Abstract
Metal ion interactions with DNA have far-reaching implications in biochemistry and DNA nanotechnology. Ag+ is uniquely interesting because it binds exclusively to the bases rather than the backbone of DNA, without the toxicity of Hg2+. In contrast to prior studies of Ag+ incorporation into double-stranded DNA, we remove the constraints of Watson-Crick pairing by focusing on homo-base DNA oligomers of the canonical bases. High resolution electro-spray ionization mass spectrometry reveals an unanticipated Ag+-mediated pairing of guanine homo-base strands, with higher stability than canonical guanine-cytosine pairing. By exploring unrestricted binding geometries, quantum chemical calculations find that Ag+ bridges between non-canonical sites on guanine bases. Circular dichroism spectroscopy shows that the Ag+-mediated structuring of guanine homobase strands persists to at least 90 °C under conditions for which canonical guanine-cytosine duplexes melt below 20 °C. These findings are promising for DNA nanotechnology and metal-ion based biomedical science.
The long-standing biochemical interest in metal-DNA interactions now extends into the field of DNA nanotechnology1, where incorporation of strongly bound metal cations promises to realize more robust, diversely functional DNA-based materials2,3,4,5. This potential stimulated the recent development of artificial bases that form metal-mediated pairs bridged by Ag+, Cu2+ and Hg2+ 6,7,8,9. The identification of more diverse metal cation pairings of the natural bases could remove the need to incorporate expensive synthetic elements and amplify the impact of such metal-mediated pairings. Ag+ is uniquely interesting because it exhibits unusually specific interactions with DNA, binding exclusively to natural bases rather than the negatively charged phosphate backbone (Hg2+ also associates exclusively with the bases, but is far more toxic)5. The potential that Ag+-DNA interactions hold for nanotechnology is already exemplified by the fluorescent, DNA-stabilized silver clusters10,11 used recently in novel chemical and biochemical sensing schemes12. These nano-optical, DNA based materials are known to incorporate Ag+ as well as neutral silver atoms13, indicating that Ag+-DNA interactions are key to stabilizing the fluorescent clusters. In addition to the previously known bridging of cytosine (C) bases by Ag+ 14,15, the formation of fluorescent silver clusters on homo-base guanine (G) strands of DNA and RNA16 and the Ag+ induced dimerization of individual modified guanine bases in non-aqueous solution17 suggests that Ag+ might also bridge G bases in DNA oligomers. In biochemistry, the strong interactions of G bases with Pt2+ are thought to be key to important chemotherapy drugs18,19. Thus the discovery of stable Ag+-G binding might find use in treatment of disease associated with mutations. Ag+-base interactions may also underlie the antimicrobial action of silver nanoparticles20. The known biochemical roles of metal cations suggest that a better understanding of how Ag+ binds to the natural DNA bases could aid future development of disease therapeutics.
Despite this compelling potential, surprisingly little is known about how Ag+ interacts with the natural bases when DNA is not conformationally constrained by the canonical Watson-Crick (WC) hydrogen bonding of adenine (A) to thymine (T) and C to G. Early studies of biological double-stranded (ds) DNA could not unravel the diversity of base-Ag+ interactions due to the mixed A, C, G and T composition21,22,23,24,25,26,27. More recent studies of synthetic dsDNA used the insertion of single-base mismatches to examine which bases could be bridged by Ag+, when subject to the conformational constraints of the WC-paired surroundings14,28,29. In addition to C-Ag+-C, evidence for a C-Ag+-A pair has been reported30, while the recent experimental literature is conflicting on the possibility of C-Ag+-T pairing28,29, and mostly silent regarding Ag+ interactions with G31. Prior computational studies of Ag+-base interactions have been challenged by the plethora of possible binding geometries, especially for G32,33,34,35,36, and have focused largely on binding geometries compatible with WC-like structure.
Although many prior studies have focused on Ag+ incorporation into a canonical dsDNA environment, there is no a priori reason to assume that base pairing by hydrogen bonding will persist in the presence of Ag+. The dominant mode of binding must depend on the affinities and geometries of Ag+-base interactions relative to WC pairing. Here we remove the constraint of WC pairing by focusing on homo-base deoxyoligonucleotides and mixtures of these AN, CN, GN and TN strands, for strand lengths N = 6 to N = 20 bases (Fig. 1a), at neutral pH. We use relative abundances in electrospray ion mass spectrometry (ESI-MS) to determine the ordering of Ag+ affinities to each homo-base strand. The competition of Ag+-mediated base pairing with WC pairing is tested by studies of the mixed complementary strands. Through quantum chemical calculations that explore an unrestricted space of binding geometries, we find that the order of binding energies (BE) for the most stable Ag+-bridged homobase complexes agree with abundance trends in ESI-MS data. Strikingly, experiments show that GN strands form fully Ag+-bridged duplexes, GN-(Ag+)N-GN, that are more stable than the WC paired CN-GN duplex.
Figure 1.
