Abstract
Background
Gastric cancer (GC) remains to have a poor prognosis via diverse process of cancer progression. Dihydropyrimidinase-like 3 (DPYSL3) is a cell adhesion molecule that has been reported to be involved in the metastatic process of tumor cells. The aim of this study was to identify a novel clinically-relevant biomarker of GC.
Methods
Expression analysis of DPYSL3 mRNA and protein levels was conducted using GC cell lines and 238 pairs of surgically resected gastric tissues. Correlations between expression status of DPYSL3 and clinicopathological parameters were investigated.
Results
DPYSL3 mRNA expression levels positively correlated with those of potentially interacting genes (VEGF, FAK and EZR) in GC cell lines. GC tissues from tumors with distant metastases (stage IV cancer) showed elevated expression levels of DPYSL3 mRNA. The DPYSL3 staining intensity in immunochemical staining was consistent with the mRNA expression patterns in GC tissues. High DPYSL3 mRNA expression in GCs was significantly associated with more malignant phenotypes and was an independent prognostic factor. Moreover, patients with high DPYSL3 mRNA expression had a significantly shorter recurrence free survival after curative resection. In subgroup analysis based on tumor histology, similar tendency was observed between patients with differentiated and undifferentiated GCs.
Conclusions
Expression status of DPYSL3 in GC tissues may represent a promising biomarker for the malignant behavior of GC.
Keywords: Gastric cancer, Dihydropyrimidinase-like 3, Expression, Prognosis, Biomarker
Background
Gastric cancer (GC) is the second leading cause of global cancer mortality, accounting for 700,000 deaths annually [1],[2]. More than 70% of countries worldwide have a mortality-to-incidence ratio greater than 0.8, suggesting that prevention of late presentation and modified treatment strategies are required to improve clinical outcomes [3]. In particular, distant metastases including peritoneal dissemination have been recognized as important prognostic determinants for GC patients [4],[5]. Identifying genes relevant to the malignant behavior of GC could aid clinicians in tailoring treatments by identifying high-risk patients and proposing novel molecular targets [6].
Recently, technological advances such as microarrays and next-generation sequencing have allowed for the exhaustive genomic characterization of malignancies, enhancing our understanding of cancer initiation and progression [7]-[9]. With these techniques, numerous genetic and epigenetic alterations relevant to gastric carcinogenesis and GC progression have been reported [10]. However, understanding the clinical significance of individual genes remains insufficient, despite the accumulating array data.
Dihydropyrimidinase-like 3 (DPYSL3) is a cell-adhesion molecule [11],[12] and actively expressed in normal tissues of cardiac myocytes, brain, pineal body, retina and smooth muscle, and moderately expressed in various tissues including gastric tissues [13]. DPYSL3 has been reported to be involved in the metastatic process of tumor cells [14],[15]. Gao et al. conducted expression and functional analyses of DPYSL3 in prostate cancer and found that DPYSL3 is a metastasis suppressor that is inversely associated with the expression of vascular endothelial growth factor (VEGF) [14]. In contrast, Kawahara et al. reported that DPYSL3 facilitates pancreatic cancer cell metastasis via a strong interaction with other cell adhesion factors, including ezrin (EZR), focal adhesion kinase (FAK) and c-SRC [15]. Thus, DPYSL3 has attracted attention as a metastatic modulator; however, the role of DPYSL3 expression in GC initiation and progression has not been investigated.
Here, we focused on DPYSL3 as a potential facilitator of malignant behavior in GC. The aim of this study was to evaluate the clinical significance of DPYSL3 expression in GC.
Material and methods
Ethics
This study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki‐Ethical Principles for Medical Research Involving Human Subjects and has been approved by the Institutional Review Board of Nagoya University, Japan. Written informed consent for usage of clinical samples and data, as required by the institutional review board, was obtained from all patients.
