Abstract
Low efficiency and significant toxicity are the main obstacles to successful gene delivery. We have developed a cationic reduction-responsive vector based on a disulfide cross-linked stearylated polyarginine peptide modified with histidine (C-SHR) for DNA delivery. The structure of the C-SHR was characterized, and the in vitro and in vivo transfection efficiency and cytotoxicity of C-SHR/plasmid DNA complexes were examined. Compared with non-cross-linked stearylated polyarginine peptide (SHR), C-SHR increased the intracellular uptake and dissociation behavior of the complexes. In addition, the gene transfection efficiency of C-SHR/plasmid DNA complexes in HEK293 and HeLa cells was improved and was comparable with that of bPEI-25K/plasmid DNA complexes, and the cytotoxicity of C-SHR was significantly less than that of bPEI-25K. Importantly, the in vivo gene transfection efficiency of C-SHR/plasmid DNA complexes was five fold higher than that of SHR/plasmid DNA complexes, suggesting that C-SHR is an efficient non-viral vector for DNA delivery.
Keywords: polyarginine, histidine, stearyl, reduction-responsive, peptide, DNA delivery
Introduction
Gene therapy offers new hope for curing human diseases, including cancer.1,2 Tremendous efforts have been made to develop viral and non-viral vectors for gene delivery.3 Although viral vectors provide efficient gene delivery, their immunogenicity, potential for recombination in vivo, and virus-induced chromosomal integration have become obstacles to their further application.4,5 Non-viral vectors do not come with these concerns, but have relatively low gene delivery efficiency and high cytotoxicity.6 To improve the efficiency of gene delivery, more effort should be made to improve cell uptake, endosomal escape, and cytoplasmic DNA dissociation when developing non-viral vectors.
In this study, we developed a disulfide cross-linked stearylated polyarginine peptide modified with histidine (C-SHR) for delivery of DNA. Polyarginine, as a cell-penetrating peptide, can help enhance intracellular uptake through interactions of the guanidine group with the sulfate, carbonate, and phosphate of the proteoglycans on the cellular surface.7,8 In particular, its ability to condense DNA with the positive charge and control DNA release make it possible to use polyarginine as an intracellular delivery vector for plasmid DNA (pDNA).9 In addition, histidine has a proton sponge effect, thus making it possible to enhance the endosomal escape rate by increasing the pH of the endosomes, leading to osmotic swelling and bursting.10,11 It was reported that stearylation could improve intracellular uptake and endosomal escape.12–14 A stearyl moiety could increase the buffering capacity of histidine, thus increasing the efficiency of transfection by the hydrophobic moiety of stearylation on the N-terminus of the peptide and facilitating absorption of the complex through the membrane.15,16 Additionally, the disulfide bonds in C-SHR could be cleaved rapidly by thiol-disulfide exchange reactions with intracellular reducing molecules, especially with glutathione.17–19 The glutathione concentration in the cytosol of tumor cells is several times higher than that of normal cells. Thus, reduction-responsive vectors are available for tumor-specific drug delivery. We therefore hypothesized that C-SHR could be used as a DNA delivery vector to increase DNA binding capacity, enhance cell uptake, improve endosomal escape, and promote DNA release into the cytoplasm and nucleus, thus improving gene transfection efficiency (Figure 9). For this purpose, we first synthesized and characterized C-SHR, examined the cytotoxicity and transfection efficiency of C-SHR/pDNA in HEK293 and HeLa cells, and finally evaluated the gene transfection efficiency in vivo.
Figure 9.
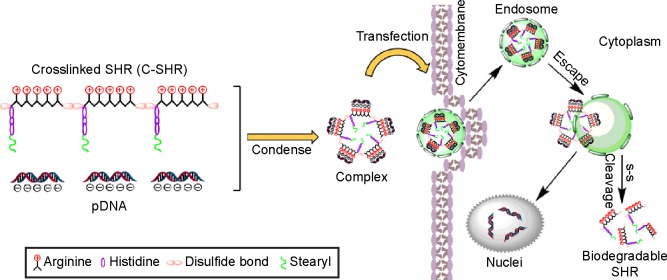
Illustration of the structures, condense process, and intracellular uptake of C-SHR/pDNA complexes.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; pDNA, plasmid DNA.
Materials and methods
Materials
Materials used in this study were l-histidine hydrochloride, l-arginine, stearic acid, and hydrogen peroxide (30% w/v; Sangon Biotech, Shanghai, People’s Republic of China); polyethylenimine (PEI, branched, molecular weight 25 kDa), Lipofectamine™ 2000, l-buthionine-sulfoximine, 4′,6-diamidino-2-phenylindole dihydrochloride, l-cysteine hydrochloride monohydrate, ethidium bromide, dithiothreitol (DTT, Sigma-Aldrich, St Louis, MO, USA); a luciferase assay kit (Promega, Madison, WI, USA); pDNA (pGL3 and pEGFP) (Shanghai Innovation Biotechnology Co Ltd, Shanghai, People’s Republic of China); enhanced bicinchoninic acid protein assay kit (Beyotime, Nanjing, People’s Republic of China); Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum, YOYO-1 (Y3601) and penicillin–streptomycin solution 5 kU/mL (Life Technologies, Carlsbad, CA, USA); a Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies Inc, Nanjing, People’s Republic of China); and HEK293 cells and HeLa cells (Cell Culture Center of the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences, Shanghai, People’s Republic of China). All other reagents were of analytical grade. All animal experiments were performed in accordance with the ethics and regulations of animal experiments of Second Military Medical University (Shanghai, People’s Republic of China).
Cell lines and cell culture
HeLa cells (a human cervical carcinoma cell line, American Type Culture Collection, Manassas, VA, USA) and HEK293 cells (a human embryonic kidney cell line, American Type Culture Collection) were grown in DMEM supplemented with 10% fetal bovine serum and antibiotics (100 U/mL penicillin and streptomycin) at 37°C under a 5% CO2 atmosphere.
