Abstract
Kinesins are a superfamily of microtubule-based molecular motors that perform various transport needs and have essential roles in cell division. Among these, the kinesin-5 family has been shown to play a major role in the formation and maintenance of the bipolar mitotic spindle. Moreover, recent work suggests that kinesin-5 motors may have additional roles. In contrast to most model organisms, the sole kinesin-5 gene in Caenorhabditis elegans, bmk-1, is not required for successful mitosis and animals lacking bmk-1 are viable and fertile. To gain insight into factors that may act redundantly with BMK-1 in spindle assembly and to identify possible additional cellular pathways involving BMK-1, we performed a synthetic lethal screen using the bmk-1 deletion allele ok391. We successfully knocked down 82% of the C. elegans genome using RNAi and assayed viability in bmk-1(ok391) and wild type strains using an automated high-throughput approach based on fluorescence microscopy. The dataset includes a final list of 37 synthetic lethal interactions whose further study is likely to provide insight into kinesin-5 function.
Subject terms: Mitosis, RNAi, Caenorhabditis elegans, High-throughput screening
Background & Summary
The mitotic spindle consists of microtubule filaments that are organized into a bipolar configuration by a variety of microtubule-associated proteins (MAPs) and their regulators. Of the MAPs with motor activity, members of the kinesin-5 family play a central role. With four identical motor domains configured like a ‘dumbbell’1, kinesin-5 is capable of cross-linking and sliding apart antiparallel microtubules2, thereby generating an outward force on the spindle. In mammals, kinesin-5 is essential to separate centrosomes early in mitosis to form the bipolar spindle3, while in S. cerevisiae and Drosophila S2 cells kinesin-5 activity is used later in mitosis to elongate the anaphase spindle4,5. By contrast, the C. elegans kinesin-5 homolog BMK-1 acts as a break for spindle elongation in anaphase of the first embryonic division6. In this context, the cross-linking activity of BMK-1 likely resists the strong cortical forces in the one-cell embryo that pull the two spindle poles apart. Thus, while sequence conservation suggests that the basic activity of kinesin-5 is the same in all species, considerable variation exists in the requirement for this activity in mitosis. In addition, the dispensability of kinesin-5 for successful mitosis in C. elegans may hint at redundant mechanisms for spindle assembly in this organism.
Several lines of evidence suggest that kinesin-5 also has multiple roles beyond mitosis. Mammalian Eg5 is a regulator of axonal growth7,8 and has been shown to associate with ribosomes to enhance the efficiency of translation9, while C. elegans BMK-1 was identified in a genome-wide screen for genes that regulate germ cell apoptosis10.
To gain insight into factors that may act redundantly with kinesin-5 in spindle assembly in C. elegans and to uncover possible additional functions of kinesin-5, we conducted a genome-wide synthetic lethal RNAi screen using animals harbouring the genetic deletion allele bmk-1(ok391). The viability and fertility of bmk-1(ok391) animals is indistinguishable from wild type controls, making this genetic background ideal for a synthetic lethal screen. Knockdown by RNAi was carried out by bacterial feeding using the Ahringer genome-wide library11 adapted to a 96-well liquid format. We marked animals with green fluorescent protein in the pharynx (myo-2::gfp) for detection and accurate counting with an automated fluorescence microscopy system, in which adult worms (parents) and larvae (progeny) could be distinguished by the size of their pharynx. This allowed determination of ‘reproductive fitness’, defined as the number of progeny per parent. The primary screen consisted of two technical replicates per bacterial clone (16757 clones) expressing a gene-specific dsRNA. Genes that when knocked down significantly decreased reproductive fitness in bmk-1(ok391) animals relative to control animals were considered hits. These were selected for secondary screening (Fig. 1), which consisted of 3 biological replicates, each carried out in 2 technical replicates. Data analysis and validation steps generated a list of 37 genes that display synthetic lethality with bmk-1(ok391) (Data Records 1 and 2). The list contains genes implicated in both embryonic and post-embryonic development (Table 1).
Figure 1. Experimental workflow.