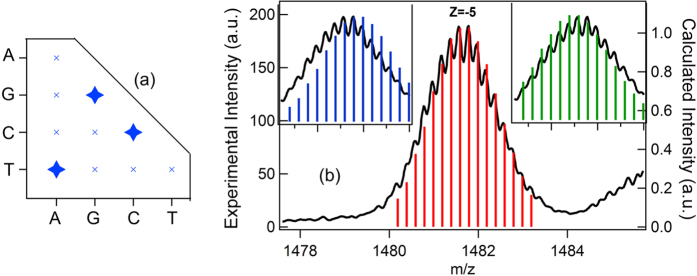
(a) Schematic of the homo-base strand types and combinations studied. Stars denote the detected Ag+-bridged duplexes. (b) Example of isotope peak envelope resolved in MS for C11-(Ag+)11-C11. Black lines: data. The total mass of the ionized species (m) is given by m = mDNA + mAgNAg – npr, where mDNA is the mass of the unionized DNA strand, mAgNAg is the mass of the total silver content, and npr is the number of protons removed by negative mode electrospray ionization. The charge state, z (negative) of the ionized species is z = QAg/e – npr, where QAg is the total charge associated with the silver content. Bars show the calculated isotope peak patterns for a net charge on the silver content of QAg = +10e (blue), +12e (green)(insets) and +11e (red) associated with the silver atom content. The best fit at a charge of +11e confirms that all of the attached silver atoms are cations, Ag+.
Results and Discussion
Detection of (Ag+)N-DNA products by ESI-MS
We use high-resolution, negative ion ESI-MS to determine the composition of complexes formed by Ag+ attachment to the homo-base oligomers. ESI-MS is a powerful tool for detecting non-covalently bound molecules to DNA in solution37 and for investigating solution binding stoichiometries to DNA38. Our use of high-resolution MS resolves the isotope peak envelopes (Fig. 1b), which enables determination of absolute composition, not just the ratio of silver cations per base (stoichiometry). This is important for unambiguous determination of strand dimerization by Ag+. By fitting the calculated isotope peak envelope to the MS data13,39,40, we find that all of the attached silver atoms are cationic (Ag+), as expected.
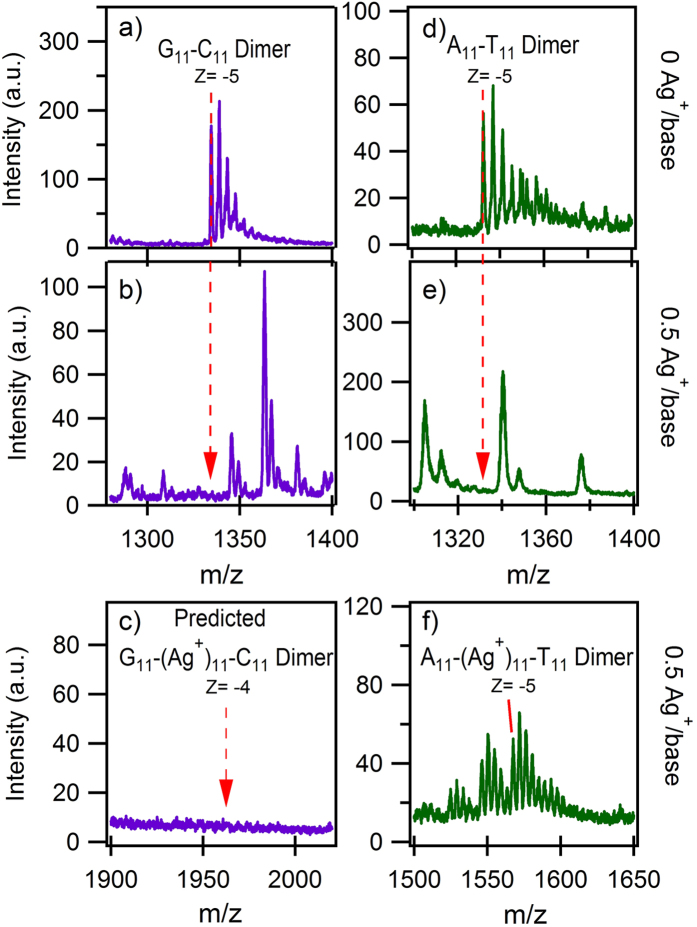
To investigate the possible disruption of WC pairing by Ag+, we combined C11 with G11, and A11 with T11, at 40 μM/strand in 10 mM ammonium acetate (pH 7). Mass spectra of the mixture of C11 and G11 strands (Fig. 2a) show the expected peaks for the WC-paired C11-G11 duplex, with additional peaks for the individual C11 and G11 strands (Fig. S1). After addition of 0.5 Ag+ per base, the C11-G11 duplexes entirely vanish from the mass spectra (Fig. 2b). New peaks appear for Ag+-decorated strand monomers, C11-(Ag+)N and G11-(Ag+)N; and for Ag+-paired homoduplexes, C11-(Ag+)N-C11 and G11-(Ag+)N-G11. Strikingly, there were no detectable C11-(Ag+)N-G11 heteroduplex or triplex products (Fig. 2c). If present, such products are at too low concentration to produce detectable ion currents, while the Ag+-paired homoduplexes are present in concentrations that result in high ion currents. We infer that the binding of Ag+ in G11 or C11 duplexes is more stable than canonical WC pairing of G11 to C11, and also more stable than C11-(Ag+)N-G11 pairing. For A11-T11 (Fig. 2d), WC-paired strands were undetectable after addition of 0.5 Ag+ per base (Fig. 2e), but heteroduplex A11-(Ag+)N-T11 products were detected (Fig. 2f) as well as A11-(Ag+)N. There were no detectable homoduplex A11-(Ag+)N-A11 or T11-(Ag+)N-T11 products.
Figure 2.
