Abstract
Severe influenza disease strikes otherwise healthy children and remains unexplained. We report compound heterozygous null mutations in IRF7, which encodes the transcription factor interferon regulatory factor 7, in an otherwise healthy child who suffered life-threatening influenza during primary infection. In response to influenza virus, the patient’s leukocytes and plasmacytoid dendritic cells produced very little type I and III interferons (IFNs). Moreover, the patient’s dermal fibroblasts and induced pluripotent stem cell (iPSC)–derived pulmonary epithelial cells produced reduced amounts of type I IFN and displayed increased influenza virus replication. These findings suggest that IRF7-dependent amplification of type I and III IFNs is required for protection against primary infection by influenza virus in humans. They also show that severe influenza may result from single-gene inborn errors of immunity.
Both seasonal and pandemic influenza viruses typically cause self-limiting respiratory disease but occasionally cause life-threatening acute respiratory distress syndrome (ARDS). The frequency of severe disease depends on the viral strain (1). Known host risk factors to severe influenza consist of a few acquired comorbidities, such as chronic pulmonary disease (2, 3). The pathogenesis of most cases of life-threatening influenza remains unexplained, especially among children (4). The lack of severe influenza in patients with known primary immunodeficiencies, including inborn errors of T and/or B cell immunity that predispose to a variety of related infections such as severe parainfluenza (5), is also unexplained (6). Only haploinsufficiency for GATA2, resulting in abnormal hematopoietic cell development, was reported in a few patients with severe influenza and other infections (7). Monogenic inborn errors of immunity may underlie life-threatening, isolated diseases in otherwise healthy children during primary infection by a few other viruses (8). We therefore hypothesized that severe influenza striking otherwise healthy children may also result from single-gene inborn errors of immunity.
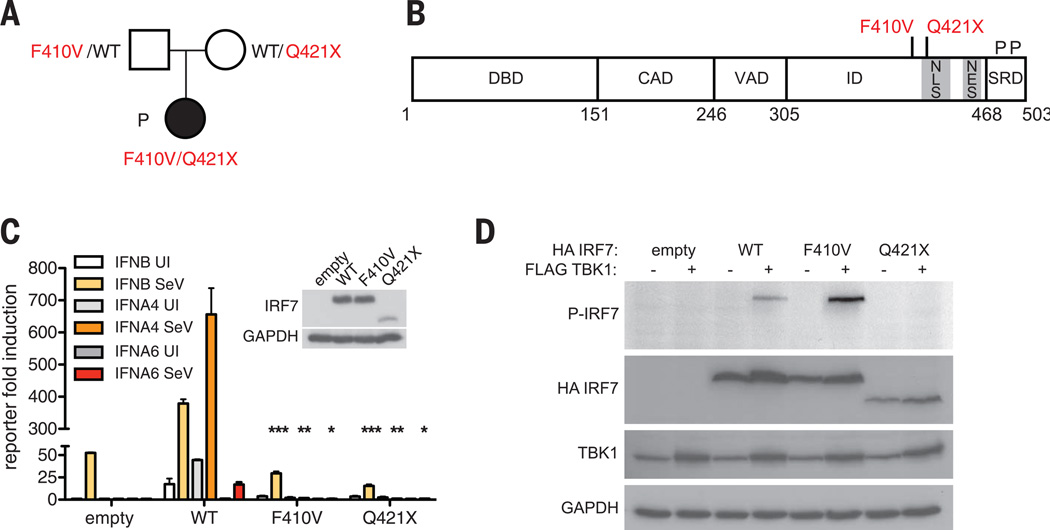
We performed whole-exome sequencing (WES) in a 7-year-old girl (“P”), one of 22 individuals sequenced (including only three children <5 years old) proven to have developed influenza in the course of primary infection (table S1). P suffered life-threatening ARDS during infection with laboratory-confirmed pandemic H1N1 (pH1N1) 2009 influenza A virus (IAV) in January 2011 at the age of 2.5 years, prior to any influenza vaccination. Serum drawn shortly after hospitalization showed protective antibody titers against A/Netherlands/602/2009 (H1N1) IAV (fig. S1A) but not against A/Perth/16/2009 (H3N2) or B/Brisbane/60/2008 (fig. S1B), indicating that this was her first encounter with IAV. She did not suffer from severe infections caused by other viruses (supplementary case report and fig. S1C). The patient had no known comorbidity and no detectable immunological abnormalities suggestive of any T or B cell deficit (table S2). She was born to nonconsanguineous parents of French descent (Fig. 1A and fig. S2A). WES analysis of the trio revealed, and Sanger sequencing confirmed, two compound heterozygous IRF7 mutations—p.Phe410Val (F410V) and p.Gln421X(Q421X)—with each parent being heterozygous for a single mutated allele (Fig. 1A and fig. S2B), which defined the best candidate genotype in this patient (9).