Synthesis and characterization of polymers
Firstly, histidine-arginine peptide (H3CR5C, HR) was synthesized using the F-moc-solid phase peptide synthesis method.20 Secondly, we coupled stearyl with the N-terminal of the HR to obtain stearyl HR (SHR) using the same synthesis method as for HR. The products were then purified by reverse-phase high-performance liquid chromatography. Next, SHR (50 mg) was mixed with various molar ratios of l-cysteine hydrochloride monohydrate (Table 1) and dissolved in 9.5 mL of phosphate-buffered saline (PBS, pH 7.4). H2O2 solution (1%, 0.5 mL) was added into the mixed solutions as a cross-linked catalyst under stirring at room temperature for 12 hours. Finally, the solution was dialyzed in distilled water for 6 hours using a dialysis membrane (molecular weight cutoff 3500, Pierce Co, Rockford, IL, USA), followed by lyophilization to obtain C-SHR with different molecular weights (C-SHR-1, C-SHR-2, C-SHR-3, C-SHR-4). The molecular weights of SHR and C-SHR were determined by HLC-8220 gel permeation chromatography (TOSOH Corporation, Tokyo, Japan). The synthesized polymers were also characterized by 1H-NMR at 600 MHz (Varian Inc, Palo Alto, CA, USA) in DMF-D7.
Table 1.
Synthetic conditions and corresponding molecular weight of C-SHR
| Polymers | SHR (mg) | Cysteine (mg) | Feed ratioa | Mw (kDa)b |
|---|---|---|---|---|
| SHR | – | – | – | 1.683 |
| C-SHR-1 | 50 | 1.44 | 2.5 | 8.9 |
| C-SHR-2 | 50 | 0.72 | 5.0 | 15 |
| C-SHR-3 | 50 | 0.36 | 10.0 | 29 |
| C-SHR-4 | 50 | 0.24 | 15.0 | 39 |
Notes:
Molar ratio of SHR to cysteine.
Data obtained by gel permeation chromatography.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; Mw, molecular weight.
Preparation and characterization of complexes
pGL3 was used as a model plasmid. C-SHR was mixed with pGL3 plasmid (60 μL per 2 μg DNA) at an N/P ratio of 0.5, 1, 2.5, 5, 10, and 15 in PBS (pH 7.4). The mixture was vortexed and incubated for 1 hour at room temperature before use. The particle size and zeta potential of the complex were further determined by dynamic light scattering (Zetasizer Nano ZS90, Malvern Instruments, Malvern, USA). The morphology of the C-SHR-3/pGL3 complex at an N/P ratio of 10 was examined under a transmission electron microscope (Hitachi, Tokyo, Japan) at an acceleration voltage of 75 kV.
Agarose gel electrophoresis
The ability of the polymers to condense pGL3 plasmid was determined by agarose gel electrophoresis. The C-SHR/pGL3 complex was prepared at an N/P ratio of 0–2. After 30 minutes of incubation, the complex was electrophoresed at 100 V for 15 minutes on agarose gels (1.0%, w/v), then stained with Gelred™ fluorescent dye and Tris-acetate-EDTA buffer for 1 hour at room temperature. To compare the stability of the complexes under reductive environments, C-SHR-3/pGL3 and SHR/pGL3 complexes at an N/P ratio of 5 in the presence of heparin or at an N/P ratio of 0–2 in the presence of DTT (3 mg/mL) were analyzed using agarose gel electrophoresis. The location of pGL3 bands was visualized under ultraviolet light.
Cellular uptake assay
The cellular uptake efficiency of C-SHR/pGL3 complexes was investigated using flow cytometry. HEK293 cells were seeded in 12-well plates at a density of 3×105 cells per well and incubated for 24 hours. Before preparation of the C-SHR/pGL3 complexes, pGL3 was labeled with YOYO-1 for 10 minutes. After replacing the culture medium, 50 μL of PBS solution containing C-SHR/pGL3-YOYO-1 complexes was added to HEK293 cells. After 3 hours of incubation, the cells were trypsinized, centrifuged, washed, and resuspended in PBS. The fluorescent intensity of cells was analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA). bPEI-25k/pGL3-YOYO-1, C-HR-3/pGL3-YOYO-1, and SHR/pGL3-YOYO-1 complexes were used as controls. The experiment was replicated three times.
Confocal laser scanning microscopic observation
For study by confocal laser scanning microscopy, HEK293 cells were seeded in a 24-well plate with a glass cover slip at the bottom at a density of 1×105 cells per well and incubated for 24 hours. The culture medium was then replaced with DMEM containing C-SHR/pGL3-YOYO-1 complexes. After 3 hours of transfection, the medium was replaced and the culture was expanded for an additional 1 hour or 13 hours. The cells were then rinsed three times with PBS, fixed in 4% paraformaldehyde, treated with 4′,6-diamidino-2-phenylindole dihydrochloride for staining of the nucleus, sealed with mounting medium, and imaged with a confocal laser scanning microscope (Olympus, Tokyo, Japan). bPEI-25K/pGL3-YOYO-1, C-HR-3/pGL3-YOYO-1, and SHR/pGL3-YOYO-1 complexes were used as controls.
Gene transfection assay
The gene transfection efficiency of the C-SHR/pDNA complexes was evaluated in HEK293 and HeLa cells using pEGFP and pGL3 as report genes. Briefly, HEK293 and HeLa cells were seeded in 24-well plates at a density of 2×105 and 6×104 cells per well, respectively. After 24 hours of incubation, the culture medium was removed. Cells were added with fresh culture medium containing C-SHR/pEGFP complexes with different N/P ratios at 37°C and incubated for another 3 hours. The culture medium was then replaced and cell culture was expanded. After an additional 21 hours of incubation, expression of EGFP in the transfected cells was observed by fluorescent microscopy (Olympus) and quantitated by flow cytometry. SHR/pEGFP, C-HR-3/pEGFP, and bPEI-25K/pEGFP complexes were used as controls. The experiment was repeated three times.
The luciferase assay for C-SHR/pGL3 complexes in cells was conducted according to the manufacturer’s protocol (Promega). The relative luciferase light units were normalized to the amount of cellular protein, which was determined using a bicinchoninic acid protein assay kit (Beyotime). The luciferase gene expression level was expressed as relative luciferase light units per milligram of protein. bPEI-25K/pGL3, SHR/pGL3, and C-HR-3/pGL3 complexes were used as controls. The experiment was repeated three times.
