Abstract
Hepatitis C virus (HCV) is a global health burden with an estimated 170–200 million peoples chronically infected worldwide. HCV infection remains as an independent risk factor for chronic hepatitis, liver cirrhosis, hepatocellular carcinoma, and a major reason for liver transplantation. Discovery of direct acting antiviral (DAA) drugs have shown promising results with more than 90% success rate in clearing the HCV RNA in patients, although long-term consequences remain to be evaluated. microRNAs (miRNAs) are important players in establishment of HCV infection and target crucial host cellular factors needed for productive HCV replication and augmented cell growth. Altered expression of miRNAs is involved in the pathogenesis associated with HCV infection by controlling signaling pathways such as immune response, proliferation and apoptosis. miRNA is emerging as a means of communication between various cell types inside the liver. There is likely possibility of developing circulating miRNAs as biomarkers of disease progression and can also serve as diagnostic tool with potential of early therapeutic intervention in HCV associated end stage liver disease. This review focuses on recent studies highlighting the contribution of miRNAs in HCV life cycle and their coordinated regulation in HCV mediated liver disease progression.
Keywords: Circulatory miRNAs, HCV, Interferon signaling, Liver disease, MicroRNA
Introduction
Hepatitis C virus (HCV) is a hepatotropic, enveloped, single stranded and positive sense RNA virus belongs to family flaviviridae and genus hepacivirus. The viral genome contains 5′ and 3′ untranslated regions (UTR) that are important for viral replication and translation. An internal ribosome entry site (IRES) directs the synthesis of a single precursor polyprotein of approximately 3010 amino acids, which is cleaved by viral and cellular proteases into three structural (core, E1 and E2) and seven non-structural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins. Core protein forms the capsid, which is surrounded by a lipid bilayer containing the glycoproteins, E1 and E2.1 These viral proteins both singly or in coordinated manner interact with host cellular factors and regulate various signaling pathways to facilitate virus mediated persistent infection.2
HCV is a major cause of chronic liver disease, mostly asymptomatic in nature. Majority of infected patients, approximately 80%, develop persistent chronic infection and are at high risk for liver cirrhosis and hepatocellular carcinoma (HCC). An estimated 170–200 million peoples worldwide are infected with hepatitis C virus3 and about 2.7–3.9 million peoples are living with HCV infection in the United States.4 In addition, HCC and cirrhosis have been increasing among persons infected with HCV.5, 6 Advances in anti-HCV therapy have moved into a new era with interferon (IFN)-free regimens. Treatment options are evolving and the once difficult to treat genotype (genotype 1) has shown significant improvements in sustained virologic response (SVR) rates. All-oral, IFN-free combinations of drugs are expected to cure more than 90% of infections.7, 8, 9 Direct-acting antivirals (DAAs) target non-structural proteins of HCV resulting in the termination of viral replication. Several antiviral drugs targeting viral and host factors essential for productive HCV infection are depicted in Fig. 1. The first NS3/4A protease inhibitors boceprevir (Merck, Whitehouse Station, NJ) and telaprevir (Vertex Pharmaceuticals, Cambridge, MA) have shown improved rate of SVR in genotype 1 HCV infected patients in compared with standard pegylated interferon (PEG-IFN) and ribavirin (RBV) therapy, but their toxicities combined with PEG-IFN and RBV limited their overall efficacy. Simeprevir (Janssen Pharmaceuticals, Titusville, NJ), faldaprevir, and asunaprevir are second-wave, first-generation NS3/4A inhibitors that have already been or will soon be approved. Second-generation protease inhibitors are in clinical trials. Daclatasvir (BMS-790052, Bristol-Myers Squibb Company) is the first approved DAA belonging to the class of NS5A replication complex inhibitors. The potency of daclatasvir is very high, and this drug is an important component of combination regimens for all genotypes. Sofosbuvir, the first approved NS5B polymerase inhibitor (Gilead Sciences, Foster City, CA) has shown high potency and lower drug resistance in clinical trials. DAA drugs as anti-HCV therapeutics is highly encouraging.7, 8 However, the efficacy of these new therapeutic options for cirrhotic patients, the most-difficult-to-treat population and long-term follow-up data will be needed to confirm the excellent outcome of SVR and liver pathogenesis. An alternative or complementary approach to treat HCV infection is by targeting host factors that support viral life cycle. Identifying host factors have emerged as a promising alternative since they will have a high barrier for viral resistance and can be broadly effective against all HCV genotypes.10 Several host factors were identified by using high throughput gene silencing screening approaches that helps in HCV entry, replication, assembly and release. The most promising host factors are miR-122, cyclophilin A (Cyp A), scavenger receptor (SR)-BI and phosphatidylinositol-4-kinase III α. Drugs targeting HCV entry factors, such as SR-BI may prevent the initiation of new infection. Cyclophilins are important host factors required for viral replication and Cyp A has been shown to interact with NS5A.10, 11 The liver specific miR-122 binds to the 5′ UTR of the HCV genome and helps in viral replication and enhances viral protein synthesis.12, 13, 14, 15 Drugs targeting host factors, such as small molecule inhibitor of SR-BI (ITX 5061) and cyclophilin inhibitors, alisporivir (Debio-025, Novartis) are in clinical trials.7, 10, 11 Anti-HCV therapeutics targeting miR-122 showed reduction in HCV viremia with no evidence of viral resistance and minimal side effects in chimpanzees chronically infected with hepatitis C.16 In addition, Miravirsen (a locked nucleic acid-modified antisense oligonucleotide for miR-122) showed prolonged dose-dependent reductions in HCV RNA levels in chronic HCV genotype 1 infected patients in Phase IIa clinical trials by Santaris Pharma (Copenhagen, Denmark).17 These alternative approaches targeting host factors in combination with current DAA drugs will help in strengthening the host's innate immunity as well as interfere with host factors required for HCV induced pathogenesis.