Fig. 1. Autosomal recessive IRF7 deficiency from compound heterozygous mutations.
(A) Familial segregation of IRF7 mutations in a nonconsanguineous French family. (B) Schematic illustration of IRF7A featuring DNA binding domain (DBD), constitutive activation domain (CAD), virus-activated domain (VAD), inhibitory domain (ID), and signal response domain (SRD). A potential nuclear localization signal (NLS) lies between amino acids 417 and 440, and the nuclear export signal (NES) between amino acids 448 and 462. Phosphorylation sites (P) Ser477 and Ser479 lie at the C terminus. Mutations are shown in red. (C) Wild-type, F410V, or Q421X IRF7 activation of IFNB, IFNA4, or IFNA6 promoter-driven reporter assay. Cells are uninfected (UI) or infected with SeV. Means ± SD of three independent experiments are shown. *P < 0.01, **P < 0.005, ***P < 0.001 as determined by t test. (D) Phosphorylation of HA-tagged wild-type (WT), F410V, or Q421X IRF7 coexpressed with FLAG-tagged TBK1 as assessed by Western blot with phospho-specific IRF7 antibody (P-IRF7); GAPDH was used as a loading control. This result is representative of two experiments.
Interferon regulatory factor 7 (IRF7) is a transcription factor that amplifies interferon (IFN) production in response to viruses (10–12). Specifically, IRF7 is involved in the amplification of mouse and human type I (13 IFNA, IFNB, IFNE, IFNK, IFNW) and type III (IL29, IL28A, IL28B) IFN genes (11–13). The missense F410V substitution is predicted to be damaging and absent in public databases. The nonsense Q421X (Fig. 1B and fig. S2B) is found as a heterozygous variant in 1 out of 118,062 chromosomes of the Exome Aggregation Consortium (ExAC) cohort, yielding a minor allele frequency of 0.000008. There are currently no homozygous or compound heterozygous nonsynonymous mutations found in our in-house (table S3) and public databases (ExAC). Thus, autosomal recessive IRF7 deficiency by compound heterozygosity may underlie severe influenza in this child.
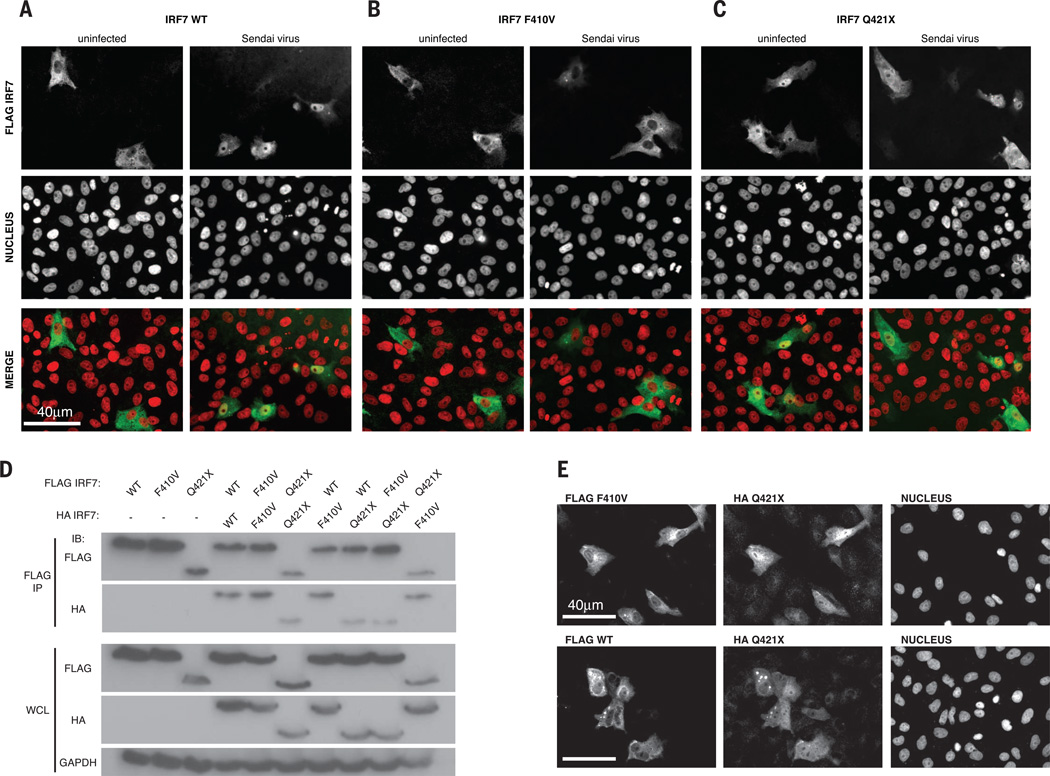
Each mutation was loss-of-function in reporter assays driven by IFNB, IFNA4, or IFNA6 promoters (Fig. 1C) (11). None of the other five heterozygous IRF7 variants tested was loss-of-function (table S4 and fig. S3). IRF7 can be activated by RIG-I recognition of IAV genomic RNA, resulting in C-terminal serine phosphorylation by the IKK-related kinases TBK1 and IKK-ε (11, 