Abstract
Objectives
Identification of patient subpopulations susceptible to develop myocardial infarction (MI) or, conversely, those displaying either intrinsic cardioprotective phenotypes or highly responsive to protective interventions remain high-priority knowledge gaps. We sought to identify novel common genetic variants associated with perioperative MI in patients undergoing coronary artery bypass grafting using genome-wide association methodology.
Setting
107 secondary and tertiary cardiac surgery centres across the USA.
Participants
We conducted a stage I genome-wide association study (GWAS) in 1433 ethnically diverse patients of both genders (112 cases/1321 controls) from the Genetics of Myocardial Adverse Outcomes and Graft Failure (GeneMAGIC) study, and a stage II analysis in an expanded population of 2055 patients (225 cases/1830 controls) combined from the GeneMAGIC and Duke Perioperative Genetics and Safety Outcomes (PEGASUS) studies. Patients undergoing primary non-emergent coronary bypass grafting were included.
Primary and secondary outcome measures
The primary outcome variable was perioperative MI, defined as creatine kinase MB isoenzyme (CK-MB) values ≥10× upper limit of normal during the first postoperative day, and not attributable to preoperative MI. Secondary outcomes included postoperative CK-MB as a quantitative trait, or a dichotomised phenotype based on extreme quartiles of the CK-MB distribution.
Results
Following quality control and adjustment for clinical covariates, we identified 521 single nucleotide polymorphisms in the stage I GWAS analysis. Among these, 8 common variants in 3 genes or intergenic regions met p<10−5 in stage II. A secondary analysis using CK-MB as a quantitative trait (minimum p=1.26×10−3 for rs609418), or a dichotomised phenotype based on extreme CK-MB values (minimum p=7.72×10−6 for rs4834703) supported these findings. Pathway analysis revealed that genes harbouring top-scoring variants cluster in pathways of biological relevance to extracellular matrix remodelling, endoplasmic reticulum-to-Golgi transport and inflammation.
Conclusions
Using a two-stage GWAS and pathway analysis, we identified and prioritised several potential susceptibility loci for perioperative MI.
Keywords: SURGERY, GENETICS
Strengths and limitations of this study.
This is the first genome-wide association study of perioperative myocardial infarction, using prospective cohorts of cardiac surgical patients, standard definitions of the primary phenotype and full adjustment for non-genetic risk factors.
We conducted comprehensive and complementary single marker and pathway-based genome-wide association analyses.
The study is powered to detect relatively large effect sizes.
Rare genetic variant effects not analysed.
Predominantly Caucasian cohort, thus findings cannot be generalised to other populations.
Introduction
Despite advances in surgical techniques and pharmacological therapy, the incidence of myocardial infarction (MI) after coronary artery bypass grafting (CABG) remains as high as 19%, and is associated with increased mortality and long-term morbidity.1 Strategies to identify subpopulations of patients at risk for developing large myocardial infarcts on the one hand, or those displaying an intrinsic cardioprotective state on the other hand, remain high-priority knowledge gaps and could inform selection of more specific protective agents.2
The evidence for heritability of MI is striking, supported both by family studies and, recently, by a number of well powered and replicated genome-wide association studies (GWAS), which primarily implicate common genetic variants at the 9p21 locus in multiple racial groups.3–6 However, although family-based methods are not practical for studying perioperative MI (PMI), its genetic basis is strongly suggested by several observations, including wide variability in incidence and severity that is poorly explained by clinical and procedural risk factors, different racial susceptibility profiles and results from preclinical animal models. Indeed, extensive genetic variability has been found in biological pathways implicated in the pathophysiology of postoperative MI, such as the complex acute inflammatory response to cardiac surgery. Mounting evidence for heritability of a proinflammatory state suggests that individual genetic history may also significantly modulate the magnitude of postoperative inflammatory response after cardiac surgery.7 Yet only a few studies have identified allelic associations with altered susceptibility to myocardial ischaemia-reperfusion injury in cardiac surgical populations, all based on a candidate gene association approach.8–12 Thus, the overall influence of common genetic variation on the incidence of PMI remains poorly understood.
Recently, integrated testing of genes involved in the same biological pathway has emerged as an alternative strategy for evaluating the combined effects of multiple genetic variants with small effect size on a disease phenotype.13–15 Given the polygenic nature of disease susceptibility, this approach is increasingly being used to identify groups of gene variants with shared cellular function that are enriched for disease, while also improving the statistical power of GWAS. In this study, we adopted this strategy by first employing genome-wide association methodology to identify common genetic variants associated with PMI after CABG, followed by pathway-based analyses to uncover biological mechanisms of relevance to PMI.
Methods
The study design and reporting of the results follow the recommendations by ‘Strengthening the Reporting of Genetic Association Studies’ (STREGA).16 We performed a joint two-stage17 GWAS combined with a pathway analysis approach.