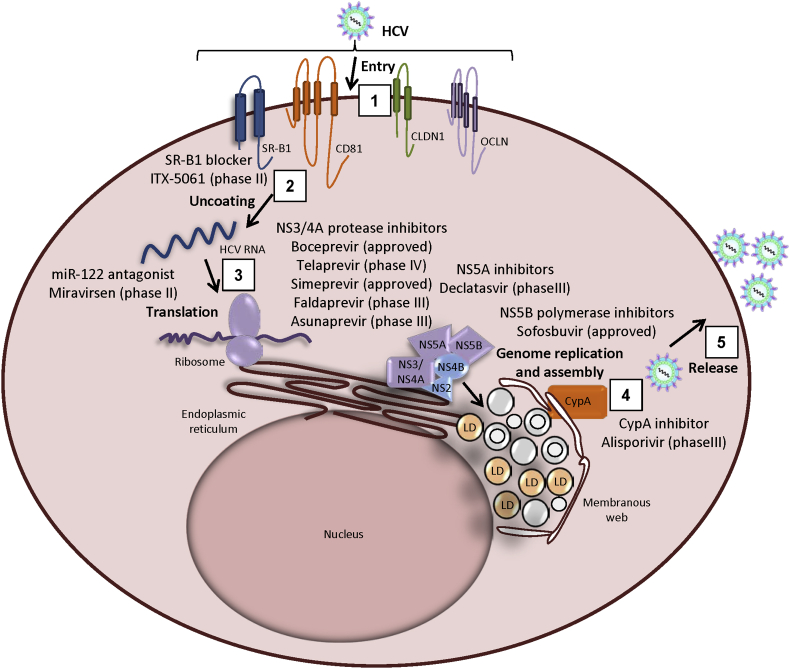
Figure 1.
Anti-HCV drugs targeting viral life cycle. (1) HCV viral particle interacts with several entry factors such as, SR-B1, CD81, CLDN1 and OCLN to enter the hepatocytes. Entry inhibitor, ITX-5061 inhibits the uptake of viral particles through SR-B1. (2) Once inside the cell, virus releases its genome by the process of uncoating. (3) The viral RNA is translated and the resulting polyprotein is processed at the endoplasmic reticulum. NS3/4A protease inhibitors inhibit viral protein synthesis and impair interferon signaling pathway. (4) Replication takes place in ER-derived membrane compartment, called as membranous web composed of single-, double-, multi-membraned vesicles and lipid droplets. miR-122 binding to viral genome enhances viral replication and translation. Drugs targeting NS5A and NS5B block viral replication. CypA interacts with NS5A and required for viral replication. Nucleocapsid is formed after interaction of core and NS5A protein with lipid droplets. The p7, NS2 and NS3-NS4A proteins are also involved in coordination of virus assembly. (5) HCV virion morphogenesis is coupled to the very-low density lipoproteins (VLDL) secretory pathway, and virus particles are released as lipoviroparticles (LVPs). Viral proteins and host factors that are the targets of direct-acting antivirals (DAAs) in advanced clinical development are indicated in the legend. CLDN1, claudin 1; CypA, cyclophilin A; miR, microRNA; OCLN, occludin; SR-B1, scavenger receptor class B member 1; LD, lipid droplets.
MicroRNA biogenesis and its regulation
MicroRNAs (miRNAs) were discovered in 1993 during a developmental timing experiment in the nematode Caenorhabditis elegans. Currently, human miRNA family has expanded to 2588 mature miRNAs (miRBase v21.0; http://www.mirbase.org/) and in silico prediction estimates that approximately 60% of human message RNA (mRNA) could be targets of miRNA. These miRNAs account for only 1% of the human genome. miRNAs are highly conserved in nearly all organisms and constitute a class of non-coding RNAs, about 18–22 nucleotides long and play a crucial role in the regulation of gene expression.18, 19 Genes encoding miRNAs are transcribed by RNA polymerase II and form transcripts as primary miRNAs (pri-miRNAs). pri-miRNAs are processed by ribonuclease Drosha to produce precursor miRNAs (pre-miRNAs) which is exported into the cytoplasm and cleaved by the ribonuclease Dicer to produce mature, single stranded miRNAs.19, 20, 21 Once synthesized, mature miRNA binds to two proteins, GW182 and Argonaute/EIF2C (AGO) family proteins and forms a complex called miRNA induced silencing complex (miRISC) and mediate the target mRNA recognition (Fig. 2). miRNA regulation takes place at multiple steps, including their transcription, their processing by Drosha and Dicer, their loading onto AGO proteins and miRNA turnover.20, 21 miRNA transcription is controlled by RNA Pol II-associated transcription factors and epigenetic regulators. Transcription factors, such as p53, MYC, ZEB1 and ZEB2, and myoblast determination protein 1 (MYOD1) positively or negatively regulate miRNA expression. Epigenetic control, such as DNA methylation and histone modifications also contribute to miRNA gene regulation. miRNA identify target mRNA through specific base-pairing interactions between the 5′ end of miRNA and sites within coding region and UTRs especially 3′ UTR of mRNAs. The domain at the 5′ end of miRNAs that spans from nucleotide position 2 to 7 is crucial for target recognition and has been termed the ‘miRNA seed’. The downstream nucleotides of miRNA (particularly nucleotide 8 and less importantly nucleotides 13–16) also contribute to base pairing with the targets. miRNAs with almost identical sequences at their 5′ ends forms miRNA seed families and they share targets. For example, miR-17, miR-20 and miR-106 belong to the same family by sharing a common seed sequence and they target a common gene, such as the cyclin-dependent kinase inhibitor 1A (CDKN1A; also known as p21). Moreover, 64% of the human miRNAs are part of multimember seed families and therefore, co-expression of seed-related miRNAs induces a stronger downregulation of their common targets.22 miRNA inhibits the target gene expression either by mRNA degradation or translational repression. The incomplete complementary binding leads to repression of translation or deadenylation of the target mRNA, whereas a complete complementary binding leads to degradation of the target mRNA. miRNA promotes mRNA cleavage by inducing deadenylation or suppressing protein synthesis by repressing the translation initiation at the cap recognition or inducing ribosomes to drop off prematurely.19, 20, 21, 23 Paradoxically, miRNA can also activate gene expression by targeting gene regulatory sequences. miR-10a interacts with the 5′ UTR of mRNAs encoding ribosomal proteins to enhance their translation.24 A putative target site for miR-373 has been identified in the promoter of E-cadherin and miR-373 overexpresssion has been shown to induce E-cadherin expression in prostate cancer cell line.25 In another report, miR-369-3 is shown to be involved in the recruitment of Ago and fragile X mental retardation related protein 1 (FXR1) genes and enhances the translation of tumor necrosis factor (TNF) mRNA during cell cycle arrest.26 A combinatorial nature of miRNA regulation i.e., each miRNA regulates hundreds of different mRNAs allow miRNA to be a part of complex regulatory networks in controlling gene expression in almost every biological process including development, immune response, aging, cell proliferation and apoptosis.