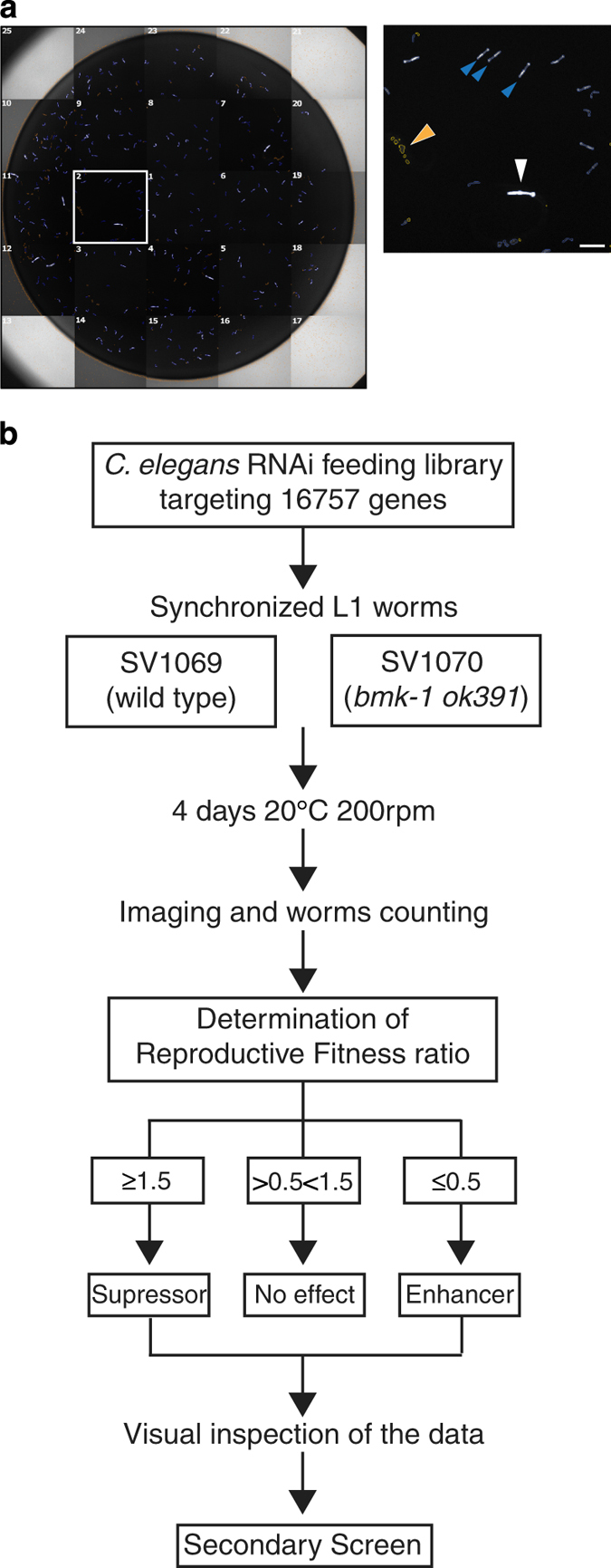
(a) Example of a mosaic reconstruction of an imaged well of the screen. In the magnification of field 2, the white arrowhead shows the pharynx of a parent worm, blue arrowheads show pharynxes of progeny, and the orange arrowhead points to excluded objects. Scale bar is 150 μm. (b) Flowchart of screening protocol. The primary screen was done in two technical replicates. The secondary screen followed the same protocol as the primary screen but used three biological replicates with two technical replicates each.
Table 1. Final hit list.
| hit plate | well | Cosmid(s) | Primary targets | Secondary targets | Gene(s) name(s) |
N2 (wild type)
|
SV1005 (kinesin-5 mutant)
|
||
|---|---|---|---|---|---|---|---|---|---|
| Embryonic | Post-embryonic | Embryonic | Post-embryonic | ||||||
| Table summarizing the genes that genetically interact with the worm kinesin-5 and the observed developmental defects upon dsRNA treatment. Description of the abbreviations can be found at www.RNAi.org15. | |||||||||
| 1 | B08 | R06C7.10 | myo-1 | Gro | Gro | ||||
| 1 | B11 | F36H2 | F36H2.1 | tat-5 | Emb | Emb | |||
| 1 | C09 | C39E9+B0564 | C39E9.14 | dli-1 | Emb | Adl+Ste+Bag | Emb | Ste+Pvl | |
| 1 | C11 | F46A9+F30A10 | F46A9.4 | F46A9.5 | skr-2+skr-1 | Gro | Gro | ||
| 1 | D05 | W09C3+T03F1 | W09C3.6 | T03F1.5 | gsp-3+gsp-4 | Gro | Gro | ||
| 1 | D07 | W01A8 | W01A8.4 | nuo-6 | Lva | Lva | |||
| 1 | D11 | C17E4 | C17E4.5 | pabp-2 | Gro+Lva | Gro+Lva | |||
| 1 | E11 | C41G7+M01E11 | C41G7.2 | M01E11.6 | klp-16+klp-15 | Emb | Gro | Emb | Gro |
| 2 | B11 | T26E3 | T26E3.3 | par-6 | Emb | Ste | Emb | Ste | |
| 2 | E11 | T08B2 | T08B2.8 | T08B2.7 | mrpl-23+ech-1.2 | Gro | Gro | ||
| 3 | C04 | F26E4 | F26E4.8 | tba-1 | Emb | Emb | |||
| 3 | C10 | Y71F9AM | Y71F9AM.4 | Y71F9AM.7 | cogc-3+Y71F9AM.7 | Ste+Sck | Ste+Sck | ||
| 3 | F11 | F52C6+M7 | M7.1 | F52C6.12 | ubc-2(let-70)+F52C6.12 | Emb | no defect | Emb | no defect |
| 4 | E05 | F37C12 | F37C12.3 | F37C12.3 | Gro | Gro | |||
| 4 | E08 | B0285 | B0285.1 | cdtl-7 | Lva | Lva | |||
| 4 | F03 | T12G3 | T12G3.5 | mrpl-51 | Gro+Lva | Gro+Lva | |||
| 4 | H05 | B0280 | B0280.9 | B0280.9 | Gro+Ste+Sck | Gro+Ste+Sck | |||
| 5 | C11 | W09H1+several other | several his- genes | Emb | Emb | ||||
| 5 | F02 | F09E5 | F09E5.1 | pkc-3 | Emb | Emb | |||
