Abstract
Hepatitis C virus (HCV) chronically infects 130-170 million people worldwide and is a major public health burden. HCV is an RNA virus that infects hepatocytes within liver, and this infection is sensed as non-self by the intracellular innate immune response to program antiviral immunity to HCV. HCV encodes several strategies to evade this antiviral response, and this evasion of innate immunity plays a key role in determining viral persistence. This review discusses the molecular mechanisms of how the intracellular innate immune system detects HCV infection, including how HCV pathogen-associated molecular patterns are generated during infection and where they are recognized as foreign by the innate immune system. Further, this review highlights the key innate immune evasion strategies used by HCV to establish persistent infection within the liver, as well as how host genotype influences the outcome of HCV infection. Understanding these HCV-host interactions is key to understanding how to target HCV during infection and for the design of more effective HCV therapies at the immunological level.
Keywords: HCV, RIG-I, MAVS, interferon, mitochondrial-associated membranes
Introduction
Hepatitis C virus (HCV) is an RNA virus that chronically infects 130-170 million people worldwide, with 3-4 million new infections per year.1 HCV infects hepatocytes within the liver, and persistent infection by HCV in the liver leads to varying stages of liver disease, including fibrosis, cirrhosis, and hepatocellular carcinoma.1 Every year, more than 350,000 people die from HCV-related liver diseases. Until recently, the standard of care for HCV was treatment with pegylated interferon-α and ribavirin and this was only effective in about 50% of infected patients.2 The efficacy of HCV therapy has increased dramatically in recent years with the advent of direct acting antivirals, including the newly developed HCV protease and polymerase inhibitors. 3 While treatment with these therapies can lead to a successful treatment outcome up to 70% of the time, these therapies often have significant side effects, are by and large genotype specific, and can cause viral resistance to emerge.4 Currently, there is no vaccine for HCV.5
The host defenses that initially sense HCV infection take place within an arm of the immune system termed the antiviral innate immune response. This immune response is triggered in a cell intrinsic manner when pattern recognition receptors (PRRs) within the infected cell sense the virus as non-self or foreign, and trigger downstream signaling cascades that activate immunity. This antiviral response is the first line of defense against viral infection, and not only can it be directly antiviral by acting to suppress viral replication and spread to other cells, but this innate immune response is also required for programming functional adaptive immune responses and therefore coordinates the entire host immune response to infection. The importance of innate immunity in control of viral infection is underscored by the fact that many viruses, including HCV, have evolved ways to inactivate various innate immune signaling factors.6
Interestingly, approximately 20-30% of people who are infected with HCV clear the virus during the acute stage of infection, while 70-80% develop a chronic, life-long infection.1 The mechanisms that underlie these differences are still not fully understood, but likely reflect a complex interplay between the virus and the host at the level of the immune response. Therefore, a detailed understanding of what makes up an effective immune response to HCV and how HCV counteracts this immune response will be essential for developing new antiviral strategies, with the ultimate goal of reducing the disease burden of HCV, including preventing liver disease and cancer. This review will focus on recent advances in how HCV activates and then subsequently evades the antiviral innate immune response to establish a productive infection. In addition, this review will discuss how the host genetic background influences the antiviral response to HCV in both natural and treatment-induced clearance.
How is HCV sensed as non-self?
The innate immune response to RNA viruses is composed of three main classes of PRRs, termed the RIG-I (retinoic acid-inducible gene I)-like receptors (RLRs), the toll-like receptors (TLRs), and the nucleotide oligomerization domain-like receptors (NLRs).7 These proteins sense specific features called pathogen-associated molecular patterns (PAMPs) within the viruses or present during viral infection. Following PAMP recognition, PRRs signal through various downstream molecules to activate transcription factors that drive the expression of antiviral genes and various cytokines, such as the type I and III interferons and IL-1β. Secretion of these cytokines from the infected cell activates signaling in a paracrine and/or autocrine fashion to establish the full antiviral state.
HCV is an enveloped, positive-strand RNA virus of the genus hepacivirus and a member of the Flaviviridae family. HCV isolates have been classified into 7 major genetic groups, referred to as genotypes, with sequence diversity of greater than 30%.8; 9 HCV replicates as a quasispecies population and it is thought that this contributes to viral persistence because it enables the virus to quickly mutate to escape neutralizing antibodies, preventing an effective antibody response.10 The HCV virion, which is coated with host lipoprotein, is comprised of the viral E1 and E2 glycoproteins surrounding the nucleocapsid core. This lipoprotein-coated virion interacts with several host cell entry factors in a sequential fashion for entry into the hepatocyte via receptor-mediated endocytosis followed by fusion in the early endosome.11 Following HCV entry, the viral RNA genome of 9.6 kilobases is released into the cytoplasm. From there, and in association with the rough ER, this RNA is translated from an internal ribosome entry site (IRES) into a single polyprotein. This polyprotein is then co- and post-translationally cleaved into the structural (core, E1, and E2) and non-structural (p7, NS2, NS3, NS4A, NS5A, and NS5B) proteins of the virus by host proteases and two virally-encoded proteases.12 HCV replication induces a rearrangement of intracellular membranes into a structure called the “membranous web”. Viral RNA replication takes place in association with these intracellular membranes, and many of the HCV proteins themselves are membrane associated.13 HCV RNA replication, catalyzed by the viral RNA-dependent RNA polymerase NS5B, produces an antigenomic RNA that serves as a template for the production of more positive sense genomic viral RNA. These new viral genomes are then packaged into a nucleocapsid through interactions with several HCV proteins at the lipid droplet and subsequently at ER membranes in close proximity to these sites. HCV assembly is closely coupled to the host cell lipid synthesis pathway and utilizes this pathway for entry into the secretory pathway and eventual release of a lipoprotein-coated virion from the infected cell.14; 15
HCV can be sensed by all three of these classes of PRRs (RLRs, TLRs, and NLRs; see Fig. 1). The best described antiviral sensor protein for HCV is RIG-I. RIG-I is a cytosolic RNA helicase that belongs to the mammalian RLR family, which also includes MDA5 (melanoma differentiation-associated protein 5) and LGP2 (laboratory of genetics and physiology 2). RIG-I has three major domains, including a C-terminal domain (often referred to as the repressor domain), a central DExD/H box RNA helicase domain, and two CARD domains at the N-terminus.16; 17 The stimulatory ligands for RIG-I have been well-characterized (reviewed in18; 19; 20), and consist of RNA containing a 5’ triphosphate (5’-ppp) moiety and/or having double stranded structure.21; 22 The C-terminal domain of RIG-I selectively binds to the 5’-ppp, a distinguishing feature of non-self RNA.23; 24
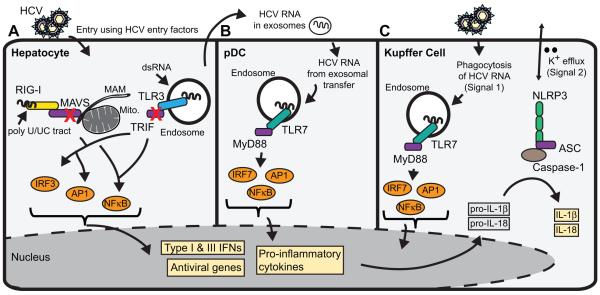
Figure 1. Innate immune sensing of HCV.
