Papillomaviruses (PVs) are ubiquitous, highly diverse, DNA viruses that have been isolated from all four classes of Tetrapoda, including most mammals as well as birds, turtles and snakes [1-3]; their origin predates the existence of modern humans [4-7]. Observational accounts of warts from ancient Greece and Rome describe condylomatous lesions on the skin and genitals and they presumed that genital warts were associated with promiscuous sexual behavior. Warts in general were surmised to be transmissible [8]. Manifestations of animal papillomaviruses have historically been documented in myths and paintings, particularly of the ‘jackalope’. This animal does not exist, but likely represents a case of mistaken identity as a result of a papillomavirus infection that produced cornified growths (i.e., horns) on jackrabbits. Historical records and myths provide evidence for the antiquity of papillomaviruses and the diseases they cause. In 1842, the Italian physician Dr. Rigoni-Stern was the first to hypothesize cervical cancer might be linked with sexual behavior. He observed that cervical cancer was disproportionally higher in prostitutes and married women, than nuns and virgins, implying the causative agent was likely sexually transmitted [9]. Dr. Rigoni-Stern’s observation would be validated almost 150 years later; today human papillomaviruses (HPVs) are known to be among the most commonly sexually transmitted infections (STIs), and infection by specific high-risk (HR) HPV types are known to be the etiological agents of cervical cancer [10-13].
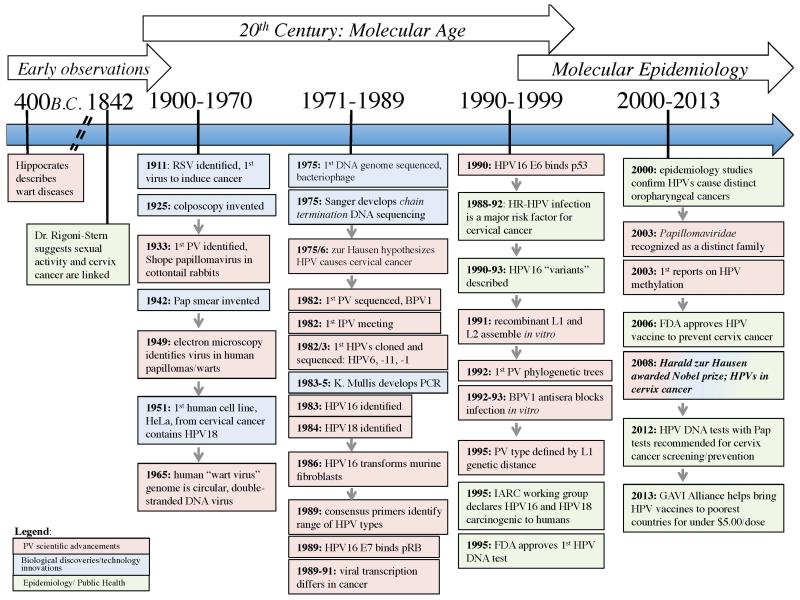
In 1911, Francis Peyton Rous famously demonstrated that filtered tumor-cell extract obtained from chicken carcinomas and transplanted to naïve chickens promoted sarcoma growth of a virulent nature, identifying the first oncogenic virus i.e. Rous sarcoma virus. Almost 25 years later, Dr. Rous and Dr. Richard Shope identified the first papillomavirus, known as Shope papillomavirus or cottontail rabbit PV (CRPV), from warty growths on cottontail rabbits. They went on to demonstrate that transmission of CRPV exhibited neoplastic potential in domestic rabbits [14,15]. Approximately 75 years later, Professor zur Hausen was awarded a Nobel Prize for the discovery of human papillomaviruses causing cervical cancer [16]. The discovery that HPVs are major contributors to cancer and represent highly adaptive, carcinogenic viral pathogens causing essentially all of cervical cancer and approximately 20% of head and neck cancers has energized the research and medical communities [13]. See Figure 1.
Figure 1. History of HPV, Cervix Cancer and Technological Advances.
The timeline displays the landmarks in Papillomavirus (PV) biology and clinical discovery. The color of the boxes serves to distinguish events and/or discoveries relating to basic PV scientific advancements in red, biological discoveries/technology innovations in blue, or epidemiology/public health in green.
Cervical cancer ranks 3rd amongst cancers affecting women worldwide and 2nd in developing countries [13]. Women in developing countries account for 85% of the global incidence of cervical cancer. Incidence rates are nearly double in developing compared to developed countries, 17.8% and 9.0% respectively. This difference is thought to be largely due to the implementation of early diagnostic screening methods, which have reduced the risk of cervical cancer associated with persistent HPV infection if left untreated. Yet, cervical cancer is still responsible for 275,000 deaths per year [17].
HPV infection by any of the 150 identified HPV types is not sufficient to cause cancer. All genital oncogenic types belong to the genus Alphapapillomavirus, which is currently comprised of 62 known HPV genotypes that infect the mucosal epithelium. Further classification beyond the genus level is required to distinguish the 13-15 HR-HPV types that are associated with oncogenic risk. It is well established that phylogenetic analysis clusters HPV taxa by host cell tropism (e.g., skin vs. genitalia), degree of oncogenic potential, and morphology of clinical lesions [1,18]. Progression to cancer is rare. The malignant potential of specific HPVs likely results from niche adaptation (i.e., evolution of an organism/virus to a specific biological/anatomic ecosystem on the body) of PVs, as cancer is undesirable for both the virus and its host. The majority of HPV infections are cleared within 6-10 months. Persistent infections with HR-HPVs are a critical risk factor for development of HPV associated precancer and cancer [19-21]. Delineating the differences intrinsic to these HR-HPV genotypes, compared to the majority of HPV types that lack oncogenic potential, will help to elucidate the genetic basis of such carcinogenic properties, thus, contributing to a better understanding of the biological mechanisms exploited by the virus to facilitate cancer development. Epidemiology studies provide a platform for obtaining viral isolates used to investigate the dynamic relationship of HPV genotype differences and clinical disease. The recent explosion in DNA sequencing technologies will continue to revolutionize methods of HPV detection [22] contributing to a better understanding of HPV biology, and the development of new therapies against HPV-associated cancers.