To assess whether the disulfide bond could contribute to the high gene transfection efficiency of the C-SHR/pEGFP complexes, HEK293 cells were seeded in 24-well plates at a density of 2×105 cells per well for 24 hours and then incubated in the presence of different concentrations of buthionine sulfoximine (a glutathione depleting agent) for 6 hours prior to transfection. The C-SHR-3/pEGFP and SHR/pEGFP complexes were then added at an N/P ratio of 10. Three hours later, the culture medium was replaced and the cell culture was expanded for 21 hours. EGFP expression in the transfected cells was observed by fluorescent microscopy. The experiment was replicated three times.
Cytotoxicity assay
The cytotoxicity of the polymers on HEK293 and HeLa cells was evaluated by CCK-8 assay. HEK293 and HeLa cells in 100 μL of DMEM containing 10% fetal bovine serum were seeded in 96-well plates at a density of 8,000 cells per well and incubated at 37°C under an atmosphere of 5% CO2. After 24 hours, the cells were transfected as described above, with C-SHR/pGL3 complexes at an N/P ratio of 10. After 3 hours of transfection, cells were washed with PBS and cultured in 100 μL of DMEM containing 10% fetal bovine serum for another 21 hours. The medium was then replaced by 90 μL of fresh complete medium and 10 μL of CCK-8 solution. The cells were incubated for an additional 4 hours. The absorbance of each well was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). The absorbance of untreated cells was set at 100%, and cell viability was expressed as the percentage relative to the absorbance of the untreated cells. bPEI-25K/pGL3 and SHR/pGL3 complexes were used as controls. The experiment was repeated four times.
In vivo transfection
A HeLa tumor xenograft model was established to evaluate the gene transfection efficiency of C-SHR-3/pGL3 complexes in vivo. Nine female Balb/c nude mice (aged 4–5 weeks) weighing 19–20 g were purchased from the Department of Experimental Animals, Second Military Medical University (Shanghai, People’s Republic of China), and housed in standard conditions with access to standard food and water. The mice were subcutaneously injected with 5.0×106 HeLa cells. Ten days after inoculation, the mice were randomized into three groups containing three mice each. Next, 100 μL of C-SHR-3/pGL3 in 5% glucose solution (212 μg C-SHR-3 combined with 20 μg pGL3, N/P ratio 10) was injected into the tumor at a pGL3 dose of 1 mg/kg every day for 3 days.21 A group treated with SHR/pGL3 complexes in 5% glucose solution was used as the control. Forty-eight hours after the final injection, d-luciferin potassium salt (200 μL, 150 mg/kg) was injected intraperitoneally into the mice which were anesthetized by pentobarbital sodium (70 μL, 0.02 mg/mL). Luminescent images were recorded using an IVIS imaging system (Xenogen IVIS-200, Caliper Life Sciences, Hopkinton, MA, USA). Tumor tissue specimens were harvested and homogenized. Luciferase assays were performed as previously described. All animal experiments were performed in accordance with the ethics and regulations of animal experiments of Second Military Medical University.
Statistical analysis
The Student’s unpaired t-test was used for the statistical analysis. Differences were considered to be statistically significant at P<0.05. The results are shown as the mean ± standard deviation.
Results
Characterization of C-SHR
The peptide HR was synthesized with high purity (>95%) and a precise molecular weight of 1,416.65 Da. The stearyl moiety was further grafted onto the N-terminal of HR using the solid-phase peptide synthesis method (Figure 1A). The mass-average molecular weights (Mws) of the polymers are summarized in Table 1. The Mw of C-SHR increased with the molar ratio of SHR to cysteine increasing and the Mw of C-SHR was significantly enhanced when compared with that of the original monomer SHR (1,683.65 Da).
Figure 1.
Synthesis and 1H-NMR determination of C-SHR (A) Synthesis of C-SHR-3 (n=3) and (B) 1H-NMR spectra of C-SHR-3 in DMF-D7 at 600 mHz.
Note: (a) Signal a was assigned to the proton of stearyl moiety, (b) signal b was attributed to arginine and (c) signal c was from protons of imidazole in histidine.
Abbreviations: Arg, arginine; His, histidine; HR, histidine-arginine; C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; NMR, nuclear magnetic resonance.
Formation of C-SHR was verified by 1H-NMR (Figure 1B). The peak at δ 0.07–0.6 was assigned to the proton of the stearyl moiety (signal a). The peak at δ 0.7–1.4 was attributed to arginine (signal b) and the peak at δ 6.7–7.4 was from protons of imidazole in histidine (signal c). Both 1H-NMR and gel permeation chromatography data demonstrated that C-SHR was successfully synthesized.
Characterization of C-SHR/pGL3 complexes
An appropriate size and zeta potential are important for efficient gene transfection.22 In this study, the particle size and zeta potential of C-SHR/pGL3 complexes with different N/P ratios are shown in Figure 2. As shown in Figure 2A, all complexes carried a positive surface charge with a zeta potential ranging from 30 to 40 mV when the N/P ratio was higher than 2.5. Figure 2B shows that the particle size of the C-SHR/pGL3 complexes was less than 300 nm at an N/P ratio higher than 10. When the N/P ratio was higher than 2.5, most C-SHR/pGL3 complexes had a higher zeta potential and a smaller particle size than the SHR/pGL3 complexes. In addition, the particle size and zeta potential were dependent on the degree of cross-linking of C-SHR. A higher degree of cross-linking induced a higher zeta potential and a smaller particle size of C-SHR. Among the C-SHR with different Mw, C-SHR-3/pGL3 was found to have an appropriate particle size and zeta potential (Figure 2C and D) at an N/P ratio of 10, with a particle size and zeta potential of 111.5±14.8 nm and 34.3±2.86 mV, respectively. Transmission electron microscopy showed that the C-SHR-3/pGL3 complexes formed a compact nanostructure, with a size of about 100 nm at an N/P ratio of 10 (Figure 2C). Thus, C-SHR-3/pGL3 complexes were selected for further study.
Figure 2.
Characterization of polymer/pGL3 complexes.
Notes: Zeta potential (A) and particle size (B) of polymer/pGL3 complexes at various N/P ratios. (C) Transmission electron micrograph of C-SHR-3/pGL3 complexes at an N/P ratio of 10. (D) Surface zeta potential distribution of C-SHR-3/pGL3 at an N/P ratio of 10. Data are shown as the mean ± standard deviation (n=3).
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide.