Effects of Ag+ on solutions of mixed C11 and G11 (a)-(c), and solutions of mixed A11 and T11 (d)-(f), at 40 μM per strand. (a) Mass spectra (MS) of the C11-G11 mixture at 0 Ag+/base and (b) 0.5 Ag+/base. Dashed line: m/z for the WC paired G11-C11 duplex, present in the absence of Ag+ (a) but undetectable after adding Ag+(b). (The peak envelope to higher m/z in (a), (d) and (f) is attachment of Na+ from residual salts). (c) No G11-(Ag+)N-C11 products were detected, exemplified by the absence of G11-(Ag+)11-C11 (expected at dashed line). (d) MS of A11-T11 mixture at 0 Ag+/base and (e) 0.5 Ag+/base. Dashed line: m/z for the WC paired product showing no detectable signal after adding Ag+. (f) A11-(Ag+)N-T11 products did form, exemplified by A11-(Ag+)11-T11 (dashed line). Additional, unlabeled peaks in (b) and (e) are various (Ag+)N-DNA products (see Fig. S1).
Measurements on every strand combination (Fig. 1a) found A11-(Ag+)N-T11 as the only detectable Ag+-bound heteroduplex. Apparently the favored mode of attachment of Ag+ to A11 is incompatible with homoduplex duplex formation under these solution conditions. Other, less stable Ag+-bridged heteroduplexes may exist but be reduced to undetectable levels by formation of C11-(Ag+)N-C11, G11-(Ag+)N-G11 and A11-(Ag+)N instead.
To better understand the patterns of Ag+ attachment, we investigated the products formed on all individual strands at Ag+/base ratios of 0.5, 0.75 and 1.0. Figure 3a-l show the integrated counts (IC) measured for the highest abundance charge state, zmax, of the strand monomer (zmax = −3 or −4) and duplex (zmax = −5 or −6) products. Figure 3m,n show full spectra for C11 and G11 at 1 Ag+/base (Figs. S2-S4 show all other full spectra). The IC provide a semi-quantitative comparison of the relative abundances of Ag+-DNA products (IC do not give quantitative product yields due to dependence of count rates on z and the possibility of unbinding events during ESI). The small shifts in Ag+/strand stoichiometry from 0.75 to 1.0 Ag+/base suggest that the silver binding to DNA is near saturation. For T11 (Fig. 3a–c), the dominant product at all Ag+/base was the bare strand. A11 formed a wider range of strand monomer products, with A11-(Ag+)3 dominant (Fig. 3d–f). It appears that A11 binds Ag+ more stably than T11, as expected at neutral pH41.
Figure 3.
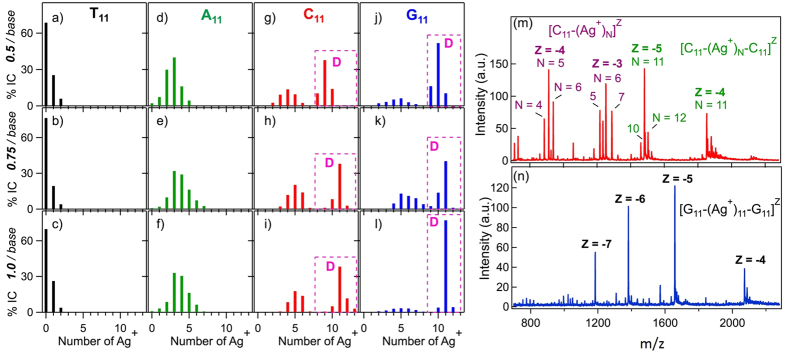
(a-l) Percentage of the total integrated counts (%IC) for each detected Ag+-bearing DNA product plotted versus number of attached Ag+. Top, middle and bottom rows: solutions with 0.5, 0.75 and 1 Ag+/base. Product yields are qualitatively different depending on base type. The boxed peaks labeled “D” are Ag+-paired duplexes, containing two copies of the strand. All other peaks correspond to strand monomers. Data are for the highest abundance charge state of each product. (a-c) T11 solutions exhibit the bare strand as the major product. (d-f) A11 shows a range of Ag+ attachment to strand monomers and no duplexes. (g-i) C11 exhibits strand monomer and duplex products. (j-l) G11 products are heavily biased to duplexes. The duplex with 11 bridging Ag+, G11-(Ag+)11-G11, is the overwhelming majority product at 1 Ag+/base (l). (m,n) Mass spectra of C11 and G11 solutions at 1 Ag+/base, corresponding to % IC plots i) and l). Major peaks are labeled.
For C11 (Fig. 3g–i,m), dominant products at the higher Ag+ concentrations were C11-(Ag+)11-C11, corresponding to Ag+ bridging each pair of C bases, and the strand monomer product C11(Ag+)5. The presence of the Ag+-bound C11 duplexes (dashed boxes labeled “D”, Fig. 3g–i) agrees with previous circular dichroism (CD) studies that suggested Ag+-induced dimerization of a C8 strand42.
Mass spectra for G11 (Fig. 3j–l,n) exhibit narrower product distributions than for C11. The fully Ag+-bridged duplex, G11-(Ag+)11-G11, dominates overwhelmingly at 1 Ag+/base (Fig. 3l,n). Small amounts of strand monomer G11-(Ag+)N products are still detectable, but in much reduced abundances relative to strand monomer Ag+-C11 products. Results were similar for CN and GN strands with N = 6 and 20 (Fig. S5).
