-derived HBV particles have demonstrated that isolated SERPINB3 protein is able to bind preS1 encoded sequence HBV surface protein, allowing virus entry into human hepatocytes and also peripheral blood mononuclear cells, underlying its potential biological role in HBV infection [33–35].
This serpin induces also cell proliferation and deregulation of adhesion processes, leading to epithelial-mesenchymal transition (EMT) with increased invasiveness potential [36]. In addition, it has been reported that it induces TGF-β expression [37,38] and promotes fibrogenesis in experimental models [39].
In addition, SERPINB3 may enhance its oncogenic potential through inhibition of several tumor suppressive miRNAs [40] and could play a role in the development of cancer phenotype.
More recently, it has been reported that increased SERPINB3 expression leads to inhibition of protein turnover, unfolded protein response, activation of NF-kB and is essential for Ras-mediated cytokine production and tumour growth [41].
Homology
SERPINB3 and SERPINB4 isoforms, also known as squamous cell carcinoma antigen 1 and 2 (SCCA1 and SCCA2) are highly homologous, 91% identical at the amino acid level [42], share conserved tertiary structure, and use a unique conformational rearrangement for their inhibitory activity [43].
However, SERPINB3 and SERPINB4 show distinct properties and substrates: SERPINB3 inhibits papain-like cysteine proteinases, cathepsins K, L, and S while SCCA2 inhibits chymotrypsin-like serine proteinases, cathepsin G and mast cell chymase [44][22]. In mouse, SCCA locus (SERPINB3 and SERPINB4) was expanded and contained four genes, Serpinb3a, -b3b, -b3c, and -b3d, and three pseudogenes, Serpinb3-ps1, -ps-2, and -ps-3. Percentage protein sequences identity: 55–59%) [45].
Moreover similarity-to-human data found for SERPINB3 was found in: worm (Caenorhabditis elegans), fruit fly (Drosophila melanogaster), mosquito (Anopheles gambiae), chicken (Gallus gallus), mouse (Mus musculus), rattus (Rattus norvegicus) and chimpanzee (Pan troglodytes).
Mutations
Maps of different Single-nucleotide polymorphisms (SNP) in human SERPINB3 are reported in Fig. 2.
Only SNP rs3180227 has beeen well characterised [46]. This polymorphic variant (also known as SCCA-PD) presents the 351G/A mutation in the variable reactive site loop (RSL) of the protein (GenBank accession number: AY190327). The prevalence of SCCA-PD was 24% in the normal population, while in patients with cirrhosis it was 45%, supporting the hypothesis of a higher contribution of this isoform to liver disease progression. In addition, the specific amino acid change detected in the reactive center of this SNP might confer a different biological behavior to the serpin improving the antiprotease activity of SERPINB3 [47].
Implicated in
Hepatocellular carcinoma
Several papers have documented the key role of SERPINB3 in Hepatocellular Carcinoma (HCC).
SERPINB3 is almost undetectable in normal liver but it is expressed in HCC cells and in cells of highly dysplastic nodules and hepatocytes of peri- tumoral cirrhotic tissue, suggesting that SERPINB3 expression may represent a relatively early event in hepatocarcinogenesis [48].
SERPINB3 has been proposed as a serological biomarker that, alone or in combination with α- fetoprotein, may improve the sensitivity of HCC diagnosis [49, 50].
Circulating SERPINB3-IgM immuno-complexes have been described in cirrhotic patients at higher risk of HCC development [20] and in patients with HCC diagnosis [49]
Moreover, it has been reported that human hepatoma cells, stably transfected in order to over- express SERPINB3, exhibited a significant increase in proliferation rate and unequivocal evidence of invasiveness and Epithelial to Mesenchimal Transition, then potentially acting as a putative paracrine mediator able to favor cancer cell growth and metastatic invasiveness [36].
In addition a very recent study revealed that high levels of SERPINB3 were detectable in 22% of HCCs specimens from cirrhotic patients and were found to be significantly associated with early tumor recurrence, then representing a candidate prognostic marker able to identify the subset of most aggressive HCCs [51].