Figure 3 shows the results of agarose gel electrophoresis for the pDNA binding affinity for SHR and C-SHR-3. pGL3 mobilization was totally retarded at an N/P ratio of 1 for C-SHR-3/pGL3 complexes, and thoroughly retarded at an N/P ratio of 2 for SHR/pGL3 complexes, indicating that the binding affinity of SHR was improved by cross-linking with the disulfide bond. To verify this finding, DTT (a reducing agent) was used to break the disulfide bond.23 As was shown in Figure 3, C-SHR-3/pGL3 complexes showed a weaker pGL3 binding affinity in the presence of DTT due to depolymerization of C-SHR-3. Therefore, we hypothesized that the disulfide bond could be reduced in the reductive environment of the cytoplasm, which could weaken the binding affinity between pGL3 and C-SHR-3.24 The character of the disulfide bond was relatively stable in the extracellular space but was not stable under the intracellular homeostasis, making it an effective way to enhance nanoparticle formation and cytoplasm-sensitive dissociation.25
Figure 3.
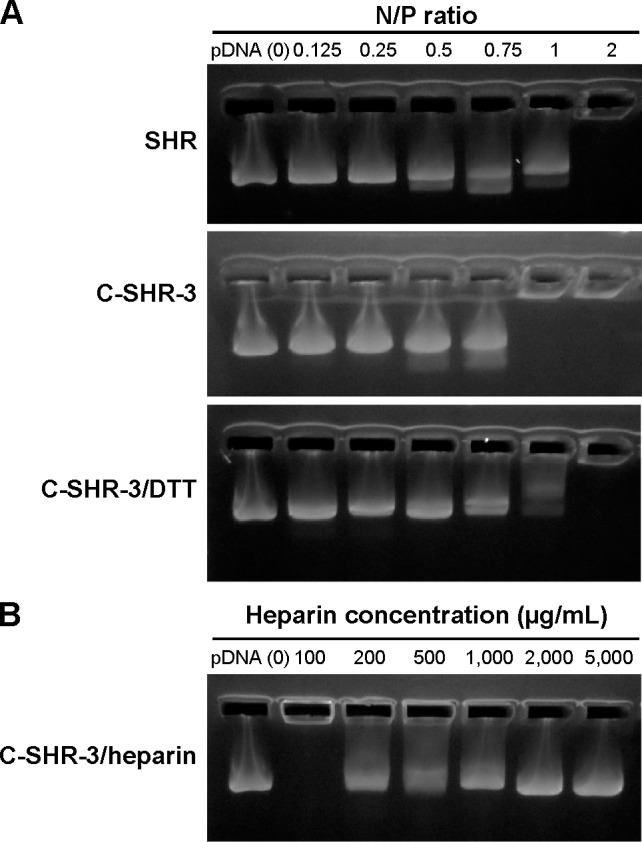
Agarose gel electrophoresis assay.
Notes: (A) Agarose gel electrophoresis of pGL3 binding affinity for SHR and C-SHR-3 at various N/P ratios. (B) Heparin were used to evaluate the release of pGL3 from C-SHR-3/pGL3 complexes at an N/P ratio of 5.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; DTT, dithiothreitol; pDNA, plasmid DNA.
To measure the ability of pGL3 releasing from C-SHR-3/pGL3 complexes, a heparin (a highly sulfated glycosaminoglycan with a strong negative charge) replacement test was conducted.26 As shown in Figure 3, C-SHR-3/pGL3 complexes were resistant to replacement of 100 μg/mL heparin without pGL3 release. With increasing amounts of heparin, pGL3 started being replaced and was completely replaced at a concentration of 2,000 μg/mL.
Cellular uptake
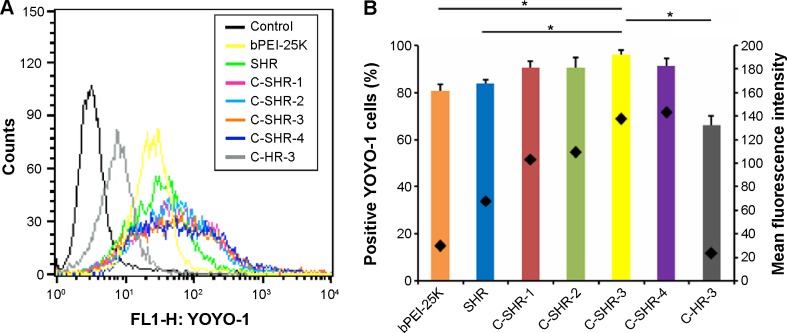
Cellular uptake is an important factor in gene delivery, and is closely related to final transfection efficiency. Here, cellular uptake of the complexes was examined for C-SHR with HEK293 cells by flow cytometry. YOYO-1-labeled C-SHR/pGL3 complexes were prepared by mixing C-SHR and YOYO-1-labeled pGL3 at an N/P ratio of 10. YOYO-1 fluorescence intensities of HEK293 cells were then evaluated. As shown in Figure 4A and B, cellular uptake efficiency of the complexes for SHR and C-SHR was significantly higher than that for bPEI-25K and C-HR-3 in HEK293 cells. The mean fluorescence intensity of C-SHR-3/pGL3 complexes in cells was approximately 5.84-fold higher than that of C-HR-3/pGL3 complexes (P<0.05), indicating that the hydrophobic moiety with stearic acid enhanced the membrane affinity of the cellular uptake of polymer/pDNA complexes. In addition, the significantly improved uptake of C-SHR/pGL3 complexes indicated that the transfection efficiency was high. Moreover, increased formation of disulfide bonds enhanced the cellular uptake of pGL3 in HEK293 cells. The mean fluorescence intensity of the C-SHR-1/pGL3, C-SHR-2/pGL3, C-SHR-3/pGL3, and C-SHR-4/pGL3 complexes was approximately 1.52-fold, 1.61-fold, 2.03-fold, and 2.11-fold that of the SHR/pGL3 complexes, respectively, indicating that reduction-responsiveness improved uptake of the complexes (P<0.05). In addition, there was no significant difference in cellular uptake between the four C-SHR/pGL3 complexes (P>0.05).
Figure 4.
Intracellular uptake of pGL3-YOYO-1 complexes based on different polymers at an N/P ratio of 10 in HEK293 cells after 3 hours of transfection.
Notes: (A) Flow cytometry figures and (B) mean fluorescence intensity and the percentage of positive YOYO-1 cells. Diamonds (♦) indicate mean fluorescence intensity (B). Data are shown as the mean ± standard deviation (n=3). *P<0.05.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; PEI, polyethylenimine.