Hallmarks of HCV infection can be sensed by pattern recognition receptors (PRRs) in a number of cell types within the liver to activate innate immunity. (A) HCV infection in hepatocytes is sensed by RIG-I, which detects the poly U/UC tract present in the 3’end of HCV RNA, and by TLR3, which detects dsRNA replication intermediates or HCV RNA that has been endocytosed from dying cells. These PRRs signal downstream to activate the innate immune response program, including the production of IFNβ, IFNλ, and pro-inflammatory cytokines, which act in an autocrine and paracrine manner to establish the full innate immune response. The red “X” indicates points of regulation of these signaling pathways by the HCV NS3/4A protease. (B) HCV is sensed in plasmacytoid dendritic cells (pDCs) by TLR7-mediated recognition of HCV RNA in the endosome. While pDCs themselves are not productively infected with HCV, the viral RNA is taken up by the pDC following exosomal transfer of viral RNA from productively infected hepatocytes. C) HCV can also be sensed in Kupffer cells, the liver resident macrophages, by the inflammasome signaling complex. Kupffer cells phagocytose HCV RNA (Signal 1) for TLR7-dependent signaling that activates the transcription of the pro-forms of IL-1β and IL-18, while potassium efflux (Signal 2) sensed by NLRP3 drives caspase 1 processing of IL-1β and IL-18 into the mature form for secretion and full activation of the inflammasome.
RLR recognition of HCV
HCV activates the RIG-I pathway at very early times after infection25; 26, and RIG-I activation attenuates HCV replication.27 HCV RNA physically binds to RIG-I27; 28, and the HCV PAMP sensed by RIG-I contains a multi-motif signature consisting of poly U/UC region located within the 3’NTR of the virus, along with a 5’-ppp.28; 29 Recent work has further shown that the 34 nucleotide poly-uridine core within the poly U/UC region is a key RNA sequence motif for recognition of the HCV PAMP by RIG-I.30 The poly U/UC region of the HCV genome is highly conserved among HCV genotypes. It is also essential for HCV replication31; 32; 33, and therefore the HCV RNA sequence in this region is likely evolutionarily constricted and unable to evolve to evade detection by RIG-I. It is likely because of this fact that HCV has other mechanisms to inactivate RIG-I pathway signaling (see below).
It is not yet known how exactly the 5’-ppp and poly U/UC region interact to form the HCV PAMP during an actual HCV infection or when this PAMP would be presented to RIG-I. It could be that known long range or “kissing loop” interactions between the 5’ and 3’ ends of the HCV genome bring the 5’-ppp in close proximity to the poly U/UC region for presentation to RIG-I.31; 33 Alternatively, the anti-genomic strand of HCV may contain a 5’-ppp, and dsRNA intermediates formed during HCV replication between this strand and the genomic strand could present a multi-component PAMP signature to RIG-I.28 Both the exact timing and subcellular location of this PAMP detection by RIG-I during HCV infection are questions that remain to be answered. For example, is the incoming full-length genomic HCV RNA sensed by RIG-I? Is the HCV RNA in viral nucleocapsids the first RIG-I ligand, similar to influenza A virus, vesicular stomatitis virus, and bunyaviruses?34 Or is some level of HCV RNA replication a pre-requisite for RIG-I sensing, as has been proposed for some aspects of influenza A virus sensing by RIG-I?35; 36
When the HCV PAMP binds to RIG-I, it promotes a RIG-I conformational change that relieves the autorepression formed through the C-terminal domain16; 22 to promote its activation and oligomerization.16; 37 The crystal structures of RIG-I have shown that this conformational change following 5’-ppp or dsRNA binding allows the helicase domain to make direct contacts with viral RNA38; 39; 40, possibly scanning the PAMP via ATP hydrolysis41 for specific activating sequences, such as the poly U/UC tract found in HCV RNA. This conformational change in RIG-I release the CARD domains for ubiquitination by TRIM2542, and interaction with 14-3-3ε to promote association with intracellular membranes43 for translocation to the mitochondrial-associated ER membrane (MAM)44 to interact with MAVS, the RIG-I signaling adaptor protein. This interaction of RIG-I with MAVS results in assembly of a signalosome complex that activates effector molecules, including the transcription factors IRF3 and NFκB, to drive downstream innate immune signaling.