Lung cancer
Elevated levels of Squamous cell carcinoma antigen (SCC-Ag) is secreted by non-small cell lung tumours (NSCLC) and can be detected in serum [52].
It has been reported that preoperative SCC-Ag level in serum and its postoperative decrease have prognostic significance in NSCLC [53].
Moreover, in another study, tumor transcriptome analysis has revealed the predictive and prognostic impact of lysosomal protease inhibitors (SERPINB3 and cystatin C) with clinical response in platinum-based chemotherapy – treated in NSCLC patients [54]. These molecules potentially represent novel targets for NSCLC therapeutics.
Ovarian cancer
In a model of human epithelial ovarian cancer (EOC) using chickens, the most relevant animal model, SERPINB3 mRNA was induced in cancerous, but not normal ovaries, and it was abundant only in the glandular epithelium of cancerous ovaries of chickens.
In addition, strong expression of SERPINB3 protein was reported as prognostic factor for platinum drug resistance and for poor progression- free survival in patients with EOC [32].
Breast cancer
Elevated expression of SERPINB3 is associated with both high grade and advanced stage human breast carcinomas. In addition, it has been reported that SCCA-positive breast tumors have a worse clinical outcome, including decreased overall survival and recurrence free survival [17].
Furthermore, SERPINB3 positivity predicted poor survival in patients treated with anthracycline-based chemotherapy [55].
The implication of SERPINB3 in breast cancer may provide a novel diagnostic approach that will help to understand the initiation and advancement of breast cancer and provide new therapeutic options.
Hepatoblastoma
Hepatoblastoma (HB) is the most common liver malignancy in early childhood and it is considered an embryonal tumour of the liver.
According to a recent paper, SERPINB3 was overexpressed in 79% of the cases of HBs. Moreover, by immunohistochemistry SERPINB3 expression was found mainly in the embryonic, blastemal, small cell undifferentiated (SCUD) components of HB. High SERPINB3 reactivity was also detected in neoplastic cell clusters of portal vein tumour thrombosis. Furthermore a direct correlation was observed between SERPINB3 gene expression and tumour extension, suggesting that this serpin might help in defining the risk profile of children affected by this neoplasm [14].
Autoimmune disorders
Alteration in serpin function was shown to associate with deregulation of cell survival as well as with some autoimmune traits, meaning that people carrying serpin dysfunction often display an altered immune response. A recent study explored SERPINB3 expression in patients with impaired immune response to assess the potential involvement of SERPINB3 in the deregulation of B-cell reactivity.
Although serpins mainly act at the intracellular level, membrane-bound expression of SERPINB3 was recently demonstrated also on peripheral blood mononuclear cells, especially on CD27+ B cells. Interestingly, SERPINB3 was found to be absent on autoimmune diseases as SLE (systemic lupus erythematosus) CD27+ B lymphocytes, consistent with its expression being suppressed by high levels of type I interferon, which is a typical finding in SLE [13].
Since SERPINB3 displays an antiapoptotic behavior, alterations in its expression might contribute to the apoptotic deregulation seen in SLE, thereby increasing the autoantigen burden [56]. Then, SERPINB3 expression and CD27 positivity were found to be directly related, suggesting that this serpin might also be implicated in normal B cell activation. It has to be noted that the peripheral B cell repertoire and particularly CD27+ B cell number is heterogeneously altered in SLE [57].
In summary, these results may suggest a role for SERPINB3 in maintaining immune homeostasis, and that the impairment in serpin function may contribute to the development of autoimmune disorders.
Lung fibrosis
Idiopathic pulmonary fibrosis (IPF), with its histopathological signature of usual interstitial pneumonia is a chronic progressive disorder of interstitial lung disease of unknown etiology with a poor prognosis.
It has been reported overexpression of SERPINB3 in lung tissue of IPF patients compared with other forms of interstitial lung diseases and normal lungs. In IPF, SERPINB3 was abnormally secreted by metaplastic epithelial cells other than bronchial cells where it is normally expressed [37].