HPV Genome Characteristics
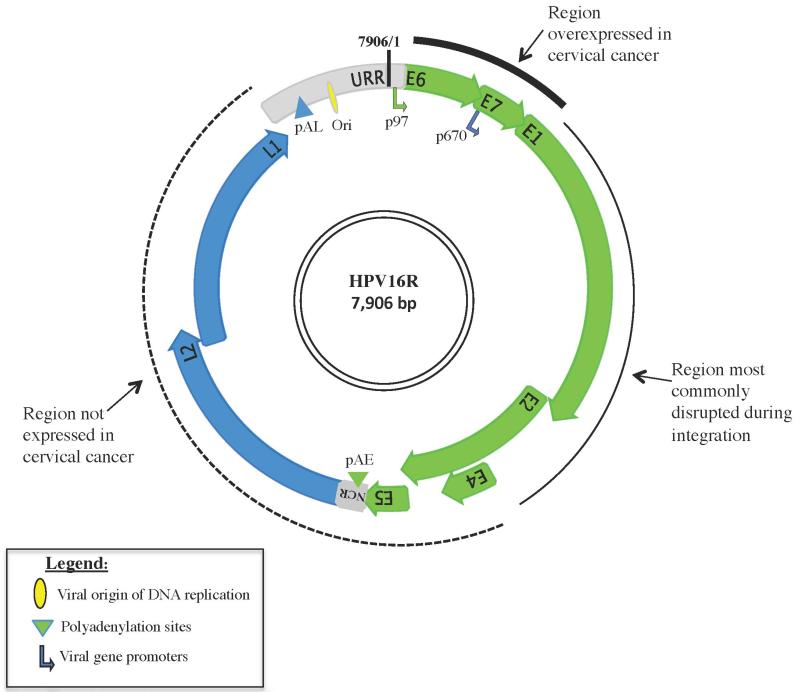
HPV is a circular, non-enveloped, double stranded DNA (dsDNA) virus approximately 8 kb in size that infects basal keratinocytes. Upon infection, the virus exists as an autonomous episome in the host-cell nucleus. The viral life cycle is mediated by a series of viral-host interactions, which govern viral transcription, virion production and eventual clearance in the majority of infections [23,24]. See Figure 2.
Figure 2. Schematic of HPV16R Genome.
The HPV16 genome displayed shows the general organization of the HPV open reading frames (ORFs) and regulatory regions. Early (E) genes and related transcriptional regulatory motifs are shown in green; late (L) genes and related transcriptional regulatory are shown in blue; and the 2 noncoding regions are shown in grey, the smaller region between E5 and L2 is termed the NCR (non-coding region), and the larger region between L1 and E6 is known as the URR (Upstream Regulatory Region). The URR contains regulatory elements including the viral DNA origin of replication (Ori) depicted by a yellow oval and the early gene promoter, p97 depicted by a green arrow. The blue arrow in the E7 ORF depicts late gene expression regulated by the late promoter, p670. Polyadenylation sites termed early (pAE) or late (pAL) are shown as green or blue triangles, respectively. Arrowheads at the tip of each ORF depict their overlap. The thick solid line represents genomic regions commonly overexpressed in cancer, the thin black line represents genomic regions frequently disrupted by integration, and the dashed line represents the genomic region rarely expressed in cancer.
The structure and function of the HPV genome is conserved throughout Papillomaviridae and is broadly divided into 3 general components (1) early gene region, denoted by “E” consists of 6 open reading frames (ORFs): E6, E7, E1, E2, E4, and E5. The early genes E1, E2, E6 and E7 are generated as a polycistronic transcript with splicing under control of the early gene promoter. Several additional early proteins E3, E5 and E8 have also been identified, but their expression is not uniformly observed throughout the Papillomaviridae. Viral transcripts can undergo extensive alternative splicing, contributing to the intricate balance between viral and host-regulated transcription (for review see [25]). The early genes code for non-structural proteins that function in viral replication, adaptation of the cellular milieu for viral activities, transactivation of viral transcription and cellular transformation and proliferation; (2) late gene region, denoted by “L” consists of the L2 and L1 ORFs. L1 and L2 encode the structural proteins, the major and minor capsid proteins, respectively. The L2 ORF encodes for group specific epitopes whereas the L1 ORF contains type specific protein domains; (3) Upstream Regulatory Region (URR) is the noncoding region between the end of the L1 ORF and the E6 start codon, comprising ~10% of the genome. The URR contains DNA recognition sites for both viral and host transcription factors and regulates early gene transcription, viral amplification and cellular tropism. The URR contains a keratinocyte-specific enhancer region proximal to the early gene promoter (p97), which highlights the significance of host cell tropism to viral gene expression and life cycle. A smaller non-coding region (NCR) located between the E5 stop and L2 start codons, harbors a highly conserved early polyadenylation signal (pAE) required for gene expression from the early promoter, including alternatively spliced early transcripts and their gene products [25]. Both the NCR and the URR display variability, useful in assessing genetic heterogeneity [26]. Increased understanding of the inherent differences between HPV species that contribute to viral function is predicated on genomic analysis. Great strides have been made since the inception of clinical HPV molecular biology in the 1970’s [7,27-29].
The viral proteins E1 and E2 function in viral genome replication and are dependent on the host DNA polymerase and replication machinery. E1 functions as a helicase and is capable of melting dsDNA for strand separation, prior to polymerization, and harbors ATPase activity. The E2 ORF encodes for a viral-DNA binding transcription factor. E1 and E2 form heterodimers at the viral origin of replication to initiate bi-directional genome synthesis. Recently, E1 and E2 have shown to function in the early induction of the DNA Damage Response (DDR) pathway contributing to a permissive environment for viral genome replication. E1 can induce beaks in the host dsDNA that activates the ATM (ataxia-telangiectasia mutated) DDR pathway, signaling cell cycle arrest [30]. The URR contains 4 highly conserved E2 binding sites that differentially regulate viral replication and early gene transcription. E2-dependent down-regulation of the early promoter activity helps the virus maintain a low profile during its early life cycle stages. In high-grade neoplastic lesions/cancer the E2 ORF can be disrupted through integration into the host genome. Integration results in the loss of E2 dependent early promoter regulation, a ramification of which can be overexpression of E6 and E7 [23,24,31]. The viral proteins E6, and E7, function as oncogenes in the HR alpha-HPVs. HR-HPV E6 and E7 disrupt cell cycle regulation in upper epithelial cells, (stratum spinosum), which normally exit cell cycle to terminally differentiate. Viral mediated inactivation of key cell-cycle regulators, known tumor suppressors, e.g., p53 and pRB (by E6 and E7, respectively) are distinct to certain alpha-PV types, although not specific to those causing cancer [32]. The E5 ORF encodes for a transmembrane protein that probably contributes to cell signaling [33].