The relative abundances in Fig. 3 reflect the partitioning of Ag+ between the solvent and the various Ag+-DNA products. The hydrated state of the Ag+ in solution is the same in all cases. If the hydrated Ag+ has lower free energy than the Ag+-DNA complexes, the bare DNA strand will be the major product. This is the case for T11, for which the bare strand comprises ~70% of all products. For A11 the bare strand is still detectable, but as only 1-2% of all products. This indicates that the hydrated Ag+ is no longer the thermodynamically favored state and consequently, that the complexes of Ag+ with A11 have lower free energy than the complexes of Ag+ with T11. For C11 and G11, the bare strand is undetectable, indicating a further lowering in free energy of the complexes of Ag+ with C11 and G11. The fully Ag+-bridged GN-(Ag+)N-GN duplex appears to be the most stable of all Ag+-GN complexes. The higher presence of duplex relative to monomer strand products for GN than for CN may indicate a reduced propensity for strand self-folding around Ag+ for the larger G base. To our knowledge this is the first study to detect Ag+-mediated pairing of guanine bases, a possibility that has not been investigated previously without the imposition of structural constraints from canonical WC pairing.
Thermal stability of silver mediated DNA homobase duplexes
To investigate thermal stability we carried out circular dichroism (CD) studies of C6 and G6. The solutions contained 1 Ag+/base (pH 7) at DNA concentrations of 17 μM. Silver mediated homoduplex products remain abundant at this concentration in mass spectra (Figure S6). In the absence of Ag+, the C6 solution (Fig 4a; black dashed line) shows the expected peak structure for predominantly unstructured cytosine oligonucleotides43,44. The peak structure of the G6 solution (Fig. 4b) indicates some presence of parallel G-quadruplex type structures45,46. With Ag+, the CD spectra are dramatically altered by base-Ag+ interactions (Fig. 4). Remarkably, the Ag+-imposed structure persists to the highest temperature we investigated, 90 °C (red curves). For comparison, the nearest-neighbor two-state model calculated melting temperature (mfold) for the C6-G6 duplex formed by canonical WC pairing is 16 °C for the low ionic conditions in Fig. 4. Apparently Ag+-bridging of G to G and C to C bases is much more stable than canonical WC pairing. This discovery of highly stable, silver mediated pairing of G bases, in addition to the previously known C-Ag+-C pairing, has promise to broaden the range of applications for DNA nanotechnology, which is currently limited by the low thermal melting temperatures imposed by Watson-Crick pairing, and may also impact development of disease therapeutics based on metal-DNA interactions.
Figure 4.
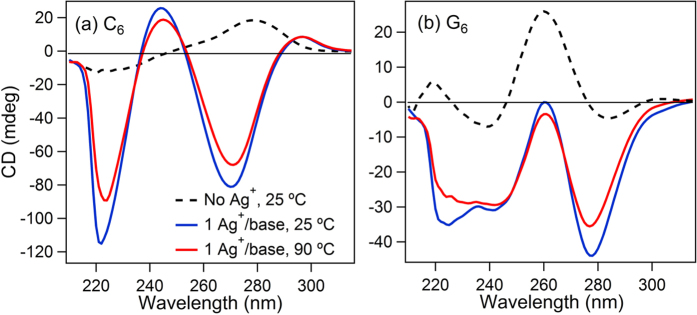
Circular dichroism (CD) spectra of C6 (a) and G6 (b). Dashed black lines: data for the bare strands at 25 °C. Blue lines: with 1 Ag+/base at 25 °C. Red lines: with 1 Ag+/base at 90 °C. The restructuring of the CD spectra upon addition of Ag+ reflects the reconfiguration of the DNA by incorporation of Ag+. Persistence of the dichroic peaks to the highest temperature accessible experimentally shows that this structural change is thermally robust.
Quantum chemical calculations of binding strengths and geometries
For theoretical investigation of the geometries and stabilities of Ag+-mediated base pairing, we consider two bases and a silver atom in vacuum with the charge of the entire system set to +1. To calculate the electronic ground state we used density functional theory with a real space basis and the projector-augmented wave method47. The exchange correlation functional PBE+TS09 was chosen to account for van der Waals dispersion interactions48,49. The grid spacing was 0.18 Å and the calculation was spin-polarized. Per atom, the electronic configuration of the valence electrons are H(1s1), O(2s22p4), C(2s22p2), N(2s22p3), Ag(4p64d105s1), including a scalar relativistic correction and a frozen core. We also report CAM-B3LYP binding energies (BE) calculated with the Gaussian code50 and a LANL2DZ/ECP basis set for the silver atom and 6-311+G(d,p) for the rest.
Starting from many initial geometries (SI), a global search is performed via force optimization using the Hessian matrix (BFGS algorithm in ASE) until the residual force is below 0.02 eV/Å. To verify that the relative ordering of silver-mediated BE is invariant with functional, CAM-B3LYP/6-311+G(d,p) was used. The same ordering as PBE+TS09 is obtained (SI).