14–20). Wild-type IRF7 was phosphorylated upon overexpression of TBK1, as was F410V (Fig. 1D). The truncated Q421X product lacks the C-terminal serine residues and was not phosphorylated (Fig. 1D). Phosphorylation induces IRF7 nuclear accumulation and transcription of type I and III IFNs (11, 14–16). Wild-type IRF7 accumulated in the nuclei of all transfected Vero cells by 8 hours post-infection (hpi) with Sendai virus (SeV) (Fig. 2A), whereas F410V was cytoplasmic (Fig. 2B) and Q421X was nuclear with or without SeV infection (Fig. 2C). This suggested that F410V disrupts a potential nuclear localization signal (amino acids 417 to 440) and was consistent with Q421X missing the nuclear export signal (amino acids 448 to 462) (15).
Fig. 2. P’s IRF7 alleles are loss-of-function by different mechanisms.
(A to C) Localization of FLAG-tagged wild-type (A), F410V (B), or Q421X (C) IRF7 in uninfected or Sendai virus–infected Vero cells by immunofluorescence imaging. This result is representative of two experiments. (D) Wild-type and mutant IRF7 dimerization by immunoprecipitation (IP) with antibody to FLAG followed by Western blot with antibodies to FLAG and HA.WCL, whole-cell lysate. (E) Localization of FLAG F410V (top) and FLAG wild-type IRF7 (bottom) cotransfected with HA Q421X IRF7 in Vero cells as assessed by immunofluorescence imaging. This result is representative of two experiments.
The two mutant IRF7 alleles led to loss of function by different mechanisms: F410V did not accumulate in the nucleus despite phosphorylation, whereas Q421X resided in the nucleus without phosphorylation. The mutant IRF7 proteins could homo- or heterodimerize (Fig. 2D), which suggests that coexpression might enable nuclear unphosphorylated Q421X to shuttle phosphorylated cytoplasmic F410V to the nucleus, where IRF7 mutant heterodimers could up-regulate IFNs. However, there was no nuclear relocalization of F410V or the wild type in the presence of Q421X (Fig. 2E). Consistently, there was no rescue in reporter assays when the two mutants were coexpressed (fig. S4A). Moreover, neither allele was dominant negative in terms of IRF7 or IRF3 function (fig. S4, B to D). This is consistent with the lack of infectious phenotype, including severe influenza, in P’s heterozygous parents. Overall, the patient’s severe influenza may result from lack of functional IRF7 homodimers, or IRF3-IRF7 heterodimers, or both.