Sample collection
Ten GC cell lines (H111, KATOIII, MKN1, MKN28, MKN45, MKN74, NUGC2, NUGC3, NUGC4 and SC-6-LCK) were obtained from the American Type Culture Collection (Manassas, VA, USA) or Tohoku University, Japan and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum in 5% CO2 at 37°C. Primary GC tissues and corresponding normal adjacent tissues were collected from 238 patients undergoing gastric resection for GC without neoadjuvant therapy at Nagoya University Hospital between 2001 and 2012. The collected tissue samples were immediately flash-frozen in liquid nitrogen and stored at −80°C until RNA extraction. Approximately 5 mm2 was extracted from each tumor sample, avoiding necrotic tissue by gross observation and only samples confirmed to comprise more than 80% tumor components by H&E staining were included in this study. Corresponding normal adjacent gastric mucosa samples were obtained from the same patient and were collected > 5 cm from the tumor edge [16]. The specimens were classified histologically using the 7th edition of the Union for International Cancer Control (UICC) classification [17]. To evaluate whether the expression status of DPYSL3 differed by tumor histology, patients were categorized into two histological subtypes; differentiated (papillary, well differentiated and moderately differentiated adenocarcinoma) and undifferentiated (poorly differentiated adenocarcinoma, signet ring cell and mucinous carcinoma) tumors. Since 2006, adjuvant chemotherapy using S-1 (an oral fluorinated pyrimidine) has been applied to all UICC stage II–IV GC patients unless contraindicated by the patient’s condition [18],[19].
Expression analysis of DPYSL3 mRNA
DPYSL3 mRNA expression levels of 10 GC cell lines and 238 primary GC tissues and corresponding normal adjacent tissues were analyzed through quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) with an ABI StepOnePlus Real-Time PCR System (Perkin-Elmer, Applied Biosystems) as described previously [20],[21]. The primer sequences used in this study are listed in Additional file 1: Table S1. In clinical samples, mean expression level of DPYSL3 mRNA were compared between GC tissues and corresponding normal adjacent tissues. Additionally, expression level of DPYSL3 mRNA in GCs was compared with each patient subgroup based on UICC stage to investigate the oncological role of DPYSL3.
Expression of genes potentially interacting with DPYSL3
The expression status of VEGF, EZR, FAK and c-SRC, which have previously been reported to be genes that potentially interact with DPYSL3[14],[15], was evaluated in GC cell lines through qRT-PCR. Primers specific for VEGF, EZR, FAK and c-SRC are listed in Additional file 1: Table S1.
Immunochemical staining
DPYSL3 protein localization was determined by immunochemical staining using 54 representative formalin-fixed and paraffin-embedded sections of well-preserved GC tissue as described previously [22],[23] with a mouse monoclonal antibody against DPYSL3 (LS-C133161, LifeSpan BioSciences, Seattle, WA, USA) diluted 1:150 in antibody diluent (Dako, Glostrup, Denmark). Staining patterns were compared between GCs and the corresponding normal adjacent tissues, and the intensity of DPYSL3 protein expression was graded depending on the percentage of stained cells as follows: no staining, minimal (<20%); focal (20 – 60%); and diffuse (>60%) [24],[25]. To avoid subjectivity, the specimens were randomized and coded before analysis by two independent observers blinded to the status of the samples. Each observer evaluated all specimens at least twice to minimize intra-observer variation [26].
Evaluation of clinical significance of DPYSL3 expression
Patients were stratified into two groups divided by the median value of DPYSL3 mRNA expression level in cancerous tissues of the all analyzed patients; high DPYSL3 expression (higher than the median value) and low DPYSL3 expression (the median value or lower). Correlations between the pattern of DPYSL3 mRNA expression and clinicopathological parameters were evaluated. Outcome analyses including disease specific survival rate, recurrence free survival rate and multivariate analysis were performed in 169 patients who underwent curative surgery (i.e. stage I - III). Additionally, the prognostic impact of DPYSL3 mRNA expression was assessed in each patient subgroup based on tumor differentiation.
Statistical analyses
The relative mRNA expression levels (DPYSL3/GAPDH) between the two groups were analyzed using the Mann–Whitney U test. The strength of a correlation between two variables was assessed by the Spearman's rank correlation coefficient. The χ2 test was used to analyze the association between the expression status of DPYSL3 and clinicopathological parameters. Disease specific and recurrence free survival rates were calculated using the Kaplan–Meier method, and the difference in survival curves was analyzed using the log-rank test. We performed multivariable regression analysis to detect prognostic factors using the Cox proportional hazards model, and variables with a P value of < 0.05 were entered into the final model. All statistical analyses were performed using JMP 10 software (SAS Institute Inc., Cary, NC, USA). P < 0.05 was considered significant.