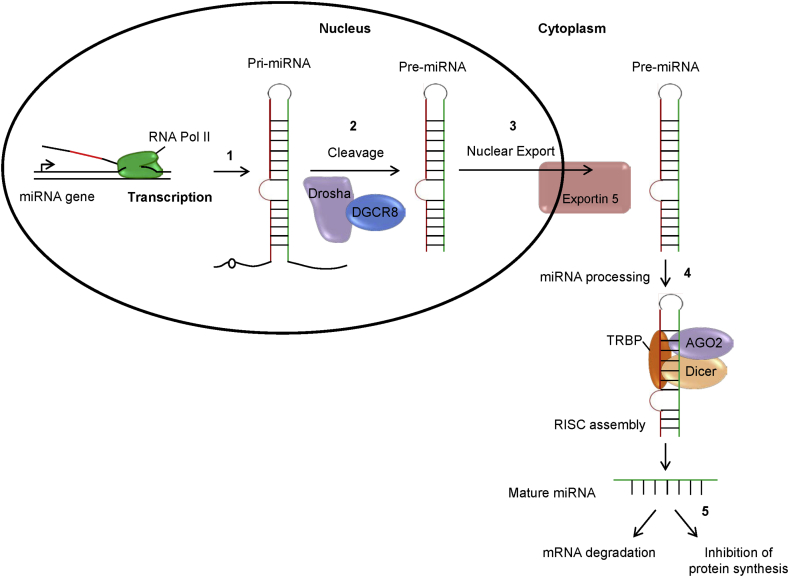
Figure 2.
miRNA biogenesis and regulation of gene expression. MicroRNAs (miRNAs) are small non-coding RNA synthesized from protein coding genes or introns with the help of RNA polymerase II. (1) First, the miRNA gene transcribed to a primary long transcript with stem loop structure as pri-miRNA. (2) This pri-miRNA is processed by RNase III family of enzymes, Drosha with the help of double stranded RNA binding protein, DGCR8 and produce small ∼70-nucleotide precursor hairpin structure as precursor miRNA (pre-miRNA). (3) Pre-miRNA then transported to the cytoplasm with the help of exportin5 protein. (4) Pre-miRNA was further cleaved by Dicer together with transactivation-responsive (TAR) RNA-binding protein TRBP, in the cytoplasm and generate a ∼20-bp miRNA/miRNA* duplex. Following processing, one strand of the miRNA/miRNA*duplex (the guide strand) is preferentially loaded into the miRNA-induced silencing complex (miRISC) containing argonaute 2 (AGO2) and form mature miRNA. (5) Then, the mature miRNA targets specific messenger RNA (mRNA) at the seed region that lead to either mRNA degradation or inhibition of protein synthesis.
Many miRNA genes are located in chromosomal regions frequently involved in chromosomal alterations such as deletion or amplification during tumor development.27 Therefore, it is not surprising that dysregulation of miRNA networks have been associated with cancer progression. Virus-host interactions also involve several regulatory steps to control gene expression and one of them is changes in cellular miRNA expression profiles. Cellular miRNAs control protein expression that may influence cellular tropism of viruses, modulate viral infectivity, and play a crucial role in inducing appropriate antiviral immune responses.28 Several RNA viruses have evolved mechanisms to degrade, boost, or hijack cellular miRNAs to benefit the viral life cycle.29 HCV infection exerts a profound effect on the expression of cellular miRNAs.30 Some of the cellular miRNAs affect viral replication directly by binding to the viral genome or indirectly by targeting host factors.
