Abstract
The protein mucin1 (MUC1) is an attractive target for cancer biomarkers because it is overexpressed in most adenocarcinomas. In this study, we exploited a MUC1-binding aptamer (AptMUC1) as a targeting agent for nanoparticle-based imaging systems coupled with laser desorption/ionization mass spectrometry (LDI-MS). We found that AptMUC1-conjugated gold nanoparticles immobilized, through hydrophobic and π–π interactions, on graphene oxide (AptMUC1–Au NPs/GO) bound effectively to MUC1 units on tumor cell membranes. The ultrahigh density and high flexibility of AptMUC1 on the GO surface enhanced the platform’s cooperative and multivalent binding affinity for MUC1 on cell membranes. After we had labeled MUC1-overexpressing MCF-7 cells (human breast adenocarcinoma cell line) with AptMUC1–Au NPs/GO, we used LDI-MS to monitor Au cluster ions ([Aun]+; n = 1–3), resulting in the detection of as few as 100 MCF-7 cells. We also employed this AptMUC1–Au NPs/GO–LDI-MS system to analyze four different MUC1 expression cell lines. In addition, the AptMUC1–Au NPs/GO platform could be used further as a labeling agent for tumor tissue imaging when coupled with LDI-MS. Thus, Apt–Au NPs/GO can function as a highly amplified signal transducer through the formation of large Au clusters ions during LDI-MS analysis.
Biological imaging enables the morphological features of organs to be correlated with pathological symptoms; this process is important for finding biomarkers and subsequently using them for the diagnosis of diseases1. Among the different technologies used for tissue imaging, mass spectrometry (MS) for molecular imaging has attracted a great deal of interest for the chemical characterization of biological molecules and for the real-time identification of tissues in biological and clinical applications3. An attractive feature of MS-based tissue imaging is that it can be performed without using fluorescent probes or radioactive labels3. In addition, MS imaging (MSI) is a highly sensitive, rapid, and multiplexed technology that enables the simultaneous identification of a broad range of complex biomolecules in cell and tissue samples. To date, secondary ion MS, matrix-assisted laser desorption/ionization (LDI) MS, laser ablation electrospray ionization MS, and desorption electrospray ionization MS have been the major techniques used for MSI3. Mass spectrometric analysis using nanomaterials and nanostructured substrates has become a popular technique because of the versatility of the nanomaterials and nanosubstrates7. Recently, nanoparticles, including those prepared from gold, silver, and carbon nanotubes, have been employed as matrixes for LDI-based MSI analyses to achieve higher spatial resolution, lower background noise, and higher sensitivity2. Therefore, analyses of the distributions of specific molecular makers through simple nanoparticle-assisted LDI (NALDI) are in high demand for tissue imaging. Nevertheless, the drawback of MSI coupled with NALDI is the lack of sufficient specificity.
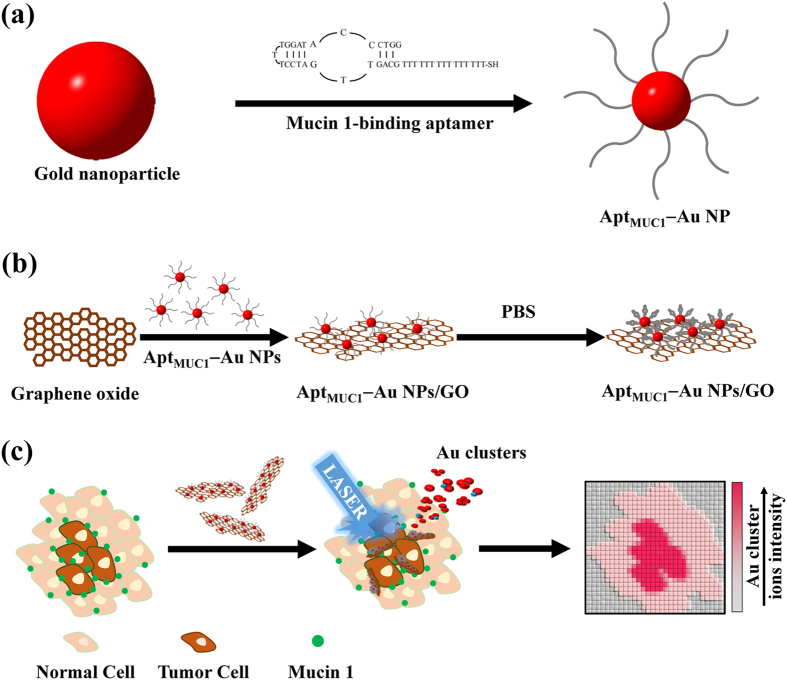
In this study, we found that graphene oxide (GO) modified with Mucin1 (MUC1) binding aptamer–conjugated gold nanoparticles (AptMUC1–Au NPs/GO) coupled with LDI-MS is a facile platform for the detection of tumor cells and for tissue imaging (Fig. 1). MUC1 is a large transmembrane glycoprotein (250–500 kDa) of the mucin family; it consists of a varying amplified repeat sequence of 20 amino acid residues that is rich in serine, threonine, and proline residues in its extracellular domain, a hydrophobic membrane-spanning domain of 31 amino acid residues, and a cytoplasmic tail comprising 69 amino acid residues3. The overexpression (>10-fold) of MUC1 is often associated with a variety of malignant tumors, including breast, gastric, colorectal, lung, prostate, ovarian, pancreatic, and bladder carcinomas4, making MUC1 an ideal target molecule for tumor labeling and chemotherapeutics. AptMUC1 (for its sequence, see the Method section) binds to synthetic peptides (Kd: ca. 0.1 nM) containing MUC1 tandem repeated sequences of the extracellular domain5. In addition, AptMUC1 has been employed to recognize and bind to native MUC1 at the surfaces of MCF-7 breast cancer cells6. In this study, we used LDI-MS analysis to demonstrate that the nanocomposite AptMUC1–Au NPs/GO binds selectively to MCF-7 cells. In addition, we employed AptMUC1–Au NPs/GO in combination with LDI-MS, monitoring the Au cluster ions ([Aun]+; Fig. 1c), for tumor tissue imaging. We conclude that the binding of Apt–Au NPs/GO with tumor cells can be transduced to highly amplified Au cluster ion signals during LDI-MS analysis.