The E5 ORF defined by the presence of a start and stop codon exists in subsets of the human alpha-PVs; the HR-HPV species, the alpha-10 species and other PVs including Bovine papillomavirus 2 (BPV2) implicated in urinary cancer (cattle). At the nucleotide level, E5 is not highly conserved. The E5 protein is tied with late gene viral life-cycle events and interacts with EGF (epidermal growth factor) and PDGF (platelet derived growth factor) to influence cellular proliferation [34]. The integrity of the E5 ORF provides another example of the differences revealed through studying HPV genotyping and phylogeny [35]. The late genes, L1 and L2 function in virion maturation and orchestrate virion self-assembly that packages the genome for release in the upper epithelia. During late stages of precancer/cancer L1 and L2 proteins are not expressed. This initially made early studies on HPV identification difficult and required the use of extracted DNA from warts to be obtained for analysis. Virus-like particles (VLPs) made from HR-HPV L1 readily self assemble and induce a neutralizing antibody response. This is the basis for the two prophylactic vaccines currently available.
Viral Identification and Classification: Phylogeny and Taxonomy
HPV genome heterogeneity was apparent during the initial studies examining viral isolates obtained from dysplastic skin lesions (cutaneous or genital). By the late 1970’s, an unknown viral agent was known to be the causative agent implicated in the development of multiple wart types, condyloma acuminata (genital warts) and possibly cervical cancer. Attempts to identify the viral agent from lesions was hampered by the fact that DNA probes often did not cross react with DNA extracts from the different warts, implicating the presence of diverse viruses [36]. DNA extracted from warts at similar anatomical sites had similar restriction enzyme digestion patterns and cross-hybridization results. Furthermore, transplantation across host species (i.e., human to rabbit) repeatedly failed [29,37]. Understanding the delicate balance of HPV-host interactions proved fundamental to addressing the clinical significance of HPV related disease.
Human papillomaviruses are extremely ancient viruses and related PVs have been identified in most nonhuman primates, including humans’ most recent common ancestors [38-40]. Sequence and phylogenetic analysis of nonhuman primate PVs isolated from the cervicovaginal area revealed genomic similarity to the alpha-HPVs. Several non-human primate PVs identified from rhesus macaques (Macaca mulatta), cynomolgus macaques (Macaca fascicularis), and olive baboons (Papio hamadryas anubis) comprise the alpha-12 species. These phylogenetically related alpha-PV types can induce epithelial dysplasia of varying degrees, resembling the high-grade cervix intraepithelial neoplasia (CIN) associated with persistent HR mucosal-HPV infections, specifically within the alpha-9 species, which includes HPV16 [5,6,41]. Experimental transmission of MfPV-3 (Macaca fascicularis PV-3) from a naturally infected female to naive female macaques was associated with the development of cervical intraepithelial neoplasia [42]. These nonhuman primate PVs are more similar to the alpha-9 HPV species, than more distantly related HPV types, such as the alpha-10 species, suggesting the mechanisms governing cellular transformation result from a common ancestral trait that predates human/primate divergence.
Diversification of HPVs occurred prior to the evolution of Homo sapiens approximately 200,000-150,000 years ago [43]. The extensive diversification and demographic range of HPVs reflect human dispersal and population expansion [4], and may intuitively suggest that HPVs have had to undergo a high rate of mutations to adapt to such diverse hosts; however, this is not the case. Rates of nucleotide mutations are remarkably slow in the virus, arising at a rate of approximately 10−8-10−7 nucleotides per year [44].
HPV characterization in population-based studies affords the unique opportunity to study hominid evolution through a viral lens to better understand virus-host interactions as they pertain to immune system surveillance and cancer progression [5,6,35]. The unique coupling of the PV-host dependent lifecycle has selected for specialized viral adaptation to specific host niches, coincident with the ability of the virus to evade a host immune response. These features have enabled PVs to capitalize on their hosts, and exploit global diversity to thrive throughout evolutionary time. Genital HPV infections are commonly dependent on sexual transmission. This exemplifies one way the virus invests in its existence; they have hijacked the most fundamental aspect of host species success, reproductive fitness. Post-infection, viral gene expression is tightly regulated such that viral gene expression is dependent on host epithelial differentiation for near exclusive activation of either the early or late gene promoters driving gene expression. Such adaptation tactics suggests a commensal virus-host relationship, yet HPV dependent malignancies defy this viewpoint. Molecular epidemiological studies assessing phylogenetic association of HPVs based on oncogenic risk support specific biological and pathological traits distinct to HPV genera, species and types [35]. HPV16 is unique in its ability to establish persistence that is highly associated with neoplastic progression, both of the cervix and in head and neck carcinogenesis. The human alpha-PVs exhibit agreement between (clinical) natural history studies and HPV phylogeny and taxonomy, providing evidence to support carcinogenicity is an evolved trait most probably related to niche adaptation [35,45-47].
HPV Classification
Methods to culture HPV in vitro or produce infectious virus through xenotropic models are not efficient, and do not provide a robust method for identification and characterization of HPV types [48]. Furthermore, a lack of a robust, consistent antibody response in infected individuals limits the use of antibody titer and serology for PV taxonomy [48]. Historically, HPV has been identified from biopsies of warts or lesions and classified by comparison to known types by restriction endonuclease cleavage patterns and/or DNA-DNA or DNA-RNA blot hybridization. Such methods had innate flaws for characterizing PVs; specifically there was no quantifiable means of comparison amongst HPV types from different lesions, titer of virus in lesions was not available, and cross-hybridization was difficult to explain except for association by anatomical site [29,37]. The appreciation for a genomic, DNA-sequence based classification system was agreed upon in the PV community by the late 1980’s. This system has facilitated the rapid advances in DNA technologies from PCR to the Next-Gen era upon us.
Today, PCR based amplification of DNA obtained from clinical samples is common. As the realization that phylogeny and genotyping methods validated one other, the notion that only a limited set of HPV types were associated with cancer spurred the need for highly specific assays that could discriminate HPV types. Consensus PCR primers targeting highly conserved regions within the L1 ORF, such as the MY09/11 PCR assay or the GP5+/GP6+ PCR assays are typically used for HPV identification [49]. Review of HPV detection methods have been recently published [50]. In addition, sequencing of the PCR amplicons for alignment to known HPV type(s) facilitate classification of genotype by nucleotide identity and identification of novel types.
PVs belong to the family Papillomaviridae and were given this status by recognition of a genome-based, DNA sequence system for classification [51]. DNA sequencing provides a quantifiable means to catalog nucleotide heterogeneity, affording the classification and taxonomy of HPV to genera, species and variant lineages [26,52]. Classification of HPV genera and species was recognized by the PV Working Group at the 14th International Papillomavirus (IPV) conference in Quebec 1995, later adapted by the International Committee on the Taxonomy of Viruses (ICTV) [1,18,48,53]. Variant lineage classification is a more recent development within the PV community that will become increasingly more relevant as high resolution techniques, such as next-generation sequencing (NGS) generate a plethora of PV sequencing data that need to be coherently analyzed, named and correlated with phenotype and geographic locations.