| 5 | F08 | Y49E10+Y51H7C+ Y9D1A+Y46H3C+etc | Gro | Gro | |||||
| 6 | C03 | several | F54D5.11 | T01G5.8 | Emb | Gro+Rup | Emb | Gro+Rup+Pat | |
| 6 | C06 | F01F1 | F01F1.7 | ddx-23 | Emb | Lva | Emb | Lva | |
| 6 | C09 | F58A4 | F58A4.8 | tbg-1 | Emb | Ste+Pvl | Emb | ||
| 6 | D07 | Y43C5A | Y43C5A.6 | rad-51 | no defect | no defect | |||
| 7 | A10 | H06I04 | H06I04.3 | H06I04.3 | Gro | Gro+Ste | |||
| 7 | E09 | M04D8 | M04D8.6 | xbx-3 | no defect | ||||
| 7 | H04 | F42A8+C06C3 | C06C3.1 | mel-11 | Adl+Ste+Bag | Ste+Sck | |||
| 7 | H12 | E04A4 | E04A4.7 | cyc-2.1 | Gro | Lva | |||
| 8 | A02 | K08F11 | K08F11.4 | yars-1 | Gro | Gro+Lva | |||
| 9 | H10 | YAC Y38F2AL | Y38F2AL.4 | vha-3 | Lva | >Gro | |||
| 9 | H12 | Y6E2A | Y6E2A.9 | sfxn-1.3 | Gro+Lva | no defect | |||
| 11 | B02 | Y82E9BR | Y82E9B5.3 | Y82E9BR.3+Y82E9BR.27+Y82E9BR.30 | Gro | >Gro | |||
| 11 | B07 | F26H11 | F26H11.1 | kbp-3 (knl) | Emb | Ste | Emb | Ste+Adl+Gro+Pvl | |
| 11 | D03 | F11A3+C50F4 | F11A3.2 | F11A3.2 | Gro | Ste+Lva | |||
| 12 | H02 | T27B1 | T27B1.2 | ztf-19/pat-9 | Emb | Emb | Pat+Adl+Rup+Dpy | ||
| 12 | F04 | T27B1 | T27B1.2 | ztf-19/pat-9 | Emb | Ste+Sck | Emb | ||
| 13 | D05 | F25G6 | F25G6.2 | symk-1 | Emb | no defect | Emb | Gro+Sck+Adl | |
| 13 | H02 | F49C12 | F49C12.13 | vha-17 | Gro | ||||
The RNAi dataset presented here points to cellular pathways beyond spindle assembly in which C. elegans kinesin-5 might play a role. Furthermore, the data on reproductive fitness in the wild type strain N2 may serve as a useful reference for other genome-wide RNAi screens in C. elegans. Finally, we hope that our adaptation of 96-well liquid format screening12 to an automated pipeline with a microscope image-based read-out will encourage others to execute similar large-scale screens using this approach.
Methods
Strains and culture
Worms were cultured on nematode growth media (NGM) plates seeded with OP50 bacteria. Strains used in this study include: N2 wild type Bristol variant, SV1005 bmk-1(ok391) V backcrossed 8 times with N2, SV1069 myo-2::GFP backcrossed 4 times with N2 and SV1070 myo-2::GFP; bmk-1(ok391) V from crossing SV1005 with SV1069. Worm strains were grown at 16 or 20 °C. Newly generated strains will be donated and available through the Caenorhabditis Genetics Center (CGC).