The PRR MDA5 is also a sensor of RNA virus infection and in many ways quite similar to RIG-I; it has an equivalent domain architecture with two N-terminal CARD domains, a central DExD/H box RNA helicase domain, and a C-terminal domain.45 To date, there is no described role for MDA5 in sensing of HCV RNA, but it is quite possible that it could play a role in the antiviral response triggered by HCV infection. Similar to RIG-I, MDA5 also signals through MAVS to drive innate immune signaling. However, MDA5 does have a distinct role from RIG-I in the antiviral response.46 The viral ligand for MDA5 is long higher order dsRNA or dsRNA replication intermediates.21; 47; 48; 49; 50 Interestingly, during infection of West Nile virus, an RNA virus also in the Flaviviridae, both RIG-I and MDA5 contribute to the antiviral response. In this case, RIG-I is important early after infection, while MDA5 is important for signaling during later times of infection.51 It may be that innate immune signaling to HCV is orchestrated in a similar fashion, with differing contributions of RIG-I and MDA5 to the antiviral response via sensing of different PAMPs presented at different times by HCV as it establishes a productive infection. While in vitro transcribed HCV RNA itself does not require MDA5 to activate signaling to IFN-β28, dsRNA replicative intermediates of HCV that accumulate at later times during infection could be bona fide MDA5 ligands. Importantly, expression of the paramyxovirus V protein, known to block MDA5 signaling, can enhance HCV replication, strongly suggesting a role for MDA5 in antiviral immunity to HCV during a productive infection.52
TLR recognition of HCV
HCV infection is also monitored in the host by the TLRs. These receptors can recognize either viral nucleic acid (TLR3, TLR7, TLR9) or protein (TLR2 and TLR4) PAMPs. HCV proteins can be sensed by TLR253; 54, likely on the cell surface, and HCV infection can induce expression of TLR4.55 Intracellularly, TLR3 and TLR7 have both been shown to play roles in sensing of HCV RNA (Fig. 1).56; 57; 58; 59; 60; 61 The endosomal protein TLR3 senses HCV RNA for the activation of IFN-β and other proinflammatory cytokines. TLR3 is expressed in liver cell types that would be relevant for HCV infection, including hepatocytes and Kupffer cells, the liver-resident macrophages.59; 60; 61 Activation of TLR3 inhibits HCV replication, suggesting that TLR3 is part of the antiviral response to HCV infection.59; 61 TLR3 signals are transduced through TIR-domain containing adapter-inducing IFN-β (TRIF) leading to activation of the transcription factors IRF3 and NFκB for induction of innate immunity. The HCV PAMPs for TLR3 are comprised of dsRNA replicative intermediates that form over the course of a productive infection.59 Additional HCV PAMPS for TLR3 could come following uptake of HCV RNA from the extracellular milieu (possibly from dying infected cells) by scavenger receptors or through autophagic processes.62; 63 This uptake could lead to viral PAMP presentation to TLR3 in the endosome in uninfected cells setting up an antiviral state even in uninfected hepatocytes. HCV sensing by TLR7 occurs in both plasmacytoid dendritic cells (pDCs) and Kupffer cells, leading to production of IFN or activation of the inflammasome (Fig. 1B-C; see below).56; 57; 58
HCV activation of NLRs and the inflammasome
A more recently discovered sensing pathway in the innate immune response to RNA viruses is governed by NLRs and results in activation of the inflammasome.64 The inflammasome is a multi-protein complex made up of a sensor protein (for RNA viruses, this is NLRP3), the adaptor protein ASC, and the cellular protease caspase 1. Activation of the NLRs triggers the production and secretion of the proinflammatory cytokines IL-1β and IL-18. NLRP3 inflammasome activation requires two main triggers. The first (Signal 1) is a priming event involving transcriptional upregulation of the pro-forms of IL-1β and IL-18, The second trigger (Signal 2) results in activation of caspase 1 to process IL-1β and IL-18 into their mature forms for secretion from the cell. The nature of these two activating signals is diverse, suggesting that there is no single specific activator for NLRP3.65 For influenza, Signal 1 is sensing of the viral RNA by TLR7, and Signal 2 is activated by the viral M2 ion channel protein.66 Additional triggers that could play a relevant role during viral infection included mitochondrial reactive oxygen species and potassium efflux.67; 68 We now know that HCV also activates the inflammasome, resulting in IL-1β and IL-18 secretion and induction of a proinflammatory response.57; 69; 70 HCV induction of the inflammasome occurs in Kupffer cells, which are not productively infected by HCV but rather are able to phagocytose HCV to drive TLR7 signaling leading to transcriptional upregulation of the pro-form of IL-1β (see Fig. 1C). Potassium efflux drives caspase 1 processing of IL-1β into the mature form.57 It is still unknown how potassium efflux is activated in response to HCV exposure of Kupffer cells, but it is interesting that UV-inactivated HCV also induces the inflammasome, suggesting a non-replication mechanism of action.57 The activation of IL-1β by HCV likely contributes to the liver inflammation that is observed in chronically infected hepatitis C patients. IL-1β has been shown to inhibit replication of HCV RNA71, and so production of IL-1β during HCV infection could also be playing a direct antiviral role. Interesting, IL-18 activation has also been reported during acute cases of HCV infection72, and so it still remains to be determined how IL-18 activation by the inflammasome would contribute liver inflammation in chronically infected patients, but the combination of IL-18 and IL-1β could program hepatic stellate cells towards fibrogenesis.
Other HCV sensors that drive innate immunity
Another recently described sensor protein for HCV is the well-known antiviral protein protein kinase R (PKR). PKR is a dsRNA binding protein that upon activation uses its kinase domain to phosphorylate the α subunit of the eukaryotic translation initiation factor 2 (eIF2α) to inhibit translation of host mRNAs. HCV evades this host shutoff and the actions of PKR as its RNA uses an IRES for cap-independent translation.73; 74; 75; 76 Recently PKR was shown to bind to HCV RNA at very early times after infection (ie at 2-6hr, prior to RIG-I sensing) and signal to activate ISGs and IFN-β, independent of the kinase activity of PKR.25 This signaling induces protein-protein interactions between PKR with MAVS, which has previously been described as a signaling adaptor protein for PKR.25; 77; 78; 79 It has been suggested that PKR sensing of HCV RNA induces a proviral state due to negative regulation of RIG-I via the induction of specific genes that negatively regulate RIG-I (e.g. ISG15).25 However, more work is required to fully understand the cross-talk between PKR and RIG-I for control of HCV replication and innate immune induction during HCV infection. In addition, while the PAMP for PKR is the highly structured IRES region in the genome73; 76, it is still unknown how and where this PAMP is presented to PKR for signaling.