HPV Genome “Typing”
The L1 gene is highly conserved and provides the basis for HPV genotyping. An HPV “type” is designated when the nucleotide sequence of the L1 ORF from the cloned viral genome is more than 10% dissimilar to all known types [1].
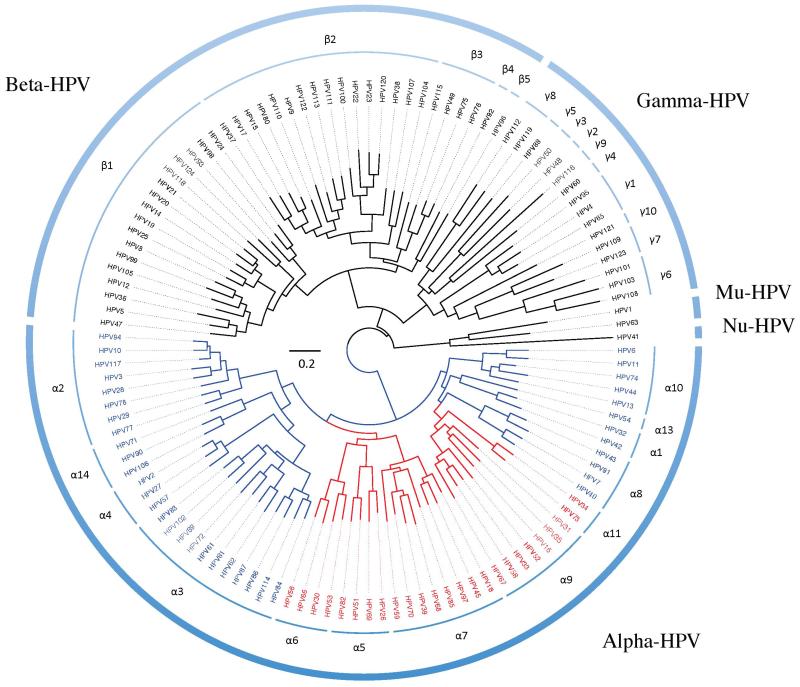
Nucleotide differences across HPV genotypes are correlated with the viral lineages based on evolution without significant, if any, recombination and these correlated changes are the result of lineage fixation [54]. Whole genome sequencing established L1 sequence identity as representative of complete genome variation due to its high conservation [1,47]. Nucleotide sequences are used to build phylogenetic trees used for HPV taxonomy. Phylogenetic analysis suggests the underlying relationship between the biological observations identified within this heterogeneous group of viruses: including host cell tropism (mucosal or cutaneous), carcinogenic risk and associated pathology [35]. Currently, over 150 HPV types have been formally identified and predominantly cluster to 3 main genera. (1) Alphapapillomaviruses are primarily isolated from the genital, mucosal epithelium and are the overwhelming cause of anogenital cancer; (2) Betapapillomaviruses are primarily isolated from cutaneous, skin lesions. The Beta-PVs include HPV types frequently associated with the rare genetic disorder, epidermodysplasia verruciformis (EV), which predisposes individuals to develop HPV-associated cutaneous, scaly wart-like squamous cell carcinomas (SCC). Many beta-HPV types were originally identified in isolates obtained from EV patients, previously termed HPV-EV types and include HPV5 and HPV8 both members of the beta-1 species, identified in approximately 90% of EV-related cutaneous SCC. Less prevalent EV-types extend to the beta-2 species (HPV38) and beta-3 species (HPV49). These types are also found associated with malignancy in immunocompromised hosts, and are less prevalent in the general population [55]; (3) Gammapapillomavirus are primarily isolated from cutaneous epithelia, some form cutaneous lesions histologically defined by the presence of homogenous intracytoplasmic inclusion bodies [1,56]. Both Gammapapillomaviruses and Betapapillomaviruses have been identified in oral samples suggesting they have an expanded tropism including the oral cavity [57]. See Figure 3. Thus, the tropism of these later genera have to be reconsidered in light of the new information. This also demonstrates that not testing a specific anatomic site (e.g., the oral cavity) for HPV (using methods to detect the gamut of types) doesn’t mean the virus is not there.
Figure 3. Human papillomavirus phylogenetic tree clustering the majority of HPVs into three genera, Alphapapillomavirus, Betapapillomavirus and Gammapapillomavirus.
A maximum likelihood (ML) tree was constructed using RAxML v7.2.8.27 [77] and an alignment of the nucleotide sequences of the L1 open reading frame (ORF) of 120 published HPV types. HPV species groups were generally classified according to the classification system for PVs by Bernard and colleagues [18]. All high-risk mucosal HPV types cluster within the genus Alphapapillomavirus, highlighted in red. The clades in blue represent mucosal HPV types with low risk or no risk to cervical cancer. The scale bar represents nucleotide change of 0.2 per site.
Alpha-HPVs Phylogenetic association with cancer risk
Phylogenetic analysis based on the sequence of the HPV L1 ORF has been the standard for genomic analysis and type classification [1,18]. Observations of phylogenetic incongruence within the alpha-PVs are shown by comparison of trees built utilizing either the early or late regions of the genome. This results in differences regarding the monophyletic origin of the oncogenic alpha-HPV types. Trees generated from the early genes or the complete genome cluster alpha-PVs by associated oncogenic risk as a monophyletic clade. Phylogeny based on late gene regions (L1 and L2) does not support a monophyletic origin for the 5 oncogenic alpha-HPV species (alpha-5, alpha-6, alpha-7, alpha-9, alpha-11). This phylogenetic incongruence is suggestive of genomic distinctions within the alpha-PVs that are exemplified by examining differences inherent to the alpha-9 (HPV16-related) and alpha-7 (HPV18-related) species that differ in biology and pathological outcome. HPV16 and HPV18 are highly prevalent, oncogenic types implicated in 70% of cervix cancer, as members of different species groups they exhibit different biological niches, cellular tropisms and manifest as precancerous and cancerous lesions differently. HPV16 targets the cutaneous squamous epithelia abundant in the ectocervix and predominantly causes cervical squamous cell carcinoma (SCC) that evolves through differing grades of squamous epithelial neoplasia observed by histological/cytological screens [58]. HPV16 also infects the squamous epithelia of the oropharynx and is identified in oropharynx cancer (targeting the regions of the throat including the base of the tongue, the soft palate and the tonsils). Additionally, HPV16 is also implicated as causal for cancers of the vulva, vagina, penis and anus [17]. Conversely,HPV18 disproportionately targets glandular, mucin-secreting columnar epithelial cells found primarily in the endocervix, and is over represented in the development of cervical adenocarcinoma (AdCa)[35,45,59,60].