Direct interaction of cellular miRNAs to the HCV genome
Recent studies have identified several miRNAs as key players in virus-host interactions, regulating virus replication and pathogenesis during HCV infection. The role of miR-122 in HCV infection was first demonstrated by sequestration of endogenous miR-122 that led to a substantial reduction in HCV RNA abundance.12 miR-122 binds directly to 5′ UTR of the virus genome at two adjacent sites in association with Ago2. It forms an oligomeric complex in which one miR-122 molecule binds to the 5′ UTR of HCV RNA with 3′ overhanging nucleotides, masking the 5′ terminal sequences from nucleolytic degradation, thereby promoting viral RNA stability and propagation of HCV genome.14, 31 Furthermore, specific internal nucleotides as well as 3′ terminal nucleotides in miR-122 were absolutely required for maintaining HCV RNA.31 Recent study also demonstrated that miR-122 protects HCV RNA from 5′ decay by targeting 5′ exonuclease Xrn1.32 HCV genome harbors two more miR-122 target sites, one in the variable region of the 3′UTR and the other in the NS5B coding region. However, miR-122 binding to NS5B and 3′UTR impairs HCV RNA replication and translation.33 Exogenous expression of miR-122 allows efficient HCV RNA replication and/or infectious virion production in non-permissive cell line.34, 35, 36 Besides miR-122, other miRNAs such as, miR-448, miR-196, miR-199a, let-7b and miR-181c have been reported to interact directly with HCV genome, however, upon binding to HCV RNA, they inhibit HCV replication (Fig. 3). Overexpression of miR-448 and miR-196 were able to substantially attenuate viral replication by directly targeting Core and NS5A coding region of the HCV genome, respectively.37 Overexpression of miR-199a inhibited HCV replication in cells bearing HCV-1b or -2a genome length replicon on binding to stem-loop II region of 5′UTR of HCV genome.38 Let-7b was also reported to directly target HCV genome and elicits anti-HCV activity.39 Mutational analysis identified let-7b binding sites at the coding sequences of NS5B and 5′-UTR of HCV genome that were conserved among various HCV genotypes. We have recently showed that miR-181c is a novel miRNA that binds to E1 and NS5A regions of HCV genome and overexpression of miR-181c reduces the viral replication.40 However, miR-181c binding efficiency to genotype 1a and genotype 2a specific HCV genome is different. Thus, further work is needed to understand the association between different genotypes of HCV genome and miRNAs.
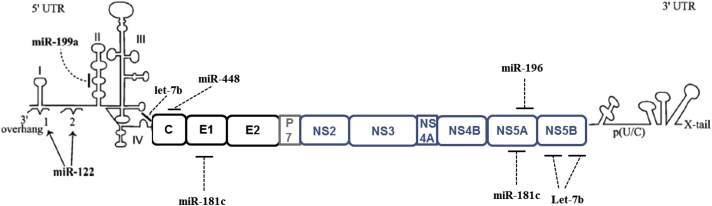
Figure 3.
Cellular miRNAs targeting HCV genome. The binding of two copies of miR-122 within the 5′ UTR of the HCV genome enhances viral replication and translation. The binding of miR-199a at 5′UTR inhibits viral replication. Let-7b interacts with viral genome at two different locations, 5′UTR and NS5B coding region. miR-448 targets Core and miR-196 targets NS5A of the HCV genome. miR-181c inhibits viral replication by targeting E1 and NS5A regions of viral genome.
Cellular miRNAs that regulates HCV replication by targeting interferon signaling pathway
HCV has developed several strategies to evade the IFN signaling pathway to facilitate its own replication.41 HCV evades type I IFN pathway by inducing miRNAs that regulate the expression of target genes involved in innate immune response to viral infections. The direct correlation of cellular miRNA in regulating type I IFN signaling pathway to promote HCV replication was demonstrated by our group. We established that HCV induces the expression of miR-130a to evade IFN response by targeting IFITM1. Our study demonstrated that miR-130a expression was upregulated in liver biopsy from HCV infected patients as well as in HCV infected hepatocytes in vitro.42 Knockdown of miR-130a enhances IFITM1 expression that possesses anti-HCV activity.43 During IFN treatment, IFITM1 accumulates at hepatic tight junctions in the liver of HCV-infected patients and then, interacts with HCV co-receptors, including CD81 and occludin, to disrupt the process of viral entry.44 Similar observation of reduced HCV RNA copies was reported in anti-miR-130a transfected virus infected cells.45 Subsequently, miR-130a expression was correlated with genes involved in transforming growth factor beta (TGF-β) signaling pathway and found to be reduced in HCV infection and upregulated on IFN treatment.45 However, another study reported that overexpression of miR-130a inhibited HCV replication by restoring the expression of endogenous IFN-α and IFN-β and interferon stimulated genes, MxA, ISG15 and USP18 respectively, in TLR3 and RIG-I deficient hepatocytes.46 Upregulated miR-21 suppressed MyD88 and IRAK1 expression in HCV infected hepatocytes, which subsequently repressed type I IFN effector gene expression and the type I IFN-mediated antiviral response, thereby promoting viral replication.47 In a separate study, upregulation of miR-758 expression was reported in HCV infected patients and shown to be negatively correlated with decrease in TLR3 and TLR7 expression levels and thereby, reduced IFN-α and IFN-β production to impair innate immune response.48 Overexpression of miR-122 has also been associated with inhibition of IFN signaling pathway. Silencing of miR-122 enhances IFN-induced ISRE activity by decreasing expression of SOCS3. This decrease in SOCS3 level was also regulated by enhanced methylation at SOCS3 gene promoter, implicating additional mechanism of inhibition of HCV replication using antisense oligonucleotides of miR-122.49 Recently, SOCS1 and SOCS3 were identified as the targets of miR-221 and overexpression of miR-221 was shown to accelerate anti-HCV effect of IFN-α in HCV infected hepatocytes.50 IFN-α treatment also modulates HCV-specific miRNAs expression in hepatocytes. miR-324-5p and miR-489 were shown to be upregulated in the presence of IFN-α while differential expression of miR-30c and miR-130a were observed between HCV-infected Huh7.5 cells treated with or without IFN-α.45 miR-30 cluster targets SOCS1 and SOCS3 genes that act as negative regulators of cytokine signaling. Specifically, SOCS1 and SOCS3 inhibit JAK tyrosine kinase activity and STATs in the JAK-STAT signaling pathway suggesting that IFN-α induced miRNAs modulates gene expression in HCV infected hepatocytes.45 IFN-β treatment of Huh7 cells showed an upregulation of miR-142-3p and miR-128a, and these miRNAs were downregulated in HCV replicon-expressing cells.51 IFN-β induced miRNAs, in conjunction with downregulation of miR-122, were also studied to prevent HCV replication. Introduction of anti-miRs against miR-196, miR-296, miR-351, miR-431 and miR-448, with and without the inclusion of miR-122 mimic, attenuated the IFN-β mediated reduction of viral RNA by ∼75%.37 In a recent study, a set of 750 miRNAs expression profiles was generated in response to IFN-α and interleukin (IL)-28B treatment to hepatocytes. Let-7b was shown to inhibit HCV replication and viral protein translation by targeting host factor insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1). Furthermore, inhibition of let-7b attenuated the anti-HCV effects of IFN-α and IL-28B.52 We recently observed that miR-373 was upregulated during HCV infection and negatively regulated type I IFN signaling pathway by suppressing JAK1 and IRF9 (unpublished observation). Together, these observations suggest that type I and type III IFNs induced host miRNAs directly involved in regulation of HCV replication and infectivity along with targeting host factors resulting in additional mode of virus restriction.