Figure 1.
Schematic representation of (a) the preparation of MUC1-binding aptamer–modified gold nanoparticles (AptMUC1–Au NPs) and (b) their conjugation to graphene oxide (AptMUC1–Au NPs/GO) for (c) tumor tissue imaging by monitoring Au cluster ions when coupled with laser desorption/ionization mass spectrometry.
Results and Discussion
LDI-MS of AptMUC1–Au NPs/GO
We synthesized GO from graphite (7–11 μm) using the modified Hummers method7. Atomic force microscopy (AFM) and transmission electron microscopy (TEM) images revealed that the average size of a single-layer GO (thickness: ca. 1.4 nm) was approximately 235 nm (see Supplementary Fig. S1a and S1b). The 5´-thiolated AptMUC1, which comprised the functionality of a MUC1-binding aptamer (25-mer) and a polythymidine (T15) linker, was bound to the surfaces of the Au NPs (diameter: ca. 13 nm) through Au–S bonding, resulting in the preparation of AptMUC1−Au NPs (ca. 50 AptMUC1 molecules per Au NP). The 25-mer AptMUC1 contains three Watson–Crick pairs, three consecutive mispairs, and four Watson–Crick pairs capped by a TTT triloop motif (Fig. 1a); their base-paired nucleotides melt cooperatively at approximately 47 °C. The TTT triloop structure is the primary binding site for MUC1 protein5. AptMUC1–Au NPs/GO was prepared through simple mixing of AptMUC1–Au NPs (1.0 nM) with GO (0.25 g L–1) in 5 mM sodium phosphate (pH 7.4). AptMUC1–Au NPs were absorbed on GO with stabilization mainly through multivalent nucleobase–graphene π–π stacking between the linear AptMUC1 and GO (Fig. 1b), although hydrophobic and hydrogen bonding interactions between AptMUC1 and GO could not be excluded9. The conformation of AptMUC1 was predominantly a stretched linear structure on the Au NP surfaces in a solution of low ionic strength (5 mM sodium phosphate, pH 7.4). Linear single-stranded DNA interacts with graphene more strongly than do hairpin and double-stranded DNA1. After the AptMUC1–Au NPs had self-assembled on GO, we added phosphate-buffered saline (PBS; 10 mM sodium phosphate, 140 mM NaCl, 2.70 mM KCl; pH 7.6) to the AptMUC1–Au NPs/GO solution to induce folding of the AptMUC1 units into hairpin structures on the Au NP surfaces. As a result, the AptMUC1 molecules could be arranged in an appropriate orientation and conformation for the binding of MUC1.
TEM imaging of AptMUC1–Au NPs/GO revealed that the Apt–Au NPs had assembled homogeneously on the GO (see Supplementary Fig. S1c). UV–Vis absorption and DLS measurements (see Supplementary Fig. S2) revealed that AptMUC1–Au NPs/GO dispersed well (no aggregation) in PBS solution. We used LDI to further study the formation efficiency of Au cluster ions from AptMUC1–Au NPs and AptMUC1–Au NPs/GO (Fig. 2). The photoabsorption of the Au NPs (13 nm) induced the desorption and ionization of the surface Au atoms under SmartBeam laser irradiation (355-nm Nd:YAG; 100 Hz; pulse width: 6 ns; 2.84 × 104 W cm–2; beam diameter: ca. 40 μm). Photothermal evaporation and Coulomb explosion are two well-established mechanisms for the fragmentation of metallic NPs under pulsed laser illumination2. The Coulomb explosion mechanism involves the ejection of a large number of electrons to generate multiple ionized metallic clusters/nanoparticles3. In contrast, the photothermal mechanism suggests that the degree of surface evaporation is highly determined by laser-induced thermal energy transfer into the lattice system3. The formation of Au cluster ions from Au NPs under pulsed laser irradiation is very sensitive to the composition, size, and surface properties of the particles and the wavelength, intensity, and pulse width of the laser5. Similar to our previous findings5, the formation efficiency of the Au cluster ions [Aun]+ (n = 1–3) from the Au NPs decreased to 16.7% after the Au NPs had been modified with AptMUC1 (Figs. 2A, B). The Au NPs may have transferred their absorbed energy to the surface AptMUC1 molecules, decreasing the lattice temperature (Tl) of the Au NPs, after laser excitation5. As a result, the evaporation or ionization of the surface Au atoms/ions into the gas phase was suppressed. In addition, the dense mantle of AptMUC1 units may have strongly inhibited electron ejection from the Au NPs, decreasing the electron temperature (Te) of the Au NPs; therefore, the explosive fragmentation-induced formation of Au cluster ions was strongly inhibited3.
Figure 2.