Patient populations
The stage I cohort (discovery cohort) comprised 1493 ethnically diverse subjects who underwent an isolated CABG with cardiopulmonary bypass for the first time and were enrolled between 2002 and 2003 in the Institutional Review Board (IRB) approved Genetics of Myocardial Outcomes and Graft Failure (GeneMAGIC) ancillary substudy of the multicentre Project of Ex-vivo Vein Graft Engineering via Transfection (PREVENT-IV).18 Of those, 1433 patients met eligibility requirements after applying quality control criteria and excluding patients with missing genotypes or phenotypic information.
In stage II, we tested the top genetic variants in an expanded data set in which an additional 622 patients of self-reported European ancestry were added to the discovery data set, leading to a total of 2055 patients. The additional patients underwent CABG with cardiopulmonary bypass between 1997 and 2006 as part of the Perioperative Genetics and Safety Outcomes Study, an IRB-approved longitudinal study at the Duke University Medical Center.11 12
Definition of PMI
PMI was defined according to the universal definition of MI19 as an elevation in the plasma level of creatine kinase MB isoenzyme (CK-MB) that was >10 times the upper limit of normal, as measured by a core laboratory within 24 h after surgery, and that was not attributable to an intervening clinical event or preoperative MI (adjudicated by the PREVENT-IV Clinical Events Committee).18
Genotyping and quality controls
Genomic DNA was isolated from whole blood or saliva using standard procedures. Genotyping in both cohorts was performed on the Illumina Human610-Quad BeadChip at the Duke Genomic Analysis Facility. Sample and genotype quality control of data flow included assessment of call rates, gender check, cryptic relatedness, SNP missingness and the Hardy-Weinberg equilibrium, as previously described.20 We used principal components derived from the EIGENSTRAT21 method to control for population stratification (see online supplementary methods).
Statistical analysis
Univariate regression analysis was performed to test differences in demographic, clinical and procedural characteristics between patients with and without postoperative MI; statistically significant covariates were subsequently used to adjust genetic association tests. Genome-wide association analyses performed in the stage I cohort used multivariable logistic regression models implemented in PLINK 1.07, assuming an additive genetic model, and including significant clinical covariates and the top 10 principal components to adjust for population stratification (see online supplementary table S1). Statistical significance for stage I analyses was a priori arbitrarily defined as a two-tailed p<0.001, to balance between the overly conservative Bonferroni correction and type II error, given that we had an a priori defined replication data set to obviate type I error. In stage II analyses, the same clinical covariate and principal component adjustments (re-estimated for the expanded data set) were applied, with statistical significance defined as a Bonferroni adjusted p value of 0.05/number of SNPs identified in stage I, to control the overall type I error rate. Haplotype association analysis was performed in the stage II cohort for genes tagged by the significant SNPs, adjusting for the same clinical covariates and principal components. Secondary analyses were performed for the top SNPs identified in the stage II cohort, using the continuous phenotype of CK-MB values and a dichotomised extreme CK-MB phenotype, which included participants within the first and fourth quartiles of the CK-MB distribution. For CK-MB, a linear regression model was applied with adjustment for the same set of covariates as in the primary analysis. For the extreme CK-MB subset, we reassigned trait status as ‘0’ for participants within the first CK-MB quartile, and ‘1’ for participants within the fourth CK-MB quartile from the stage II cohort. Logistic regression with the same set of covariates was performed. Finally, we employed pathway analysis to prioritise association results and provide biological interpretation, using functional ontology enrichment analysis tools implemented in MetaCore (GeneGO, St. Joseph, MI; see online supplementary methods).
Results
PMI was observed in 112 of 1433 patients (7.8%) and in 225 of 2055 patients (10.9%) in stage I and II cohorts, respectively (see online supplementary table S2). Aortic cross-clamp time, number of coronary artery grafts, and procedures in addition to CABG were significantly associated with PMI in stage I analyses. Stage II analyses showed that extracardiac arteriopathy and year of surgery also played significant roles. All the clinical variables identified above were included in multivariable logistic regression models to adjust the genetic association tests for potential confounders (see online supplementary table S2).
After applying quality control criteria (see online supplementary methods: A.1. Quality control of data flow), 534 390 markers were analysed for association with PMI in stage I. While none of the SNPs reached genome-wide significance (figure 1), 521 SNPs met the a priori defined discovery threshold of p<0.001 (minimum p=2.76×10−6, rs2044061 on chromosome 8) and were subsequently analysed in stage II (see online supplementary table S10). A quantile-quantile (Q-Q) plot of observed versus expected p values showed that the population substructure was well adjusted for (see online supplementary figure S2).
Figure 1.