Comparison of phylogenetic trees generated from either early or late gene sequences result in different phylogenies for distinguishing higher-level taxa [45]; the clades defining species groups remain intact. Yet, the nodes defining a clades’ most recent common ancestor (MRCA) appear differently depending on the genes used for the phylogenetic tree construction, reflecting a degree of phylogenetic incongruence. This hints at the occurrence of early selection events likely driven by ecological niche adaptation. Such that early and late genes are regulated through distinct promoters, that are strictly governed by the availability of host proteins differentially expressed within the stratified epithelia [24,45]. Incongruence may also result from disproportionate selective pressures on the two genomic regions. Incongruence does not likely result from an early viral recombination event, as similar genome characteristics are maintained across diverse hosts throughout evolutionary time, such as humans and non-human primates, supporting the clonal nature of viral expansion as opposed to recombination [35,45].
Variant Lineages: Model for Recent HPV speciation
Classification below the species level is not formally recognized by the ICTV [18,26,40]. Evidence from large epidemiological studies identifying HPV genomic heterogeneity and associated pathologies [43,47,61] support the need for distinction of taxa below the HPV-type level and will likely be updated soon (Burk, R. et al., 2013, in press) [52]. The term HPV subtype has become obsolete. It previously referred to isolates that exhibited 2-10% differences in nucleotide identity compared to its closest known type and/or differences in restriction enzyme cleave patterns [1]. As better systems for identification and classification of HPVs emerged subtypes are now considered variant lineages.
Variant Lineages and Sublineages: Updates on current Nomenclature Guidelines
HPV variant lineages represent viral isolates that exhibit genomic nucleotide differences of 1% to 2%, as compared to their prototype, or reference genome [5,6,26,47,61]. Viral variants share a most recent common ancestor (MRCA) that is unique to the specific HPV type. HPV variants are common, and may differ in risk for development cervical precancer (CIN2/3) or cancer. Initial studies examining HPV16 and HPV18 intratypic variation aimed to discern the contribution of variant lineages to geographic differences in virus distribution [5,6,62]. Variants of both HPV16 and HPV18 were initially classified by sequencing the URR, as this non-coding region exhibits greater nucleotide variability than protein coding regions, and is a valuable tool for identifying stable nucleotide polymorphisms, the basis for HPV classification. However, unlike type identification that is sufficiently designated by sequencing the L1 ORF, complete genome sequencing is required to identify HPV variants since the distribution of differences is not evenly spaced across the genome. In addition, a variant nomenclature system based on the complete genome permits the identification and quantification of nucleotide polymorphisms using different regions the genome. Evidence supports that certain genomic regions exhibit greater heterogeneity than others.
HPV replication depends on host DNA replication machinery, proofreading capabilities by host DNA polymerases maintain a slow rate of mutation within the viral genome at approximately 10−8 to 10−7 nucleotide substitutions per year [4]. Intratypic HPV genetic variation results from random nucleotide polymorphisms or insertions/deletions (indels) acquired through genetic drift or natural selection that become fixed over time. Stable acquisition over time, of these nucleotide changes, eventually leads to PV speciation, through a process termed lineage fixation [54] and is further supported by a lack of evidence for viral recombination events [4-6,46]. Furthermore, the stable acquisition of polymorphisms among isolates of the same HPV type (sharing a MRCA) eventually leads to type speciation, characterized by genomic nucleotide identity that is less than 90% and occurs over millions of years [43]. To this end, evidence of prehistoric human population bottlenecks is reflected through inter- and intra- typic PV genomic diversity. Distinction below the species level (PV type) is common within the PV research community, and is useful for physicians, researchers and epidemiologists investigating HPV variants for association with geographical host population and viral genetic changes that confer variable phenotypic outcomes, including varying pathological manifestations such as cancer. At present, guidelines for variant HPV lineage classification are beginning to be formally established [26,52](Burk, R. et al., 2013, in press). A formal classification and nomenclature system to describe intratypic HPV variants, at the lineage or sublineage level, will undoubtedly become increasingly useful as the future of HPV genomics expands to include data obtained from next-generation sequencing and metagenomic studies, and will facilitate a better system for cataloguing genotype-phenotype changes.
Variant Lineages and Sublineages: Identification and Clinical Relevance
Variant lineage classification is based on isolates of a known type that have had their complete genome sequenced and reveal genetic heterogeneity of at least 1% and less than 10%, based on multiple sequence alignments to the prototype (first characterized genome of a given type). Parameters for classification have been recently established through phylogenetic analysis on isolates of the alpha-9 species and HPV6 and HPV11 (including HPV16, HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67) (Burk, R. et al., 2013, in press) [26,52]. A divergence rate of 1% was determined to conservatively designate variant lineages. Similarly, pairwise nucleotide identity differences in the range of 0.5%- 1% define a type sublineage. Nomenclature for lineage and sublineage is based on an alphanumeric system wherein the prototype “reference” genome is always designated with an “A”. If sublineages for the given type are present, the prototype reference sequence is designated as “A1” (See Table 1).
Table 1. HPV Taxa Definition and Nomenclature Overview.
The table lists the current taxonomic distinctions used for classification of PVs illustrated by the HPV16 reference genome. The defining genomic characteristics refer to the aligned nucleotide sequence identity differences based on either the sequence of the L1 ORF or complete genome (CG), these values are used to distinguish HPV taxa. Proper taxonomic orthography is shown for the HPV16R genome. The inclusion of lineage and sublineage designation is not currently recognized by the ICTV, but represents important taxa distinction recognized by the PV community.
| Taxa | Defining genomic characteristics | Region | Example (HPV16 reference genome) |
|---|---|---|---|
| Family | genome size ~8kb, distinct genome organization | L1 | Papillomaviridae |
| Genus | > 40% difference between genera | L1 | Alphapapillomavirus |
| Species | 30 - 40% difference to other types | L1 | Alphapapillomavirus-9 |
| Type | > 10% difference to known types | L1 | Human papillomavirus 16 (HPV16) |
| Lineage | 1 - 10% difference to same type | CG^ | variant lineage “A” |
| Sublineage | 0.5 - 1% difference to same type | CG^ | variant sublineage “A1” |
CG refers to the complete genome sequence used for identification and nomenclature
Variant lineages from many clinically relevant alpha-HPVs are known. Variants have been characterized for the majority of the high risk mucosal HPV types: HPV16 [5,62,63], HPV18 [6,43], HPV types 31, 33, 35, 52, 58, 67 [26]; other alpha-HPV variants that have been identified include members of the: alpha-4 species HPV2, HPV27 and HPV57 [64] and two types from the low risk alpha-10 species, specifically HPV6 and HPV11 [52] associated with genital warts have been extensively examined.