Results
Expression of DPYSL3 and potentially interacting genes in GC cell lines
The relative mRNA expression levels of DPYSL3 and its potential interacting genes in GC cell lines are shown in Figure 1A. There were large differences in mRNA expression level of DPYSL3 and other genes among GC cell lines. DPYSL3 expression levels positively correlated with those of VEGF, FAK and EZR, while no interaction was observed with c-SRC (Figure 1B).
Figure 1.
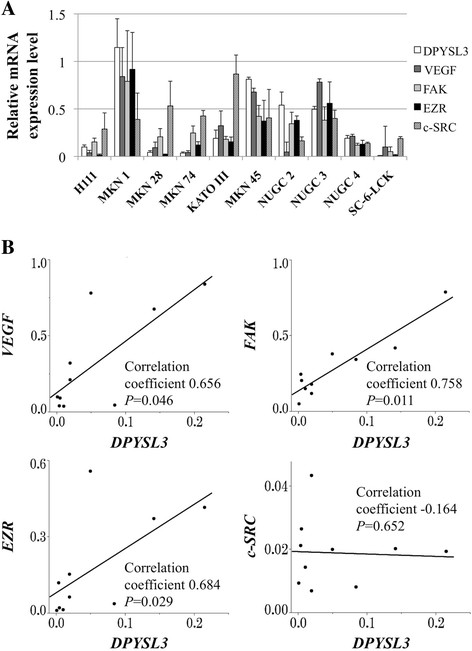
Expression profile of GC cell lines. (A) Expression status of DPYSL3 and potentially interacting genes in GC cell lines. Differential mRNA expression in GC cell lines was observed. Error bars indicated standard deviation among three biological replicates. (B) Correlative analysis between the mRNA expression levels of DPYSL3 and those of VEGF, FAK, EZR and c-SRC.
Patient characteristics
The patient ages ranged from 20 to 84 years (65.3 ± 11.7 years, mean ± standard deviation), and the male:female ratio was 179:59. Pathologically, 139 patients were diagnosed with undifferentiated GC and 99 with differentiated GC. According to the 7th edition of the UICC classification, 58, 40, 71 and 69 patients were in stages I, II, III and IV, respectively. Sixty of the 69 stage IV patients were diagnosed as stage IV due to positive peritoneal lavage cytology, localized peritoneal metastasis or distant lymph node metastasis including para-aortic lymph nodes. Eight patients in stage IV had synchronous liver metastasis one had lung metastasis, and they underwent gastrectomy with the purpose of controlling tumor bleeding or obstruction to the passage of food.
Expression status of DPYSL3 mRNA in 238 clinical GC samples
Elevation of the mean expression level of DPYSL3 mRNA was observed in GC tissues compared with the corresponding normal adjacent tissues (Figure 2A). When subdividing patients by UICC stage, DPYSL3 expression levels were significantly higher in stage IV patients than in stage I-III patients, indicating that DPYSL3 may promote distant metastasis (Figure 2B).
Figure 2.
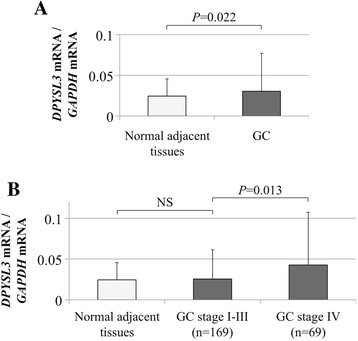
Expression status ofDPYSL3in clinical specimens. (A) GC tissues showed higher mean expression levels of DPYSL3 mRNA than corresponding normal adjacent tissues. (B) After subdividing patients according to UICC staging, GC tissues from patients with stage IV GC showed the highest DPYSL3 mRNA expression levels compared with corresponding normal adjacent tissues and those from patients with stage I-III GC. NS, not significant.