Cellular miRNAs in response to IFN therapy and HCV infection
Increasing evidence suggests that miRNAs have a profound impact on host defense to HCV infection and clinical outcome of standard HCV therapy. miRNA expression profiles were examined to identify the miRNAs associated with the standard treatment (IFN-α with RBV) to chronic hepatitis C (CHC) patients. Expression levels of 9 miRNAs (upregulated: miR-27b, miR-122, miR-378 and miR-422b; downregulated: miR-18a, miR-34b, miR-143, miR-145 and miR-652) were significantly different in the SVR and non-responder (NR) groups, suggesting that expression pattern of these hepatic miRNA are associated with the therapeutic outcome in CHC patients.53 The expression level of hepatic miR-122 was reportedly associated with early response to IFN treatment. HCV infected patients who did not respond to therapy had significantly lower miR-122 levels as compared to responder.54 In addition, an association between miR-122, IFN-λ3 polymorphism and outcome to IFN therapy was examined in chronic HCV patients. HCV infected patients with IFN-λ3 CC genotype, favorable outcome to therapy, were associated with increased hepatic expression of miR-122. Interestingly, during PEG-IFN/RBV therapy, patients with the IFN-λ3 CC genotype had a more rapid early HCV viral decline than patients with CT/TT genotype and a stronger association of miR-122 was observed with complete early virologic response (cEVR) than with SVR. This implies that miR-122 expression may play an important role during early viral decline induced by the innate immune response.55 In a recent study, the kinetics of serum levels of miR-122 was determined during IFN therapy. miR-122 expression in sera remained low in SVR, but increased to baseline levels in patients not responding or showing relapse to therapy, suggesting that serum miR-122 levels reflect the response rate to IFN/RBV therapy in chronic HCV patients.56 Higher levels of miR-27a displayed favorable response to IFN-α and RBV combination therapy.57 Serum levels of miR-181a were significantly elevated in SVR patients following treatment compared to NR patients and treatment-naïve SVRs.58 IFN-λ3 expression is strongly associated with favorable response to IFN-α and RBV in HCV infected patients. However, HCV infection induces the expression of miR-208b and miR-499a-5p, which in turn downregulates IFN-λ3 expression. The unfavorable IFN-λ3 polymorphism at rs4803217 SNP: T genotype was targeted by miR-208b and miR-499a-5p and decreases IFN-λ3 mediated antiviral response. Inhibition of miR-208b and miR-499a-5p represents a therapeutic option to improved response rate to IFN-α and RBV with unfavorable IFN-λ3 genotype.59 Hepatic miR-155 expression was significantly elevated in HCV-infected patients. Lower viral load in sera and SVR to standard treatment was found to be associated with elevated levels of miR-155 in HCV infected patients. In addition, miR-155 was significantly elevated in HCV infected patients harboring the favorable IL-28B CC allele (rs1279860).60 Treatment-naïve patients with chronic HCV infection were shown to have higher expression of miR-155 in their circulating monocytes as compared to individuals who cleared HCV infection after therapy, suggesting a possible correlation between increased miR-155 in monocytes and HCV viral presence and/or replication.61 Expression profiling of peripheral blood mononuclear cells (PBMC) associated miRNAs was also compared between responder and non-responders to PEG-IFN/RBV antiviral therapy for chronic HCV patients. Circulating PBMC associated miR-125b was a predictive marker for SVR with significantly reduced levels of miR-125b in responders versus non-responders.62
Cellular miRNAs in HCV mediated liver disease progression
HCV modulates the expression of miRNAs to regulate critical gene networks that help in determining viral load, clearance and HCV related disease progression. Recently, in vivo kinetics of hepatic and serum miR-122 and viral replication was studied in HCV infected chimpanzees to understand the complexities of the virus-host interaction during the acute phase of HCV infection. It was observed that during acute infection (first 4 weeks) there was rise in hepatic miR-122 levels indicating the significance of miR-122 in viral replication. However, in later time points (10–14 weeks), an inverse correlation between hepatic miR-122 and HCV RNA in the liver and serum was noted. Subsequently, there is rise of hepatic miR-122 level during HCV clearance and alanine leucine transaminase (ALT) normalization suggesting a tri-phasic relationship among hepatic miR-122 expression and viral RNA levels during HCV infection.63 Apart from regulating viral replication, miR-122 is also involved in cell cycle progression in hepatoma cell line by regulating viral translation.64 Ectopic expression of miR-122 promotes HCV IRES dependent translation during the G₀ and G₁ phase of cell cycle.64 Alcohol consumption increases viral replication by regulating miR-122 and cyclin G1 expression and use of miR-122 inhibitor has been reported to prevent alcohol-induced increase in HCV RNA and protein levels.65 In addition, alcohol consumption enhances miR-122 expression by increasing the expression levels of GW182 and heat shock protein 90 further promoting HCV replication.66 Overexpression of miR-122 also decreases HCV entry into hepatocytes through down-regulation of occludin.67 Apart from directly targeting the viral genome, miR-196a targets Bach1, a repressor of anti-oxidative and anti-inflammatory heme oxygenase1 (HMOX1) and inhibits HCV RNA and NS5A protein expression in subgenomic replicon.68 In addition, higher HMOX1 expression was correlated with higher expression of Bach1 and miR-122 in chronic HCV patients.69 Several miRNA profiling studies have demonstrated alterations in miRNAs expression and identified miRNA-mRNA regulatory networks during HCV infection.51, 70, 71, 72 Differential upregulation of hsa-miR-130a, hsa-miR-130b, hsa-miR-298, hsa-miR-193a-5p and hsa-miR-371-5p were observed in HCV Con1 replicon in comparison to control cells. These miRNAs have been associated with cell growth by targeting genes PPARG, IRF1 and STAT3 in HCV infected cells.71 Differential expression of miRNAs such as, miR-24, miR-149, miR-638 and miR-1181 were also identified following HCV infection and are involved in HCV entry, replication and propagation.72