Figure 5 shows the calculated ground state geometries for the Ag+-bridged bases. Bond lengths and dihedral angles are specified in the SI, along with the geometries of higher-lying structures. For homo-base pairs bridged by Ag+, the calculated BE, defined as the sum of the energy of fragments minus the energy of the complex, are 129.7, 126.2, 111.9, and 91.5 kcal/mol for G-Ag+-G, C-Ag+-C, A-Ag+-A and T-Ag+-T, respectively. These BE are all higher than those for attachment of Ag+ to the individual bases, calculated in prior work to be to be 77.08, 71.16, 60.30 and 51.19 kcal/mol for the most stable G-Ag+, C-Ag+, A-Ag+ and T-Ag+ complexes, respectively (in the most stable C-Ag+ and G-Ag+ configurations, the silver coordinates to two binding sites on one base51). With respect to prior work with PBE only functional, the BE is very close and ordering is the same. The pairing of bases by Ag+ and the attachment of Ag+ to individual bases show the same base-dependent ordering of BE, G ~ C > A > T.
Figure 5.
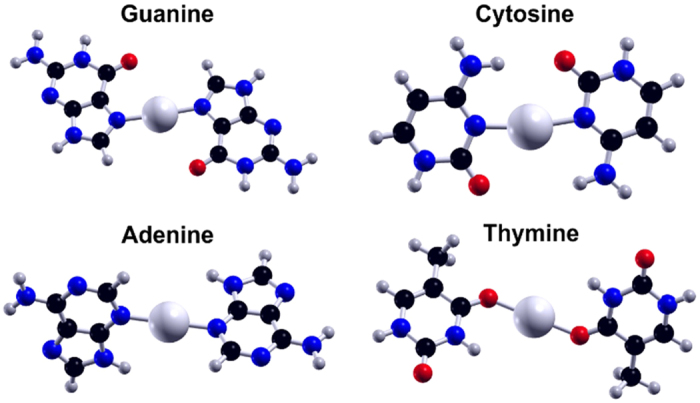
Calculated ground state geometries of Ag+-mediated homo-base pairs. Binding energies decrease in the order G > C > A > T (see text). G-Ag+-G and C-Ag+-C are planar, while A-Ag+-A and T-Ag+-T are non-planar.
In the absence of interfering steric factors, the higher BE for base-Ag+-base pairing than for individual base-Ag+ binding should result in higher yields of Ag+-bridged duplexes than strand monomer products with Ag+. However, experimentally we observe Ag+-bridged homoduplexes only for GN and CN (Fig. 3; dashed boxes labeled “D”). The calculated ground state structures are consistent with this experimental observation. The bases in the G-Ag+-G ground state are very nearly coplanar, with dihedral angle θ = 181.2°, and close to co-planar for C-Ag+-C (θ = 171.9°), as required to permit base-stacking interactions in Ag+-paired DNA strands containing multiple bases. In contrast, for A-Ag+-A and T-Ag+-T the ground state structures have highly non-coplanar bases with θ = 101.6° and 140°, respectively. Such a twisted geometry would disrupt base-stacking and sterically hinder the formation of Ag+-bridged strand homoduplexes of A and of T bases, consistent with the absence of these products in data (Fig. 3). We expect that the small (~10°) calculated rotation of the base planes in C-Ag+-C may account for the relatively high presence of strand monomer complexes, CN-(Ag+)m, in Fig. 3. Overall, the stoichiometric abundances of Ag+ bound to bases in Fig. 3 agree with the ordering of the calculated BE, G ~ C > A > T.
Our calculations find that in G-Ag+-G, Ag+ binds through the N7 atom of G, a site that does not engage in WC pairing (Fig. 5; see Fig. S7 for site numbering). This can account for the non-detection of G-Ag+-G pairing in prior studies of single G base mismatches within otherwise WC-paired duplexes, in which the canonically-paired surroundings restricted presentation of the mismatch bases28. In C-Ag+-C, we find that Ag+ bridges the N3 atom of the C bases. This site also participates in WC pairing, consistent with the observation of Ag+-pairing of C base mismatches in earlier work on WC duplexes28. For both G-Ag+-G and C-Ag+-C, hydrogen bonding helps stabilize the complexes. We note that G-Ag+-C has a comparable predicted BE (SI), with nearly co-planar bases. Presumably factors that are beyond the present calculations, specifically base stacking and solvation, cause the preferential formation of G11-(Ag+)11-G11 and C11-(Ag+)11-C11 rather than C11-(Ag+)11-G11 (Fig. 2).
Our calculations for binding energy (BE) contribute to the enthalpy only while unaccounted entropic costs of complex formation also contribute to relative solution abundances. For canonical WC duplexes, it is well-known that the entropic costs of duplex formation substantially compensate the enthalpic contributions to the free energy52,53; however the free energy and the enthalpy correlate, as evidenced by higher melting temperatures for canonical G-C rich duplexes compared to A-T rich duplexes. Because the BE of the Ag+-bridged bases are substantially higher than for canonical WC pairing (25.5 kcal/mol for G-C and 13.5 kcal-mol for A-T with PBE+TS09, respectively), we expect that the relative solution free energies of base-Ag+-base products will have the same ordering as the calculated BE if the calculated ground state geometries are consistent with formation of helices and the hydration free energies of the Ag+-bridged duplexes are similar to each other (as is the case for canonical duplexes of varying composition)54. For the homo-base, Ag+-bridged duplexes, relative experimental abundances (G ~ C > A > T) are consistent with the ordering of the calculated BE. The narrower product distribution for G-Ag+-G complexes (which are heavily dominated by G11-(Ag+)11-G11) than for C-Ag+-C complexes may reflect greater solvent stabilization associated with the higher solvent accessible area for the larger G base, and greater stacking tendencies.