Detection of DPYSL3 protein
Representative cases with each staining grade in GC tissues are shown in Figure 3A. Diffuse staining of DPYSL3 protein in the cytoplasm of cancerous cells was observed, whereas cells in the adjacent normal adjacent tissue had less staining. Generally, the expression patterns of DPYSL3 protein detected by IHC were consistent with the qRT-PCR data. When grading the staining intensity of the cancerous cells, patient numbers 8, 19, 15 and 12 were categorized as no staining, minimal, focal and diffuse, respectively. A positive correlatin between the DPYSL3 staining grade and mRNA expression levels in GC tissues was confirmed (Figure 3B).
Figure 3.
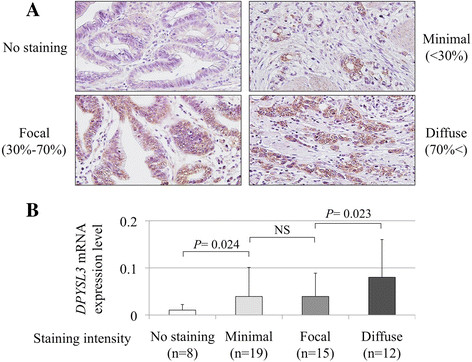
Detection ofDPYSL3protein. (A) Representative cases of each DPYSL3 staining intensity; no staining, minimal, focal and diffuse (400× magnification). (B) A positive correlation was observed between the expression level of DPYSL3 mRNA and the staining intensity in GC tissues.
Prognostic impact of expression status of DPYSL3 in gastric tissues
Correlations between expression status of DPYSL3 mRNA and clinicopathological parameters were evaluated in 238 patients with GC. High expression level of DPYSL3 mRNA in GCs was significantly associated with more aggressive phenotype including pT4, invasive growth, lymph node metastasis, positive peritoneal lavage cytology, and UICC stage IV, and but not tumor location (Table 1).
Table 1.
Association between expression level of DPYSL3 mRNA and clinicopathological parameters in 238 patients
| Variables | High DPYSL3 mRNA in GC tissue (n) | Low DPYSL3 mRNA in GC tissue (n) | P -value |
|---|---|---|---|
| Age | 0.793 | ||
| < 65 year | 51 | 49 | |
| ≥ 65 year | 68 | 70 | |
| Gender | 0.453 | ||
| Male | 87 | 92 | |
| Female | 32 | 27 | |
| Carcinoembryonic antigen (ng/ml) | 0.415 | ||
| ≤ 5 | 93 | 98 | |
| > 5 | 26 | 21 | |
| Carbohydrate antigen 19–9 (IU/ml) | 0.504 | ||
| ≤ 37 | 95 | 99 | |
| > 37 | 24 | 20 | |
| Tumor location | 0.769 | ||
| Entire | 12 | 8 | |
| Upper third | 24 | 27 | |
| Middle third | 37 | 35 | |
| Lower third | 46 | 49 | |
| Tumor size (mm) | 0.090 | ||
| < 50 | 48 | 61 | |
| ≥ 50 | 71 | 58 | |
| Tumor depth (UICC) | <0.001* | ||
| pT1-3 | 51 | 77 | |
| pT4 | 68 | 42 | |
| Histology | 0.098 | ||
| Papillary | 1 | 1 | |
| Well differentiated | 4 | 10 | |
| Moderately differentiated | 33 | 48 | |
| Poorly differentiated | 74 | 56 | |
| Signet ring cell | 5 | 2 | |
| Mucinous | 2 | 2 | |
| Differentiation | 0.006* | ||
| Differentiated | 39 | 60 | |
| Undifferentiated | 80 | 59 | |
| Lymphatic involvement | 0.016* | ||
| Absent | 11 | 24 | |
| Present | 108 | 95 | |
| Vessel invasion | 0.036* | ||
| Absent | 44 | 60 | |
| Present | 75 | 59 | |
| Infiltrative growth type | <0.001* | ||
| Invasive growth | 55 | 28 | |
| Expansive growth | 64 | 90 | |
| Lymph node metastasis | <0.001* | ||
| Absent | 32 | 57 | |
| Present | 87 | 62 | |
| Peritoneal lavage cytology | 0.001* | ||
| Negative | 84 | 104 | |
| Positive | 35 | 15 | |
| UICC stage | 0.032* | ||
| I - III | 77 | 92 | |
| IV | 42 | 27 |
Abbreviations: UICC Union for International Cancer Control. *Statistically significant (P < 0.05).