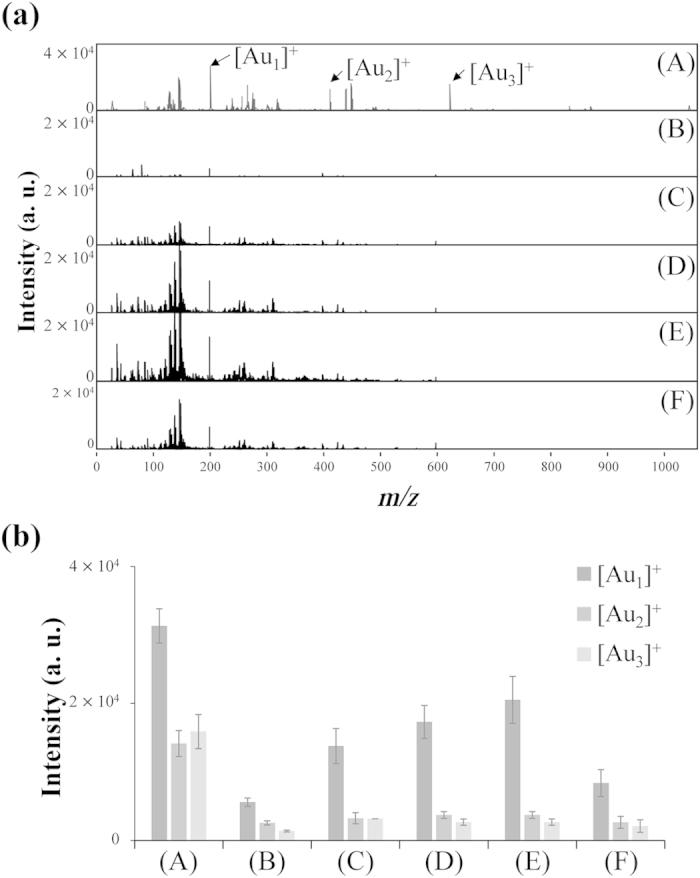
(a) LDI-MS spectra of (A) Au NPs (1.0 nM) and AptMUC1–Au NPs (1.0 nM) in the (B) absence and (C–F) presence of GO at concentrations of (C) 0.0025, (D) 0.025, (E) 0.25, and (F) 2.5 g L–1. (b) Intensity of [Aun]+ signals obtained, under LDI, from (A) Au NPs (1.0 nM) and AptMUC1–Au NPs (1.0030 nM) in the (B) absence and (C–F) presence of GO at concentrations of (C) 0.0025, (D) 0.025, (E) 0.25, and (F) 2.5 g L–1. Signals at m/z 196.97, 393.93, and 590.90 are assigned to [Au1]+, [Au2]+, and [Au3]+ ions, respectively. A total of 500 pulsed laser shots was applied to accumulate the signals from five LDI-targeted positions at a laser power density of 2.84 × 104 W cm−2. Peak intensities are plotted in arbitrary units (a. u.). The error bars in (b) represent the standard deviations of five repeated measurements.
We found that the formation of Au cluster ions from AptMUC1–Au NPs was enhanced in the presence of GO. As revealed in Fig. 2B–F, the intensities of the Au cluster ions in the LDI-MS spectra increased upon increasing the concentration of GO (0–0.25 g L−1). Because of their high specific surface areas (ca. 2600 m2 g–1), high UV-region absorption coefficients, and energy transduction capabilities, graphene and GO have been used recently as nanomaterials in surface-assisted LDI-MS for the analysis of oligonucleotides, proteins, lipids, and small molecules0. We suspect that energy (heat) transfer from GO to the AptMUC1–Au NPs may have been the main reason for the enhanced formation of [Aun]+ cluster ions; GO-assisted electron ejection from the AptMUC1–Au NPs may also have contribute to the higher formation efficiency of the [Aun]+ cluster ions. The aggregation of AptMUC1–Au NPs (1.0 nM; ~0.014 g L–1)/GO resulted in a slight decrease in the signal intensity of [Aun]+ when the concentration of GO was greater than 0.25 g L–1 (Fig. 2F). Aggregation occurred when the GO concentration was >0.25 g/L presumably because of the relative low number of adsorbed AptMUC1–Au NPs on each GO and therefore, their stability in PBS solution decreased (see Supplementary Fig. S3 and Fig. S4). In addition, a low ratio (<0.06) of the concentrations of AptMUC1–Au NPs (1.0 nM; ~0.014 g L–1) to GO (>0.25 g L–1) may have resulted in the formation of crosslinks (AptMUC1–Au NPs/GO), thereby inducing aggregation of the nanocomposites. Such aggregation of AptMUC1–Au NPs/GO nanocomposites made it difficult to fragment, desorb, and ionize the surface Au atoms/ions on the Au NPs. The combination of AptMUC1–Au NPs/GO composites and LDI-MS monitoring of the [Au1]+ signals allowed detection of the AptMUC1–Au NPs at concentrations as low as 10 fM. In contrast, the equivalent concentration from the UV–Vis absorption measurements, monitoring the surface plasmon resonance (SPR) absorption of the Au NPs at 520 nm, was approximately 10 pM (see Supplementary Fig. S5). In addition, the Au ion–driven signals arising from the high surface ratio of Au atoms/ions on Au NPs are much stronger than those from proteins in the LDI-MS analysis because of the low ionization/desorption efficiencies of proteins. Therefore, we suspected that our AptMUC1–Au NPs/GO would be an exceptional labeling agent for the analysis of marker proteins in tumor cells when coupled with LDI-MS.
Detection of tumor cells by AptMUC1–Au NPs/GO–LDI-MS
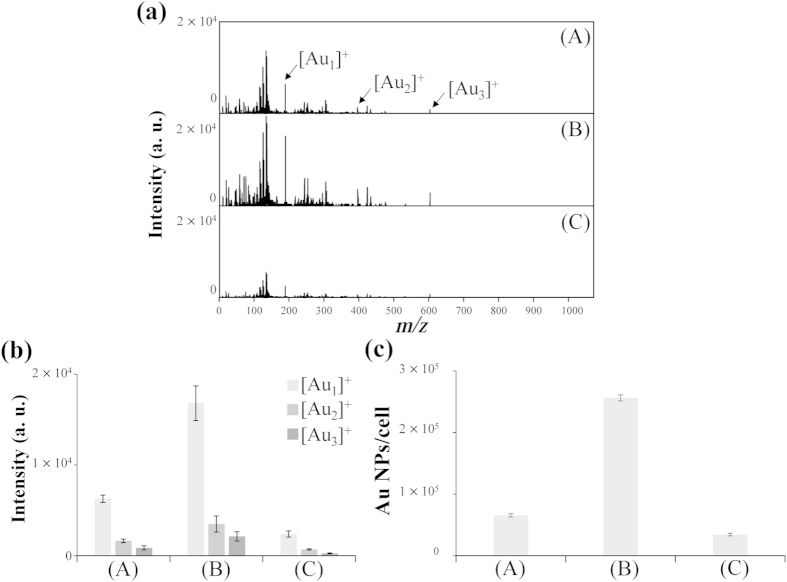
As revealed in Fig. 3a, b, the [Aun]+ signals obtained using AptMUC1–Au NPs/GO, prepared from AptMUC1–Au NPs (1.0 nM) and GO (0.25 g L–1), when coupled with LDI-MS for the detection of MUC1-overexpressing MCF-7 cells were approximately three times stronger than those obtained using AptMUC1–Au NPs. In a control experiment, polythymine (T45)-conjugated Au NPs provided low-intensity [Aun]+ signals, compared with those from AptMUC1–Au NPs and AptMUC1–Au NPs/GO, when analyzing MCF-7 cells. The detection of MCF-7 cells using AptMUC1–Au NPs/GO was superior to that using AptMUC1–Au NPs, presumably because the former displayed a higher formation efficiency of Au cluster ions and/or stronger binding affinity toward MUC1 on the cell membrane. In a quantitation analysis performed using inductively coupled plasma mass spectrometry (ICP-MS), we found that the number of Au NPs bound or uptaken by MCF-7 cells from AptMUC1–Au NPs/GO was approximately four times higher than that from AptMUC1–Au NPs (Fig. 3c), revealing that AptMUC1–Au NPs/GO had relatively stronger binding affinity toward MUC1 on the cell membrane. The signal enhancement obtained by LDI-MS (~3-fold) was lower than that of by ICP-MS (~4-fold) probably due to the incomplete fragmentation, desorption, and ionization of AptMUC1−Au NPs to Au cluster ions in the LDI-MS analysis. The ultrahigh density of AptMUC1 on the surface of the Au NPs and the ultrahigh density of AptMUC1–Au NPs on the GO surface provided a high local concentration