Moreover, mice transgenic for human SERPINB3, showed higher TGF-β expression and more extended pulmonary fibrosis than controls [58].
In addition, it has been reported that SERPINB3 immunocomplexed is increased in scleroderma patients with lung fibrosis [59]
Cholesteatoma
Cholesteatoma is a destructive and expanding growth consisting of keratinizing squamous epithelium in the middle ear and/or mastoid process.
Recent data suggest that SERPINB3, STAT3, cathepsin K, and cathepsin L are associated with the proliferation and growth of cholesteatoma and that these proteins may be influential factors in cholesteatoma growth [60].
Skin disease
Many intrinsic and extrinsic factors are associated with the stratum corneum (SC) barrier disruption. In the study of Katagiri C, it has been reported a high correlation between SERPINB3 and transepidermal water loss (TEWL). This finding was confirmed by means of a barrier disruption study with a topical oleic acid treatment: subjects with high levels of SERPINB3 readily developed impaired SC barrier function. Furthermore, SERPINB3 content showed a very high correlation with the number of parakeratotic cells in the cornified layer of the skin. These findings indicate that SERPINB3 can be considered a marker of parakeratosis and may play an inhibitory role in the process of nuclear digestion [61].
Psoriatic Skin
It has been also reported that Cathepsin L (a target of SERPINB3) is one of the lysosomal acid proteinases recently identified in psoriatic epidermis [62] together with various proteinases including tryptase, chymase, and cathepsin G (targets of SERPINB4: see homology in previous paragraph) released by degranulation in psoriatic lesion [63].
Takeda A, et al., have shown that SERPINB3/4 isoforms (SERPINB3 and SERPINB4) are indeed predominantly present along the periphery of the intercellular space in the upper spinous cell layer of psoriatic epidermis from patients with a high serum SERPINB3/4 level. In addition, SERPINB3/4 immunoreactivity was detected around the degranulated cells near the dermo-epidermal junction as well as in the granules of filtrated cells. Furthermore, strong positive staining for SERPINB3/4 was also found in nuclei of granular layer cells and a considerable number of cells in the elongated rete rete ridges of psoriatic epidermis. In particular, SERPINB3 mRNA was ubiquitously expressed in all normal skin, and significantly overexpressed in psoriatic skin tissues. On the other hand, SERPINB4 mRNA expression was specific for psoriatic skin tissues, while it was absent in normal epidermis [64].
Asthma related pathology
A protective role of SERPINB3 in asthma was initially suggested by a microarray analysis of human bronchial epithelial cell cultures after stimulation with IL-4 and IL-13 [65]. This serpin may exert its protective role by inhibiting endogenous proteases associated with the inflammatory response.
Further studies indicated that SERPINB3 serum levels were increased in patients with bronchial asthma, asthma exacerbation and in patients with allergic rhinitis [66–68].