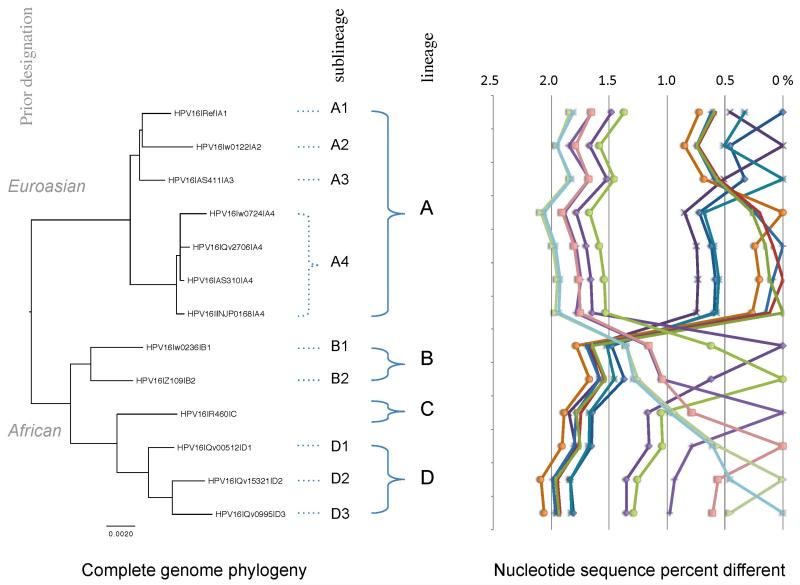
Phylogenetic analysis of HPV16 variation revealed variants reflective of human dispersal out of Africa and divergence into the 3 major human races, Africans, Caucasians and Asians. Initial phylogenetic studies identified 4 major HPV16 variants representative of host geographic origin. The major HPV16 variants were broadly termed “European” now lineage “A” or “non-European” now lineages “B, C and D”. The non-European lineages are more heterogeneous and included two African HPV16 lineages, African-1 and African-2 (B and C, respectively), and Asian-American variants (lineage D) [5,54,59,65]. These variants display phylogenetic congruence with host ethnicity and geographic origin [5]. However, with the recent update to nomenclature the designation of variant lineage simply (for the case of HPV16 example) as non-European or European is misleading and overlooks details within the viral genomes (see Figure 4). Previously termed HPV16 variants have thus been re-named according to the updated variant nomenclature guidelines whereby, European variants are classified as the “A” lineage, further resolved to four sublineages (A1, A2, A3, A4). The HPV16 non-European variants are now recognized as three distinct lineages with the appropriate sublineage designation: HPV16B (previously Af-1) contains sublineages B1, and B2; HPV16C, (previously Af-2); and HPV16D (previously NA1, AA1, AA2) contains sublineages D1, D2 and D3. This update helps to resolve and term the viral distinctions below the type level and is important since many of the nucleotide changes are correlated within taxa. See Figure 4.
Figure 4. HPV16 variant tree topology and pairwise comparisons of individual complete genomes.
A maximum likelihood (ML) tree was inferred from a global alignment of 13 complete nucleotide sequence genomes of HPV16 using RAxML v7.2.8 [77]. Distinct variant lineages (i.e., termed A/B/C/D) and sublineages (e.g., termed A1/A2/A3/A4) were classified according to the topology and nucleotide sequence differences from > 1% to < 10%, and > 0.5% to < 1%, respectively [18,26]. The percent nucleotide sequence differences were calculated for each isolate compared to all other isolates based on the nucleotide sequences (complete genome) and are shown in the panel to the right of the phylogeny. Values for comparison from an isolate are connected by lines and the comparison to self is indicated by the 0% difference point. Symbols and colored lines are used to distinguish each isolate. The scale bar at the bottom of the tree represents nucleotide change of 0.002 per site. Multiple isolates for the HPV16 A4 sublineage are shown to highlight the intra-sublineage relationship, note the clustering in the right hand graph that depicts nucleotide sequence differences.
The Future of HPV Genomics: Biomedical technology advancements and biomarker prediction
Global epidemiological studies now permit access to vast numbers of clinical HPV samples to better understand the natural history of HPV infections [47] and the biological and clinical ramifications [47,66]. The risk associated with a persistent HR infection, such as HPV16 is significantly associated with neoplastic progression. However, determinants of viral persistence and clearance are not well understood. Recent evidence suggests differences in the duration of persistence for different HPV types and/or lineages associated with clinical outcomes [47,67,68]. Understanding of HPV genomics and classification will help further characterize host/viral components involved in viral persistence and/or clearance [69,70]. This may contribute to the design of new treatments and therapeutics targeting those most at risk for cancer. On a broader scale, it may also help delineate which infections require treatment versus those that naturally regress, having implications on the financial burden associated with cancer treatment globally.
Rapid technology advances during the past decade have, and will continue to, advance our understanding of PV genomic heterogeneity that contributes to pathological clinical outcomes. Next-generation sequencing (NGS) provides increased resolution for the detection and classification of viral DNA, at the single molecule level, and will enhance methods for identification of new HPV taxa that may have previously fallen under the limit of detection by current methods [71]. This will facilitate a more complete view of Papillomaviridae diversity and should contribute to an improved understanding of the mechanisms of HPV associated malignancy [72,73]. Increased use of novel genotyping methods will improve the efficiency of identifying HPV DNA from an array of samples obtained through large cohort studies. The explosion in data generated by NGS techniques reinforces the need for current, widely accepted methods to characterize the results. This is highlighted by recent reports describing an extensive array of novel Betapapillomavirus and Gammapapillomavirus types, which are currently difficult to classify given the extensive diversity of these types and the lack of a simple method for their characterization in a standard clinical assay.
New codes in the HPV genome
The appreciation of an additional “epigenetic code” (i.e., CpG sites that can be methylated) in the HPV genome has energized new and future studies on understanding the significance of information. HPV epigenetics is a burgeoning field (for review see [74]). Accounts of CpG methylation of viral DNA, predominately from HPV16 and HPV18 were initiated at the turn of the 21st century and identified regions of viral CpG methylation by methylation-specific restriction endonuclease maps. Different cleavage patterns from viral isolates obtained from women with different stages of HPV associated CIN have been observed. The onset of technological advances within the past two decades have generated methods for more accurately identifying and quantitating DNA methylation. CpG sites are highly conserved amongst HPV species. CpG sites appear throughout the genome and exhibit varied methylation states and likely play a physiological role in the viral life cycle. Large epidemiological studies that aim to elucidate the oncogenic properties of HPV have demonstrated that methylation at specific CpG sites within the viral genome is predictive of clinical outcome, such as precancer (CIN2/3) or cancer [74-76]. Specifically, CpG sites found within the HPV16 L1 ORF are highly predictive of CIN2/3 [75]. Interestingly, the alpha-7 species contain higher numbers of CpG sites relative to the alpha-9 species. The development of high-resolution methods for HPV identification and classification should lead to the discovery of additional coded information in the HPV genome beyond nucleotide, amino-acid, CpG and DNA-binding protein regions.