of flexible AptMUC1 ligands, enhancing the cooperative and multivalent binding affinity toward MUC1 units on the cell membrane2. The nanosheet structure of GO, allowing ready adhesion to the plasma membrane and high uptake by mammalian cells through phagocytosis and clathrin-mediated endocytosisi5, may have contributed to the strong affinity of AptMUC1–Au NPs/GO toward MCF-7 cells. In addition, high density of Au NPs on GO can generate greater number of Au cluster ions for signal enhancement. Our AptMUC1–Au NPs/GO–LDI-MS system could detect as few as 100 MCF-7 cells when monitoring the [Au1]+ signals (see Supplementary Fig. S6). Compared to other mass spectrometry based tumor cell analysis techniques8, our AptMUC1–Au NPs/GO–LDI-MS platform is relatively simple, rapid, and selective and shows great legibility for tumor cells quantification.
Figure 3.
(a) LDI-MS spectra recorded using (A) AptMUC1–Au NPs (1.0 nM), (B) AptMUC1–Au NPs/GO ([AptMUC1–Au NPs] = 1.0 nM; [GO] = 0.25 g L–1), and (C) T45–Au NPs/GO [(T45–Au NPs) = 1.0 nM; (GO) = 0.25 g L–1] a probes for the detection of 105 MCF-7 cells. (b) Peak intensities of [Aun]+ ions obtained from (A) AptMUC1–Au NPs, (B) AptMUC1–Au NPs/GO, and (C) T45–Au NPs/GO after labeling of MCF-7 cells. (c) Concentrations of Au NPs accumulated in MCF-7 cells, as determined using ICP-MS. Other conditions were the same as those described in Fig. 1.
Specificity of AptMUC1–Au NPs/GO
We further applied our AptMUC1–Au NPs/GO–LDI-MS platform to analyze other types of cells, namely MCF-10A (normal mammary epithelial cells), MDA-MB-231 (breast adenocarcinoma cells), and 293T (transformed embryonic kidney cells). Among the four tested cell lines, the highest-intensity [Au1]+ signals arose from the MCF-7 breast cancer cell line (see Supplementary Fig. S7a and S7b), consistent with our results from ICP-MS analysis (see Supplementary Fig. S7c). Notably, the sample preparation for ICP-MS analysis required relatively complicated and tedious pretreatment steps. We also observed that the intensity of the [Au1]+ signals increased upon increasing the ratio of MCF-7 to MCF-10A cells in a co-culture system containing these two cell types (see Supplementary Fig. S8). MUC1 is widely expressed by normal epithelial cells (e.g., MCF-10A); this expression increases significantly when the cells become malignant, such as in breast, pancreatic, or ovarian cancer4. MUC1 is overexpressed in approximately 90% of human breast cancers; the expression of membrane-associated MUC1 in MDA-MB-231 cells is, however, lower than that in MCF-7 cells9. The low level of MUC1 expression in some breast adenocarcinoma cells is associated with decreased cytokeratin expression and increased vimentin expression0. In fact, the MUC1 protein is expressed aberrantly in many solid tumor cell lines1. For example, the 293T cell line is MUC1-negative because it does not expresses endogenous MUC11. Our results from AptMUC1–Au NPs/GO–LDI-MS analysis platform (Fig. S7) are consistent with these reports and the results of the western blotting analysis (see Supplementary Fig. S9)1. MUC1 is associated with cellular transformation and tumorigenicity; it is considered as both a potential cancer biomarker and an attractive target for cancer immunotherapy4. Therefore, we suspect that our proposed assay will have practical applications in the detection of MUC1 expression in tumor cells, offering high accuracy and reliability with comprehensive information for the early detection of MUC1-related cancers.
Analysis of MUC1 expression
Furthermore, we applied our AptMUC1–Au NPs/GO–LDI-MS platform to the analysis of MCF-7 cells after treatment with apigenin (4´,5,7-trihydroxyflavone; 0–75 μM) for 24 h. Apigenin, a natural polyphenol product belonging to the flavone class, is abundant in some fruits (e.g., cherries, apples, grapes) and vegetables (e.g., parsley, Chinese cabbage, bell peppers, celery). Apigenin has been studied widely for its anti-inflammatory, antioxidant, and antitumor properties; it can inhibit tumor cell invasion and metastases by regulating protease production2. Recently, it was demonstrated that apigenin can inhibit mucin gene expression and the production of mucin protein in epithelial and tumor cells3. Using our AptMUC1–Au NPs/GO–LDI-MS detection system, we also observed an apigenin-induced downregulation of MUC1 expression: the intensity of the [Au1]+ signals decreased upon increasing the concentration of apigenin used for treatment of the MCF-7 cells (Fig. 4a). This downregulation of MUC1 in MCF-7 cells was consistent with the results of western blotting (Fig. 4c), revealing that our LDI-MS–based assay has great potential for use in the analysis of membrane protein expression levels after labeling with functional Au NPs. Compared with western blotting analysis, our assay is relatively simple, sensitive, and capable of high throughput.