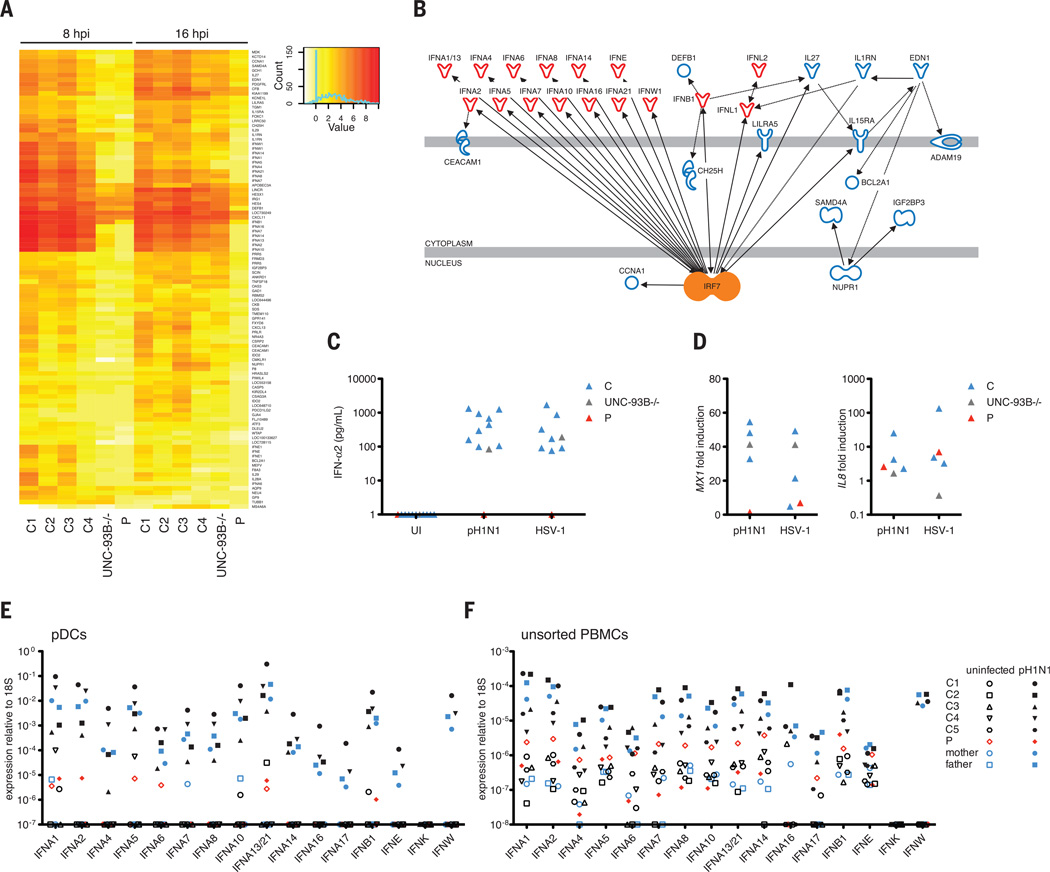
We investigated the genome-wide impact of IAV infection in peripheral blood mononuclear cells (PBMCs) by microarray, quantitative polymerase chain reaction (qPCR), and enzyme-linked immunosorbent assay (ELISA). At baseline, PBMCs from P displayed significant down-regulation of innate immune genes (fig. S5). We observed a robust induction of type I IFN genes IFNA14, -16, -7, -2, -10, -13, and -21 (in order of relative change); IFNE; and IFNW1, as well as type III IFN genes IL29 and IL28A, in healthy donors but not in P, at 8 and 16 hpi (Fig. 3A). Among IFN genes, the only exception was a factor of 2 induction of IFNB in P, albeit this was less than in controls by a factor of 4. Ingenuity pathway analysis software independently predicted IRF7 as an upstream regulator of these genes (P = 2.02 × 10−13) (Fig. 3B). IFN-stimulated genes (ISGs) known to inhibit IAV replication—MX1 (21), RSAD2 (22), BST2 (23), and SERPINE1 (24)—were up-regulated normally in P, perhaps via stimulation by IFN-β. Her PBMCs displayed a profound or complete defect of IFN-α2 production after infection with 11 other viruses or stimulation with Toll-like receptor (TLR) agonists (fig. S6). IFN-β and IFN-λ1 levels were impaired but less so than IFN-α, whereas IL-6 production was normal (fig. S6, B to D). Overall, we observed an overwhelming and selective defect of type I and III IFN induction in P’s PBMCs.