Chronic HCV infection is closely associated with hepatic inflammation, fibrosis, steatosis, liver cirrhosis and HCC. Chronic HCV infection induced liver fibrosis is mediated by upregulation of TGF-β.73 TGF-β signaling activates hepatic stellate cells (HSCs) to induce extracellular matrix production. In HCV-infected patients and in a mouse carbon tetrachloride fibrosis model, expression levels of miR-21 were higher and found to be positively correlated with fibrotic stage.74 miR-21 was shown to target SMAD7, a negative regulator of TGF-β signaling, leading to increased fibrogenesis.74 In HCV infected patients, lower expression levels of miR-29 was observed in liver and overexpression of miR-29 inhibits viral RNA in HCV infected hepatocytes.75 Inhibition of miR-29 was also linked with activation of HSCs and collagen synthesis.75 Recently, upregulation of miR-200c was linked with hepatic fibrosis in HCV patients. miR-200c reduces the expression of Fas associated phosphatase 1 (FAP1) and subsequent activation of Src kinase signaling increases the expression of collagen and fibroblast growth factor involved in fibrosis.76 Reduced expression of miR-449a and miR-107 was observed in chronic HCV patients but not in alcoholic and non-alcoholic liver diseases by genome wide miRNA analyses.77, 78 miR-449a regulates the expression of YKL40 by targeting NOTCH signaling pathway following HCV infection.77 Elevated levels of YKL40 were observed in chronic HCV patients and YKL40 is known to promote the synthesis of extracellular matrix and fibrosis. Downregulation of miR-107 and miR-449a modulates the expression of CCL2 chemokine by targeting components of the interleukin-6 receptor (IL-6R) complex in patients with HCV related liver disease. miR-449a and miR-107 target IL-6R and JAK1 respectively and inhibit IL-6 signaling and impair STAT3 activation in human hepatocytes.78 Collectively, these reports suggest the role of miRNAs in targeting genes involved in linking inflammation and fibrosis in chronic HCV infection. The incidence of hepatic steatosis is higher in patients infected with genotype 3a of HCV.79 The role of miR-27 was implicated in HCV associated steatosis by two separate groups. In the first study, miR-27a was reported to targets lipid metabolism related transcription factor, RXRα and lipid transporter, ABCA1 resulting in lipid accumulation and HDL synthesis in virus infected hepatocytes.57 In a second study, miR-27b also induced lipid droplet accumulation in HCV infected hepatocytes by targeting peroxisome proliferator-activated receptor (PPAR-α) and angiopoietin-like protein 3 (ANGPTL3).80 miR-27a repression increased the cellular lipid content, decreased the buoyant density of HCV particles and increased viral replication and infectivity.57 However, miR-27a expression was induced by HCV infection and HCV replication was hampered by miR-27a reflecting a negative feedback role in HCV infection. In a separate study, upregulation of miR-136 and downregulation of miR-126 and miR-181a was observed in liver biopsies of HCV infected patients with different genotypes compared with noninfected individuals. Interestingly, miR-122 and miR-126 expression levels were correlated with viral load in sera and miR-136 and miR-122 correlated with the presence of steatosis.81
CHC is a major risk factor associated with HCC.82 miRNA dysregulation has been linked with initiation and progression of HCC,83, 84, 85 however, the role of miRNAs in HCV-related HCC is poorly understood. The identification of HCC related miRNA signatures is of great value for the early diagnosis of HCC, prior to the onset of disease in HCV-positive patients. HCV modulates miRNA expression to facilitate hepatocyte growth towards tumorigenesis by regulating various signaling pathways. We have observed reduced expression of miR-181c at the transcriptional level in HCV infected hepatocytes. miR-181c targets HOXA1 to promote hepatocyte growth by modulating Stat3 and Stat5 expression during HCV infection.40 miR-155 expression levels were markedly increased in patients infected with HCV. Overexpression of miR-155 promoted the cell proliferation through activation of β-catenin and a concomitant increase in cyclin D1, c-myc, and survivin. miR-155 also reduces the expression of adenomatous polyposis coli (APC), a negative regulator of Wnt signaling, to promote hepatocyte proliferation and tumorigenesis.86 In contrast, miR-141 inhibits the expression of tumor suppressor gene, DLC-1 (a Rho GTPase-activating protein) to enhance viral replication and tumorigenesis in HCV-infected primary human hepatocytes.87 HCV core protein is a known oncogene involved in HCV mediated tumorigenesis. In a recent study, core overexpression significantly reduces miR-152 expression in HepG2 cells. miR-152 directly target Wnt1, a activating ligand for β-catenin pathway therefore, inhibiting Wnt signaling.88 Reduced expression of miR-491 promotes tumorigenesis by inhibiting phosphoinositol-3 kinase (PI3K)-Akt prosurvival pathway in chronic HCV infection. miR-192/miR-215 and miR-491 are capable of enhancing HCV replication in replicon cells.89 Limited studies are available addressing the role of miRNA expression in HCV associated HCC. Differential miRNA expression from formalin fixed paraffin embedded HCV infected HCC specimens indicated 10 upregulated and 19 downregulated miRNAs.90 Another study in HCV infected patients, 13 miRNAs were shown to be downregulated and were predicted to target genes related to immune response, antigen presentation, cell cycle, proteasome, and lipid metabolism signaling pathways.91 However, validation of these miRNAs and their predicted targets are needed to elucidate the conclusive role of particular miRNA in HCV related HCC.