References
- 1.Silverman GA, Whisstock JC, Askew DJ, et al. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61:301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeshima N, Suminami Y, Takeda O, et al. Expression of mRNA of SCC antigen in squamous cells. Tumour Biol. 1992;13:338–342. doi: 10.1159/000217784. [DOI] [PubMed] [Google Scholar]
- 3.Schneider SS, Schick C, Fish KE, et al. A serine proteinase inhibitor locus at 18q21.3 contains a tandem duplication of the human squamous cell carcinoma antigen gene. Proc Natl Acad Sci U S A. 1995;92:3147–3151. doi: 10.1073/pnas.92.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontisso P, Calabrese F, Benvegnu L, et al. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br J Cancer. 2004;90:833–837. doi: 10.1038/sj.bjc.6601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed ST, Darnell JE., Jr Serpin B3/B4, activated by STAT3, promote survival of squamous carcinoma cells. Biochem Biophys Res Commun. 2009;378:821–825. doi: 10.1016/j.bbrc.2008.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannito S, Turato C, Paternostro C, et al. Hypoxia up-regulates SERPINB3 through HIF-2alpha in human liver cancer cells. Oncotarget. 2015;6:2206–2221. doi: 10.18632/oncotarget.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remold-O’Donnell E. The ovalbumin family of serpin proteins. FEBS Lett. 1993;315:105–108. doi: 10.1016/0014-5793(93)81143-n. [DOI] [PubMed] [Google Scholar]
- 8.Hunt LT, Dayhoff MO. A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor. Biochem Biophys Res Commun. 1980;95:864–871. doi: 10.1016/0006-291x(80)90867-0. [DOI] [PubMed] [Google Scholar]
- 9.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 10.Schick C, Pemberton PA, Shi GP, et al. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 11.Kato H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. 1996;16:2149–2153. [PubMed] [Google Scholar]
- 12.Cataltepe S, Gornstein ER, Schick C, et al. Co-expression of the squamous cell carcinoma antigens 1 and 2 in normal adult human tissues and squamous cell carcinomas. J Histochem Cytochem. 2000;48:113–122. doi: 10.1177/002215540004800112. [DOI] [PubMed] [Google Scholar]
- 13.Vidalino L, Doria A, Quarta SM, et al. SERPINB3 expression on B-cell surface in autoimmune diseases and hepatitis C virus-related chronic liver infection. Exp Biol Med (Maywood) 2012;237:793–802. doi: 10.1258/ebm.2012.012024. [DOI] [PubMed] [Google Scholar]
- 14.Turato C, Buendia MA, Fabre M, et al. Over-expression of SERPINB3 in hepatoblastoma: A possible insight into the genesis of this tumour? Eur J Cancer. 2012;48 doi: 10.1016/j.ejca.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Villano G, Turato C, Quarta S, et al. Hepatic progenitor cells express SerpinB3. BMC Cell Biol. 2014;15:5-2121-15-5. doi: 10.1186/1471-2121-15-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uemura Y, Pak SC, Luke C, et al. Circulating serpin tumor markers SCCA1 and SCCA2 are not actively secreted but reside in the cytosol of squamous carcinoma cells. Int J Cancer. 2000;89:368–377. doi: 10.1002/1097-0215(20000720)89:4<368::aid-ijc9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Catanzaro JM, Guerriero JL, Liu J, et al. Elevated expression of squamous cell carcinoma antigen (SCCA) is associated with human breast carcinoma. PLoS One. 2011;6:e19096. doi: 10.1371/journal.pone.0019096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Falco S, Ruvoletto MG, Verdoliva A, et al. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J Biol Chem. 2001;276:36613–36623. doi: 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- 19.Katagiri C, Nakanishi J, Kadoya K, et al. Serpin squamous cell carcinoma antigen inhibits UV-induced apoptosis via suppression of c-JUN NH2-terminal kinase. J Cell Biol. 2006;172:983–990. doi: 10.1083/jcb.200508064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pontisso P, Quarta S, Caberlotto C, et al. Progressive increase of SCCA-IgM immune complexes in cirrhotic patients is associated with development of hepatocellular carcinoma. Int J Cancer. 2006;119:735–740. doi: 10.1002/ijc.21908. [DOI] [PubMed] [Google Scholar]