Fig. 3. Impaired IRF7-dependent innate immunity in leukocytes and pDCs.
(A) The top 5% of genes, in PBMCs infected with pH1N1 at MOI (multiplicity of infection) = 2, whose relative changes were >2 (up or down) in controls and ≤1 in the patient, P, as analyzed by gene array. (B) Causal network analysis of the differentially regulated genes. (C) IFN-α production at 24 hpi with pH1N1 IAV or HSV-1 in pDCs from four healthy controls, P, and an UNC-93B–deficient individual (UNC-93B−/−), a control for TLR responses. (D) MX1 and IL8 were measured in cells from (C) by qPCR. (E and F) The expression of all indicated type I IFN genes was measured by qPCR in purified pDCs (E) and unsorted PBMCs (F) separately infected with pH1N1 IAV at MOI = 1. The probe for IFNA13 also detects IFNA21 mRNA. All data shown in (C) to (F) are representative of two independent experiments.
In mice and humans, IRF7 is constitutively expressed in plasmacytoid dendritic cells (pDCs), resulting in pDCs being the major type I IFN-producing cells (25, 26). We measured IFN-α2 production in response to pH1N1 infection in P’s pDCs (Fig. 3C), which were found at normal frequency (table S5). We observed no IFN-α2 production in pDCs from P at 24 hpi with pH1N1 and herpes simplex virus–1 (HSV-1) (Fig. 3C). MX1 induction was abolished, while that of IL8 was normal (Fig. 3D). The patient’s heterozygous mother produced IFN-α2 like controls (fig. S7A). We quantified the induction of all 20 human type I and type III IFN genes by qPCR in purified pDCs and unsorted PBMCs after infection with pH1N1 (Fig. 3, E and F). At 8 hpi, P was deficient for type I IFN (including the 13 IFNA, IFNE, IFNK, and IFNW) and type III IFN (IL29, IL28A, and IL28B) genes in both cell preparations (Fig. 3, E and F, and fig. S7B). IFNB was, however, mildly induced in P’s pDCs (Fig. 3E), and the induction of MX1 in P at 8 hpiwas normal (fig. S7C). Collectively, a small amount of IRF7-independent IFN-β triggered early ISG up-regulation at 8 hpi in pDCs, but the IRF7-dependent amplification of type I and III IFNs was lacking for sustained ISG induction at 24 hpi.
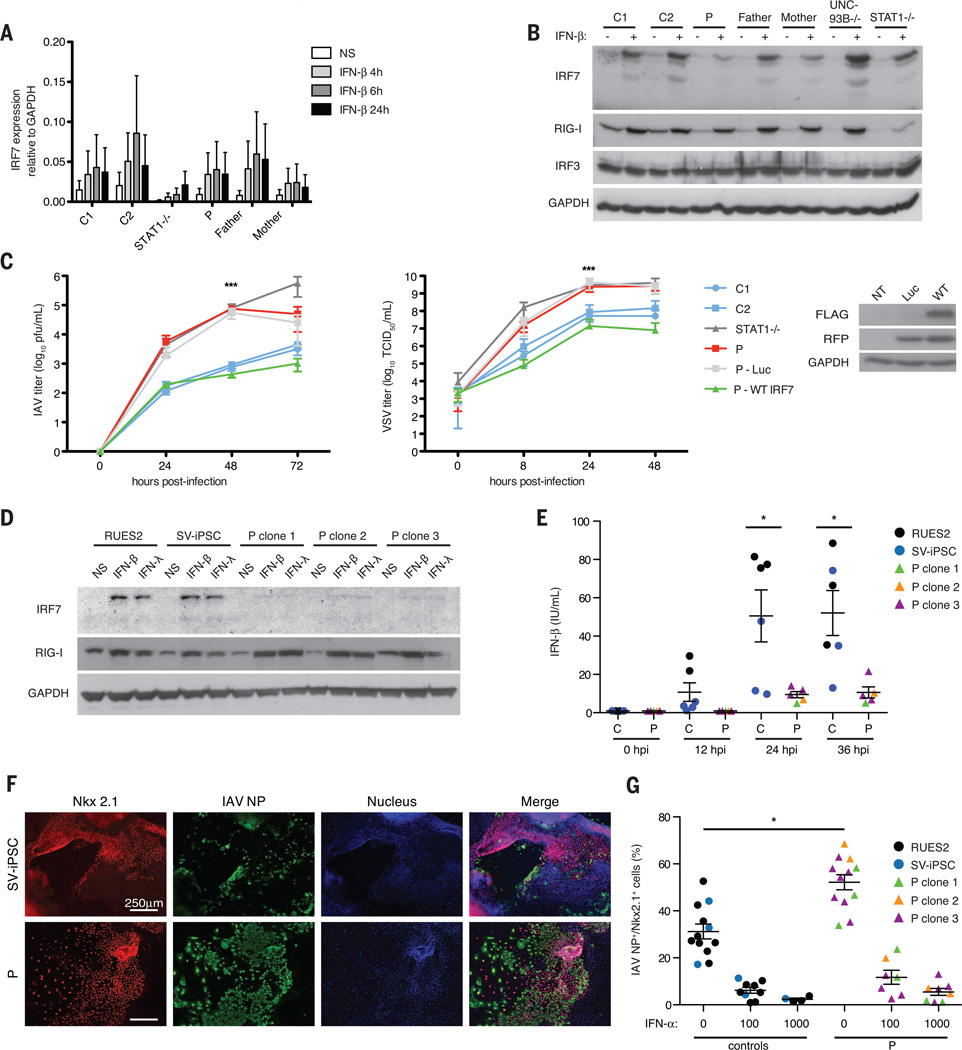