Circulatory miRNAs as biomarkers in HCV infection
One of the major challenges in HCV research is the detection of early stage liver disease which will allow for rapid intervention and improved outcome of antiviral treatment. Non-invasive or minimally invasive methods need to be developed to evaluate disease severity and the likelihood of disease progression. Circulating miRNAs might have potential value as biomarkers for detection and as predictive marker for liver disease progression in HCV infection. Circulating miRNAs are released in extracellular space and carried in various forms, including in association with Ago2, exosomes or HDL in circulation.92, 93 The biological function of these circulating miRNA is still unknown in HCV infection. However, circulating miRNA might provide a means of communication to neighboring recipient cells and influence gene expression on target cells. To determine the potential of developing circulating miRNA as predictive biomarker in HCV related liver disease, we performed serum/plasma specific miRNA array and observed that several circulating miRNAs were significantly upregulated in sera of HCV infected patients as compared to healthy controls.94 We observed increased expression of miR-20a and miR-92a in sera specific to HCV associated liver disease but not in sera of patients with non-HCV related liver disease. Subsequently, we observed that elevated levels of miR-20a were positively correlated with disease severity in HCV infected patients, however, miR-92a expression was reduced with higher grade of fibrosis in HCV infected patients.94 Longitudinal sample analyses suggested that miR-20a expression remained higher when disease progresses from acute to chronic infection whereas, miR-92a expression declines in response to spontaneously resolved infection. miRNA profiling was also performed to identify the expression of 940 human miRNAs in the serum of HCV infected individuals. Serum levels of miR-134, miR-320c and miR-483-5p were significantly upregulated in HCV infected patients.95 Serum levels of miR-122 were correlated with disease parameters in patients with CHC by several groups. The higher levels of miR-122 and miR-192 were observed in sera from patients with CHC and in other etiologies associated with liver injury as compared to sera from healthy controls.96, 97, 98, 99, 100 The serum level of miR-122 and miR-21 strongly correlates with serum ALT levels and higher necroinflammatory activity in the liver of CHC patients suggesting their potential as better serum biomarker over ALT in predicting the presence of chronic HCV infection,56, 96, 97, 100, 101 although the specificity of miR-122 for HCV mediated liver disease is controversial. Reduced levels of miR-122 were observed in both serum and liver samples from patients with CHC with advanced fibrosis. During fibrosis progression, the number of hepatocytes decreases and numbers of lymphocytes, Kupffer cells, and HSCs increases. This may explain the reduced amount of miR-122 with hepatocyte loss at advanced stage of fibrosis.55, 102 miR-221 was found to be upregulated in the serum of CHC patients.50 miR-181a expression was significantly upregulated in the serum of HCV patients compared to the healthy controls.58 The serum miR-181a expression level in HCV patients is inversely correlated with the level of viremia, as well as liver enzymes (ALT, AST). Levels of miR-125b and miR-146a were also increased in the serum of CHC patients compared to healthy controls.61
Circulating miRNAs in urine were also examined for developing screening methods. Expression of three upregulated miRNAs, miR-625, miR-532 and miR-618, were evaluated as non-invasive biomarkers for early diagnosis of HCC among high-risk HCV positive patients. Elevated expression of miR-625, miR-532 and miR-618 were observed in 56%, 62.5% and 72% of HCC-post HCV positive patients, respectively. In addition, miR-516-5p and miR-650 were found to be down-regulated in 50% and 72% of HCC-post HCV positive patients, respectively. Differential expressions of these miRNAs were predicted to possibly target genes related to HCC development and progression in high risk HCV patients.103 Further studies are warranted to understand the function of these extracellular circulating miRNAs during HCV infection.
Conclusion
Cross-talk between miRNA and HCV is an emerging area of research. Understanding the potential of manipulating miRNAs as a therapeutic option either alone or in combination with currently available drugs to treat HCV infected patients is still in preliminary stage. Direct involvement of miR-122 in regulating viral replication and pathogenesis has led to the development of the very first miRNA based antiviral therapy. Therapeutic silencing of miR-122 showed long lasting antiviral activity in treatment of naïve HCV patients and no off-target effects in clinical trials. HCV infection modulates several miRNAs that regulate host immune response and cell growth interconnected with multiple signaling pathways (Fig. 4). miRNA mediated regulation of gene expression will help in identifying important signaling networks required towards HCV associated liver disease progression. Indeed, further in-depth studies are needed to identify the mechanistic insights behind modulated miRNAs by HCV infection. Therefore, the identification of miRNAs with a prominent role in HCV viral life cycle and its implication in liver disease progression is exploring a new frontier in the discovery of novel drugs against chronic HCV infections. MicroRNA in circulation is a growing area of research and there are challenges ahead in understanding the cellular origin and functional role of circulatory miRNAs in liver disease progression. Recent studies on circulating miRNAs generate a potential for development of predictive or diagnostic biomarkers to categorize patients with high risk groups for developing end stage liver disease during HCV infection. In addition, circulatory miRNAs is emerging as a tool to assess therapeutic outcome in response to therapy in HCV infected patients.
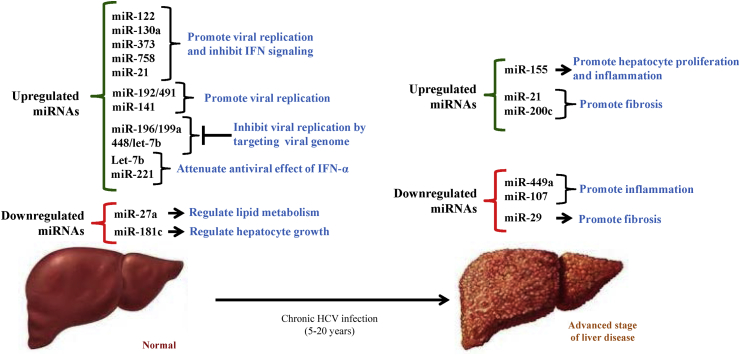
Figure 4.