High throughput RNA interference screening
Methodology overview
To perform the genome-wide RNA interference (RNAi) screen we devised an automated screening platform, based on a previously developed protocol for C. elegans screening in liquid culture using 96-well plates12. We used synchronized first larval stage (L1) animals, which we fed with bacteria from the C. elegans RNAi feeding library constructed by the Ahringer group11. We compared the viability of progeny from control and bmk-1(ok391) parents following feeding with bacteria expressing gene-specific double-stranded RNA (dsRNA). For each 96-well library plate with bacteria, we assayed two technical replicates per strain as well as negative and positive controls, positioned in column 1 (wells A1 to H1). The negative controls were empty feeding vector (L4440, a gift from Andrew Fire (Addgene plasmid # 1654)) and hil-5 dsRNA that exhibit no lethality. plk-1 dsRNA, which causes 100% embryonic lethality, was used as a control for RNAi, and dli-1 dsRNA was used as a control for enhanced lethality specifically in the bmk-1(ok391) background (hereafter referred to as positive control). Worm strains SV1069 and SV1070 express green fluorescent protein (GFP) in the pharynx from the myo-2 promoter, which allowed image acquisition and worm counting with an automated fluorescence microscope (Thermo ArrayScan VTi) (Fig. 1a). All liquid dispensing steps were carried out with the Sciclone ALH 3000 liquid handling robot (Caliper Lifesciences). L1 larvae were dispensed into each well of 96-well plates, using a MultidropTM Combi Reagent Dispenser (Thermo Scientific). For the secondary screen, bacterial clones that enhanced lethality above a certain threshold (see below) were re-assayed in three biological replicates using the same experimental procedure of the primary screen, (Fig. 1b).
Screening reagents
Ahringer RNAi feeding library with 16757 bacterial clones11 (Source BioScience, Nottingham, GB)
LB (Luria-Bertani) media plus 100 μg ml−1 Ampicillin and 12,5 μg ml−1 Tetracyclin hydrocloride
NGM agar plates seeded with Escherichia coli OP50 (available from the C. elegans Genetics center) for growing worms
IPTG 4 mM to induce lac operon controlled gene expression in bacteria
M9 buffer
Complete S medium plus 10 ml l−1 Penicillin-Streptomycin and 50 mg l−1 Nystatin
Bleach solution (10 M NaOH, 4% sodium hypochlorite)
Tween 20 (P2287, Sigma-Aldrich)
Tricaine (E10521, Sigma-Aldrich)
Tetramisole hydrochloride (T1512, Sigma-Aldrich)
Day 1: RNAi library replication
The 96-well RNAi library plates with bacterial glycerol stocks and the microtiter plates with controls were thawed at room temperature for 10 min prior to use. A 96-pin replicator (BOEKEL), with the first column of pins removed, was used to inoculate 100 μl of LB (supplemented with Ampicillin and Tetracyclin hydrocloride) in each well of a new 96-well plate with bacteria. Positive and negative controls were then added to the first column in each 96-well plate. The first column of every library plate was replicated into a separate 96-well plate for screening. Bacteria were grown overnight (ON) at 37 °C.
Day 2: Preparation of bacteria and worms
5 μl of the bacterial ON cultures were used to inoculate 400 μl of LB (supplemented with Ampicillin and Tetracyclin hydrocloride) in a 2-ml 96-well plate, followed by ON incubation in a shaking incubator at 220 r.p.m. at 37 °C. Gravid adult worms were washed off NGM agar plates in M9 buffer, pelleted by centrifugation for 1 min at 1,350 r.p.m. and subjected to bleach solution for 5–10 min with occasional vortexing until only embryos remained. Embryos were pelleted by centrifugation for 1 min at 1,350 r.p.m. and resuspended in M9 buffer three times to remove bleach. Embryos were allowed to hatch ON in M9 buffer in a shaking incubator at 200 r.p.m. at 20 °C.
Day 3: RNAi feeding
L1 worms were resuspended at a concentration of approximately seven larvae per 10 μl of M9 buffer. Tween 20 was added to the worms to a final concentration of 0,01% to avoid worms sticking to the plastic tubing during dispensing. IPTG was added to each well of the bacterial cultures to a final concentration of 4 mM to induce dsRNA transcription during 1 h incubation at 37 °C. Bacteria was then pelleted by spinning at 4,000 r.p.m. for 5 min and subsequently resuspended in 400 μl of Complete S medium plus 4 mM IPTG. 40 μl of each bacterial culture were transferred to flat-bottomed 96-well plates followed by 10 μl of M9 buffer containing the L1 worms. The plates were incubated at 20 °C for 4 days with shaking at 200 r.p.m., allowing sufficient time for the L1 worms to grow to adults and lay eggs, and for the eggs to hatch and develop into larvae.