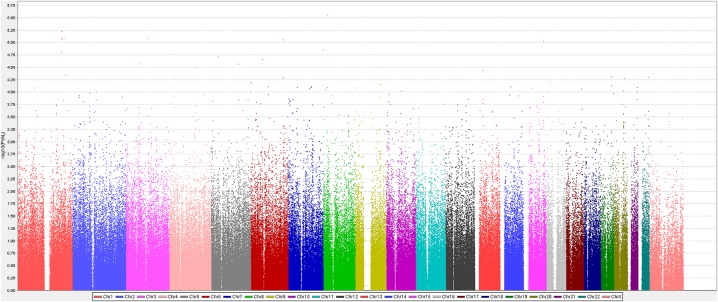
Manhattan plot of genome-wide association with perioperative myocardial infarction in stage I analysis. The x axis represents the genome in physical order (coloured by chromosome); the y axis showing –log10(p) for all single nucleotide polymorphisms (SNPs). None of the SNPs reached genome-wide significance (p<9.09×10−8), but 521 SNPs met the prespecified discovery threshold p<0.001 for inclusion in stage II analyses (minimum p=2.76×10−6, rs2044061 on chromosome 8).
In stage II analyses, eight of the 521 SNPs met the Bonferroni correction threshold (p<9.6×10−5 for 521 SNPs). The top 2 SNPs (rs10454444 and rs10913237), located in the pregnancy-associated plasma protein A2 gene (PAPPA2), were in high linkage disequilibrium, with p values for stage II of 2.43×10−6 (OR 0.46; 95% CI 0.33 to 0.63) and 2.47×10−6 (OR 0.46; 95% CI 0.33 to 0.63), respectively. In addition, one intronic SNP in histone deacetylase-4 (HDAC4), two in the SEC24 family, member D (SEC24D, a member of the cytoplasmic coat protein complex II, COPII), and two located in intergenic regions were associated with PMI (table 1).
Table 1.
Top eight SNP associated with postoperative myocardial infarction
| Stage I: discovery data set* |
Stage II: expanded data set* |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
MAF |
||||||||||
| Chr | SNP | Base pair | Gene symbol | Controls N=1321 |
Cases N=112 |
OR (95% CI) | p Value | Controls N=1830 |
Cases N=225 |
OR (95% CI) | p Value |
| 1 | rs10489478 | 176 566 350 | PAPPA2 | 0.18 | 0.09 | 0.41 (0.25 to 0.66) | 2.67×10−4 | 0.18 | 0.11 | 0.47 (0.34 to 0.65) | 4.70×10−6 |
| 1 | rs10913237 | 176 630 416 | PAPPA2 | 0.18 | 0.09 | 0.39 (0.24 to 0.64) | 1.85×10−4 | 0.19 | 0.11 | 0.46 (0.33 to 0.63) | 2.47×10−6 |
| 1 | rs10454444 | 176 649 181 | PAPPA2 | 0.18 | 0.09 | 0.39 (0.24 to 0.64)1 | 1.81×10−4 | 0.19 | 0.11 | 0.46 (0.33 to 0.63) | 2.43×10−6 |
| 2 | rs10 200 850 | 240 234 901 | HDAC4 | 0.06 | 0.13 | 2.22 (1.42 to 3.46) | 4.34×10−4 | 0.06 | 0.11 | 2.23 (1.57 to 3.17) | 8.18×10−6 |
| 4 | rs4834703 | 119 691 624 | SEC24D | 0.09 | 0.18 | 2.27 (1.54 to 3.34) | 3.13×10−5 | 0.09 | 0.16 | 1.98 (1.47 to 2.66) | 6.80×10−6 |
| 4 | rs6822035 | 119 710 421 | SEC24D | 0.29 | 0.40 | 1.73 (1.29 to 2.33) | 2.59×10−4 | 0.28 | 0.36 | 1.65 (1.32 to 2.05) | 9.55×10−6 |
| 12 | rs2303970 | 74 471 340 | TRHDE|LOC552889 | 0.48 | 0.35 | 0.58 (0.43 to 0.78) | 3.51×10−4 | 0.48 | 0.36 | 0.62 (0.50 to 0.76) | 8.89×10−6 |
| 13 | rs609418 | 37 417 427 | RFXAP | SMAD9 | 0.25 | 0.35 | 1.73 (1.28 to 2.33) | 3.14×10−4 | 0.25 | 0.33 | 1.67 (1.34 to 2.09) | 5.81×10−6 |
*Adjusted for clinical characteristics including extracardiac arteriopathy, recent myocardial infarction, procedure other than CABG only, year of surgery, number of diseased vessels and aortic cross-clamp time.
HDAC4, histone deacetylase-4; PAPPA2, pregnancy-associated plasma protein A2; RFXAP, regulatory factor X-associated protein; SEC24D, SEC24-related protein D; SMAD9, mother against decapentaplegic homolog 9; TRHDE, thyrotropin-releasing hormone-degrading ectoenzyme; intergenic region is expressed by two franking genes with ‘|’ in between.
CABG, coronary artery bypass grafting; Chr, chromosome; MAF, minor allele frequency (based on the discovery data set); SNP, single nucleotide polymorphisms.