- 21.Suminami Y, Nawata S, Kato H. Biological role of SCC antigen. Tumour Biol. 1998;19:488–493. doi: 10.1159/000030042. [DOI] [PubMed] [Google Scholar]
- 22.Schick C, Kamachi Y, Bartuski AJ, et al. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- 23.Suminami Y, Nagashima S, Vujanovic NL, et al. Inhibition of apoptosis in human tumour cells by the tumour-associated serpin, SCC antigen-1. Br J Cancer. 2000;82:981–989. doi: 10.1054/bjoc.1999.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidalino L, Doria A, Quarta S, et al. SERPINB3, apoptosis and autoimmunity. Autoimmun Rev. 2009;9:108–112. doi: 10.1016/j.autrev.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Ciscato F, Sciacovelli M, Villano G, et al. SERPINB3 protects from oxidative damage by chemotherapeutics through inhibition of mitochondrial respiratory complex I. Oncotarget. 2014;5:2418–2427. doi: 10.18632/oncotarget.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng B, Matoba Y, Kumagai T, et al. Crystal structure of SCCA1 and insight about the interaction with JNK1. Biochem Biophys Res Commun. 2009;380:143–147. doi: 10.1016/j.bbrc.2009.01.057. [DOI] [PubMed] [Google Scholar]
- 27.Murakami A, Suminami Y, Hirakawa H, et al. Squamous cell carcinoma antigen suppresses radiation-induced cell death. Br J Cancer. 2001;84:851–858. doi: 10.1054/bjoc.2000.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ullman E, Pan JA, Zong WX. Squamous cell carcinoma antigen 1 promotes caspase-8-mediated apoptosis in response to endoplasmic reticulum stress while inhibiting necrosis induced by lysosomal injury. Mol Cell Biol. 2011;31:2902–2919. doi: 10.1128/MCB.05452-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim W, Kim HS, Jeong W, et al. SERPINB3 in the chicken model of ovarian cancer: a prognostic factor for platinum resistance and survival in patients with epithelial ovarian cancer. PLoS One. 2012;7:e49869. doi: 10.1371/journal.pone.0049869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto K, Kiyoshima T, Matsuo K, et al. Effect of SCCA1 and SCCA2 on the suppression of TNF-alpha-induced cell death by impeding the release of mitochondrial cytochrome c in an oral squamous cell carcinoma cell line. Tumour Biol. 2005;26:165–172. doi: 10.1159/000086949. [DOI] [PubMed] [Google Scholar]
- 31.Collie-Duguid ES, Sweeney K, Stewart KN, et al. SerpinB3, a new prognostic tool in breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2012;132:807–818. doi: 10.1007/s10549-011-1625-9. [DOI] [PubMed] [Google Scholar]
- 32.Lim W, Kim HS, Jeong W, et al. SERPINB3 in the chicken model of ovarian cancer: a prognostic factor for platinum resistance and survival in patients with epithelial ovarian cancer. PLoS One. 2012;7:e49869. doi: 10.1371/journal.pone.0049869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Falco S, Ruvoletto MG, Verdoliva A, et al. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J Biol Chem. 2001;276:36613–36623. doi: 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- 34.Pontisso P, Morsica G, Ruvoletto MG, et al. Hepatitis B virus binds to peripheral blood mononuclear cells via the pre S1 protein. J Hepatol. 1991;12:203–206. doi: 10.1016/0168-8278(91)90939-9. [DOI] [PubMed] [Google Scholar]
- 35.Ruvoletto MG, Tono N, Carollo D, et al. Surface expression of squamous cell carcinoma antigen (SCCA) can be increased by the preS1(21–47) sequence of hepatitis B virus. J Gen Virol. 2004;85:621–624. doi: 10.1099/vir.0.19130-0. [DOI] [PubMed] [Google Scholar]