Gene transfection efficiency
Gene transfection efficiency in HEK293 and HeLa cells was evaluated by luciferase protein expression assays, using pGL3 and pEGFP as the model plasmid. bPEI-25K and Lipofectamine 2000 were used as controls. It seemed that the gene transfection efficiency was dependent on the N/P ratio, which could be observed for both HEK293 and HeLa cells (Figure 5A and B). Gene transfection efficiency in the C-SHR group was significantly higher than that in the SHR and C-HR-3 groups at all N/P ratios (P<0.05), probably due to the reduction-responsiveness of the C-SHR and stearyl moiety. It is noteworthy that C-SHR showed optimal gene transfection efficiency at an N/P ratio of 10, which has similar effects with with Lipofectamine 2000. Among the C-SHRs with different degrees of cross-linking, C-SHR-3 showed the maximum luciferase gene transfection rate at an N/P ratio of 10, approaching the high level of transfection by the bPEI-25K control on HEK293 cells. Additionally, the pEGFP transfection efficiency of the C-SHR/pEGFP complexes in HEK293 cells was assayed at various N/P ratios of 5, 7.5, 10, and 15. SHR/pEGFP, C-HR-3/pEGFP, bPEI-25K/pEGFP, and Lipofectamine 2000/pEGFP complexes were used as controls. As shown in Figure 5C, C-SHR-3/pEGFP complexes showed markedly higher EGFP fluorescence intensities than SHR/pEGFP, C-SHR-1/pEGFP, C-SHR-2/pEGFP, C-SHR-4/pEGFP, and C-HR-3/pEGFP complexes at N/P ratios of 5, 7.5, and 10. The fluorescence intensity of C-SHR-3/pEGFP complexes at an N/P ratio of 10 was comparable with that of bPEI-25K/pEGFP and Lipofectamine 2000/pEGFP complexes. In addition, the EGFP fluorescence intensity transfected by C-SHR-3/pEGFP complexes at an N/P ratio of 10 in HeLa cells was also higher than that with SHR/pEGFP, C-SHR-1/pEGFP, C-SHR-2/pEGFP, and C-SHR-4/pEGFP complexes (Figure 5D). Flow cytometry assay about positive EGFP cells and mean fluorescence intensity on HEK293 cells also confirmed this tendency (Figure 5E), which is in good agreement with the results of the luciferase gene expression assay. In addition, buthionine sulfoximine, a glutathione depleting agent, was used to inhibit the disulfide bond in C-SHR-3/pGL3 complexes during gene transfection. As shown in Figure 5F, EGFP expression in the C-SHR-3/pGL3 complexes significantly decreased when the buthionine sulfoximine concentration increased from 0 to 1,000 μM, indicating that the disulfide bond was essential for efficient gene transfection of C-SHR-3/pGL3 complexes. Importantly, C-SHR-3 could condense DNA and promote the cellular uptake of DNA effectively. Their reduction-responsive ability facilitated the unpacking of DNA. These findings may explain the high efficiency of gene transfection of C-SHR-3/pDNA complexes.
Figure 5.
In vitro transfection efficacy of polymer/pDNA complexes.
Notes: Luciferase assay of polymer/pGL3 complexes on HEK293 cells (A) and HeLa cells (B). Fluorescent images of transfection efficiency of polymer/pEGFP complexes at an N/P ratio of 10 in HEK293 cells (C) and HeLa cells (D). Quantitative measurements of transfection efficiency for polymer/pEGFP complexes at an N/P ratio of 10 in HEK293 cells using flow cytometry (E). Fluorescent images of transfection efficiency for C-SHR-3/pEGFP at an N/P ratio of 10 in the presence of BSO in HEK293 cells (F). Diamonds (♦) indicate mean fluorescence intensity (E). Data are shown as the mean ± standard deviation (n=3). *P<0.05. Scale bar, 250 μm.
Abbreviations: BSO, l-buthionine-sulfoximine; C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; PEI, polyethylenimine; RLU, relative light units; pDNA, plasmid DNA.
Intracellular distribution
Figure 6 shows the intracellular distribution of the complexes in HEK293 cells at 1 hour and 13 hours post transfection. As seen in Figure 6A, all the YOYO-1 labeled pGL3 (pGL3-YOYO-1, green) transfected by the vectors was distributed mainly in the cytoplasm at 1 hour post transfection. The fluorescence intensity of pGL3-YOYO-1 transfected by C-SHR-3 was higher than that of SHR/pGL3-YOYO-1, bPEI-25K/pGL3-YOYO-1, and C-HR-3/pGL3-YOYO-1. After 13 hours of transfection, the pGL3-YOYO-1 transfected by C-SHR-3 was evenly dispersed in both the cytoplasm and nucleus with high fluorescence intensity, indicating that pGL3-YOYO-1 successfully escaped from the endosome and entered the nucleus. However, pGL3-YOYO-1 transfected by SHR, bPEI-25K, and C-HR-3 was not uniformly dispersed in the cytoplasm by 13 hours post transfection (Figure 6B).
Figure 6.