We investigated the impact of IRF7 deficiency on cell-intrinsic, nonhematopoietic immunity, using P’s SV40-immortalized fibroblasts (F-SV40). Basal and IFN-β–induced IRF7 mRNA expression in F-SV40 from P was normal (Fig. 4A). In contrast, IRF7 protein expression was diminished, even after IFN-β treatment (Fig. 4B).We observed approximately 2-log higher titers of IAV at 48 hpi relative to healthy controls (Fig. 4C). Stable transfection of wild-type IRF7 complemented this phenotype (Fig. 4C). Wild-type IRF7 similarly rescued enhanced replication of vesicular stomatitis virus (VSV) (Fig. 4C). Highly pathogenic avian H5N1 IAV also replicated to high titers in P’s F-SV40, indicating that the phenotype was not IAV strain–specific (fig. S8A). Further, treatment with exogenous IFN-α2b protected the fibroblasts from IAV and VSV replication (fig. S8, B and C). IFN-β production by fibroblasts was impaired after IAV infection (fig. S8D); however, it was normal after stimulation with extracellular or intracellular polyinosine-polycytidine (synthetic double-stranded RNA) (fig. S8E). This is consistent with intact IRF3-dependent signaling (fig. S4D) and detectable IFN-β production in P’s PBMCs (fig. S6A).
Fig. 4. IRF7-dependent intrinsic immunity is required for control of IAV infection.
(A) IRF7 mRNA induction at indicated time points after IFN-β treatment in F-SV40. Means ± SD of three replicates are shown. (B) IRF7 expression in F-SV40 measured byWestern blot 18 hours after treatment with IFN-β. (C) Virus titers in F-SV40 from P stably transfected luciferase (Luc) or wild-type IRF7 with an internal ribosome entry site–expressed red fluorescent protein, after infection with pH1N1 (MOI = 10) or VSV (MOI = 3). Means ± SD for pH1N1 IAV (n = 3) and VSV (n = 7) are shown. ***P < 0.001 between controls and P as determined by t test. (D) IRF7 expression in IFN-β– or IFN-λ–treated PECs derived from healthy control ESCs (RUES2), SV iPSCs, or three individual clones (clones 1, 2, and 3) of P’s iPSCs as detected by Western blot. (E) IFN-β production in PECs infected with A/PR/8/34-GFP as measured by ELISA at indicated time points. Means ± SD of two independent experiments are shown. *P < 0.05 as determined by t test. (F) Staining of IAV nucleoprotein (NP) (green) and Nkx2.1 (red) in PECs derived from SV-iPSC control and P infected at MOI = 1 with pH1N1 IAV. Cells derived from a single representative clone of P’s iPSCs are shown. (G) Percentage of Nkx2.1+ cells scored as positive for IAV NP, in PECs untreated or treated with IFN-α (100 or 1000 U/ml) for 18 hours and infected with pH1N1 for 24 hours. Means ± SD of two independent experiments are shown. *P < 1 × 10−6 as determined by χ2 analysis.