For the heterobase Ag+ duplexes, the lowered symmetry in steric properties adds additional complexity. The data for mixed A11-T11 (Fig. 2 and S1b) show T-Ag+-A heterobase complexes but no T or A homobase duplexes. For T-Ag+-T, this is consistent with the lower calculated BE (91.5 kcal/mol) than for T-Ag+-A (102 kcal/mol); however for A-Ag+-A, the calculated BE (111.9 kcal/mol) is ~10 kcal/mol higher than for the hetero-base complex. We expect that the non-planar calculated structure of A-Ag+-A (SI) may be destabilizing relative to A-Ag+-T when stacking and hydration are included, accounting for the absence of A-Ag+-A in the data (A-Ag+-T is also calculated to be non-planar, but given the small size of the T base there may still be A stacking). In the case of the mixed C11-G11 (Fig. 2), the predicted BE of G-Ag+-C (130.95 kcal/mol) is similar to G-Ag+-G (129.72 kcal/mol) and C-Ag+-C (126.18 kcal/mol) but the heterobase complexes are not detectable in the data for mixed G11 and C11 strands. The differences in hydration and stacking free energies may be what is dictating the preferential formation of homobase complexes. We also note that previous experimental work has detected C-Ag+-G pairings31, but only within hydrogen-bonded triplexes stabilized by multiple inter-strand T-A-T triplet pairings, with facing C bases embedded in the T-rich regions and facing G bases in A-rich regions. The homobase oligomers studied here provide an entirely different context.
In conclusion, our experimental studies of homo-base strands of DNA have identified an unanticipated Ag+-mediated pairing of guanine bases in homobase oligonucleotides. This discovery of the highly stable, silver-mediated pairing of G bases expands the diversity of known nontoxic metal-mediated interactions with natural DNA bases. Our complementary calculations do not constrain Ag+ to attach at base sites that correspond to WC pairing. Instead the unrestricted binding configurations identify that the most stable Ag+ attachment in G-Ag+-G pairs is to base sites that do not engage in WC pairing. Our results suggest that in mixed base, double stranded DNA with long enough runs of consecutive C or G bases, addition of sufficient Ag+ should reconfigure WC pairing to more stable Ag+-mediated base pairing. This expansion of the known interactions between silver cations and DNA bases paves the way for more robust DNA nanotechnology and for potential applications in biomedical science.
Methods
DNA preparation
DNA oligonucleotides were synthesized by Integrated DNA Technologies with standard desalting. We solvent-exchanged the strands to remove residual salts. All solutions used RNase/DNase-free distilled water (Life Technologies). AgNO3 was analytical grade (Sigma-Aldrich). Before each analysis, strands with and without Ag+ were annealed to 90 °C for 5 minutes and allowed to cool slowly to ambient temperature.
Mass spectrometry experiments
Samples for ESI-MS used 80 μM single-stranded DNA concentrations in 10 mM ammonium acetate buffer. Solutions of DNA with Ag+ contained a ratio of 0.5, 0.75 or 1.0 AgNO3 per base, as detailed in the main text. The oligonucleotide solutions were injected into the MS (Waters QTOF2) at 10 μL/min in ESI negative mode with a 2 kV capillary voltage, 30 V cone voltage and 10 V collision energy. The signal was integrated over approximately 5 minutes.
Circular dichroism experiments
Samples for CD experiments used 17 μM single-stranded DNA concentrations in 7.5 mM MOPS buffer (pH = 7.0) containing approximately 2.5 mM Na+. All measurements were collected on an Aviv 202 circular dichrometer. For the measurements at 90 °C, the samples were heated at a rate of 3 °C/min and allowed 10 minutes to equilibrate at 90 °C before taking the full spectra. Blanks containing the appropriate concentration of buffer were collected before and after the samples, averaged and then subtracted from the sample spectra to correct for background signal.
Author Contributions
S.M.S. and E.G.G. conceived and designed the experiments. L.E.L. and O.L.A. conceived and designed the calculations, with initial suggestions from E.G.G. S.M.S. carried out the experiments and J.P. provided additional expertise in ESI-MS. S.M.S. and E.G.G. analyzed and interpreted data and wrote the experimental parts of the manuscript. L.E.L. and O.L.A. analyzed the computational information and wrote the theoretical parts of the manuscript. All authors reviewed the manuscript.
Additional Information
How to cite this article: Swasey, S. M. et al. Silver (I) as DNA glue: Ag+-mediated guanine pairing revealed by removing Watson-Crick constraints. Sci. Rep. 5, 10163; doi: 10.1038/srep10163 (2015).
Supplementary Material
Acknowledgments
This work was supported by NSF-CHE-1213895 and Academy of Finland projects 279240 and 251748. Computational resources were provided by Finland’s IT Center for Science (CSC). We thank Stacy Copp for helpful conversations.