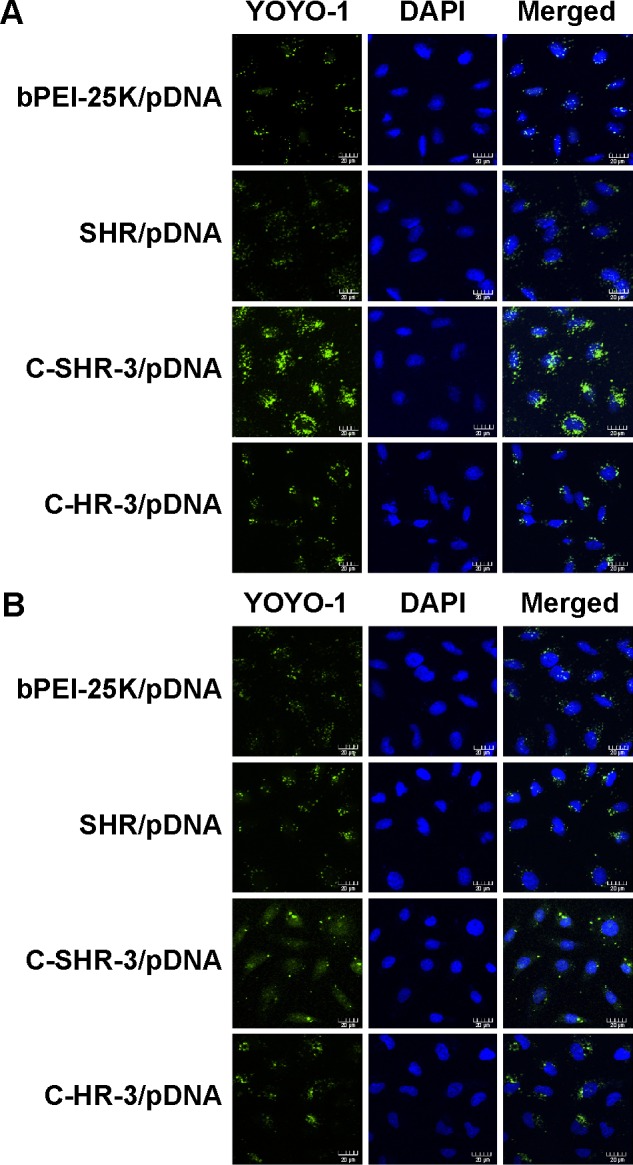
Confocal laser scanning micrographs of YOYO-1 labeled pGL3 (green) on HEK293 cells at 1 hour (A) and 13 hours (B) post transfection. The cells were transfected by polymer/pGL3-YOYO-1 complexes (1 μg/mL pGL3), respectively. The nuclei were stained with DAPI (blue). Scale bar, 20 μm.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; PEI, polyethylenimine; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; pDNA, plasmid DNA.
Cytotoxicity
The cytotoxicity of the C-SHR/pGL3 complexes in HEK293 and HeLa cells is shown in Figure 7. As shown in Figure 7, C-SHR/pGL3 and SHR/pGL3 complexes showed significantly lower cytotoxicity than the bPEI-25K/pGL3 complexes (P<0.05). No significant cytotoxicity was observed over the 21-hour period of transfection with C-SHR/pGL3 and SHR/pGL3 complexes at concentrations of 5–60 μg/mL, indicating their favorable biocompatibility with HEK293 and HeLa cells. However, cell viability decreased to approximately 20% in both cell lines when compared with untreated cells for bPEI-25K/pGL3 complexes at a concentration of 20 μg/mL. In addition, cross-linking did not increase the cytotoxicity of the polymers. C-SHR/pGL3 complexes showed no significant cytotoxicity with high molecular weight and charge density.
Figure 7.
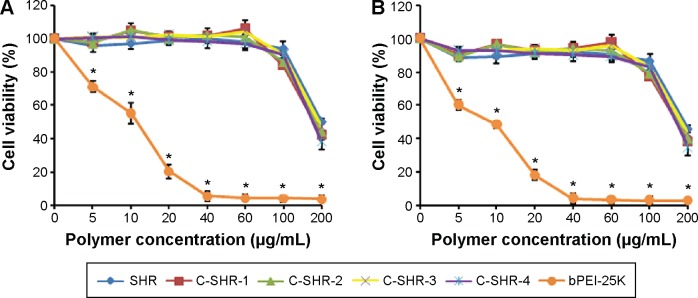
Cytotoxicity of polymer/pGL3 complexes on HEK293 cells (A) and HeLa cells (B) for 24 hours. Data are shown as the mean ± standard deviation (n=3). *P<0.05.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; PEI, polyethylenimine.
In vivo gene transfection efficiency
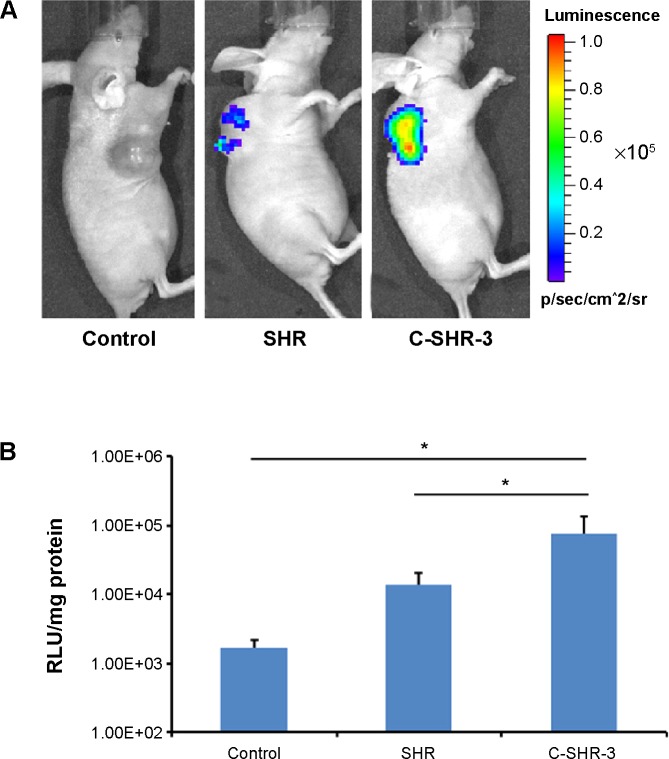
For the in vivo study, a HeLa xenograft model was established on the right back of each mouse. C-SHR-3 and SHR complexes containing pGL3 were injected into the tumor once a day for 3 days. A group injected with 5% glucose solution was used as the control. The results showed that the luminescence intensity of the group that received the C-SHR-3/pGL3 complexes was 5.28-fold that of the groups that received the SHR/pGL3 complexes (Figure 8A and B, P<0.05). The in vivo gene transfection study further suggested that C-SHR could be a potential gene delivery vector.
Figure 8.
In vivo gene transfection of polymer/pGL3 complexes on a HeLa tumor xenograft model.
Notes: (A) Luminescent images of mice after injection of polymer/pGL3 complexes. (B) Quantitative comparison of luciferase activity for polymer/pGL3 complexes with a luminometer. Data are shown as the mean ± standard deviation (n=3). *P<0.05.
Abbreviations: C-SHR, disulfide cross-linked stearylated polyarginine peptide modified with histidine; SHR, non-cross-linked stearylated polyarginine peptide; RLU, relative light units.