References
- 1.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Herbst LH, Lenz J, Van Doorslaer K, et al. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology. 2009;383:131–5. doi: 10.1016/j.virol.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Lange CE, Favrot C, Ackermann M, Gull J, Vetsch E, Tobler K. Novel snake papillomavirus does not cluster with other non-mammalian papillomaviruses. Virology journal. 2011;8:436. doi: 10.1186/1743-422X-8-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard HU. Coevolution of papillomaviruses with human populations. Trends in microbiology. 1994;2:140–3. doi: 10.1016/0966-842x(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 5.Ho L, Chan SY, Burk RD, et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. Journal of virology. 1993;67:6413–23. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong CK, Chan SY, Campo MS, et al. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. Journal of virology. 1993;67:6424–31. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.zur Hausen H, de Villiers EM. Human papillomaviruses. Annual review of microbiology. 1994;48:427–47. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 8.Burns DA. ‘Warts and all’--the history and folklore of warts: a review. Journal of the Royal Society of Medicine. 1992;85:37–40. doi: 10.1177/014107689208500113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowson KE, Mahy BW. Human papova (wart) virus. Bacteriological reviews. 1967;31:110–31. doi: 10.1128/br.31.2.110-131.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffman M, Wentzensen N. Human papillomavirus infection and the multistage carcinogenesis of cervical cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22:553–60. doi: 10.1158/1055-9965.EPI-12-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 12.Jemal A, Simard EP, Dorell C, et al. Annual Report to the Nation on the Status of Cancer, 1975-2009, Featuring the Burden and Trends in Human Papillomavirus (HPV)-Associated Cancers and HPV Vaccination Coverage Levels. Journal of the National Cancer Institute. 2013;105:175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 14.Shope RE, Hurst EW. Infectious Papillomatosis of Rabbits: With a Note on the Histopathology. The Journal of experimental medicine. 1933;58:607–24. doi: 10.1084/jem.58.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rous P, Beard JW. The Progression to Carcinoma of Virus-Induced Rabbit Papillomas (Shope) The Journal of experimental medicine. 1935;62:523–48. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 17.Cancer WIICoHaC . Summary Report 2010. 2010. Human Papillomavirus Related Cancers in World. [Google Scholar]
- 18.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–9. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz N, Bosch FX, de Sanjose S, et al. The causal link between human papillomavirus and invasive cervical cancer: a population-based case-control study in Colombia and Spain. International journal of cancer Journal international du cancer. 1992;52:743–9. doi: 10.1002/ijc.2910520513. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. Journal of the National Cancer Institute. 2008;100:513–7. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny L, Kuhn L, Hu CC, Tsai WY, Wright TC., Jr. Human papillomavirus-based cervical cancer prevention: long-term results of a randomized screening trial. Journal of the National Cancer Institute. 2010;102:1557–67. doi: 10.1093/jnci/djq342. [DOI] [PubMed] [Google Scholar]
- 22.Barzon L, Militello V, Lavezzo E, et al. Human papillomavirus genotyping by 454 next generation sequencing technology. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2011;52:93–7. doi: 10.1016/j.jcv.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiology and molecular biology reviews: MMBR. 2004;68:362–72. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clinical science. 2006;110:525–41. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 25.Johansson C, Schwartz S. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nature reviews Microbiology. 2013;11:239–51. doi: 10.1038/nrmicro2984. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PloS one. 2011;6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klug A, Finch JT. Structure of Viruses of the Papilloma-Polyoma Type. I. Human Wart Virus. Journal of molecular biology. 1965;11:403–23. doi: 10.1016/s0022-2836(65)80066-3. [DOI] [PubMed] [Google Scholar]
- 28.Klug A, Finch JT. Structure of viruses of the papilloma-polyoma type. IV. Analysis of tilting experiments in the electron microscope. Journal of molecular biology. 1968;31:1–12. doi: 10.1016/0022-2836(68)90050-8. [DOI] [PubMed] [Google Scholar]
- 29.Gissmann L, Pfister H, Zur Hausen H. Human papilloma viruses (HPV): characterization of four different isolates. Virology. 1977;76:569–80. doi: 10.1016/0042-6822(77)90239-2. [DOI] [PubMed] [Google Scholar]
- 30.Reinson T, Toots M, Kadaja M, et al. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. Journal of virology. 2013;87:951–64. doi: 10.1128/JVI.01943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegde RS. The papillomavirus E2 proteins: structure, function, and biology. Annual review of biophysics and biomolecular structure. 2002;31:343–60. doi: 10.1146/annurev.biophys.31.100901.142129. [DOI] [PubMed] [Google Scholar]
- 32.Fu L, Van Doorslaer K, Chen Z, et al. Degradation of p53 by human Alphapapillomavirus E6 proteins shows a stronger correlation with phylogeny than oncogenicity. PloS one. 2010:5. doi: 10.1371/journal.pone.0012816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SL, Mounts P. Transforming activity of E5a protein of human papillomavirus type 6 in NIH 3T3 and C127 cells. Journal of virology. 1990;64:3226–33. doi: 10.1128/jvi.64.7.3226-3233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang ES, Nottoli T, Dimaio D. The HPV16 E5 protein: expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology. 1995;211:227–33. doi: 10.1006/viro.1995.1395. [DOI] [PubMed] [Google Scholar]
- 35.Schiffman M, Herrero R, Desalle R, et al. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.zur Hausen H, Meinhof W, Scheiber W, Bornkamm GW. Attempts to detect virus-secific DNA in human tumors. I. Nucleic acid hybridizations with complementary RNA of human wart virus. International journal of cancer Journal international du cancer. 1974;13:650–6. doi: 10.1002/ijc.2910130509. [DOI] [PubMed] [Google Scholar]
- 37.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. The EMBO journal. 1984;3:1151–7. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Ranst M, Fuse A, Sobis H, et al. A papillomavirus related to HPV type 13 in oral focal epithelial hyperplasia in the pygmy chimpanzee. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 1991;20:325–31. doi: 10.1111/j.1600-0714.1991.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 39.Chan SY, Bernard HU, Ratterree M, Birkebak TA, Faras AJ, Ostrow RS. Genomic diversity and evolution of papillomaviruses in rhesus monkeys. Journal of virology. 1997;71:4938–43. doi: 10.1128/jvi.71.7.4938-4943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, van Doorslaer K, DeSalle R, et al. Genomic diversity and interspecies host infection of alpha12 Macaca fascicularis papillomaviruses (MfPVs) Virology. 2009;393:304–10. doi: 10.1016/j.virol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergin IL, Bell JD, Chen Z, et al. Novel genital Alphapapillomaviruses in baboons (Papio hamadryas Anubis) with cervical dysplasia. Veterinary pathology. 