- 36.Quarta S, Vidalino L, Turato C, et al. SERPINB3 induces epithelial-mesenchymal transition. J Pathol. 2010;221:343–356. doi: 10.1002/path.2708. [DOI] [PubMed] [Google Scholar]
- 37.Calabrese F, Lunardi F, Giacometti C, et al. Overexpression of squamous cell carcinoma antigen in idiopathic pulmonary fibrosis: clinicopathological correlations. Thorax. 2008;63:795–802. doi: 10.1136/thx.2007.088583. [DOI] [PubMed] [Google Scholar]
- 38.Turato C, Calabrese F, Biasiolo A, et al. SERPINB3 modulates TGF-beta expression in chronic liver disease. Laboratory Investigation. 2010;90 doi: 10.1038/labinvest.2010.55. [DOI] [PubMed] [Google Scholar]
- 39.Lunardi F, Villano G, Perissinotto E, et al. Overexpression of SERPIN B3 promotes epithelial proliferation and lung fibrosis in mice. Lab Invest. 2011;91:945–954. doi: 10.1038/labinvest.2011.1. [DOI] [PubMed] [Google Scholar]
- 40.Turato C, Simonato D, Quarta S, et al. MicroRNAs and SerpinB3 in hepatocellular carcinoma. Life Sci. 2014;100:9–17. doi: 10.1016/j.lfs.2014.01.073. [DOI] [PubMed] [Google Scholar]
- 41.Catanzaro JM, Sheshadri N, Pan JA, et al. Oncogenic Ras induces inflammatory cytokine production by upregulating the squamous cell carcinoma antigens SerpinB3/B4. Nat Commun. 2014;5:3729. doi: 10.1038/ncomms4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suminami Y, Kishi F, Sekiguchi K, et al. Squamous cell carcinoma antigen is a new member of the serine protease inhibitors. Biochem Biophys Res Commun. 1991;181:51–58. doi: 10.1016/s0006-291x(05)81380-4. [DOI] [PubMed] [Google Scholar]
- 43.Stein PE, Leslie AG, Finch JT, et al. Crystal structure of ovalbumin as a model for the reactive centre of serpins. Nature. 1990;347:99–102. doi: 10.1038/347099a0. [DOI] [PubMed] [Google Scholar]
- 44.Takeda A, Yamamoto T, Nakamura Y, et al. Squamous cell carcinoma antigen is a potent inhibitor of cysteine proteinase cathepsin L. FEBS Lett. 1995;359:78–80. doi: 10.1016/0014-5793(94)01456-b. [DOI] [PubMed] [Google Scholar]
- 45.Askew DJ, Askew YS, Kato Y, et al. Comparative genomic analysis of the clade B serpin cluster at human chromosome 18q21: amplification within the mouse squamous cell carcinoma antigen gene locus. Genomics. 2004;84:176–184. doi: 10.1016/j.ygeno.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Turato C, Ruvoletto MG, Biasiolo A, et al. Squamous cell carcinoma antigen-1 (SERPINB3) polymorphism in chronic liver disease. Digestive and Liver Disease. 2009;41 doi: 10.1016/j.dld.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Turato C, Biasiolo A, Pengo P, et al. Increased antiprotease activity of the SERPINB3 polymorphic variant SCCA-PD. Exp Biol Med. 2011;236 doi: 10.1258/ebm.2011.010229. [DOI] [PubMed] [Google Scholar]
- 48.Guido M, Roskams T, Pontisso P, et al. Squamous cell carcinoma antigen in human liver carcinogenesis. J Clin Pathol. 2008;61:445–447. doi: 10.1136/jcp.2007.051383. [DOI] [PubMed] [Google Scholar]
- 49.Beneduce L, Castaldi F, Marino M, et al. Squamous cell carcinoma antigen-immunoglobulin M complexes as novel biomarkers for hepatocellular carcinoma. Cancer. 2005;103 doi: 10.1002/cncr.21106. [DOI] [PubMed] [Google Scholar]
- 50.Giannelli G, Fransvea E, Trerotoli P, et al. Clinical validation of combined serological biomarkers for improved hepatocellular carcinoma diagnosis in 961 patients. Clin Chim Acta. 2007;383:147–152. doi: 10.1016/j.cca.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 51.Turato C, Vitale A, Fasolato S, et al. SERPINB3 is associated with TGF-beta1 and cytoplasmic beta-catenin expression in hepatocellular carcinomas with poor prognosis. Br J Cancer. 2014;110:2708–2715. doi: 10.1038/bjc.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mino N, Iio A, Hamamoto K. Availability of tumor-antigen 4 as a marker of squamous cell carcinoma of the lung and other organs. Cancer. 1988;62:730–734. doi: 10.1002/1097-0142(19880815)62:4<730::aid-cncr2820620415>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 53.Vassilakopoulos T, Troupis T, Sotiropoulou C, et al. Diagnostic and prognostic significance of squamous cell carcinoma antigen in non-small cell lung cancer. Lung Cancer. 2001;32:137–144. doi: 10.1016/s0169-5002(00)00225-7. [DOI] [PubMed] [Google Scholar]