Most of the research that has addressed HCV activation of IFN has focused the mechanisms leading to activation of type I IFN. However, increasingly type III IFN (the IFNλ’s) has been shown to be an important antiviral cytokine for HCV. Not only is it directly antiviral towards HCV80, but genome wide association studies have found single nucleotide polymorphisms (SNPs) near the gene encoding the IFNL3 cytokine that can predict whether one will have an acute or chronic HCV infection81; 82, as well as the outcome of HCV therapies.82; 83; 84; 85 The type III IFNs are comprised of the three closely related cytokines, IFNL1, IFNL2, and IFNL3 (also known as IL29, IL28A, and IL28B). Additionally a fourth type III IFN, IFNL4, has also recently been discovered (and SNPs in IFNL4 have also been described to be associated with natural and treatment-induced HCV clearance), although much of the biology governing IFNL4 remains to be determined.86; 87 Similar to type I IFN, signaling by the type III IFNs upon receptor engagement activates the JAK/STAT signaling pathway for the formation of the ISGF3 transcription complex (made of up IRF-9 and STAT1/STAT2 heterodimers) and transcriptional activation of very similar ISG profiles (reviewed in88). However, the receptor for type III IFN is different from that of type I IFN, and is comprised of the IL10R2 and IFNLR1 receptor, which have a more limited cellular distribution profile than IFNAR, the receptor for type I IFN.89 This limited receptor distribution profile likely contributes to a more refined, local antiviral action of the type III IFNs. HCV infection of primary hepatocyte cultures does induce type III IFN90; 91; 92, but not much is known about the sensing pathways and transcription factors that contribute to this activation during HCV infection. While the HCV poly U/UC PAMP, the HCV 3’UTR, as well as poly I:C, can signal to various members of the type III IFN family through IRF3 and NFκB dependent signaling pathways in different cell types91; 93; 94, the actual HCV PAMP generated during HCV infection that triggers type III IFN, as well as the timing of this activation, differential induction of the members of the type III IFN family, and relevant cell types, are still unknown. The presence of the unfavorable SNP near IFNL3 does appear to result in less expression of IFNL3 within the liver, peripheral blood mononuclear cells, blood dendritic cell antigen 3+ dendritic cells that accumulate in the liver, and whole blood84; 85; 95; 96; 97; 98, suggesting that IFNL3 is required for optimal HCV clearance and therapy responses. However, a functional SNP that governs regulation of IFNL3, as well as the mechanism for this regulation, have yet to be identified. Further, it remains to be determined why IFNL3 itself would be the key IFNλ that governs natural and treatment-induced responses to HCV infection.99 Answers to these questions will produce great advances in our understanding of how to make a productive immune response to HCV and in the design of new therapies for HCV.
The above described antiviral sensing pathways for HCV likely all contribute to a functional immune response to HCV, including priming of an adaptive immune response, important for ultimate viral clearance.100; 101 However, we know very little about how these sensor proteins cooperate to induce the antiviral response, the timing of their activation, as well as the most relevant cell types of action. In addition, in most cases the natural HCV PAMP for these sensors is unknown. The most authentic HCV PAMP may in fact be HCV RNA in complex with viral RNA binding proteins.102 HCV infection takes place within hepatocytes located within the liver. Other cell types within the liver, including pDCs58; 94, Kupffer cells (described above57), and stellate cells103 can induce type I and III IFNs and other proinflammatory cytokines during HCV infection through uptake of viral RNA and/or cross-talk with hepatocytes, for example by exosomal PAMP carriers56, and cytokine production by these cells undoubtedly influences the outcome of HCV infection. Further, it is possible that other as yet undiscovered RNA, protein, and metabolite-sensing pathways contribute to the antiviral response to HCV. For example, membrane perturbations that occur during the HCV entry process could activate some level of innate immune signaling, as has been described for virus-like particles and other enveloped viruses.104; 105
Immune effectors of HCV
Innate immunity activates a signaling cascade that induces IFN and also hundreds of ISGs, many with direct antiviral properties that control virus replication and spread.106; 107 One of the major therapies for HCV utilizes IFN-α, which can drive expression of ISGs, even in the infected patient.108 As IFN-α-based therapies are effective at eliminating HCV in about half of those infected with the virus, ISGs likely have some direct antiviral activity towards HCV in the infected hepatocyte. It is important to also remember that innate immune signaling, including IFN signaling, is involved in orchestrating functional adaptive immune responses known to control HCV infection outcome.109; 110; 111 Several recent screens have identified key ISGs with antiviral activity towards infectious HCV.112; 113; 114; 115 These studies have complemented other single and combinatorial ISG functional studies that have previously identified a range of ISGs with anti-HCV effects (reviewed in 107; 116). While many ISGs have been identified with anti-HCV activity, the antiviral mechanism of action towards HCV for many of these ISGs has yet to be discovered, and at times has been conflicting based on the experimental cell lines and assays used to asses activity.116 Several ISGs, including PKR, Viperin, and the IFITM family of proteins, have consistently been identified as having anti-HCV activity. In particular, Viperin, which blocks viral replication through interactions with NS5A 117; 118; 119, and also IFITM1, which prevents HCV entry by blocking the function of the HCV entry factors120 have very strong anti-HCV activity. It is unlikely that any single ISG is the “key anti-HCV ISG” that dictates viral clearance during either natural infection or in response to IFN-based therapy112, but rather it is likely the combinatorial action of these ISGs that, in concert with the adaptive immune response, dictate the outcome of the infection.