Next, outcome analysis was carried out for 169 patients who underwent curative surgery. Patients with high expression level of DPYSL3 mRNA in GCs (n = 84) were more likely to have a shorter disease specific survival than those with low expression level of DPYSL3 mRNA (n = 85; the 5-year survival rates were 61% and 77%, respectively, P = 0.010; Figure 4A). Multivariate analysis identified high expression level of DPYSL3 mRNA in GCs as an independent prognostic factor (Table 2). Moreover, high expression level of DPYSL3 mRNA in GCs was significantly associated with shortened recurrence free survival (the 2-year survival rates were 67% in high expression group and 84% in low expression group, respectively, P = 0.015; Figure 4B).
Figure 4.
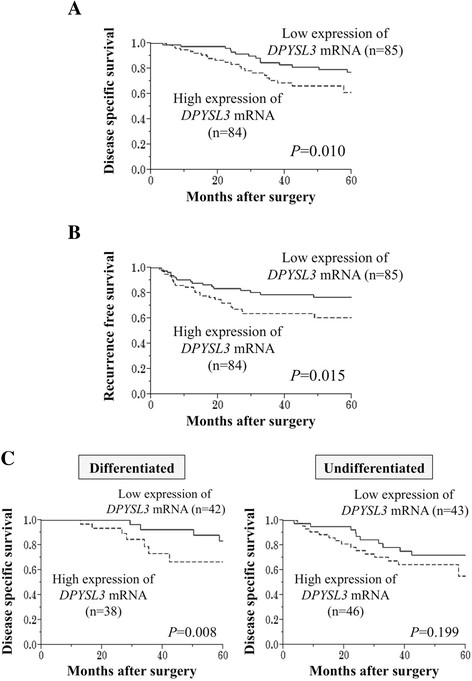
Prognostic impact ofDPYSL3mRNA expression in GC patients. (A) The high DPYSL3 mRNA expression group had significantly shorter disease specific survival than the low expression group. (B) Recurrence free survival was significantly shortened in the high DPYSL3 mRNA expression group. (C) Curves comparing the disease specific survival of patient groups categorized by expression level of DPYSL3 mRNA in each tumor differentiation.
Table 2.
Prognostic factors for disease specific survival in 169 patients who underwent curative surgery
| Variable | n | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P -value | Hazard ratio | 95% CI | P -value | ||
| Age (≥65) | 97 | 1.38 | 0.73 – 2.70 | 0.327 | |||
| Gender (male) | 128 | 1.27 | 0.60 – 2.49 | 0.517 | |||
| Tumor location (distal) | 107 | 0.42 | 0.22 – 0.78 | 0.006 | 0.53 | 0.27 – 1.05 | 0.067 |
| Carcinoembryonic antigen (>5 ng/ml) | 27 | 1.71 | 0.73 – 3.56 | 0.202 | |||
| Carbohydrate antigen 19–9 (>37 IU/ml) | 23 | 2.33 | 0.99 – 4.90 | 0.054 | |||
| Tumor size (≥50 mm) | 76 | 3.02 | 1.54 – 6.35 | 0.001 | 2.06 | 0.98 – 4.57 | 0.056 |
| Tumor depth (pT4, UICC) | 55 | 2.82 | 1.50 – 5.39 | 0.001 | 1.09 | 0.52 – 2.32 | 0.815 |
| Tumor differentiation (undifferentiated) | 89 | 1.79 | 0.93 – 3.60 | 0.081 | |||
| Lymphatic involvement | 137 | 5.70 | 1.74 – 35.2 | 0.002 | 1.12 | 0.14 – 6.12 | 0.905 |
| Vessel invasion | 83 | 4.10 | 2.02 – 9.20 | <0.001 | 2.93 | 1.31 – 7.52 | 0.008* |
| Invasive growth | 41 | 2.51 | 1.31 – 4.73 | 0.006 | 1.39 | 0.64 – 3.00 | 0.404 |
| Lymph node metastasis | 86 | 8.70 | 3.71 – 25.5 | <0.001 | 4.01 | 1.40 – 14.6 | 0.008* |
| Expression of DPYSL-3 mRNA (high) | 84 | 2.36 | 1.22 – 4.72 | 0.010 | 2.22 | 1.14 – 4.49 | 0.019* |
*Statistically significant in multivariable analysis. GC, gastric cancer; CI, confidence interval; UICC, Union for International Cancer Control.