When CK-MB was tested as a quantitative trait, all eight SNPs remained significantly associated with plasma levels of CK-MB. However, rs4834703 in SEC24D (p=7.72×10−6) and rs10200850 in HDAC4 (p=8.26×10−5) showed the strongest association, followed by the 2 SNPs in PAPPA2 (p=0.0001; table 2). When the dichotomised extreme CK-MB phenotype was studied in the stage II cohort, only the 2 SNPs located in SEC24D (rs4834703 and rs6822035, p=0.002) and rs609418 in the intergenic region between RFXAP and SMAD9 (p=0.001) remained nominally significant (table 2). Overall, the rs4834703 SNP in SEC24D consistently showed strong association signals across postoperative MI, quantitative CK-MB and the extreme CK-MB traits.
Table 2.
Association of the top eight SNP with CK-MB as a quantitative trait and as an extreme phenotype in the joint analysis data set
| MAF (Overall) | CK-MB as a quantitative trait* |
CK-MB as an extreme phenotype* |
||||||
|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Base pair | Gene symbol | β-Coefficient | p Value | OR (95% CI) | p Value | |
| 1 | rs10489478 | 176 566 350 | PAPPA2 | 0.17 | −5.40 | 2.27×10−4 | 0.77 (0.574 to 1.047) | 0.1 |
| 1 | rs10913237 | 176 630 416 | PAPPA2 | 0.18 | −5.58 | 1.30×10−4 | 0.78 (0.578 to 1.052) | 0.1 |
| 1 | rs10454444 | 176 649 181 | PAPPA2 | 0.18 | −5.59 | 1.21×10−4 | 0.77 (0.575 to 1.043) | 0.09 |
| 2 | rs10200850 | 240 234 901 | HDAC4 | 0.07 | 9.26 | 8.26×10−5 | 1.49 (0.950 to 2.326) | 0.08 |
| 4 | rs4834703 | 119 691 624 | SEC24D | 0.10 | 8.54 | 7.72×10−6 | 1.82 (1.242 to 2.654) | 0.002 |
| 4 | rs6822035 | 119 710 421 | SEC24D | 0.30 | 4.42 | 4.81×10−4 | 1.48 (1.152 to 1.903) | 0.002 |
| 12 | rs2303970 | 74 471 340 | TRHDE|LOC552889 | 0.47 | −2.93 | 0.01 | 0.84 (0.669 to 1.047) | 0.12 |
| 13 | rs609418 | 37 417 427 | RFXAP|SMAD9 | 0.26 | 4.80 | 2.26×10−4 | 1.56 (1.189 to 2.034) | 0.001 |
*Adjusted for clinical characteristics including extracardiac arteriopathy, recent myocardial infarction, procedure other than CABG only, year of surgery, number of diseased vessels, and aortic cross-clamp time using either linear regression (Β-coefficient) or logistic regression (OR, 95% CI) analyses.
HDAC4, histone deacetylase-4; PAPPA2, pregnancy-associated plasma protein A2; RFXAP, regulatory factor X-associated protein; SEC24D, SEC24-related protein D; SMAD9, mother against decapentaplegic homolog 9; TRHDE, thyrotropin-releasing hormone-degrading ectoenzyme.
CABG, coronary artery bypass grafting; Chr, chromosome; CK-MB, creatine kinase MB isoenzyme; MAF, minor allele frequency (based on the expanded data set); SNP, single nucleotide polymorphisms.
The haplotype structures of genic regions surrounding the significant SNPs in PAPPA2, HDAC4 and SEC24D are shown in online supplementary figure S3, and the results of haplotype analysis are shown in table 3 and online supplemental table S3. Haplotype analysis performed for the linkage disequilibrium blocks containing the significant markers showed that the A-A haplotype (rs6822035, rs10518325) in SEC24D had the most significant association with postoperative MI (p=5.54×10−7, OR 1.87; 95% CI 1.46 to 2.39, table 3).
Table 3.
Estimated haplotype frequencies in the SEC24D gene and results of association tests with incidence of PMI in the stage II analysis cohort (n=2055)
| Haplotype frequency |
||||||
|---|---|---|---|---|---|---|
| SNP1 | SNP2 | Haplotype | Patients with PMI (n=225) | Patients without PMI (n=1830) | OR (95% CI) | p Value |
| rs6828577 | rs6822035 | A-A | 0.3956 | 0.2874 | 1.65 (1.33 to 2.06) | 8.84×10−6 |
| rs6828577 | rs6822035 | G-C | 0.6044 | 0.7126 | Reference | |
| rs6822035 | rs10518325 | A-G | 0.03516 | 0.0528 | 0.86 (0.43 to 1.70) | 0.657 |
| rs6822035 | rs10518325 | C-G | 0.009083 | 0.02636 | 1.39 (0.67 to 2.90) | 0.378 |
| rs6822035 | rs10518325 | A-A | 0.3631 | 0.2358 | 1.87 (1.46 to 2.39) | 5.54×10−7 |
| rs6822035 | rs10518325 | C-A | 0.5927 | 0.685 | Reference | |
| rs10518325 | rs11098451 | A-A | 0.03097 | 0.06231 | 0.76 (0.48 to 1.19) | 0.225 |
| rs10518325 | rs11098451 | G-G | 0.04425 | 0.07845 | 0.87 (0.58 to 1.31) | 0.509 |
| rs10518325 | rs11098451 | A-G | 0.9248 | 0.8592 | Reference | |
Adjusted for clinical characteristics including extracardiac arteriopathy, recent myocardial infarction, procedure other than CABG only, year of surgery, number of diseased vessels, aortic cross-clamp time.