Day 7: Phenotype scoring
Prior to image acquisition, 70 μl of worm paralyzing solution (50 μl of complete S medium and 20 μl of 0,7% Tricaine+0,07% Tetramisole) was added to all wells. We imaged the entire well (25 fields per well with a 20x objective) and counted the number of adult worms (parents, bigger pharynxes) and the number of larvae (progeny, smaller pharynxes) using the Cellomics® Target Activation BioApplication (Fig. 1a, magnification).
Data collection and analysis
Quantification of number of adults
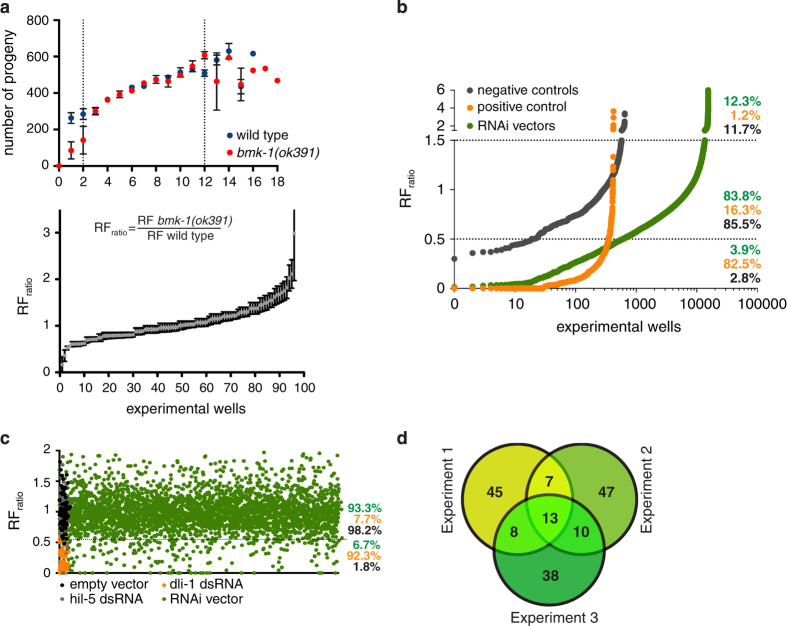
Automated image analysis was performed and the number of parent worms, which correspond to the number of L1s dispensed on day 3, was determined. Only wells with 2–12 parent worms were considered for further analysis, as the number of obtained progeny within this range is linear (Fig. 2a, top graph).
Figure 2. Genome-wide RNAi screen results.
(a, top) Determination of the ideal number of parents per well. Between 2 and 12 the number of obtained progeny is linear. Wild type strain is myo-2::GFP. bmk-1(ok391); myo-2::GFP is the mutant strain. (a, bottom) RFratio calculation and distribution of worms treated with an empty RNAi vector. Error bars represent SEM of three biological replicates, each containing two technical replicates. (b) Primary screen results. RFratio distribution of screen controls and library RNAi vectors. Dashed lines represent the thresholds for the definition of ‘enhancers’ (<0.5) and ‘suppressors’ (>1.5). Percentages represent the total number of negative (grey) and positive (orange) controls within each cut-off range. Total number of library RNAi vectors in each category is also represented by percentages (green). (c) Secondary screen results. RFratio distribution of each control or library RNAi vector across three biological replicates. Dashed line indicates the threshold value below which we considered the RNAi to ‘enhance’ bmk-1(ok391). Percentages represent the total number of negative controls (empty vector, hil-5 dsRNA, in dark grey), positive controls (orange) and library RNAi vectors (green) in each category. (d) Venn diagram showing the common and unique hits found within the biological replicates. 38 targeting sequences classify as ‘enhancers’ in at least two out of the three replicates.
Determination of reproductive fitness ratio
Reproductive Fitness (RF) is defined as the number of progeny per parent worm treated with dsRNA. It was obtained by dividing the total number of viable progeny by the number of parents for each experimental well. The Reproductive Fitness ratio (RFratio) was obtained dividing the RF of the control strain (SV1070) by the RF of the bmk-1(ok391) strain (SV1069) (Fig. 2a, bottom graph).