IAVs first target the entire respiratory tract, with pH1N1 2009 viral antigen present in type I and type II pneumocytes in humans (2, 27). We generated patient-specific pulmonary epithelial cells (PECs) from induced pluripotent stem cells (iPSCs) derived from P’s primary fibroblasts (28). IAV replication and IFN-β induction were compared in PECs derived from an embryonic stem cell line (RUES2), a SeV-reprogrammed healthy control iPSC line (SV-iPSC), and three independent IRF7-deficient iPSC clones. IRF7 expression in response to IFN-α, IFN-β, and IFN-λ1 treatment and IFN-β production in response to IAV were impaired in P lines (Fig. 4, D and E). Overall titers of IAV appeared equal regardless of IRF7 genotype, largely because of efficient replication in cells negative for Nkx2.1, a pulmonary epithelium marker in these cultures. However, when we scored the infected Nkx2.1+ cells for IAV nucleoprotein (NP) antigen (Fig. 4F), 52.4% of P’s PECs were double positive, versus 27.2% in controls (Fig. 4G). This phenotype was rescued by treatment with IFN-α2b (Fig. 4G), IFN-β, or IFN-λ1 (fig. S9). These data suggest that impaired intrinsic immunity in the pulmonary epithelium may have contributed to P’s ARDS. Impaired IFN production by P’s pDCs may also have caused disease, as evidenced by the pulmonary cell rescue with exogenous IFN-α2b.
A single-gene inborn error of IRF7 immunity can underlie life-threatening, isolated influenza in humans during primary infection, broadening the range of human infections that result from genetic lesions (8, 9, 29, 30). IRF7 deficiency disrupts the main function of pDCs, the production of antiviral IFNs. This is distinct from deficiencies of IRF8 and GATA2 that impair development of all circulating monocytes and DCs and of multiple myeloid and lymphoid subsets, respectively (7, 31, 32). IRF7 deficiency also affects cell-autonomous, intrinsic immunity in PECs. The lack of IRF7-dependent type I and III IFN amplification by pDCs, PECs, or possibly other cell types likely underlies the patient’s severe influenza. Interestingly, IRF7 is redundant for vaccine-mediated immunity to influenza viruses, as the child has been healthy for 5 years with annual influenza vaccination as the sole secondary prevention. The IRF7-deficient child, now 7 years old, also displays a narrow infectious phenotype, restricted to severe influenza, at odds with the broad role of mouse IRF7 in antiviral immunity (10, 30). Human IRF7 seems to be largely redundant in host defense against viruses. It will be important to search for deficiencies in IRF7 and related genes in children with influenza and other severe viral illnesses. Our study provides proof of principle that single-gene inborn errors of immunity can cause severe childhood influenza. IFN-based, patient-tailored therapeutic strategies could be helpful in life-threatening influenza of childhood (33, 34)
Supplementary Material
ACKNOWLEDGMENTS
We warmly thank our patient and her family. We thank S. Boucherit, T. Kochetkov, M. Fenner, M. Duchamp, A. Abhyankar, A. Belkadi, L. Shang, Y. Liang, L. Amar, and Y. Nemirovskaya for their contributions and all members of the laboratory for fruitful discussions. Plasmids used to generate recombinant influenza viruses are subject to a material transfer agreement. We thank members of the French Memo-Flu-ARDS study group (PIs: B. Autran, Inserm U945, and C.-E. Luyt, Service de réanimation médicale, Hôpital Pitié-Salpêtrière, Paris) for recruiting adult severe flu patients. Supported by National Center for Research Resources and National Center for Advancing Translational Sciences grant 8UL1TR000043; NIH grants 5R01NS072381 (J.-L.C. and L.D.N.), 5R01AI100887 (L.D.N.), and 1U19AI109945 (C.F.B.); the Rockefeller University, the St. Giles Foundation, the French National Research Agency under the “Investments for the Future” program grant ANR-10-IAHU-01, the Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (ANR-10-LABX-62-IBEID), INSERM, Paris Descartes University, and ERC grant 2010-StG-261299 (F.G.); and Center for Research on Influenza Pathogenesis and the NIAID-funded Center of Excellence for Influenza Research and Surveillance contract HHSN272201400008C (A.G.-S.). The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Raw data for the microarray analyses performed in this study are available from the public repository of GEO DataSets (accession no. GSE66486). The raw sequence data are available on the Sequence Read Archive (SRA) database: Bioproject SRP055919.