CABG, coronary artery bypass grafting; PMI, perioperative myocardial infarction; SNP, single nucleotide polymorphism.
In multivariate risk factor-adjusted stage I and II analyses, we found no evidence for association between genetic variation at the 9p21 locus and incident postoperative MI, including the rs10116277 SNP previously associated with MI and mortality in non-surgical20 and cardiac surgical8 9 cohorts (see online supplementary figure S4).
The top 10 enriched pathway maps for the stage I analysis SNPs are shown in online supplementary figure S5. The most significant were ‘Cell adhesion: extracellular matrix remodelling’ and ‘Cytoskeleton remodelling: TGF, WNT, and cytoskeletal remodelling’ (enrichment p value=1.9×10−8 and 7.3×10−6, respectively). The most significant canonical pathway maps in stage I and II comparative analyses were ‘Immune response: NFAT signalling and leucocyte interaction’ and ‘Cell adhesion: extracellular matrix remodelling’ (p=7.2×10−4 and 1.1×10−3, respectively; see online supplementary tables S4 and 5).
Discussion
We present the first report of a two-stage GWAS involving 225 PMI cases and 1830 controls. After accounting for clinical and procedural covariates, we identified eight significant SNPs mapped to three genes (PAPPA2, HDAC4, SEC24D) and two intergenic regions. The most significant association with PMI was exhibited by rs10454444 in PAPPA2 (p=2.43×10−6) in single-marker analyses, by SEC24D A-A (rs6822035, rs10518325, p=5.54×10−7) in haplotype analyses, and by SEC24D SNPs in secondary analyses when CK-MB was evaluated as a quantitative trait (rs4834703, p=2.43×10−6) or as an extreme CK-MB phenotype (rs4834703 and rs6822035, p=0.002).
Among these eight SNPs, rs4834703 in SEC24D showed the most consistently strong association with all three phenotypes evaluated in this study. The transport protein SEC24D is an integral component of cytoplasmic COPII transport machinery, a key player in vesicle trafficking of secretory proteins from the endoplasmic reticulum (ER) to the Golgi apparatus for delivery to downstream compartments. COPII is responsible for cargo sorting and vesicle morphogenesis, with roles in modulating ER exit, cell surface transport, lipid secretion and cholesterol biosynthesis, and function of G protein-coupled receptors. Conditions of ischaemia, oxidative injury or acute phase-response result in ER stress through accumulation of misfolded proteins, which leads to activation of the unfolded protein response (UPR) signalling pathway. If the protective mechanisms activated by the UPR are insufficient, cells die by apoptosis and autophagy. However, altered expression or function of SEC24 proteins could also explain ER trapping of misfolded proteins under conditions of ER stress,22 23 with SEC24D being the only isoform implicated in extracellular matrix secretion.24 Animal models of defects in SEC24D involve characteristic skeletal malformations, whereas its complete disruption results in early-embryonic lethality.25 However, there are no reports of human diseases associated with genetic variation in SEC24D.
PAPPA2 is a metalloproteinase that regulates local insulin-like growth factor (IGF) bioavailability by specifically cleaving IGF-binding protein 5 (IGFBP-5). In experimental models of myocardial ischaemia, IGFBP-5 inhibits both myocardial IGF-1, with implications for inflammation-linked angiogenesis and repair processes,26 and IGF-2, limiting its cardioprotective effects following ischaemia reperfusion.27 28 The related protein PAPPA1 cleaves IGFBP-4 and is activated and released from vulnerable atherosclerotic plaques. PAPPA1 has been extensively studied as a cardiovascular risk biomarker for the diagnosis and prognosis of acute coronary syndrome;29 30 however, the role of PAPPA2 in cardiovascular biology has not been previously reported. Consistent with our single-marker analysis findings, the GWAS pathway analysis identified significant contributions of variants in other IGF system component genes, namely IGF-2 and the IGF-1 receptor (see online supplementary figure S6 and table S5).
HDAC4, a member of the HDAC family, mediates changes in the chromatin structure by removing acetyl groups from the core histones, resulting in transcriptional repression. HDAC4 is highly expressed in the myocardium, where it plays an important role in the regulation of gene expression and is involved in myocardial cell cycle progression, differentiation and apoptosis.31 Experimental HDAC inhibition is associated with a profound reduction in ischaemia-induced myocardial cell death,31 by triggering preconditioning effects and promoting myocardial repair. Genetic variants in HDACs are important determinants of susceptibility to cardiovascular diseases,32 suggesting functional roles for HDAC4 in modulating perioperative myonecrosis, and their potential use in predicting individual patient responsiveness to HDAC inhibition.