Figure 4.
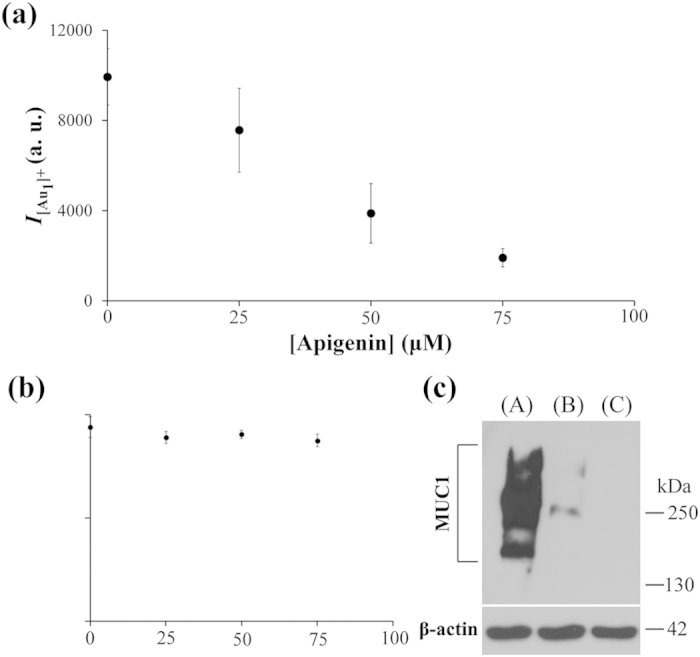
(a) Mass signal intensities of [Au+] ions (I[Au1]+) obtained when using AptMUC1–Au NPs/GO to analyze apigenin-treated (0–75 μM) MCF-7 cells (105 cells well–1). (b) Cell viability of MCF-7 cells incubated with apigenin in alpha-MEM at 37 °C under 5% CO2 for 24 h. (c) Western blotting for MUC1 in the cell lysates of (A) MCF-7 cells (105 cells well–1), (B) MCF-7 cells (105 cells well–1) after treatment with 75 μM apigenin for 24 h, and (C) 293T cells (105 cells well–1). Other conditions were the same as those described in Fig. 2.
Tissue imaging
The development of a rapid, high-throughput method for MUC1 expression imaging in a typical tissue microarray would be a significant advance over the current practice. Therefore, we further employed our AptMUC1–Au NPs/GO–LDI-MS platform for tissue microarray imaging by monitoring the [Au1]+ signals after the AptMUC1–Au NPs had targeted cells in a tissue microarray (US Biomax). The tissue microarray, comprising breast carcinoma or normal tissues (diameter: 1.5 mm) was labeled with AptMUC1–Au NPs/GO and subsequently studied using LDI-MS. Representative images for breast carcinoma and normal microarray tissues sections (see Supplementary Fig. S10) revealed that the total intensity of the [Au1]+ signals from the breast carcinoma tissue was much higher (>10-fold) than that from the normal tissue, suggesting that our AptMUC1–Au NPs/GO–LDI-MS system is capable of analyzing MUC1 expression in tissue samples. The spatial resolution of our LDI-MS imaging technique (~80 μm) is limited by laser beam diameter size (40 μm) which let the spatial resolution inferior to the current practices such as fluorescence imaging4. However, AptMUC1–Au NPs/GO–LDI-MS platform provides the great specificity and sensitivity for the molecular imaging due to the specific and strong binding of AptMUC1–Au NPs/GO to targeted molecules on cell membrane and the high signal enhancement from Au NPs.
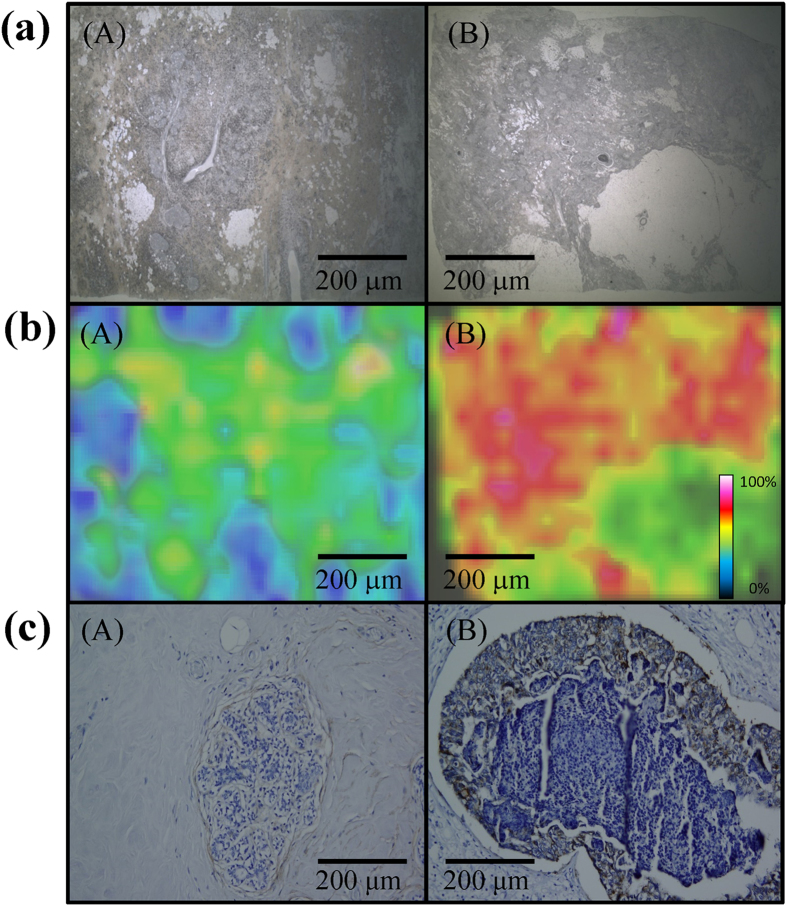
We also applied our AptMUC1–Au NPs/GO–LDI-MS platform to analyze the tissue sections (US Biomax) of normal breast tissue (normal human mammary gland; female, 35 years old) and human breast cancer tissue (human breast invasive ductal carcinoma; female, 42 years old). As indicated in Fig. 5b, MSI revealed, through monitoring of the intensity of [Au1]+ signals, a dramatic difference between the normal and tumor tissues, suggesting high sensitivity of our LDI-MS–based tumor tissue system for the diagnosis of malignancies. We used immunohistochemistry (IHC) to further validate MUC1 expression and distribution in tumor and normal breast tissue. Figure 5c presents the immunostaining images of the sections; the results are in good agreement with those obtained through LDI-MS, with the heterogeneously developed brown color being deeper in the breast tumor tissue (Fig. 5cB). These IHC validation studies confirmed that the expression pattern of MUC1 reflects the tissue distribution in the malignant breast cells.