Discussion
In the present study, we synthesized a C-SHR polymer using a F-moc-solid-phase peptide method as previously reported.20 During the synthesis process, cysteine was used to control the degree of polymerization due to its single sulfhydryl group. We also used a low concentration of H2O2 in a near-neutral pH solution as a catalyst to simply the cross-link and after-treatment process as compared with dimethyl sulfoxide (DMSO), knowing that it is difficult to remove DMSO thoroughly after dialysis.27 In addition, the cytotoxicity of the residual DMSO in cells could be avoided by using H2O2 instead of DMSO.
The cellular uptake result showed that the cellular uptake efficiency of SHR/pGL3 complexes and C-SHR/pGL3 complexes was significantly higher than that of bPEI-25K and C-HR-3 in HEK293 cells (Figure 4). The mean fluorescence intensity of C-SHR-3/pGL3 complexes in cells was approximately 5.84-fold that of C-HR-3/pGL3 complexes, probably due to the hydrophobic moiety. Zhang et al reported that a hydrophobic moiety such as stearic acid could enhance membrane affinity and greatly improve the cellular uptake of polymer/pDNA complexes mediated by clathrin and caveolin.28 Our study showed that the significantly enhanced uptake of C-SHR/pGL3 complexes further improved the efficiency of transfection (Figure 5A and B).
Moreover, the responsive sensitive attribute also improved the uptake and transfection efficacy of C-SHR/pDNA complexes. It was reported that the glutathione concentration in the cytosol of tumor cells was 100–1,000 times higher than that in normal cells.29 Kim et al reported that the disulfide bond could be cleaved rapidly via thiol-disulfide exchange reactions with intracellular reducing molecules, especially glutathione.19 Our results also demonstrated that introduced reduction sensitive disulfide bond formation in C-SHR significantly enhanced the uptake of pGL3 by approximately one- to twofold (P<0.05).
In addition, C-SHR/pDNA complexes showed significantly higher uptake and gene transfection efficiency than SHR/pDNA complexes, which are comparable with Lipofectamine 2000/pDNA complexes. The cross-linked polymer improved gene transfection, reaching a high level of transfection efficiency at an Mw of 29 kDa (Table 1 and Figure 5). Further, our in vivo study also demonstrated that the gene transfection efficiency of C-SHR-3/pGL3 complexes was 5.28-fold higher than that of SHR/pGL3 complexes (Figure 8A and B, P<0.05), which is consistent with the in vitro result.
Interestingly, C-SHR-3 with a Mw of 29 kDa showed higher in vitro gene transfection efficiency than C-SHR-1, C-SHR-2, and C-SHR-4 (Figure 5A and B), while there was no significant difference in cellular uptake between the four C-SHR/pGL3 complexes (P>0.05, Figure 4B).
Figure 6 shows the intracellular distribution by confocal laser scanning microscopy. As shown in Figure 6A, pGL3-YOYO-1 transfected by C-SHR-3 was evenly dispersed in both cytoplasm and nucleus with high fluorescence intensity, indicating that pGL3-YOYO-1 successfully escaped from the endosome and entered the nucleus. However, pGL3-YOYO-1 transfected by SHR, bPEI-25K, and C-HR-3 was not dispersed evenly in the cytoplasm at 13 hours post transfection (Figure 6B). There are two possible reasons for the improved cellular uptake and enhanced endosomal escape ability of C-SHR-3/pGL3-YOYO-1 complexes. One is related to the stronger buffering capacity provided by histidine imidazole moieties, which act as a proton sponge, preventing a lower pH milieu in the endosome, resulting in DNA release.30,31 The other reason is that the high density of hydrophobic stearyl moieties increases the cellular uptake and fusion of complexes with the endosomal membrane by breaking it to facilitate endosomal escape.32,33
Our cytotoxicity study demonstrated high biocompatibility of C-SHR/pGL3 and SHR/pGL3 complexes (Figure 7). Although the C-SHR/pGL3 complex has a higher molecular weight and charge density than the SHR/pGL3 complex, it did not induce additional cytotoxicity in cells. This may be due to the reduction-responsiveness of C-SHR. The results demonstrated that C-SHR had relatively high transfection efficiency with low cytotoxicity, suggesting that it may prove to be a promising vector for gene delivery.
Conclusion
In this study, we developed a reduction-responsive cationic stearyl polypeptide for delivery of pDNA. Low-concentration H2O2 was used as a catalyst, thus simplifying the cross-linking and after-treatment process. An agarose gel electrophoresis assay showed the stronger plasmid DNA binding affinity of C-SHR. The reduction-responsive C-SHR showed high cellular uptake, high in vitro gene transfection efficiency, and good biocompatibility as compared with bPEI-25K. The in vivo study also demonstrated the high gene transfection efficiency of C-SHR/pGL3 complexes. These results indicate that C-SHR may prove to be a promising gene delivery vector with high efficiency and low cytotoxicity.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81372762, 81272819, 81302212, 81172514, and 81101658).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J Gene Med. 2013;15(2):65–77. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 2.Lehto T, Simonson OE, Mäger I, et al. A peptide-based vector for efficient gene transfer in vitro and in vivo. Mol Ther. 2011;19(8):1457–1467. doi: 10.1038/mt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 4.de la Fuente M, Raviña M, Sousa-Herves A, et al. Exploring the efficiency of gallic acid-based dendrimers and their block copolymers with PEG as gene carriers. Nanomedicine (Lond) 2012;7(11):1667–1681. doi: 10.2217/nnm.12.51. [DOI] [PubMed] [Google Scholar]
- 5.Zarogoulidis P, Hohenforst-Schmidt W, Darwiche K, et al. 2- diethylaminoethyl-dextran methyl methacrylate copolymer nonviral vector: still a long way toward the safety of aerosol gene therapy. Gene Ther. 2013;20(10):1022–1028. doi: 10.1038/gt.2013.27. [DOI] [PubMed] [Google Scholar]