Data import, management, quality assessment and replicate summarization
Raw data, i.e., the RF for each targeted gene in the two strains, was analyzed using the software package cellHTS2 (ref. 13) implemented in Bioconductor/R. Normalization to correct for plate effects was performed by dividing each measurement by the median value across the ‘sample’ wells in each plate.
Hits selection. Primary screen
Upon determination of the RFratios, we continued with genes whose depletion led to minimally a 50% increased lethality in SV1070 when compared to SV1069 (RFratio≤0.5) (Fig. 2b). Upon visual inspection of the data, we also included genes whose depletion led to a significant (RFratio≥1.5) reduction in lethality in SV1070 when compared to SV1069. Targeted genes resulting in more than 90% lethality in the control strain were excluded, since the RFratio would be unreliable. In total 1,057 bacterial clones were selected for the secondary screen (575 enhancers and 482 suppressors) (Data Record 1).
Hits selection. Secondary screen
Using the RFratio of the positive and negative controls, we determined the percentage of expected false positives and false negatives for cut-offs ranging from 40–70%. We decided to continue with bacterial clones showing a minimum increase in lethality of 45% in SV1070 when compared with SV1069 (RFratio<0.55), since this predicts a similar percentage of false positive and false negative results across three biological replicates. 168 bacterial clones classified as hits under this criterion (Fig. 2c) (Data Record 2). Next, we built a Venn diagram with the three sets of hits from the three independent experiments and accepted as final hits genes that classified as enhancers in at least two out of the three independent experiments (Fig. 2d). All targeting vectors from the final hit list were sequenced in order to confirm the identity of the targeted gene (Table 1).
Data Records
The raw and analyzed data produced by this screen are formatted as two downloadable spreadsheets from FigShare (Data Citation 1).
Data record 1—primary screen
Primary dsRNA screen data are contained in tabs termed Controls (negative and positive controls), Samples (RNAi vectors) and ND Samples (targeting sequences for which an RFratio could not be determined for technical reasons). The tab termed ‘Selection 2nd Screen’ contains all the targeting sequences taken to the secondary screen and respective location coordinates in the arrayed hit plates (1–13).
Information regarding the column headings:
Plate
Ahringer library plate number.
Well
Well coordinates.
Description
Content description. ‘sample’, RNAi vector. ‘neg’, negative control. ‘pos’, positive control. ‘other’, embryonic lethality control.
GeneID
Gene sequence identification number/name.
Raw_r1/r2_
Raw data value for technical replicate 1 or 2 for each assayed worm strain.
RF (AVG replicates)
Average (arithmetic mean) Reproductive Fitness of replicates. Sometimes the RF of one of the two replicate could not be determined for technical reasons. In these cases the RF value corresponds to the RF of the single replicate that worked.
RFratio
Obtained dividing the RF (AVG replicates) of SV1070 by the RF (AVG replicates) of SV1069.
Classification
Controls or Samples are classified as ‘enhancer’ if RFratio is ≤0.5, ‘suppressor’ if RFratio is ≥1.5, otherwise ‘no effect’.
Data record 2—secondary screen
Secondary dsRNA screen data are included under different tabs of the spreadsheet. Content description and column details are given below:
‘Total data’
Contains the raw values and calculated RFratios for every experimental well of the three independent experiments. Column headings are the same as in Data Record 1.
‘Controls’
The RFratio for the negative and positive controls from the previous sheet was calculated for each experiment. Next, we classified each result as ‘enhancer’, ‘no effect’ or ‘suppressor’ applying different cut-offs (40%, 45%, 50%, 55%, 60%, 65%, 70%) to the RFratios. We then determined the predicted percentage of false positive and false negative results for each cut-off. To generate the final hit list, we used a 45% cut-off, which predicted a similar percentage of false positive and false negative results across the three independent experiments.
‘Samples’ (45%)
RFratio for each RNAi vector was extracted from the ‘Total data’ sheet and classified as ‘enhancer’, ‘no effect’ or ‘suppressor’ according to a 45% cut-off. Cells in red correspond to targeting sequences classifying as ‘enhancers/suppressors’ in the three independent experiments. Cells in orange correspond to targeting sequences classifying as ‘enhancers/suppressors’ in two out of the three independent experiments. ‘suppressors’ were not taken into consideration for further analysis, since they were not the aim of this screen. Nevertheless, they might represent a suppression of a RNAi phenotype by the bmk-1 loss of function.
Technical Validation
Technical and biological replicates
It was not possible to do the primary screen in multiple biological replicates due to the high cost and logistic challenges. Therefore, we used two technical replicates. For the secondary screen, three biological replicates were used, each containing two technical replicates. We used the same rational as in the primary screen, and in the end selected hits based on reproducibility.