Figure 5.
(a) Optical images of (A) normal breast and (B) breast tumor tissues. (b) LDI-MS images of the [Au1]+ intensity distributions in (A) normal breast and (B) breast tumor tissues after incubation with AptMUC1–Au NPs/GO for 1 h. (c) Immunostaining of MUC1 by MUC1 antibody (VU4H5) in (A) normal breast and (B) tumor breast tissues. The dark brown and blue colors represent the staining of MUC1 and nuclei, respectively. Parameters of LDI-MS imaging: image pixel size, 1 × 1 μm2; line distance, 50 μm; repetition rate, 100 Hz; image size, 0.8 × 0.8 mm2. Other conditions were the same as those described in Fig. 3.
In summary, we have coupled an AptMUC1–Au NPs/GO probe with LDI-MS for the detection of breast cancer cells and for tissue imaging. The formation efficiency of [Aun]+ ions from AptMUC1–Au NPs under pulsed laser irradiation was enhanced after anchoring to GO. In addition, AptMUC1–Au NPs/GO could bind specifically to MUC1 units on cell membranes. The intensities of the signals for [Aun]+ cluster ions in the mass spectra revealed the levels of MUC1 expression in tumor cells; these signals behaved as a highly amplified target-labeling indicator for the targeted tumor cells. Our probe enabled the selective and sensitive detection of as few as 100 MCF-7 cells and facilitated efficient analyses of MUC1 protein expression in four different cell lines. We also applied the AptMUC1–Au NPs/GO probe to breast tumor tissue imaging, highlighting the potential for studying various cancer tissues with different aptamer-modified Au NPs/GO. To the best of our knowledge, this study provides the first example of combining AptMUC1–Au NPs/GO with LDI-MS for tumor tissue imaging. This high-throughput LDI-MS imaging system has great potential for the diagnosis of cancer.
Methods
Materials and reagents
All thiolated oligonucleotides used in this study were purchased from Integrated DNA Technologies (Coralville, IA). Graphite powder (99%, 7–11 μM) was purchased from Alfa–Aesar (Heysham, Lancashire, UK). Sulfuric acid, phosphoric acid, hydrogen peroxide, trisodium citrate, sodium phosphate tribasic, sodium phosphate dibasic, boric acid, tetrachloroauric acid, dimethyl sulfoxide, apigenin, xylene, ethanol, 3,3-diaminobenzidine tetrahydrochloride, hematoxylin, and isopropanol were purchased from Sigma–Aldrich (Milwaukee, WI). Citric acid, calcium chloride, magnesium chloride, potassium permanganate, tris(hydroxymethyl)aminomethane (Tris), and hydrochloric acid were purchased from Mallinckrodt Baker (Phillipsburg, NJ). Alpha-MEM and fetal bovine serum were purchased from GIBCO (Campinas, Brazil). An MTT assay kit was purchased from Thermo Fisher Scientific (Waltham, MA). MUC1 (VU4H5) mouse mAb, anti-mouse IgG, and HRP-linked antibody were purchased from Cell Signaling Technology (Danvers, MA). Milli-Q ultrapure water (18 MΩ, Millipore, Billerica, MA) was used in all experiments.
Preparation of AptMUC1–Au NPs/GO
A thiol-modified MUC1-binding aptamer (5´-HS-TTT TTT TTT TTT TTT GCA GTT GAT CCT TTG GAT ACC CTG G-3´) was attached to the Au NPs following a slightly modified procedure reported elsewhere5. An aliquot of aqueous Au NP solution (980 μL) in a 1.5-mL tube was mixed with the thiol-oligonucleotide (100 μM, 20 μL) to yield final concentrations of 15 nM Au NPs and 2 μM oligonucleotides. After 2 h, the mixed solution was salt-aged in the presence of 200 mM NaCl for over 16 h. Salt-aging resulted in a greater number of thiol-modified oligonucleotides bound to the Au NPs5. The mixture was then centrifuged at an relative centrifugal force (RCF) of 30,000 × g for 20 min to remove the excess thiol-oligonucleotide. After removal of the supernatant, the oily precipitate was washed with 5.0 mM Tris–HCl (pH 7.6). After three centrifuge/wash cycles, the colloid was resuspended in 5.0 mM Tris–HCl (pH 7.6) and stored at 4 °C. The AptMUC1–Au NPs/GO nanocomposite was prepared by mixing the GO (0.25 g L–1) and AptMUC1–Au NPs (1.0 nM) in 5 mM sodium phosphate (pH 7.4). After incubation for 1 h, the AptMUC1–Au NPs/GO was transferred into PBS and incubated for another 1 h at room temperature.