- 54.Petty RD, Kerr KM, Murray GI, et al. Tumor transcriptome reveals the predictive and prognostic impact of lysosomal protease inhibitors in non-small-cell lung cancer. J Clin Oncol. 2006;24:1729–1744. doi: 10.1200/JCO.2005.03.3399. [DOI] [PubMed] [Google Scholar]
- 55.Collie-Duguid ES, Sweeney K, Stewart KN, et al. SerpinB3, a new prognostic tool in breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2012;132:807–818. doi: 10.1007/s10549-011-1625-9. [DOI] [PubMed] [Google Scholar]
- 56.Vidalino L, Doria A, Quarta S, et al. SERPINB3, apoptosis and autoimmunity. Autoimmun Rev. 2009;9:108–112. doi: 10.1016/j.autrev.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 57.Korganow AS, Knapp AM, Nehme-Schuster H, et al. Peripheral B cell abnormalities in patients with systemic lupus erythematosus in quiescent phase: decreased memory B cells and membrane CD19 expression. J Autoimmun. 2010;34:426–434. doi: 10.1016/j.jaut.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Lunardi F, Villano G, Perissinotto E, et al. Overexpression of SERPIN B3 promotes epithelial proliferation and lung fibrosis in mice. Lab Invest. 2011;91:945–954. doi: 10.1038/labinvest.2011.1. [DOI] [PubMed] [Google Scholar]
- 59.Giannelli G, Iannone F, Fransvea E, et al. Squamous cellular carcinoma immunocomplexed is increased in scleroderma patients with lung fibrosis. Clin Exp Rheumatol. 2007;25:794–795. [PubMed] [Google Scholar]
- 60.Ho KY, Huang HH, Hung KF, et al. Cholesteatoma growth and proliferation: relevance with serpin B3. Laryngoscope. 2012;122:2818–2823. doi: 10.1002/lary.23547. [DOI] [PubMed] [Google Scholar]
- 61.Katagiri C, Iida T, Nakanishi J, et al. Up-regulation of serpin SCCA1 is associated with epidermal barrier disruption. J Dermatol Sci. 2010;57:95–101. doi: 10.1016/j.jdermsci.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Kawada A, Hara K, Kominami E, et al. Processing of cathepsins L, B and D in psoriatic epidermis. Arch Dermatol Res. 1997;289:87–93. doi: 10.1007/s004030050160. [DOI] [PubMed] [Google Scholar]
- 63.Harvima IT, Naukkarinen A, Paukkonen K, et al. Mast cell tryptase and chymase in developing and mature psoriatic lesions. Arch Dermatol Res. 1993;285:184–192. doi: 10.1007/BF00372007. [DOI] [PubMed] [Google Scholar]
- 64.Takeda A, Higuchi D, Takahashi T, et al. Overexpression of serpin squamous cell carcinoma antigens in psoriatic skin. J Invest Dermatol. 2002;118:147–154. doi: 10.1046/j.0022-202x.2001.01610.x. [DOI] [PubMed] [Google Scholar]
- 65.Hansel NN, Diette GB. Gene expression profiling in human asthma. Proc Am Thorac Soc. 2007;4:32–36. doi: 10.1513/pats.200606-132JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Izuhara K. The role of interleukin-4 and interleukin-13 in the non-immunologic aspects of asthma pathogenesis. Clin Chem Lab Med. 2003;41:860–864. doi: 10.1515/CCLM.2003.130. [DOI] [PubMed] [Google Scholar]
- 67.Nishi N, Miyazaki M, Tsuji K, et al. Squamous cell carcinoma-related antigen in children with acute asthma. Ann Allergy Asthma Immunol. 2005;94:391–397. doi: 10.1016/S1081-1206(10)60993-3. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki K, Inokuchi A, Miyazaki J, et al. Relationship between squamous cell carcinoma antigen and the clinical severity of allergic rhinitis caused by Dermatophagoides farinae and Japanese cedar pollen. Ann Otol Rhinol Laryngol. 2010;119:22–26. doi: 10.1177/000348941011900104. [DOI] [PubMed] [Google Scholar]