2013;50:200–8. doi: 10.1177/0300985812439725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood CE, Chen Z, Cline JM, Miller BE, Burk RD. Characterization and experimental transmission of an oncogenic papillomavirus in female macaques. Journal of virology. 2007;81:6339–45. doi: 10.1128/JVI.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, DeSalle R, Schiffman M, Herrero R, Burk RD. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. Journal of virology. 2009;83:1443–55. doi: 10.1128/JVI.02068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rector A, Lemey P, Tachezy R, et al. Ancient papillomavirus-host co-speciation in Felidae. Genome biology. 2007;8:R57. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narechania A, Chen Z, DeSalle R, Burk RD. Phylogenetic incongruence among oncogenic genital alpha human papillomaviruses. Journal of virology. 2005;79:15503–10. doi: 10.1128/JVI.79.24.15503-15510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burk RD, Chen Z, Van Doorslaer K. Human papillomaviruses: genetic basis of carcinogenicity. Public health genomics. 2009;12:281–90. doi: 10.1159/000214919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiffman M, Rodriguez AC, Chen Z, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer research. 2010;70:3159–69. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Villiers EM. Heterogeneity of the human papillomavirus group. Journal of virology. 1989;63:4898–903. doi: 10.1128/jvi.63.11.4898-4903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qu W, Jiang G, Cruz Y, et al. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. Journal of clinical microbiology. 1997;35:1304–10. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abreu AL, Souza RP, Gimenes F, Consolaro ME. A review of methods for detect human Papillomavirus infection. Virology journal. 2012;9:262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology. 2013 doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Burk RD, Chen Z, Harari A, et al. Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages. Acta dermatovenerologica Alpina, Panonica, et Adriatica. 2011;20:113–23. [PMC free article] [PubMed] [Google Scholar]
- 53.Bernard HU. The clinical importance of the nomenclature, evolution and taxonomy of human papillomaviruses. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2005;32(Suppl 1):S1–6. doi: 10.1016/j.jcv.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 54.Chen Z, Terai M, Fu L, Herrero R, DeSalle R, Burk RD. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. Journal of virology. 2005;79:7014–23. doi: 10.1128/JVI.79.11.7014-7023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottschling M, Kohler A, Stockfleth E, Nindl I. Phylogenetic analysis of beta-papillomaviruses as inferred from nucleotide and amino acid sequence data. Molecular phylogenetics and evolution. 2007;42:213–22. doi: 10.1016/j.ympev.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Egawa K, Delius H, Matsukura T, Kawashima M, de Villiers EM. Two novel types of human papillomavirus, HPV 63 and HPV 65: comparisons of their clinical and histological features and DNA sequences to other HPV types. Virology. 1993;194:789–99. doi: 10.1006/viro.1993.1320. [DOI] [PubMed] [Google Scholar]
- 57.Bottalico D, Chen Z, Dunne A, et al. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. The Journal of infectious diseases. 2011;204:787–92. doi: 10.1093/infdis/jir383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gage JC, Schiffman M, Solomon D, et al. Risk of Precancer Determined by HPV Genotype Combinations in Women with Minor Cytologic Abnormalities. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013 doi: 10.1158/1055-9965.EPI-12-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burk RD, Terai M, Gravitt PE, et al. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer research. 2003;63:7215–20. [PubMed] [Google Scholar]
- 60.Van Ranst M, Kaplan JB, Burk RD. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. The Journal of general virology. 1992;73(Pt 10):2653–60. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- 61.Hildesheim A, Schiffman M, Bromley C, et al. Human papillomavirus type 16 variants and risk of cervical cancer. Journal of the National Cancer Institute. 2001;93:315–8. doi: 10.1093/jnci/93.4.315. [DOI] [PubMed] [Google Scholar]
- 62.Chan SY, Ho L, Ong CK, et al. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. Journal of virology. 1992;66:2057–66. doi: 10.1128/jvi.66.4.2057-2066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith B, Chen Z, Reimers L, et al. Sequence imputation of HPV16 genomes for genetic association studies. PloS one. 2011;6:e21375. doi: 10.1371/journal.pone.0021375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan SY, Chew SH, Egawa K, et al. Phylogenetic analysis of the human papillomavirus type 2 (HPV-2), HPV-27, and HPV-57 group, which is associated with common warts. Virology. 1997;239:296–302. doi: 10.1006/viro.1997.8896. [DOI] [PubMed] [Google Scholar]
- 65.Yamada T, Manos MM, Peto J, et al. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. Journal of virology. 1997;71:2463–72. doi: 10.1128/jvi.71.3.2463-2472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. International journal of cancer Journal international du cancer. 2012;131:2349–59. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 67.Castle PE, Porras C, Quint WG, et al. Comparison of two PCR-based human papillomavirus genotyping methods. Journal of clinical microbiology. 2008;46:3437–45. doi: 10.1128/JCM.00620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xi LF, Schiffman M, Koutsky LA, et al. Persistence of newly detected human papillomavirus type 31 infection, stratified by variant lineage. International journal of cancer Journal international du cancer. 2013;132:549–55. doi: 10.1002/ijc.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rositch AF, Koshiol J, Hudgens MG, et al. Patterns of persistent genital human papillomavirus infection among women worldwide: A literature review and meta-analysis. International journal of cancer Journal international du cancer. 2012 doi: 10.1002/ijc.27828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xi LF, Schiffman M, Koutsky LA, et al. Association of human papillomavirus type 31 variants with risk of cervical intraepithelial neoplasia grades 2-3. International journal of cancer Journal international du cancer. 2012;131:2300–7. doi: 10.1002/ijc.27520. [DOI] [PubMed] [Google Scholar]
- 71.Conway C, Chalkley R, High A, et al. Next-generation sequencing for simultaneous determination of human papillomavirus load, subtype, and associated genomic copy number changes in tumors. The Journal of molecular diagnostics: JMD. 2012;14:104–11. doi: 10.1016/j.jmoldx.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Foulongne V, Sauvage V, Hebert C, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PloS one. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johansson H, Bzhalava D, Ekstrom J, Hultin E, Dillner J, Forslund O. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology. 2013;440:1–7. doi: 10.1016/j.virol.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 74.Clarke MA, Wentzensen N, Mirabello L, et al. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:2125–37. doi: 10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mirabello L, Schiffman M, Ghosh A, et al. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. International journal of cancer Journal international du cancer. 2013;132:1412–22. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mirabello L, Sun C, Ghosh A, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. Journal of the National Cancer Institute. 2012;104:556–65. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]