Analysis of cells by AptMUC1–Au NPs/GO–LDI-MS
Cells were cultured on 8-mm chips in 48-well plates for 8 h at 37 °C. For co-culture experiments, MCF-10A/MCF-7 cells were mixed and seeded at different ratios. The cultured cells were labeled separately with AptMUC1–Au NPs/GO for 1 h in physiological buffer. The cell adhesive chips were then moved out and washed three times with PBS. The chips were then cast onto a stainless-steel 384-well MALDI target and air-dried at room temperature prior to LDI time-of-flight (TOF) MS measurements. MS experiments were performed in the reflectron positive-ion mode using an Autoflex III MALDI TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The samples were irradiated using a SmartBeam laser (355 nm Nd: YAG) operated at 100 Hz. Ions produced by LDI were energetically stabilized during a delayed extraction period of 30 ns and accelerated through the TOF chamber in the reflection mode prior to entering the mass-analyzer. The available accelerating voltages ranged from +20 to –20 kV. The instruments were calibrated with Au clusters, using their theoretical masses, prior to analysis. A total of 500 pulsed laser shots were applied to accumulate signals from five MALDI target positions at a power density of 2.84 × 104 W cm–2. For ICP-MS (Agilent 7700 Series, Agilent Technologies, Santa Clara, CA), the AptMUC1–Au NPs/GO-treated cells samples were prepared in 2% HNO3.
Tissue imaging
The human breast tumor and normal breast tissue microarray (BR811; 80 cases; diameter: 1.5 mm; thickness: 5 μm), human normal breast tissue section (HuFPT130), and human breast cancer section (HuCAT298) were used for tissue imaging. The procedures of tissue sample treatments were performed in accordance with the protocols approved by Animal Administration Committee of National Taiwan Ocean University. After deparaffinization, the tissue samples were labeled with AptMUC1–Au NPs/GO, prepared from 1.0 nM AptMUC1–Au NPs and 0.25 g L–1 GO, in PBS for 1 h. The slides were washed three times with PBS before they were cast onto an MTP slide adapter II MALDI plate and air-dried at room temperature prior to LDI-MS measurements. LDI-MS imaging experiments were performed in the image reflectron positive-ion mode using an Autoflex III MALDI TOF/TOF mass spectrometer. The tissue slides were irradiated using a SmartBeam laser (355 nm Nd: YAG) operated at 100 Hz. A laser spot diameter of 40 μm and a raster width of 50 μm were employed. Ions produced by LDI were energetically stabilized during a delayed extraction period of 30 ns and accelerated through the TOF chamber in the reflection mode prior to entering the mass-analyzer. The available accelerating voltages ranged from +20 to –20 kV. The instruments were calibrated with Au clusters, using their theoretical masses, prior to analysis. One spot was pulsed for a total of 500 laser shots at a power density of 2.84 × 104 W cm−2.
Please see the Supplementary Information for the details on the preparation and characterization of GO, preparation of 13-nm spherical Au NPs, cell cultures, and immunohistochemistry with horseradish peroxidase.
Author Contributions
R.C.H. and C.C.H designed the research. R.C.H. performed most of the experiments and data analysis. W.J.C. contributed to the characterization of the nanocomposites. I.P.J.L. contributed to western blotting analysis. R.C.H. and C.C.H co-wrote the paper. All authors discussed the results and commented on the manuscript.
Additional Information
How to cite this article: Huang, R.-C. et al. Multivalent Aptamer/Gold Nanoparticle-Modified Graphene Oxide for Mass Spectrometry-Based Tumor Tissue Imaging. Sci. Rep. 5, 10292; doi: 10.1038/srep10292 (2015).
Supplementary Material
Acknowledgments
This study was supported by the Ministry of Science and Technology of Taiwan under contracts NSC 101-2628-M-019-001-MY3, 102-2113-M-019-001-MY3, and 102-2627-M-019-001-MY3.
References
- Mitterhauser M. & Wadsak W. Imaging biomarkers or biomarker imaging? Pharmaceuticals 7, 765–778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver E. M. & Hummon A. B. Imaging mass spectrometry: from tissue sections to cell cultures. Adv. Drug. Deliver. Rev. 65, 1039–1055 (2013). [DOI] [PubMed] [Google Scholar]
- Norris J. L. & Caprioli R. M. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa R. & Kawasaki H. Functionalized nanoparticles and nanostructured surfaces for surface-assisted laser desorption/ionization mass spectrometry. Anal. Sci. 26, 1229–1240 (2010). [DOI] [PubMed] [Google Scholar]
- Lim A. Y., Ma J. & Boey Y. C. F. Development of nanomaterials for SALDI-MS analysis in forensics. Adv. Mater. 24, 4211–4216 (2012). [DOI] [PubMed] [Google Scholar]
- Chiang C.-K., Chen W.-T. & Chang H.-T. Nanoparticle-based mass spectrometry for the analysis of biomolecules. Chem. Soc. Rev. 40, 1269–1281 (2011). [DOI] [PubMed] [Google Scholar]
- Calavia R., Annanouch F. E., Correig X. & Yanes O. Nanostructure initiator mass spectrometry for tissue imaging in metabolomics: future prospects and perspectives. J. Proteomics 75, 5061–5068 (2012). [DOI] [PubMed] [Google Scholar]
- Jun J. H. et al. High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal. Chem. 82, 3255–3265 (2010). [DOI] [PubMed] [Google Scholar]
- Jackson S. N. et al. Imaging of lipids in rat heart by MALDI-MS with silver nanoparticles. Anal. Bioanal. Chem. 406, 1377–1386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.-W. et al. Molecular histology analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry using gold nanoparticles as matrix. Rapid Commun. Mass Spectrom. 25, 3690–3696 (2011). [DOI] [PubMed] [Google Scholar]
- Kim Y.-K. et al. Synergistic effect of graphene oxide/MWCNT films in laser desorption/ionization mass spectrometry of small molecules and tissue Imaging. ACS Nano 5, 4550–4561 (2011). [DOI] [PubMed] [Google Scholar]
- Dufresne M., Thomas A., Breault-Turcot J., Masson J.-F. & Chaurand P. Silver-assisted laser desorption ionization for high spatial resolution imaging mass spectrometry of olefins from thin tissue sections. Anal. Chem. 85, 3318–3324 (2013). [DOI] [PubMed] [Google Scholar]
- Gendler S. J. MUC1, the renaissance molecule. J. Mammary Gland Biol. 6, 339–353 (2001). [DOI] [PubMed] [Google Scholar]