- 6.Segovia N, Dosta P, Cascante A, Ramos V, Borrós S. Oligopeptide-terminated poly (β-amino ester) s for highly efficient gene delivery and intracellular localization. Acta Biomater. 2014;10(5):2147–2158. doi: 10.1016/j.actbio.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez E, Roldán M, Diez-Gil C, et al. Protein nanodisk assembling and intracellular trafficking powered by an arginine-rich (R9) peptide. Nanomedicine (Lond) 2010;5(2):259–268. doi: 10.2217/nnm.09.98. [DOI] [PubMed] [Google Scholar]
- 8.Futaki S, Hirose H, Nakase I. Arginine-rich peptides: methods of translocation through biological membranes. Curr Pharm Des. 2013;19(16):2863–2868. doi: 10.2174/1381612811319160003. [DOI] [PubMed] [Google Scholar]
- 9.Mann A, Thakur G, Shukla V, et al. Differences in DNA condensation and release by lysine and arginine homopeptides govern their DNA delivery efficiencies. Mol Pharm. 2011;8(5):1729–1741. doi: 10.1021/mp2000814. [DOI] [PubMed] [Google Scholar]
- 10.Chang KL, Higuchi Y, Kawakami S, Yamashita F, Hashida M. Development of lysine-histidine dendron modified chitosan for improving transfection efficiency in HEK293 cells. J Control Release. 2011;156(2):195–202. doi: 10.1016/j.jconrel.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Shigeta K, Kawakami S, Higuchi Y, et al. Novel histidine-conjugated galactosylated cationic liposomes for efficient hepatocyte-selective gene transfer in human hepatoma HepG2 cells. J Control Release. 2007;118(2):262–270. doi: 10.1016/j.jconrel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Nakase I, Akita H, Kogure K, et al. Efficient intracellular delivery of nucleic acid pharmaceuticals using cell-penetrating peptides. Acc Chem Res. 2012;45(7):1132–1139. doi: 10.1021/ar200256e. [DOI] [PubMed] [Google Scholar]
- 13.Hoyer J, Neundorf I. Peptide vectors for the nonviral delivery of nucleic acids. Acc Chem Res. 2012;45(7):1048–1056. doi: 10.1021/ar2002304. [DOI] [PubMed] [Google Scholar]
- 14.Mitsueda A, Shimatani Y, Ito M, et al. Development of a novel nanoparticle by dual modification with the pluripotential cell-penetrating peptide PepFect6 for cellular uptake, endosomal escape, and decondensation of an siRNA core complex. Biopolymers. 2013;100(6):698–704. doi: 10.1002/bip.22310. [DOI] [PubMed] [Google Scholar]
- 15.Tönges L, Lingor P, Egle R, Dietz GP, Fahr A, Bähr M. Stearylated octaarginine and artificial virus-like particles for transfection of siRNA into primary rat neurons. RNA. 2006;12(7):1431–1438. doi: 10.1261/rna.2252206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HY, Chen JX, Sun YX, et al. Construction of cell penetrating peptide vectors with N-terminal stearylated nuclear localization signal for targeted delivery of DNA into the cell nuclei. J Control Release. 2011;155(1):26–33. doi: 10.1016/j.jconrel.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Lin D, Jiang Q, Cheng Q, et al. Polycation-detachable nanoparticles self-assembled from mPEG-PCL-g-SS-PDMAEMA for in vitro and in vivo siRNA delivery. Acta Biomater. 2013;9(8):7746–7757. doi: 10.1016/j.actbio.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104(36):14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30(4):658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang WC. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford, UK: Oxford University Press; 2004. [Google Scholar]
- 21.Wang F, Wang Y, Wang H, Shao N, Chen Y, Cheng Y. Synergistic effect of amino acids modified on dendrimer surface in gene delivery. Biomaterials. 2014;35(33):9187–9198. doi: 10.1016/j.biomaterials.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 22.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 23.Kim JS, Oh MH, Park JY, Park TG, Nam YS. Protein-resistant, reductively dissociable polyplexes for in vivo systemic delivery and tumor-targeting of siRNA. Biomaterials. 2013;34(9):2370–2379. doi: 10.1016/j.biomaterials.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Saaranen MJ, Ruddock LW. Disulfide bond formation in the cytoplasm. Antioxid Redox Signal. 2013;19(1):46–53. doi: 10.1089/ars.2012.4868. [DOI] [PubMed] [Google Scholar]
- 25.Won YW, Yoon SM, Lee KM, Kim YH. Poly(oligo-D-arginine) with internal disulfide linkages as a cytoplasm-sensitive carrier for siRNA delivery. Mol Ther. 2011;19(2):372–380. doi: 10.1038/mt.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JL, Peng SW, Ko WH, et al. An efficient and low toxic human serum albumin conjugated polyethylenimine nano-sized complex for gene delivery. J Nanopart Res. 2014;16(9):1–17. [Google Scholar]
- 27.Won YW, Kim HA, Lee M, Kim YH. Reducible poly(oligo-D-arginine) for enhanced gene expression in mouse lung by intratracheal injection. Mol Ther. 2010;18(4):734–742. doi: 10.1038/mt.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Song J, Liang R, et al. Stearylated antimicrobial peptide melittin and its retro isomer for efficient gene transfection. Bioconjug Chem. 2013;24(11):1805–1812. doi: 10.1021/bc400053b. [DOI] [PubMed] [Google Scholar]
- 29.Cheng R, Feng F, Meng F, Deng C, Feijen J, Zhong Z. Glutathione-responsive nano-vehicles as a promising platform for targeted intracellular drug and gene delivery. J Control Release. 2011;152(1):2–12. doi: 10.1016/j.jconrel.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 30.Mann A, Shukla V, Khanduri R, Dabral S, Singh H, Ganguli M. Linear short histidine and cysteine modified arginine peptides constitute a potential class of DNA delivery agents. Mol Pharm. 2014;11(3):683–696. doi: 10.1021/mp400353n. [DOI] [PubMed] [Google Scholar]
- 31.Wen Y, Guo Z, Du Z, et al. Serum tolerance and endosomal escape capacity of histidine-modified pDNA-loaded complexes based on polyamidoamine dendrimer derivatives. Biomaterials. 2012;33(32):8111–8121. doi: 10.1016/j.biomaterials.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 32.Katayama S, Hirose H, Takayama K, Nakase I, Futaki S. Acylation of octaarginine: implication to the use of intracellular delivery vectors. J Control Release. 2011;149(1):29–35. doi: 10.1016/j.jconrel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Madani F, Abdo R, Lindberg S, et al. Modeling the endosomal escape of cell-penetrating peptides using a transmembrane pH gradient. Biochim Biophys Acta. 2013;1828(4):1198–1204. doi: 10.1016/j.bbamem.2012.12.008. [DOI] [PubMed] [Google Scholar]