HCV subversion of innate immune surveillance programs
Despite the fact that HCV is sensed by many different innate immune sensing pathways, in about 80% of those infected the immune response does not effectively clear the virus, resulting in a life-long chronic infection.121 While there could be many factors that contribute to the ability of the virus establish chronic infection, one hypothesis is that HCV evasion of host innate immune signaling contributes to viral persistence and chronicity. HCV utilizes several mechanisms to evade innate immunity. The key viral factor of the HCV immune evasion program is the viral NS3/4A protease, which consists of two proteins, NS3 and NS4A, that oligomerize to form a protein complex. NS3/4A plays many important roles in the viral life cycle, including in viral RNA replication, polyprotein processing, and viral assembly.122 To regulate innate immune signaling, NS3/4A utilizes its protease domain to cleave key innate immune signaling adaptor proteins, effectively inactivating those viral RNA detection programs. To regulate the RIG-I signaling pathway, NS3/4A targets and cleaves MAVS, preventing its dimerization and downstream signaling of innate immunity.26; 123; 124; 125; 126; 127; 128 Both NS3/4A and MAVS are associated with intracellular membranes and localized to diverse sites within the intracellular membrane network, including MAM, peroxisomes, and mitochondria.44; 129 NS3/4A cleavage of MAVS occurs at the MAM, releasing MAVS from its association with intracellular membranes and thereby eliminating RIG-I pathway signaling.44 The fact that this cleavage occurs specifically at the MAM, rather than the mitochondria, the first described subcellular location for MAVS130, suggests that the MAM-localized MAVS must drive RIG-I pathway signaling and antiviral responses during HCV infection. Cleavage of MAVS during HCV infection has been shown in vivo in patients, and patients with cleaved MAVS have lower levels of IFN pathway induction.26; 131 Because PKR can also signal through MAVS to activate a subset of innate immune genes25, it is likely that NS3/4A cleavage of MAVS also regulates PKR signaling of innate immunity. The subcellular location and mechanisms, including time of action during HCV infection, that govern this specific regulation are still unknown. In fact, it has been hypothesized that MAVS evolution has been driven by interactions with viral proteases of ancient hepaciviruses, implicating MAVS cleavage as a key component of hepacivirus pathogenesis.132 Overall, MAVS cleavage by NS3/4A during HCV infection is likely a critical component of the HCV immune evasion program to prevent induction of the antiviral response to HCV, contributing to mechanisms of viral persistence.
The HCV NS3/4A protease can also cleave TRIF133, the key adaptor for the TLR3 signaling pathway. TRIF-cleavage by NS3/4A has been shown directly in vitro, and during HCV infection TRIF protein levels decrease, most likely from destabilization of the protein after cleavage by NS3/4A.61; 133 We know much less about the molecular mechanisms regulating TRIF cleavage by NS3/4A than we do for cleavage of MAVS. Cleavage of TRIF by NS3/4A does block TLR3-dependent signaling.133 As TRIF has also been implicated as an adaptor protein for non-TLR3 dependent innate immune signaling134, it is quite possible that NS3/4A-mediated cleavage of TRIF could also regulate non-TLR3-mediated signaling.
In addition to blocking the IFN induction pathway, HCV has several strategies for evading the IFN response pathway. One of the ways that HCV blocks the IFN response pathway is through PKR activation. In cells persistently infected with HCV and treated with IFN-α, PKR kinase activation results in translational suppression of host mRNAs, including ISGs (but not HCV, where translation is driven by the IRES), thereby precluding the antiviral functions of IFN.74 Several HCV proteins have also been implicated as regulators of the IFN response pathway, but the literature on this topic is often conflicting.101; 135 It is clear that expression of HCV proteins blocks IFN signaling at the level of the JAK/STAT pathway.136 A clear understanding of how this block occurs will be useful to understanding how IFN therapy responses may be inhibited during HCV infection.
There are likely other strategies used by HCV for immune evasion yet to be discovered that play critical roles in viral infection outcomes. Additionally, there is an increasing role for miRNAs in mediating HCV innate immune evasion strategies. For example, miR-122, a miRNA critical for HCV replication137, has been suggested to shield the 5’-end of the HCV genome from RNA sensing pathways.138; 139 An HCV-induced miRNA has also been reported to be involved in evasion of IFN signaling to promote HCV replication.140 With recent advances in miRNA detection sensitivities, it is quite likely that other host miRNAs that regulate novel aspects of immunity to HCV will be described, and these discoveries will provide insights for the design of new antiviral therapies towards HCV.
IFN response to HCV
In spite of the fact that HCV has multiple strategies to antagonize innate immune signaling pathways, many patients infected with HCV display high hepatic ISG mRNA activation profiles within the liver.108; 141; 142; 143 Further, hepatitis C patients that have induced expression of ISGs prior to therapy are the ones that most often do not respond to therapy. There are many questions that remain concerning these pre-therapy induced ISGs, including how are these ISGs induced prior to therapy, what is the cell type producing the IFN that activates these ISGs, and why are they only induced in some patients? The HCV genotypes have differential response rates to IFN therapy, and infection with the difficult to treat HCV genotypes 1 and 4 leads to high pre-therapy hepatic ISG expression.95; 108; 144 Differential responses to distinct viral genotypes of HCV could be due to the variability of the viruses to regulate IFN induction and/or signaling pathways.131; 145; 146; 147; 148; 149
It has been proposed that this high pre-therapy activation of ISGs prevents type I IFN from having any functional antiviral effect because the cells become refractory to IFN signaling, perhaps through activation of specific ISGs that act as negative regulators of IFNAR signaling.150; 151 Additionally, it could be that while ISG mRNA levels are high in these patients, activation of PKR (as described above) prevents the translation of these ISGs and therefore prevents their anti-HCV effector function.74 Recent studies have implicated the specific cell-type profile of pre-therapy ISG levels as critical to predicting therapy responses, with the pre-therapy levels of ISGs in Kupffer cells being a strong predictor of therapy responses.95; 144; 152 This work suggests that functional crosstalk between Kupffer cells and hepatocytes, perhaps through induction and response to various cytokines (including type I and/or type III IFN), may be the key to a functional immune response towards HCV infection. Understanding the mechanisms that underlie the induction of ISGs in Kupffer cells during both HCV infection and therapy, and how patient IFNL3 genotype may influence these interactions, will be critical to understanding how interactions between Kupffer cells or other immune cells like pDCs and T cells contribute to eventual clearance of HCV in the infected hepatocyte.