Subgroup analysis based on tumor differentiation
The prognostic impact of DPYSL3 expression was evaluated in each patients subgroups classified by tumor differentiation. Although statistically significant difference was exhibited only in patients with differentiated GCs, similar tendency was observed between survival curves of patients with differentiated and undifferentiated GCs.
Discussion
DPYSL3, located on 5q32 and encoding a 62-kDa protein [11], has been gaining attention as a metastasis modulator [14],[15]. Interestingly, conflicting results have been reported in prostate and pancreatic cancer, implying that DPYSL3 has a diversity of functions among malignancies. In prostate cancer, the expression of both DPYSL3 mRNA and protein was inversely associated with lymph node metastasis and VEGF expression, and forced DPYSL3 expression in cell lines decreased metastasis in a mouse metastatic model [14]. Alternatively, DPYSL3 promoted adhesion and migration in pancreatic cancer cells in vitro as well as metastasis in vivo via activation of other cell adhesion genes [15]. In this study, the association between DPYSL3 expression and malignant behavior of GC was investigated.
First, the transcriptional status of DPYSL3 and potential interacting genes were evaluated in GC cell lines. The expression of DPYSL3 mRNA was heterogeneous in each GC cell line, and it showed a significant correlation with known tumor promoting factors (VEGF, FAK and EZR) [27]-[29]. These results indicated that DPYSL3 may be associated with the activation of cancer cell proliferation and metastasis, as is the case with pancreatic cancer. Following the experiments with GC cell lines, an expression analysis using surgical gastric tissues was conducted and provided important findings. DPYSL3 expression levels in patients with distant metastasis (stage IV) were significantly elevated compared with patients with localized GC (stage I-III), implying that DPYSL3 upregulation was an important determinant step in the GC progression. Consequently, high expression level of DPYSL3 mRNA in GC tissues was strongly associated with shortened survival and was identified as an independent prognostic factor. These results indicated that DPYSL3 upregulation may contribute to GC progression rather than carcinogenesis. Because DPYSL3 has been reported to play a role in cell adhesion and be a metastatic modulator, the correlations between expression status of DPYSL3 and metastasis were analyzed. Patients with upregulated DPYSL3 had a significantly higher prevalence of lymph node metastasis, overall distant metastasis and peritoneal dissemination, indicating that DPYSL3 is a metastasis facilitator of GC, and high expression of DPYSL3 may predict the metastatic behavior associated with an invasive GC phenotype. Data from the expression analysis of interacting genes also support the hypothesis that DPYSL3 has an oncogenic function in GC as with pancreatic cancer [15].
Because GC is considered a biologically heterogeneous disease, and genetic backgrounds can differ according to GC subtype [30]-[33], a subgroup analysis was conducted. Expression status of DPYSL3 was similar across tumor location (entire, upper third, middle third and lower third). In addition, patients with high expression level of DPYSL3 mRNA in GCs tended to have a shorter survival both in patient groups of differentiated and undifferentiated GCs. These findings indicated that DPYSL3 acts similarly in all types of GC.
Although further investigation will be necessary to clarify the underlying molecular mechanism that connects DPYSL3 upregulation directly to malignant behavior, our findings may offer valuable insight for the specific management of GC patients. Taken together, DPYSL3 can be used in clinical practice as follows: 1) DPYSL3 expression levels in the biopsy tissue obtained using endoscopic surveillance samples may identify patients in need of intensive systemic treatment; and 2) DPYSL3 expression levels in the surgical specimen may be useful for the prediction of an adverse prognosis, also aiding in determining an appropriate therapeutic strategy.
Conclusion
DPYSL3 acts as a facilitator of malignant behavior of GC. High expression level of DPYSL3 in GC tissues may represent a promising biomarker for the malignant behavior of GC.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MK, HO, SH, DS, HT and RH performed experiments and data analysis. DK, CT, SY, TF, GN, HS, MK, MF and YK collected cases and clinical data. MK and SN conceived and designed the study, and prepared the initial manuscript. YK supervised the project. All authors contributed to the final manuscript. All authors read and approved the final manuscript.