Controls
Negative controls were used to define the threshold for hit selection. The positive control was found in a pilot screen.
Validation of the genetic interactions
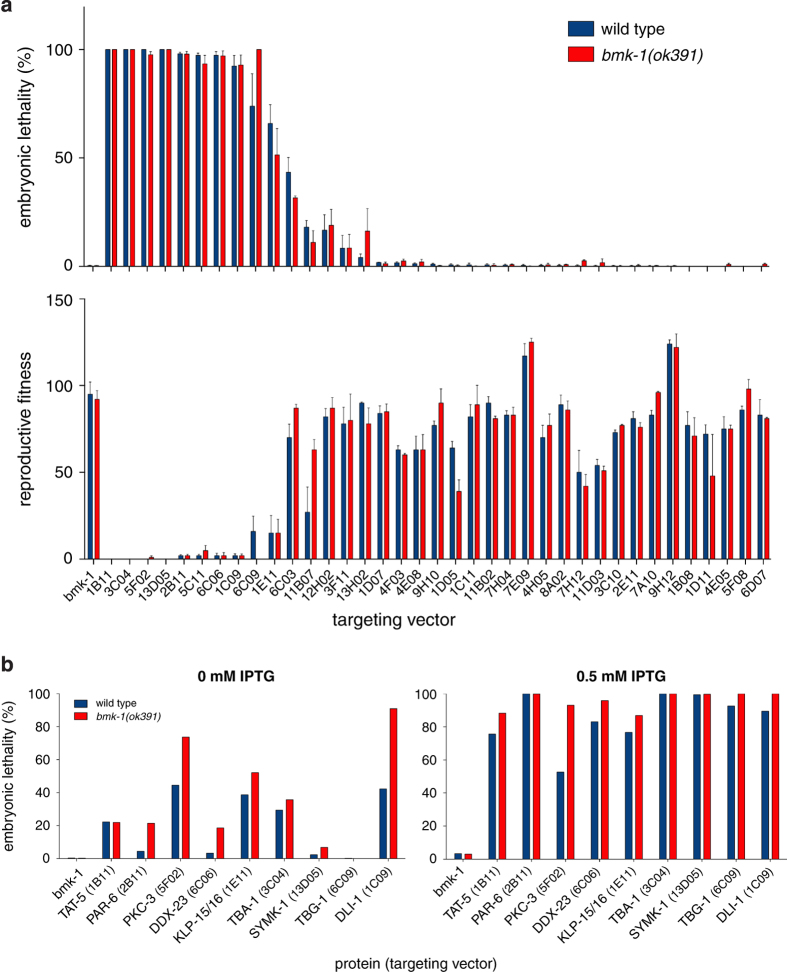
In order to confirm the lethality enhancement when the screen hits (‘enhancers’) are depleted in the bmk-1(ok391) background, we performed a classical embryonic lethality assay14. Briefly, this assay consisted of feeding wild type (N2) and bmk-1(ok391) (SV1005) L4 worms with HT115 bacteria carrying the RNAi vector and inducing the expression of dsRNA by IPTG addition (0.5 mM) on agar plates. These worm strains share the same genetic background as the strains used in the RNAi screen, with the exception that they do not express any fluorescent marker. 24 h after we started feeding the worms with dsRNA at 20 °C, we transferred them to new RNAi plates and allowed egg laying for another 24 h, after which the adult worms were removed. 24 h later embryonic lethality was accessed (Fig. 3a). Several gene knockdowns led to 100% embryonic lethality (e.g., dli-1, par-6, tbg-1) already in the wild type strain. This suggested that the knockdown of these proteins on agar plates was more penetrant than in the liquid format. Because this precluded us from detecting enhancement of lethality in the bmk-1(ok391) background, we repeated the embryonic lethality assay on agar plates in two conditions: weak RNAi (0 mM IPTG, resulting in partial depletion due to basal transcription of dsRNA in the absence of induction) and strong RNAi (0.5 mM IPTG, corresponding to maximal induction of dsRNA production). We were able to confirm an increase in embryonic lethality when the selected proteins were partially depleted (0 mM IPTG) in the bmk-1(ok391) background (Fig. 3b).
Figure 3. RNAi screen validation.
(a) Embryonic lethality assay with the screen's ‘enhancers’. N2 (blue), wild type strain. SV1005 (red), bmk-1(ok391) deletion strain. RNAi for bmk-1 was used as a negative control. Error bars represent SEM of three technical replicates. Top graph shows embryonic lethality and below the corresponding reproductive fitness (expressed in number of live progeny per parent). (b) Embryonic lethality assay upon weak (0 mM IPTG) or strong (0.5 mM IPTG) depletion of screen ‘enhancers’. Bars represent averages of two technical replicates.