One of the intergenic SNPs associated with PMI (rs609418) is located near the mother against the decapentaplegic homologue 9 (SMAD9) gene, part of the transforming growth factor β (TGF-β) signalling pathway, which is markedly activated in the infarcted myocardium. Members of the TGF-β superfamily transduce their signal from the membrane to the nucleus via a distinct combination of transmembrane receptors and downstream effectors—the SMAD proteins.33 TGF-β plays important and complex roles in regulating postinfarction inflammatory responses. In animal models, signalling through the SMAD transcription factors is associated with resolution of inflammation, repression of cytokine and chemokine gene synthesis, and protection against myocardial ischaemia-reperfusion injury.34 Consistent with this single-locus gene association result, the bone morphogenetic protein pathway, which also transduces its signals via a SMAD9-dependent cascade, was identified as one of the top-scoring pathways in our GWAS pathway analysis (p=5.2×10−3, see online supplementary table S7). Although no direct functional roles are currently attributed to this intergenic region, a query of the Regulome and Haploreg databases reports that rs609418 is located within active regulatory elements (GATA2 transcription factor binding site by ENCODE ChIP-seq and altering regulatory motifs in Gfi1 and Mef2 by the Position-Weight Matrix, see online supplementary figure S7).
Surprisingly, we have been unable to replicate previously reported associations between common genetic variants at the 9p21 locus and risk for PMI or mortality after CABG.8 9 Many different reasons may account for this, including inadequate sample size, variation in study design, differences in allele frequencies or variability in the definition of PMI phenotype.35 Indeed, our observation is consistent with other studies showing that, although genetic variants at the 9p21 locus are associated with incident coronary artery disease (CAD), they may not be associated with the actual risk of MI.36 This discrepancy between convincing associations of 9p21 with a greater burden of CAD but not with MI in the presence of underlying CAD has been further confirmed by nested case–control studies37 as well as meta-analyses.5 38 39 Taken together, in subjects with established CAD, such as those included in our study, any lack of association of the 9p21 locus with subsequent MI could have resulted from the presence of CAD in carriers and non-carriers of the risk variants at the 9p21 locus, which seems to primarily mediate an atherosclerotic phenotype.
The strengths of our study are (1) a relatively large population of cardiac surgery patients, (2) a prospective cohort design and (3) a combination of complementary single-marker and pathway-based genome-wide association analyses. This approach allowed us to identify genetic variants that carry only a small disease risk individually, but that jointly can contribute relatively large effects on PMI susceptibility. Furthermore, an application of results from pathway-based analysis may add structure to interpreting genomic data and allow exploration of cellular processes that functionally underpin the observed associations. Finally, by using this approach, replication of association findings at the gene and pathway level is much easier compared to replication at the individual SNP level.13 Of note, most of the genes identified through pathway enrichment analysis in this study encode targets for therapeutic drugs that have already been developed. Thus, by improving risk assessment and identifying allele-specific therapeutic responses, further investigation of loci and pathways prioritised in this study could yield actionable results for enhancing perioperative cardioprotection.
Several limitations are worth mentioning. Power calculations (see online supplementary results B.6.) show that, based on the current sample size and incidence of PMI, our study can detect a genotypic relative risk approximately 2 with 80% power (assuming a variant with a 10% minor allele frequency and a realistic linkage disequilibrium between the tested marker and the causal locus D′=0.8). Thus, although our study is the largest genetic association study of PMI conducted to date, it is powered to detect only common variants with relatively large effect sizes. Most published genetic association studies of PMI have reported larger effect sizes compared with ambulatory populations (OR range 1.79–3.97).8 10 11 The possibility of rare genetic variants that drive a pronounced clinical phenotype was not explored in this study, because only variants with minor allele frequencies >0.05 were assessed. Also, functional studies to further elucidate the potential biological effects of the SNPs identified were not feasible due to lack of plasma or tissue availability in the GeneMAGIC study. Finally, patients enrolled in our cohorts were predominantly Caucasian, and therefore our findings cannot be generalised to other ethnic groups.
In conclusion, we report the first GWAS in a cohort of patients at risk for MI after CABG surgery. On the basis of our integrated approach utilising primary (PMI) and secondary phenotypes (quantitative CK-MB, extreme CK-MB), single-marker analysis and pathway analysis, we identified several polymorphisms in the insulin growth factor system implicated in the regulation of extracellular matrix remodelling, as well as the ER-to-Golgi secretory pathway potentially involved in adaptive responses to ER stress. As other GWAS of PMI cohorts publish their results, we intend to collaborate on conducting a meta-analysis for this particular phenotype. While our GWAS results are intriguing, follow-up studies are needed to translate these initial findings into biological insights that could lead to predictive and therapeutic advances in perioperative care. For instance, in the regions of confirmed associations, causal variants will only occasionally be among those directly genotyped. Moreover, GWAS detect almost exclusively the effects of common SNPs, offering limited power to capture any rare and structural variants, such as insertions, deletions, inversions and translocations. Detailed sequencing may be necessary to further characterise the genetic variations in the PMI-associated regions to enhance the identification of causal variants. Furthermore, as in most cases, the functions of identified genes and their variants, as well as the mechanisms by which they may contribute to PMI pathophysiology, are largely unknown. Currently, very few existing groups can bridge from genetics to molecular biology or cell physiology and to disease or novel therapy; we propose to examine the mechanisms by which variation in the observed genes is involved in myocardial ischaemia-reperfusion injury by examining their functional effects in the human myocardium from patients undergoing cardiac surgery, as well in our previously described preclinical rodent and swine models of cardioplegic arrest and cardiopulmonary bypass.40 The use of such animal models would allow pharmacological studies targeting the identified pathways to explore the mechanism by which novel cardioprotective drugs would attenuate myocardial ischaemia-reperfusion injury, with the ultimate goal of developing personalised cardioprotective strategies.