References
- Rangnekar A. & LaBean T. H. Building DNA nanostructures for molecular computation, templated assembly, and biological applications. Acc. Chem. Res. 47, 1778–88 (2014). [DOI] [PubMed] [Google Scholar]
- Park K. S. & Park H. G. Technological applications arising from the interactions of DNA bases with metal ions. Curr. Opin. Biotechnol. 28C, 17–24 (2014). [DOI] [PubMed] [Google Scholar]
- Takezawa Y. & Shionoya M. Metal-Mediated DNA Base Pairing: Alternatives to Hydrogen-Bonded Watson-Crick Base Pairs. Acc. Chem. Res. 45, 2066–2076 (2012). [DOI] [PubMed] [Google Scholar]
- Scharf P. & Müller J. Nucleic Acids With Metal-Mediated Base Pairs and Their Applications. Chempluschem 78, 20–34 (2013). [Google Scholar]
- Berti L. & Burley G. A. Nucleic acid and nucleotide-mediated synthesis of inorganic nanoparticles. Nat. Nanotechnol. 3, 81–7 (2008). [DOI] [PubMed] [Google Scholar]
- Tanaka K. et al. Programmable self-assembly of metal ions inside artificial DNA duplexes. Nat. Nanotechnol. 1, 190–4 (2006). [DOI] [PubMed] [Google Scholar]
- Clever G. H. & Shionoya M. Metal–base pairing in DNA. Coord. Chem. Rev. 254, 2391–2402 (2010). [Google Scholar]
- Zimmermann N., Meggers E. & Schultz P. G. A novel silver(I)-mediated DNA base pair. J. Am. Chem. Soc. 124, 13684–5 (2002). [DOI] [PubMed] [Google Scholar]
- Johannsen S., Megger N., Böhme D., Sigel R. K. O. & Müller J. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nat. Chem. 2, 229–34 (2010). [DOI] [PubMed] [Google Scholar]
- Petty J. T., Zheng J., Hud N. V & Dickson R. M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 126, 5207–12 (2004). [DOI] [PubMed] [Google Scholar]
- Gwinn E. G., O’Neill P., Guerrero A. J., Bouwmeester D. & Fygenson D. K. Sequence-Dependent Fluorescence of DNA-Hosted Silver Nanoclusters. Adv. Mater. 20, 279–283 (2008). [Google Scholar]
- Yuan Z., Chen Y.-C., Li H.-W. & Chang H.-T. Fluorescent silver nanoclusters stabilized by DNA scaffolds. Chem. Commun. (2014). 10.1039/c4cc02981j. [DOI] [PubMed] [Google Scholar]
- Schultz D. et al. Evidence for rod-shaped DNA-stabilized silver nanocluster emitters. Adv. Mater. 25, 2797–803 (2013). [DOI] [PubMed] [Google Scholar]
- Ono A. et al. Specific interactions between silver(I) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 4825–7 (2008). 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- Pino G. A. Effect of Ag+ on the Excited-State Properties of a Gas-Phase (Cytosine) 2 Ag+ Complex: Electronic Transition and Estimated Lifetime. J. Phys. Chem. Lett. 6, 2295–2301 (2014). [DOI] [PubMed] [Google Scholar]
- Schultz D. & Gwinn E. Stabilization of fluorescent silver clusters by RNA homopolymers and their DNA analogs: C,G versus A,T(U) dichotomy. Chem. Commun. 47, 4715–7 (2011). [DOI] [PubMed] [Google Scholar]
- Menzer S., Hillgeris E. C. & Lippert B. Preparation, X-ray structure and solution behavior of [Ag(9-EtGH-N7)2]NO3·H2O (9-EtGH=9-ethylguanine). Inorganica Chim. Acta 210, 167–171 (1993). [Google Scholar]
- Fichtinger-Schepman A. M., van der Veer J. L., den Hartog J. H., Lohman P. H. & Reedijk J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification, and quantitation. Biochemistry 24, 707–13 (1985). [DOI] [PubMed] [Google Scholar]
- Garbutcheon-Singh K., Grant M., Harper B., Manohar M., Krause-Heuer A., Orkey N., Aldrich-Wright J., J. A.-W. Transition metal based anticancer drugs. Curr. Top. Med. Chem. 11, 521–542 (2011). [DOI] [PubMed] [Google Scholar]
- Lok C.-N. et al. Silver nanoparticles: partial oxidation and antibacterial activities. J. Biol. Inorg. Chem. 12, 527–34 (2007). [DOI] [PubMed] [Google Scholar]
- Izatt R. M., Christensen J. J. & Rytting J. H. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and and nucleotides. Chem. Rev. 5, 439–481 (1971). [DOI] [PubMed] [Google Scholar]
- DiRico D. E., Keller P. B. & Hartman K. A. The infrared spectrum and structure of the type I complex of silver and DNA. Nucleic Acids Res. 13, 251–260 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattagupta N. & Haven N. Solution structural studies of the Ag(I)-DNA complex. Nucleic Acids Res. 9, 2971–2985 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D. & Allen F. S. Electric Dichroism and Sedimentation Velocity Studies of DNA-Hg(II) and DNA-Ag(I) Complexes. Biochim. Biophys. Acta 610, 64–71 (1980). [DOI] [PubMed] [Google Scholar]
- Arakawa H., Neault J. F. & Tajmir-Riahi H. A. Silver(I) complexes with DNA and RNA studied by Fourier transform infrared spectroscopy and capillary electrophoresis. Biophys. J. 81, 1580–7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. H. & Davidson N. Spectrophotometric, Potentiometric, and Density Gradient Ultracentrifugation Studies of the Binding of Silver Ion by DNA. Biopolymers 4, 17–32 (1966). [Google Scholar]
- Nordén B., Matsuoka Y. & Kurucsev T. Nucleic acid-metal interactions: V. The effect of silver(I) on the structures of A- and B-DNA forms. Biopolymers 25, 1531–45 (1986). [DOI] [PubMed] [Google Scholar]