Conclusions
We now know that HCV is sensed as foreign or non-self by multiple arms of the innate immune response, and activation of all of these responses would be key to priming a functional adaptive response that would lead to eventual viral clearance. However, HCV antagonism of the innate immune response, both at the level of the IFN response and the IFN induction pathways, likely plays a key role in viral pathogenesis and ability of the virus to maintain a persistent, life-long infection in many who become infected. In the coming years, research aimed at understanding how the multiple arms of the innate immune response interact, as well as how the interactions between relevant cell types together drive immunity to HCV will be key to understanding what drives a protective immune response to HCV and will be required to inform the development of a vaccine for HCV. Furthermore, an understanding of how one’s genotype at IFNL3, HLA, or other loci, along with gender, age, and ethnicity, influence both spontaneous and therapy-induced HCV clearance will be important to design personalized HCV therapies in the future.
Acknowledgments
I would like to thank Dr. Courtney Wilkins, Dr. Arjun Rustagi, and John Errett for helpful suggestions and critical reading of this manuscript.
Abbreviations
- CARD
Caspase activation and recruitment domain
- eIF2α
eukaryotic translation initiation factor 2 alpha
- HCV
hepatitis C virus
- IFITM1
interferon induced transmembrane protein 1
- IFN
interferon
- IFNAR
interferon receptor
- IFNL
interferon lambda
- IL-1β
interleukin 1, beta
- IRES
internal ribosome entry site
- IRF
interferon regulatory factor
- ISG
interferon stimulated gene
- JAK
janus kinase
- LGP2
laboratory of genetics and physiology 2
- MAM
mitochondrial-associated membrane
- MAVS
mitochondrial antiviral signaling protein
- MDA5
melanoma differentiation-associated protein 5
- NFκB
nuclear factor kappaB
- NLR
Nod-like receptor
- NLRP3
Nod-like receptor protein 3
- NTR
non translated region
- PAMP
pathogen-associated molecular pattern
- pDC
plasmacytoid dendritic cell
- PKR
protein kinase R
- PRR
pattern recognition receptor
- RIG-I
retinoic acid inducible gene I
- RLR
RIG-I-like receptor
- SNP
single nucleotide polymorphism
- STAT
signal transducer and activator of transcription
- TLR
toll-like receptor
- TRIF
TIR-domain containing adapter-inducing IFN-β
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Soriano V, Peters MG, Zeuzem S. New therapies for hepatitis C virus infection. Clin Infect Dis. 2009;48:313–20. doi: 10.1086/595848. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann WP, Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2011;8:257–64. doi: 10.1038/nrgastro.2011.49. [DOI] [PubMed] [Google Scholar]
- 4.Sarrazin C, Hezode C, Zeuzem S, Pawlotsky JM. Antiviral strategies in hepatitis C virus infection. J Hepatol. 2012;56(Suppl):S88–S100. doi: 10.1016/S0168-8278(12)60010-5. [DOI] [PubMed] [Google Scholar]
- 5.Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–78. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–22. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–7. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33–53. doi: 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]
- 9.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C Virus into 7 genotypes and 67 Subtypes: updated criteria and assignment web resource. Hepatology. 2013 doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 11.Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14–9. doi: 10.1016/j.coviro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 13.Moradpour D, Gosert R, Egger D, Penin F, Blum HE, Bienz K. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 2003;60:103–9. doi: 10.1016/j.antiviral.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–49. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–7. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 18.Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–6. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 19.Rehwinkel J, Reis ESC. Targeting the viral Achilles' heel: recognition of 5'-triphosphate RNA in innate anti-viral defence. Curr Opin Microbiol. 2013 doi: 10.1016/j.mib.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlee M, Hartmann G. The chase for the RIG-I ligand--recent advances. Mol Ther. 2010;18:1254–62. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–10. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr., Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–40. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–79. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–43. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnaud N, Dabo S, Akazawa D, Fukasawa M, Shinkai-Ouchi F, Hugon J, Wakita T, Meurs EF. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog. 2011;7:e1002289. doi: 10.1371/journal.ppat.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumpter R, Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–99. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–7. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–84. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnell G, Loo YM, Marcotrigiano J, Gale M., Jr. Uridine Composition of the Poly-U/UC Tract of HCV RNA Defines Non-Self Recognition by RIG-I. PLoS Pathog. 2012;8:e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3' nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326–38. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi M, Lemon SM. 3' nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003;77:3557–68. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You S, Rice CM. 3' RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82:184–95. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. Incoming RNA virus nucleocapsids containing a 5'-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013;13:336–46. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–8. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–87. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr., Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–7. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 40.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 43.Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M., Jr. The Mitochondrial Targeting Chaperone 14-3-3epsilon Regulates a RIG-I Translocon that Mediates Membrane Association and Innate Antiviral Immunity. Cell Host Microbe. 2012;11:528–37. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–9. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 48.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–96. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–9. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–16. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, Bhatia SN, Rice CM. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–12. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang S, Dolganiuc A, Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. J Leukoc Biol. 2007;82:479–87. doi: 10.1189/jlb.0207128. [DOI] [PubMed] [Google Scholar]