- Nath S. & Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 20, 332–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C. S. M., Matthews C. S. & Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 27, 289–301 (2006). [DOI] [PubMed] [Google Scholar]
- Wu P., Gao Y., Zhang H. & Cai C. Aptamer-guided silver–gold bimetallic nanostructures with highly active surface-enhanced raman scattering for specific detection and near-infrared photothermal therapy of human breast cancer cells. Anal. Chem. 84, 7692–7699 (2012). [DOI] [PubMed] [Google Scholar]
- Marcano D. C. et al. Improved synthesis of graphene oxide. ACS Nano 4, 4806–4814 (2010). [DOI] [PubMed] [Google Scholar]
- Spiwok V., Hobza P. & Řezáč J. Free-energy simulations of hydrogen bonding versus stacking of nucleobases on a graphene surface. J. Phys. Chem. C 115, 19455–19462 (2011). [Google Scholar]
- Varghese N. et al. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem 10, 206–210 (2009). [DOI] [PubMed] [Google Scholar]
- He Y., Lin Y., Tang H. & Pang D. A graphene oxide-based fluorescent aptasensor for the turn-on detection of epithelial tumor marker mucin 1. Nanoscale 4, 2054–2059 (2012). [DOI] [PubMed] [Google Scholar]
- Wu M., Kempaiah R., Huang P.-J. J., Maheshwari V. & Liu J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir 27, 2731–2738 (2011). [DOI] [PubMed] [Google Scholar]
- Werner D. & Hashimoto S. Improved working model for interpreting the excitation wavelength- and fluence-dependent response in pulsed laser-induced size reduction of aqueous gold nanoparticles. J. Phys. Chem. C 115, 5063–5072 (2011). [Google Scholar]
- Muto H., Miyajima K. & Mafuné F. Mechanism of laser-induced size reduction of gold nanoparticles as studied by single and double laser pulse excitation. J. Phys. Chem. C 112, 5810–5815 (2008). [Google Scholar]
- Weng C.-I. et al. Detection of arsenic(III) through pulsed laser-induced desorption/ionization of gold nanoparticles on cellulose membranes. Anal. Chem. 86, 3167–3173 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Y.-C., Li Y.-J., & Huang C.-C. Information derived from cluster ions from DNA-modified gold nanoparticles under laser desorption/ionization: analysis of coverage, structure, and single-nucleotide polymorphism. Anal. Chem. 85, 1021–1028 (2013). [DOI] [PubMed] [Google Scholar]
- Kong X. & Huang Y. Applications of graphene in mass spectrometry. J. Nanosci. Nanotechnol. 14, 4719–4732 (2014). [DOI] [PubMed] [Google Scholar]
- Kawasaki H. et al. Functionalized graphene-coated cobalt nanoparticles for highly efficient surface-assisted laser desorption/ionization mass spectrometry analysis. Anal. Chem. 84, 9268–9275 (2012). [DOI] [PubMed] [Google Scholar]
- Qian K. et al. Laser engineered graphene paper for mass spectrometry imaging. Sci. Rep. 3, 1415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.-R. et al. Layer-by-layer thin film of reduced graphene oxide and gold nanoparticles as an effective sample plate in laser-induced desorption/ionization mass spectrometry. Anal. Chim. Acta 809, 97–103 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Q., Cheng M. & Jiang G. Mildly oxidized graphene: facile synthesis, characterization, and its application as matrix in MALDI mass spectrometry. Chem. Eur. J. 19, 5561–5565 (2013). [DOI] [PubMed] [Google Scholar]
- Sheng W., Chen T., Tan W. & Fan Z. H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 7, 7067–7076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Assembling DNA through affinity binding to achieve ultrasensitive protein detection. Angew. Chem. Int. Ed 52, 10698–10705 (2013). [DOI] [PubMed] [Google Scholar]
- Na H.-K. et al. Cytoprotective effects of graphene oxide for mammalian cells against internalization of exogenous materials. Nanoscale 5, 1669–1677 (2013). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Graphene microsheets enter cells through spontaneous membrane penetration at edge asperities and corner sites. Proc. Natl. Acad. Sci. U.S.A. 110, 12295–12300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Guo R. & Yan L.-T. Simulation and analysis of cellular internalization pathways and membrane perturbation for graphene nanosheets. Biomaterials 35, 6069–6077 (2014). [DOI] [PubMed] [Google Scholar]
- Heger Z. et al. Identification of estrogen receptor proteins in breast cancer cells using matrix-assisted laser desorption/ionization time of flight mass spectrometry (Review). Oncol. Lett. 7, 1341−1344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Immunomagnetic separation combined with inductively coupled plasma mass spectrometry for the detection of tumor cells using gold nanoparticle labeling. Anal. Chem. 86, 8082−8089 (2014). [DOI] [PubMed] [Google Scholar]
- Jiang F., Zhou X. Y. & Huang J. The value of surface enhanced laser desorption/ionization-time of light mass spectrometry at the diagnosis of non-small cell lung cancer: a systematic review Technol. Cancer Res. T. 13, 109–117 (2014). [DOI] [PubMed] [Google Scholar]
- Zaretsky J. Z. et al. MUC1 gene overexpressed in breast cancer: structure and transcriptional activity of the MUC1 promoter and role of estrogen receptor alpha (ERα) in regulation of the MUC1 gene expression. Mol. Cancer 5, 57 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. D. et al. Heterogeneity of MUC1 expression by human breast carcinoma cell lines in vivo and in vitro. Breast Cancer Res. Treat. 58, 255–266 (1999). [DOI] [PubMed] [Google Scholar]
- Ren C. Y., Akiyama Y., Miyake S. & Yuasa Y. Transcription factor GATA-5 selectively up-regulates mucin gene expression. J. Cancer Res. Clin. Oncol. 130, 245–252 (2004). [DOI] [PubMed] [Google Scholar]
- Shukla S. & Gupta S. Apigenin: a promising molecule for cancer prevention. Pharm. Res. 27, 962–978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Rajabi H. & Kufe D. Mucin 1 C-terminal subunit oncoprotein is a target for small-molecule inhibitors. Mol. Pharmacol. 79, 886–893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhaus K. & Ulrich Nienhaus G. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 43, 1088–1106 (2014). [DOI] [PubMed] [Google Scholar]
- Hurst S. J., Lytton-Jean A. K. R. & Mirkin C. A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 78, 8313–8318 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.