Footnotes
Contributors: MDK contributed to the analysis and interpretation of data, as well as the drafting of the manuscript. Y-JL contributed to the analysis and interpretation of data and a critical revision of the manuscript. Y-JL and Y-WL contributed to the quality control and analysis of the data. JA, MFN, PKS, DJ and JPM contributed to the acquisition of data and a critical review of the manuscript. MVP contributed to the conception and design, analysis and interpretation of data, critical manuscript review and approved the final version of the submitted manuscript.
Funding: This work was supported in part by the National Institutes of Health grants R01-HL075273 and R01-HL092071 (to MVP) and by the American Heart Association grants 0256342U and 9951185U (to JPM), and 0120492U (to MVP).
Competing interests: None.
Patient consent: Obtained.
Ethics approval: The Duke University School of Medicine Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Domanski MJ, Mahaffey K, Hasselblad V et al. Association of myocardial enzyme elevation and survival following coronary artery bypass graft surgery. JAMA 2011;305:585–91. 10.1001/jama.2011.99 [DOI] [PubMed] [Google Scholar]
- 2.Schwartz Longacre L, Kloner RA, Arai AE et al. New horizons in cardioprotection: recommendations from the 2010 National Heart, Lung, and Blood Institute Workshop. Circulation 2011;124:1172–9. 10.1161/CIRCULATIONAHA.111.032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samani NJ, Erdmann J, Hall AS et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–53. 10.1056/NEJMoa072366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488–91. 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schunkert H, Gotz A, Braund P et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 2008;117:1675–84. 10.1161/CIRCULATIONAHA.107.730614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S, Voight BF, Purcell S et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334–41. 10.1038/ng.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pankow JS, Folsom AR, Cushman M et al. Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis 2001;154:681–9. 10.1016/S0021-9150(00)00586-4 [DOI] [PubMed] [Google Scholar]
- 8.Liu KY, Muehlschlegel JD, Perry TE et al. Common genetic variants on chromosome 9p21 predict perioperative myocardial injury after coronary artery bypass graft surgery. J Thorac Cardiovasc Surg 2010;139:483–8, 88 e1–2 10.1016/j.jtcvs.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muehlschlegel JD, Liu KY, Perry TE et al. Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation 2010;122(11 Suppl):S60–5. 10.1161/CIRCULATIONAHA.109.924233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collard CD, Shernan SK, Fox AA et al. The MBL2 ‘LYQA secretor’ haplotype is an independent predictor of postoperative myocardial infarction in whites undergoing coronary artery bypass graft surgery. Circulation 2007;116(11 Suppl):I106–12. 10.1161/CIRCULATIONAHA.106.679530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podgoreanu MV, White WD, Morris RW et al. Inflammatory gene polymorphisms and risk of postoperative myocardial infarction after cardiac surgery. Circulation 2006;114(1 Suppl):I275–81. 10.1161/CIRCULATIONAHA.105.001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobato RL, White WD, Mathew JP et al. Thrombomodulin gene variants are associated with increased mortality after coronary artery bypass surgery in replicated analyses. Circulation 2011;124 (11 Suppl):S143–8. 10.1161/CIRCULATIONAHA.110.008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo L, Peng G, Zhu Y et al. Genome-wide gene and pathway analysis. Eur J Hum Genet 2010;18:1045–53. 10.1038/ejhg.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics 2008;92:265–72. 10.1016/j.ygeno.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khoriaty R, Vasievich MP, Ginsburg D. The COPII pathway and hematologic disease. Blood 2012;120:31–8. 10.1182/blood-2012-01-292086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little J, Higgins JP, Ioannidis JP et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet 2009;125:131–51. 10.1007/s00439-008-0592-7 [DOI] [PubMed] [Google Scholar]
- 17.Skol AD, Scott LJ, Abecasis GR et al. Optimal designs for two-stage genome-wide association studies. Genet Epidemiol 2007;31:776–88. 10.1002/gepi.20240 [DOI] [PubMed] [Google Scholar]
- 18.Alexander JH, Hafley G, Harrington RA et al. Efficacy and safety of edifoligide, an E2F transcription factor decoy, for prevention of vein graft failure following coronary artery bypass graft surgery: PREVENT IV: a randomized controlled trial. JAMA 2005;294:2446–54. 10.1001/jama.294.24.3108 [DOI] [PubMed] [Google Scholar]