- 54.Dolganiuc A, Oak S, Kodys K, Golenbock DT, Finberg RW, Kurt-Jones E, Szabo G. Hepatitis C core and nonstructural 3 proteins trigger toll-like receptor 2-mediated pathways and inflammatory activation. Gastroenterology. 2004;127:1513–24. doi: 10.1053/j.gastro.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 55.Machida K, Cheng KT, Sung VM, Levine AM, Foung S, Lai MM. Hepatitis C virus induces toll-like receptor 4 expression, leading to enhanced production of beta interferon and interleukin-6. J Virol. 2006;80:866–74. doi: 10.1128/JVI.80.2.866-874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–70. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, Delker DA, Jo J, Bertoletti A, Hagedorn CH, Gale M., Jr. IL-1beta Production through the NLRP3 Inflammasome by Hepatic Macrophages Links Hepatitis C Virus Infection with Liver Inflammation and Disease. PLoS Pathog. 2013;9:e1003330. doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A. 2010;107:7431–6. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55:666–75. doi: 10.1002/hep.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–35. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 61.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83:9824–34. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dansako H, Yamane D, Welsch C, McGivern DR, Hu F, Kato N, Lemon SM. Class A Scavenger Receptor 1 (MSR1) Restricts Hepatitis C Virus Replication by Mediating Toll-like Receptor 3 Recognition of Viral RNAs Produced in Neighboring Cells. PLoS Pathog. 2013;9:e1003345. doi: 10.1371/journal.ppat.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–51. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horvath GL, Schrum JE, De Nardo CM, Latz E. Intracellular sensing of microbes and danger signals by the inflammasomes. Immunol Rev. 2011;243:119–35. doi: 10.1111/j.1600-065X.2011.01050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–10. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 68.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–9. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 69.Burdette D, Haskett A, Presser L, McRae S, Iqbal J, Waris G. Hepatitis C virus activates interleukin-1beta via caspase-1-inflammasome complex. J Gen Virol. 2012;93:235–46. doi: 10.1099/vir.0.034033-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Shrivastava S, Mukherjee A, Ray R, Ray RB. Hepatitis C Virus Induces Interleukin-1beta (IL-1beta)/IL-18 in Circulatory and Resident Liver Macrophages. J Virol. 2013;87:12284–90. doi: 10.1128/JVI.01962-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu H, Liu C. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J Virol. 2003;77:5493–8. doi: 10.1128/JVI.77.9.5493-5498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chattergoon MA, Levine JS, Latanich R, Osburn WO, Thomas DL, Cox AL. High plasma interleukin-18 levels mark the acute phase of hepatitis C virus infection. J Infect Dis. 2011;204:1730–40. doi: 10.1093/infdis/jir642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. Hepatitis C virus controls interferon production through PKR activation. PLoS ONE. 2010;5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–22. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koev G, Duncan RF, Lai MM. Hepatitis C virus IRES-dependent translation is insensitive to an eIF2alpha-independent mechanism of inhibition by interferon in hepatocyte cell lines. Virology. 2002;297:195–202. doi: 10.1006/viro.2002.1455. [DOI] [PubMed] [Google Scholar]
- 76.Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–37. doi: 10.1016/j.antiviral.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 77.Kanazawa N, Kurosaki M, Sakamoto N, Enomoto N, Itsui Y, Yamashiro T, Tanabe Y, Maekawa S, Nakagawa M, Chen CH, Kakinuma S, Oshima S, Nakamura T, Kato T, Wakita T, Watanabe M. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J Virol. 2004;78:9713–20. doi: 10.1128/JVI.78.18.9713-9720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams BR. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–16. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–51. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079–89. doi: 10.1074/jbc.M804296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Gunthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Mullhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45. doi: 10.1053/j.gastro.2009.12.056. 1345 e1-7. [DOI] [PubMed] [Google Scholar]
- 83.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 84.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–4. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 86.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, Gerlach T, Malinverni R, Moradpour D, Negro F, Mullhaupt B, Bochud PY. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–16. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kelly C, Klenerman P, Barnes E. Interferon lambdas: the next cytokine storm. Gut. 2011;60:1284–93. doi: 10.1136/gut.2010.222976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–23. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–88. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park H, Serti E, Eke O, Muchmore B, Prokunina-Olsson L, Capone S, Folgori A, Rehermann B. IL-29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology. 2012;56:2060–70. doi: 10.1002/hep.25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, Shu HB, Zhong J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–8. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 94.Stone AE, Giugliano S, Schnell G, Cheng L, Leahy KF, Golden-Mason L, Gale M, Jr., Rosen HR. Hepatitis C Virus Pathogen Associated Molecular Pattern (PAMP) Triggers Production of Lambda-Interferons by Human Plasmacytoid Dendritic Cells. PLoS Pathog. 2013;9:e1003316. doi: 10.1371/journal.ppat.1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L, Papassotiropoulos A, Roth V, Heim MH. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021–31. doi: 10.1053/j.gastro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 96.Fukuhara T, Taketomi A, Motomura T, Okano S, Ninomiya A, Abe T, Uchiyama H, Soejima Y, Shirabe K, Matsuura Y, Maehara Y. Variants in IL28B in liver recipients and donors correlate with response to peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology. 2010;139:1577–85. doi: 10.1053/j.gastro.2010.07.058. 1585 e1-3. [DOI] [PubMed] [Google Scholar]
- 97.Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, Nattermann J, Oldenburg J, Sauerbruch T, Spengler U. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–65. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 98.Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M, Murata K, Fukuhara T, Matsuura Y, Hayashi N, Mizokami M, Takehara T. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-lambda in response to hepatitis C virus. Hepatology. 2013;57:1705–15. doi: 10.1002/hep.26182. [DOI] [PubMed] [Google Scholar]
- 99.Horner SM, Gale M., Jr. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–88. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–52. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 101.Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2011.00319.x. [DOI] [PubMed] [Google Scholar]