Footnotes
www.sciencemag.org/content/348/6233/448/suppl/DC1
Case Report
Materials and Methods
Supplementary Text
Figs. S1 to S9
Tables S1 to S7
References (35–50)
REFERENCES AND NOTES
- 1.Palese P, Shaw ML. In: Fields Virology. 5. Knipe D, Howley P, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1647–1689. [Google Scholar]
- 2.Shieh W-J, et al. Am. J. Pathol. 2010;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawood FS, et al. Pediatrics. 2011;128:e27–e32. doi: 10.1542/peds.2010-3343. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention, FluView: Influenza-Associated Hospitalization Surveillance Network. 2013 http://gis.cdc.gov/grasp/fluview/FluHospChars.html.
- 5.Gennery AR, Cant AJ. J. Clin. Pathol. 2001;54:191–195. doi: 10.1136/jcp.54.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Herz W, et al. Front. Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigley V, et al. J. Exp. Med. 2011;208:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casanova JL, Abel L. Annu. Rev. Genomics Hum. Genet. 2013;14:215–243. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. J. Exp. Med. 2014;211:2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda K, et al. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 11.Marié I, Durbin JE, Levy DE. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterlund PI, Pietilä TE, Veckman V, Kotenko SV, Julkunen I. J. Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 13.Sato M, et al. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 14.Caillaud A, Hovanessian AG, Levy DE, Marié IJ. J. Biol. Chem. 2005;280:17671–17677. doi: 10.1074/jbc.M411389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin R, Mamane Y, Hiscott J. J. Biol. Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- 16.Marié I, Smith E, Prakash A, Levy DE. Mol. Cell. Biol. 2000;20:8803–8814. doi: 10.1128/mcb.20.23.8803-8814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato H, et al. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehwinkel J, et al. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M, et al. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 21.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Hinson ER, Cresswell P. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Mangeat B, et al. J. Biol. Chem. 2012;287:22015–22029. doi: 10.1074/jbc.M111.319996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dittmann M, et al. Cell. 2015;160:631–643. doi: 10.1016/j.cell.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkmann M, et al. J. Immunol. 2003;170:4465–4474. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 26.Izaguirre A, et al. J. Leukoc. Biol. 2003;74:1125–1138. doi: 10.1189/jlb.0603255. [DOI] [PubMed] [Google Scholar]
- 27.Itoh Y, et al. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang SX, et al. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conley ME, Casanova JL. Curr. Opin. Immunol. 2014;30:17–23. doi: 10.1016/j.coi.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casanova JL, Abel L, Quintana-Murci L. Cold Spring Harb. Symp. Quant. Biol. 2013;78:157–172. doi: 10.1101/sqb.2013.78.019968. [DOI] [PubMed] [Google Scholar]
- 31.Hambleton S, et al. N. Engl. J. Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasquet M, et al. Blood. 2013;121:822–829. doi: 10.1182/blood-2012-08-447367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron S, Isaacs A. BMJ. 1962;1:18–20. doi: 10.1136/bmj.1.5270.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agrati C, et al. J. Infect. Dis. 2010;202:681–689. doi: 10.1086/655469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.