- Torigoe H. et al. Thermodynamic and structural properties of the specific binding between Ag+ ion and C:C mismatched base pair in duplex DNA to form C-Ag-C metal-mediated base pair. Biochimie 94, 2431–40 (2012). [DOI] [PubMed] [Google Scholar]
- Urata H., Yamaguchi E., Nakamura Y. & Wada S. Pyrimidine-pyrimidine base pairs stabilized by silver(I) ions. Chem. Commun. 47, 941–3 (2011). [DOI] [PubMed] [Google Scholar]
- Funai T. et al. Ag(I) ion mediated formation of a C-A mispair by DNA polymerases. Angew. Chemie 51, 6464–6 (2012). [DOI] [PubMed] [Google Scholar]
- Ihara T., Ishii T., Araki N., Wilson A. W. & Jyo A. Silver ion unusually stabilizes the structure of a parallel-motif DNA triplex. J. Am. Chem. Soc. 131, 3826–7 (2009). [DOI] [PubMed] [Google Scholar]
- Burda J. V, Šponer J., Leszczynski J. & Hobza P. Interaction of DNA Base Pairs with Various Metal Cations: Nonempirical ab Initio Calculations on Strcutures, Energies, and Nonadditivity of the Interaction. J. Phys. Chem. B 5647, 9670–9677 (1997). [Google Scholar]
- Brancolini G. & Di Felice R. Electronic properties of metal-modified DNA base pairs. J. Phys. Chem. B 112, 14281–90 (2008). [DOI] [PubMed] [Google Scholar]
- Marino T., Russo N., Toscano M. & Pavelka M. Theoretical investigation on DNA/RNA base pairs mediated by copper, silver, and gold cations. Dalt. Trans. 41, 1816–23 (2012). [DOI] [PubMed] [Google Scholar]
- Metal B., Li C. & Heyro J. V. Ab Initio Study of the Interaction of Guanine and Adenine with Various Mono- and Bivalent Metal Cations. J. Phys. Chem. 100, 7250–7255 (1996). [Google Scholar]
- Megger D. A., Fonseca Guerra C., Bickelhaupt F. M. & Müller J. Silver(I)-mediated Hoogsteen-type base pairs. J. Inorg. Biochem. 105, 1398–404 (2011). [DOI] [PubMed] [Google Scholar]
- Ganem B. Detection of Noncovalent Receptor-Ligand Complexes by Mass Spectrometry. J. Am. Chem. Soc. 113, 6294–6296 (1991). [Google Scholar]
- Rosu F., Gabelica V., Houssier C. & De Pauw E. Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Res. 30, e82 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszinowski K. & Ballweg K. A highly charged Ag(6)(4+) core in a DNA-encapsulated silver nanocluster. Chem. Eur. J. 16, 3285–90 (2010). [DOI] [PubMed] [Google Scholar]
- Copp S. M. et al. Magic Numbers in DNA-Stabilized Fluorescent Silver Clusters Lead to Magic Colors. J. Phys. Chem. Lett. 5, 959–963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. & Sastry M. Probing differential Ag+-nucleobase interactions with isothermal titration calorimetry (ITC): Towards patterned DNA metallization. Nanoscale 1, 122–7 (2009). [DOI] [PubMed] [Google Scholar]
- Megger D. A. & Muller J. Silver(I)-mediated cytosine self-pairing is preferred over hoogsteen-type base pairs with the artificial nucleobase 1,3-dideaza-6-nitropurine. Nucleosides. Nucleotides Nucleic Acids 29, 27–38 (2010). [DOI] [PubMed] [Google Scholar]
- Manzini G., Yathindra N. & Xodo L. E. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 22, 4634–4640 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson T., Pribylova M. & Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 278, 158–66 (2000). [DOI] [PubMed] [Google Scholar]
- Gray D. M. et al. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality 20, 431–40 (2008). [DOI] [PubMed] [Google Scholar]
- Paramasivan S., Rujan I. & Bolton P. H. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods 43, 324–31 (2007). [DOI] [PubMed] [Google Scholar]
- Enkovaara J. et al. Electronic structure calculations with GPAW: a real-space implementation of the projector augmented-wave method. J. Phys. Condens. Matter 22, 253202 (2010). [DOI] [PubMed] [Google Scholar]
- Perdew J. P., Burke K. & Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 77, 3865–3868 (1996). [DOI] [PubMed] [Google Scholar]
- Tkatchenko A. & Scheffler M. Accurate Molecular Van Der Waals Interactions from Ground-State Electron Density and Free-Atom Reference Data. Phys. Rev. Lett. 102, 073005 (2009). [DOI] [PubMed] [Google Scholar]
- Frisch M. J. et al. Gaussian 09, Revision D.01. (2009).
- Leal L. A. E. On the interaction between gold and silver metal atoms and DNA/RNA nucleobases - A comprehensive computational review of ground state properties. In Press 10.1515/ntrev-2012-0047. [DOI]
- Breslauer K. J., Frank R., Blöcker H. & Marky L. a. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. U.S.A. 83, 3746–3750 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushka J. & Goodman M. F. Enthalpy-Entropy Compensation in DNA Melting Thermodynamics. J. Biol. Chem. 270, 746–750 (1995). [DOI] [PubMed] [Google Scholar]
- Dixit S. B., Mezei M. & Beveridge D. L. Studies of base pair sequence effects on DNA solvation based on all-atom molecular dynamics simulations. J. Biosci. 37, 399–421 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.