Additional file
Supplementary Material
Primers and annealing temperature.
Contributor Information
Mitsuro Kanda, Email: m-kanda@med.nagoya-u.ac.jp.
Shuji Nomoto, Email: snomoto@med.nagoya-u.ac.jp.
Hisaharu Oya, Email: hohya@med.nagoya-u.ac.jp.
Dai Shimizu, Email: d-shimizu@med.nagoya-u.ac.jp.
Hideki Takami, Email: takamihideki@med.nagoya-u.ac.jp.
Soki Hibino, Email: sokihibino@med.nagoya-u.ac.jp.
Ryoji Hashimoto, Email: hashi64@med.nagoya-u.ac.jp.
Daisuke Kobayashi, Email: kobadai@med.nagoya-u.ac.jp.
Chie Tanaka, Email: chtanaka@med.nagoya-u.ac.jp.
Suguru Yamada, Email: suguru@med.nagoya-u.ac.jp.
Tsutomu Fujii, Email: fjt@med.nagoya-u.ac.jp.
Goro Nakayama, Email: goro@med.nagoya-u.ac.jp.
Hiroyuki Sugimoto, Email: sugi@med.nagoya-u.ac.jp.
Masahiko Koike, Email: dockoike@med.nagoya-u.ac.jp.
Michitaka Fujiwara, Email: mfuji@med.nagoya-u.ac.jp.
Yasuhiro Kodera, Email: ykodera@med.nagoya-u.ac.jp.
Acknowledgements
The authors thank Naoki Iwata for his support to collect clinical data.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan IB, Gopalakrishnan V, Ooi CH, Lee J, Qin L, Wu J, Lee M, Rha SY, Huang D, Liem N, Yeoh KG, Yong WP, Teh BT, Tan P. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut. 2013;62:22–33. doi: 10.1136/gutjnl-2011-301113. [DOI] [PubMed] [Google Scholar]
- Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, Park SR, Fujii M, Kang YK, Chen LT. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14:e535–547. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–2909. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Kanda M, Nomoto S, Okamura Y, Hayashi M, Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S, Nakao A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog. 2011;50:571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- Gonzalez CA, Agudo A. Carcinogenesis, prevention and early detection of gastric cancer: where we are and where we should go. Int J Cancer. 2012;130:745–753. doi: 10.1002/ijc.26430. [DOI] [PubMed] [Google Scholar]
- Janjigian YY, Kelsen DP. Genomic dysregulation in gastric tumors. J Surg Oncol. 2013;107:237–242. doi: 10.1002/jso.23263. [DOI] [PubMed] [Google Scholar]
- Jang BG, Kim WH. Molecular pathology of gastric carcinoma. Pathobiology. 2011;78:302–310. doi: 10.1159/000321703. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Stauffer JK, Walker RL, Meltzer P, Thiele CJ. Structure and promoter analysis of the human unc-33-like phosphoprotein gene. E-box required for maximal expression in neuroblastoma and myoblasts. J Biol Chem. 2000;275:16560–16568. doi: 10.1074/jbc.M001312200. [DOI] [PubMed] [Google Scholar]
- http://biogps.org/ http://biogps.org/ BioGPS..