- 19.Thygesen K, Alpert JS, Jaffe AS et al. Third universal definition of myocardial infarction. Circulation 2012;126:2020–35. 10.1161/CIR.0b013e31826e1058 [DOI] [PubMed] [Google Scholar]
- 20.Helgadottir A, Thorleifsson G, Manolescu A et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491–3. 10.1126/science.1142842 [DOI] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–9. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 22.Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol 2010;191:479–92. 10.1083/jcb.201004062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahyi AN, Quinton LJ, Jones MR et al. Roles of STAT3 in protein secretion pathways during the acute-phase response. Infect Immun 2013;81:1644–53. 10.1128/IAI.01332-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unlu G, Levic DS, Melville DB et al. Trafficking mechanisms of extracellular matrix macromolecules: insights from vertebrate development and human diseases. Int J Biochem Cell Biol 2014;47:57–67. 10.1016/j.biocel.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baines AC, Adams EJ, Zhang B et al. Disruption of the Sec24d gene results in early embryonic lethality in the mouse. PLoS ONE 2013;8:e61114 10.1371/journal.pone.0061114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kluge A, Zimmermann R, Munkel B et al. Insulin-like growth factor I is involved in inflammation linked angiogenic processes after microembolisation in porcine heart. Cardiovasc Res 1995;29:407–15. 10.1016/0008-6363(96)88599-3 [DOI] [PubMed] [Google Scholar]
- 27.Kluge A, Zimmermann R, Munkel B et al. Insulin-like growth factor II is an experimental stress inducible gene in a porcine model of brief coronary occlusions. Cardiovasc Res 1995;29:708–16. 10.1016/S0008-6363(96)88644-5 [DOI] [PubMed] [Google Scholar]
- 28.Vogt AM, Htun P, Kluge A et al. Insulin-like growth factor-II delays myocardial infarction in experimental coronary artery occlusion. Cardiovasc Res 1997;33:469–77. 10.1016/S0008-6363(96)00212-X [DOI] [PubMed] [Google Scholar]
- 29.Heeschen C, Dimmeler S, Hamm CW et al. Pregnancy-associated plasma protein-A levels in patients with acute coronary syndromes: comparison with markers of systemic inflammation, platelet activation, and myocardial necrosis. J Am Coll Cardiol 2005;45:229–37. 10.1016/j.jacc.2004.09.060 [DOI] [PubMed] [Google Scholar]
- 30.Conti E, Andreotti F, Zuppi C. Pregnancy-associated plasma protein a as predictor of outcome in patients with suspected acute coronary syndromes. Circulation 2004;109:e211–12; author reply e11–2 10.1161/01.CIR.0000127614.27267.8F [DOI] [PubMed] [Google Scholar]
- 31.Granger A, Abdullah I, Huebner F et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. Faseb J 2008;22:3549–60. 10.1096/fj.08-108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res 2006;98:15–24. 10.1161/01.RES.0000197782.21444.8f [DOI] [PubMed] [Google Scholar]
- 33.Bujak M, Frangogiannis NG. The role of TGF-beta signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res 2007;74:184–95. 10.1016/j.cardiores.2006.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kempf T, Eden M, Strelau J et al. The transforming growth factor-beta superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ Res 2006;98:351–60. 10.1161/01.RES.0000202805.73038.48 [DOI] [PubMed] [Google Scholar]
- 35.Lim CC, van Gaal WJ, Testa L et al. With the “universal definition,” measurement of creatine kinase-myocardial band rather than troponin allows more accurate diagnosis of periprocedural necrosis and infarction after coronary intervention. J Am Coll Cardiol 2011;57:653–61. 10.1016/j.jacc.2010.07.058 [DOI] [PubMed] [Google Scholar]
- 36.Virani SS, Brautbar A, Lee VV et al. Chromosome 9p21 single nucleotide polymorphisms are not associated with recurrent myocardial infarction in patients with established coronary artery disease. Circ J 2012;76:950–6. 10.1253/circj.CJ-11-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horne BD, Carlquist JF, Muhlestein JB et al. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet 2008;1:85–92. 10.1161/CIRCGENETICS.108.793158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan K, Patel RS, Newcombe P et al. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol 2013;61:957–70. 10.1016/j.jacc.2012.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel RS, Asselbergs FW, Quyyumi AA et al. Genetic variants at chromosome 9p21 and risk of first versus subsequent coronary heart disease events: a systematic review and meta-analysis. J Am Coll Cardiol 2014;63:2234–45. 10.1016/j.jacc.2014.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Lange F, Yoshitani K, Podgoreanu MV et al. A novel survival model of cardioplegic arrest and cardiopulmonary bypass in rats: a methodology paper. J Cardiothorac Surg 2008;3:51 10.1186/1749-8090-3-51 [DOI] [PMC free article] [PubMed] [Google Scholar]