Interestingly, 50% of the hits did not show any embryonic lethality when dsRNA was delivered by feeding on plates (Fig. 3a). We reasoned that they were selected as hits in the screen because the depletions caused a significant delay in larval development in the bmk-1(ok391) background relative to controls. As the automated analysis counted the number of progeny based on the size of their pharynxes, larvae with small pharynxes would remain undetected, lowering the reproductive fitness ratio. To test this hypothesis, we repeated the RNAi-mediated depletion of these proteins by feeding on plates and classified the phenotype of larval progeny 3 days after removal of the parent. For 14 out of 38 bacterial clones, we observed enhanced growth defects in the bmk-1(ok391) background (Table 1). We conclude that the genetic interactions uncovered in this screen include genes with roles in post-embryonic development.
Usage Notes
The RNAi screen data (Data Records 1–2) are provided for users to be able to apply their own normalisation strategies and thresholds for identifying differences between the two strains. This study focused on genes that when depleted enhanced the lethality in the worm kinesin-5 mutant bmk-1(ok391) when compared with the control strain. However, some genes showed decreased lethality when depleted in the bmk-1(ok391) background. These genes are also potential interactors of kinesin-5. Furthermore, several dsRNAs led to a significant developmental delay specifically in the bmk-1(ok391) background, which may indicate a function of kinesin-5 during development. The results obtained in this screen can be compared with data from previous RNAi screens in Caenorhabditis elegans as reported in RNAiDB (ref. 15).
Additional information
How to cite this article: Maia, A.F. et al. Genome-wide RNAi screen for synthetic lethal interactions with the C. elegans kinesin-5 homolog BMK-1. Sci. Data 2:150020 doi: 10.1038/sdata.2015.20 (2015).
Supplementary Material
Acknowledgments
We thank the members of the Medema, Kops, Lens, Boxem, The, van den Heuvel, Carvalho and Gassmann laboratories for helpful discussion. To Belen Fernandez-Garcia for helping on hit picking from the genome-wide library. To Oliver Pelz for help with web cellHTS2 application during data analysis. A.F.Maia is a FCT—Fundação para a Ciência e a Tecnologia postdoctoral fellow (SFRH/BPD/71364/2010). The R.H. Medema laboratory was supported by the Netherlands Organization for Scientific Research (ZonMw 918.46.616) and the Netherlands Genomics Initiative. S. van den Heuvel received funding from the Netherlands Organisation for Scientific Research (NWO), Chemical Sciences (CW ECHO project 711.011.010). Work on C.E. Sunkel laboratory was funded by FEDER funds through the Operational Competitiveness Programme—COMPETE and by National Funds through FCT—Fundação para a Ciência e a Tecnologia under the project FCOMP-01-0124-FEDER-019740 (PTDC/BIA-BCM/120366/2010). R.Gassmann laboratory is supported by an ERC Starting Grant (338410), an EMBO Installation Grant, and funding from the Fundação para a Ciência e a Tecnologia (IF/01015/2013/CP1157/CT0006). R.H. Medema had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Maia A. F. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1314604
References
- Kashina A. S. et al. A bipolar kinesin. Nature 379, 270–272 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L. C. et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 (2005). [DOI] [PubMed] [Google Scholar]
- Gaglio T. et al. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135, 399–414 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S. & Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70, 451–458 (1992). [DOI] [PubMed] [Google Scholar]
- Sharp D. J. et al. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125–138 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A. M., Powers J., Strome S. & Saxton W. M. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 17, R453–R454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K. A. & Baas P. W. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 178, 1081–1091 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar V. C., Ketschek A., Myers K. A., Gallo G. & Baas P. W. Kinesin-5 is essential for growth-cone turning. Curr. Biol. 18, 1972–1977 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli K. M., Jakovljevic J., Woolford J. L. & Saunders W. S. Kinesin molecular motor Eg5 functions during polypeptide synthesis. Mol. Biol. Cell 22, 3420–3430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G. et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 11, 1198–1203 (2004). [DOI] [PubMed] [Google Scholar]
- Kamath R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- Lehner B., Tischler J. & Fraser A. G. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nat. Protoc. 1, 1617–1620 (2006). [DOI] [PubMed] [Google Scholar]
- Boutros M., Brás L. P. & Huber W. Analysis of cell-based RNAi screens. Genome Biol. 7, R66 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G. & Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K. C. RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32, D 406–410 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Maia A. F. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1314604