- 102.Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo AV, Zlotnick A, Mukhopadhyay S, Ranjith-Kumar CT, Kao CC. Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS ONE. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nishitsuji H, Funami K, Shimizu Y, Ujino S, Sugiyama K, Seya T, Takaku H, Shimotohno K. Hepatitis C Virus Infection Induces Inflammatory Cytokines and Chemokines Mediated by the Cross Talk between Hepatocytes and Stellate Cells. J Virol. 2013;87:8169–78. doi: 10.1128/JVI.00974-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, Rixon FJ, Herold BC, Fitzgerald KA, Paludan SR. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–43. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noyce RS, Taylor K, Ciechonska M, Collins SE, Duncan R, Mossman KL. Membrane perturbation elicits an IRF3-dependent, interferon-independent antiviral response. J Virol. 2011;85:10926–31. doi: 10.1128/JVI.00862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–9. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–5. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walker CM. Adaptive immunity to the hepatitis C virus. Adv Virus Res. 2010;78:43–86. doi: 10.1016/B978-0-12-385032-4.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19:859–68. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Trippler M, Lohmann V, Binder M, Frese M, Bartenschlager R. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012 doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 113.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fusco DN, Brisac C, John SP, Huang YW, Chin CR, Xie T, Zhao H, Jilg N, Zhang L, Chevaliez S, Wambua D, Lin W, Peng L, Chung RT, Brass AL. A genetic screen identifies interferon-alpha effector genes required to suppress hepatitis C virus replication. Gastroenterology. 2013;144:1438–49. doi: 10.1053/j.gastro.2013.02.026. 1449 e1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao H, Lin W, Kumthip K, Cheng D, Fusco DN, Hofmann O, Jilg N, Tai AW, Goto K, Zhang L, Hide W, Jang JY, Peng LF, Chung RT. A functional genomic screen reveals novel host genes that mediate interferon-alpha's effects against hepatitis C virus. J Hepatol. 2012;56:326–33. doi: 10.1016/j.jhep.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Metz P, Reuter A, Bender S, Bartenschlager R. Interferon-stimulated genes and their role in controlling hepatitis C virus. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 117.Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, McCartney EM, Jangra RK, Lemon SM, Beard MR. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–17. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hinson ER, Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009;284:4705–12. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang S, Wu X, Pan T, Song W, Wang Y, Zhang F, Yuan Z. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J Gen Virol. 2012;93:83–92. doi: 10.1099/vir.0.033860-0. [DOI] [PubMed] [Google Scholar]
- 120.Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M., Jr. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–9. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seeff LB. The history of the "natural history" of hepatitis C (1968-2009) Liver Int. 2009;29(Suppl 1):89–99. doi: 10.1111/j.1478-3231.2008.01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J Viral Hepat. 2011;18:305–15. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 123.Foy E, Li K, Wang C, Sumpter R, Jr., Ikeda M, Lemon SM, Gale M., Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–8. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 124.Foy E, Li K, Sumpter R, Jr., Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–91. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 126.Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–22. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol. 2006;80:6072–83. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–81. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 131.Bellecave P, Sarasin-Filipowicz M, Donze O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T, Moradpour D, Heim MH. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127–36. doi: 10.1002/hep.23426. [DOI] [PubMed] [Google Scholar]
- 132.Patel MR, Loo YM, Horner SM, Gale M, Jr., Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr., Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–78. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Horner SM, Gale M., Jr. Intracellular innate immune cascades and interferon defenses that control hepatitis C virus. J Interferon Cytokine Res. 2009;29:489–98. doi: 10.1089/jir.2009.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–75. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 138.Machlin ES, Sarnow P, Sagan SM. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–8. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–6. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–44. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 142.Asselah T, Bieche I, Narguet S, Sabbagh A, Laurendeau I, Ripault MP, Boyer N, Martinot-Peignoux M, Valla D, Vidaud M, Marcellin P. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516–24. doi: 10.1136/gut.2007.128611. [DOI] [PubMed] [Google Scholar]
- 143.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ, Fried MW. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–63. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, Heathcote J, Edwards AM, McGilvray ID. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–33. doi: 10.1053/j.gastro.2009.10.046. e1-3. [DOI] [PubMed] [Google Scholar]
- 145.Donlin MJ, Cannon NA, Aurora R, Li J, Wahed AS, Di Bisceglie AM, Tavis JE. Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients. PLoS ONE. 2010;5:e9032. doi: 10.1371/journal.pone.0009032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gale M, Jr., Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–18. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Noguchi T, Satoh S, Noshi T, Hatada E, Fukuda R, Kawai A, Ikeda S, Hijikata M, Shimotohno K. Effects of mutation in hepatitis C virus nonstructural protein 5A on interferon resistance mediated by inhibition of PKR kinase activity in mammalian cells. Microbiol Immunol. 2001;45:829–40. doi: 10.1111/j.1348-0421.2001.tb01322.x. [DOI] [PubMed] [Google Scholar]
- 148.Pang PS, Planet PJ, Glenn JS. The evolution of the major hepatitis C genotypes correlates with clinical response to interferon therapy. PLoS ONE. 2009;4:e6579. doi: 10.1371/journal.pone.0006579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–10. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 150.Sarasin-Filipowicz M, Wang X, Yan M, Duong FH, Poli V, Hilton DJ, Zhang DE, Heim MH. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–51. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dill MT, Makowska Z, Duong FH, Merkofer F, Filipowicz M, Baumert TF, Tornillo L, Terracciano L, Heim MH. Interferon-gamma-stimulated genes, but not USP18, are expressed in livers of patients with acute hepatitis C. Gastroenterology. 2012;143:777–86. doi: 10.1053/j.gastro.2012.05.044. e1-6. [DOI] [PubMed] [Google Scholar]
- 152.McGilvray I, Feld JJ, Chen L, Pattullo V, Guindi M, Fischer S, Borozan I, Xie G, Selzner N, Heathcote EJ, Siminovitch K. Hepatic Cell-Type Specific Gene Expression Better Predicts HCV Treatment Outcome Than IL28B Genotype. Gastroenterology. 2012;142:1122–1131. doi: 10.1053/j.gastro.2012.01.028. e1. [DOI] [PubMed] [Google Scholar]