- Gao X, Pang J, Li LY, Liu WP, Di JM, Sun QP, Fang YQ, Liu XP, Pu XY, He D, Li MT, Su ZL, Li BY. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene. 2010;29:4555–4566. doi: 10.1038/onc.2010.213. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Hotta N, Ozawa Y, Kato S, Kano K, Yokoyama Y, Nagino M, Takahashi T, Yanagisawa K. Quantitative proteomic profiling identifies DPYSL3 as pancreatic ductal adenocarcinoma-associated molecule that regulates cell adhesion and migration by stabilization of focal adhesion complex. PLoS One. 2013;8:e79654. doi: 10.1371/journal.pone.0079654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda M, Nomoto S, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Correlations of the expression of vascular endothelial growth factor B and its isoforms in hepatocellular carcinoma with clinico-pathological parameters. J Surg Oncol. 2008;98:190–196. doi: 10.1002/jso.21095. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Gospodarowicz MK, Wittekind C. International Union Against Cancer, TNM Classification of Malignant Tumors. Wiley-Blackwell, New York; 2009. [Google Scholar]
- Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi Y, Imamura H, Higashino M, Yamamura Y, Kurita A, Arai K. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810–1820. doi: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T, Ohashi Y. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29:4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477–483. doi: 10.3892/ijo_00000448. [DOI] [PubMed] [Google Scholar]
- Inokawa Y, Nomoto S, Hishida M, Hayashi M, Kanda M, Nishikawa Y, Takeda S, Sugimoto H, Fujii T, Yamada S, Kodera Y. Detection of doublecortin domain-containing 2 (DCDC2), a new candidate tumor suppressor gene of hepatocellular carcinoma, by triple combination array analysis. J Exp Clin Cancer Res. 2013;32:65. doi: 10.1186/1756-9966-32-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu D, Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Suenaga M, Inokawa Y, Hishida M, Takano N, Nishikawa Y, Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Identification of intragenic methylation in the TUSC1 gene as a novel prognostic marker of hepatocellular carcinoma. Oncol Rep. 2014;31:1305–1313. doi: 10.3892/or.2013.2939. [DOI] [PubMed] [Google Scholar]
- Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, Asai M, Fujii T, Sugimoto H, Kodera Y. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- Loupy A, Hill GS, Suberbielle C, Charron D, Anglicheau D, Zuber J, Timsit MO, Duong JP, Bruneval P, Vernerey D, Empana JP, Jouven X, Nochy D, Legendre CH. Significance of C4d Banff scores in early protocol biopsies of kidney transplant recipients with preformed donor-specific antibodies (DSA) Am J Transplant. 2011;11:56–65. doi: 10.1111/j.1600-6143.2010.03364.x. [DOI] [PubMed] [Google Scholar]
- Kanda M, Shimizu D, Nomoto S, Hibino S, Oya H, Takami H, Kobayashi D, Yamada S, Inokawa Y, Tanaka C, Fujii T, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Clinical significance of expression and epigenetic profiling of TUSC1 in gastric cancer. J Surg Oncol. 2014;110:136–144. doi: 10.1002/jso.23614. [DOI] [PubMed] [Google Scholar]
- Hibino S, Kanda M, Oya H, Takami H, Shimizu D, Nomoto S, Hishida M, Niwa Y, Koike M, Yamada S, Nishikawa Y, Asai M, Nakayama G, Fujii T, Sugimoto H, Fujiwara M, Kodera Y. Reduced expression of DENND2D through promoter hypermethylation is an adverse prognostic factor in squamous cell carcinoma of the esophagus. Oncol Rep. 2014;31:693–700. doi: 10.3892/or.2013.2901. [DOI] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava J, Elliott BE, Louvard D, Arpin M. Src-dependent ezrin phosphorylation in adhesion-mediated signaling. Mol Biol Cell. 2005;16:1481–1490. doi: 10.1091/mbc.E04-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T, Watanabe A, Sawada H, Yamada Y, Tatsumi M, Fujimoto H, Emoto K, Nakano H. Characteristics and clinical outcome of proximal-third gastric cancer. J Am Coll Surg. 1998;187:352–357. doi: 10.1016/S1072-7515(98)00191-4. [DOI] [PubMed] [Google Scholar]
- Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano A. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. J Cell Physiol. 2007;211:287–295. doi: 10.1002/jcp.20982. [DOI] [PubMed] [Google Scholar]
- Kanda M, Shimizu D, Nomoto S, Takami H, Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, Tanaka C, Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Prognostic impact of expression and methylation status of DENN/MADD domain-containing protein 2D in gastric cancer. Gastric Cancer. 2014;ᅟ:ᅟ. doi: 10.1007/s10120-014-0372-0. Epub ahead of print, PubMed PMID: 24695972. [DOI] [PubMed] [Google Scholar]
- Wang YY, Li L, Zhao ZS, Wang YX, Ye ZY, Tao HQ. L1 and epithelial cell adhesion molecules associated with gastric cancer progression and prognosis in examination of specimens from 601 patients. J Exp Clin Cancer Res. 2013;32:66. doi: 10.1186/1756-9966-32-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and annealing temperature.