Schematic representation of HCV infection associated miRNAs in liver disease progression. Modulated expression of miRNAs regulates the expression of genes involved in several signaling pathways such as, IFN signaling, hepatocyte growth, lipid metabolism, inflammation and fibrosis linked with HCV induced liver disease.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by research grant DK081817 (RBR) and DK080812 (RR) from the National Institutes of Health and SLU Liver Center Seed Grant (RBR).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Moradpour D., Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Shulla A., Randall G. Hepatitis C virus-host interactions, replication, and viral assembly. Curr Opin Virol. 2012;2:725–732. doi: 10.1016/j.coviro.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gravitz L. Introduction: a smouldering public-health crisis. Nature. 2011;474:S2–S4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- 4.Smith B.D., Morgan R.L., Beckett G.A., Centers for Disease Control and Prevention Recommendations for the identification of chronic hepatitis C virus infection among persons born during 1945–1965. MMWR Recomm Rep. 2012;61(RR-4):1–32. [PubMed] [Google Scholar]
- 5.Kanwal F., Hoang T., Kramer J.R. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C virus infection. Gastroenterology. 2011;140:1182 e1–1188 e1. doi: 10.1053/j.gastro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas D.L. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawlotsky J.M. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology. 2014;146:1176–1192. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 8.deLemos A.S., Chung R.T. Hepatitis C treatment: an incipient therapeutic revolution. Trends Mol Med. 2014;20:315–321. doi: 10.1016/j.molmed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Schinazi R., Halfon P., Marcellin P., Asselah T. HCV direct-acting antiviral agents: the best interferon-free combinations. Liver Int. 2014;34(suppl 1):69–78. doi: 10.1111/liv.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rupp D., Bartenschlager R. Targets for antiviral therapy of hepatitis C. Semin Liver Dis. 2014;34:9–21. doi: 10.1055/s-0034-1371006. [DOI] [PubMed] [Google Scholar]
- 11.Zeisel M.B., Lupberger J., Fofana I., Baumert T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C – perspectives and challenges. J Hepatol. 2013;58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 13.Henke J.I., Goergen D., Zheng J. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts A.P., Lewis A.P., Jopling C.L. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jangra R.K., Yi M., Lemon S.M. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanford R.E., Hildebrandt-Eriksen E.S., Petri A. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen H.L., Reesink H.W., Lawitz E.J. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 18.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 21.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 22.Hausser J., Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 23.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 24.Ørom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 27.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swaminathan G., Martin-Garcia J., Navas-Martin S. RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiol Genomics. 2013;45:1035–1048. doi: 10.1152/physiolgenomics.00112.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts A.P., Lewis A.P., Jopling C.L. The role of microRNAs in viral infection. Prog Mol Biol Transl Sci. 2011;102:101–139. doi: 10.1016/B978-0-12-415795-8.00002-7. [DOI] [PubMed] [Google Scholar]
- 30.Shrivastava S., Mukherjee A., Ray R.B. Hepatitis C virus infection, microRNA and liver disease progression. World J Hepatol. 2013;5:479–486. doi: 10.4254/wjh.v5.i9.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machlin E.S., Sarnow P., Sagan S.M. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci U S A. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasheri N., Singaravelu R., Goodmurphy M., Lyn R.K., Pezacki J.P. Competing roles of microRNA-122 recognition elements in hepatitis C virus RNA. Virology. 2011;410:336–344. doi: 10.1016/j.virol.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Narbus C.M., Israelow B., Sourisseau M. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kambara H., Fukuhara T., Shiokawa M. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J Virol. 2012;86:1382–1393. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuhara T., Kambara H., Shiokawa M. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J Virol. 2012;86:7918–7933. doi: 10.1128/JVI.00567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen I.M., Cheng G., Wieland S. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami Y., Aly H.H., Tajima A., Inoue I., Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a*. J Hepatol. 2009;50:453–460. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Cheng J.C., Yeh Y.J., Tseng C.P. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621–2633. doi: 10.1007/s00018-012-0940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee A., Shrivastava S., Bhanja Chowdhury J., Ray R., Ray R.B. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol. 2014;88:7929–7940. doi: 10.1128/JVI.00787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrivastava S., Ray R.B. Hepatitis C virus infection, autophagy and innate immune response. In: Hayat M.A., editor. vol. 3. Academic Press, Elsevier; 2014. pp. 164–190. (Autophagy. Cancer, Other Pathologies, Inflammation, Immunity, Infection and Aging). [Google Scholar]
- 42.Bhanja Chowdhury J., Shrivastava S., Steele R., Di Bisceglie A.M., Ray R., Ray R.B. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol. 2012;86:10221–10225. doi: 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkins C., Woodward J., Lau D.T. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Daucher M., Armistead D., Russell R., Kottilil S. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS One. 2013;8:e55733. doi: 10.1371/journal.pone.0055733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S., Duan X., Li Y., Liu B., McGilvray I., Chen L. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat. 2014;21:121–128. doi: 10.1111/jvh.12131. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y., Chen J., Wang H. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Q., Fu S., Wang J. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Arch Virol. 2014;159:2997–3003. doi: 10.1007/s00705-014-2167-3. [DOI] [PubMed] [Google Scholar]
- 49.Yoshikawa T., Takata A., Otsuka M. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep. 2012;2:637. doi: 10.1038/srep00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu G., Yang F., Ding C.L. MiR-221 accentuates IFN׳s anti-HCV effect by downregulating SOCS1 and SOCS3. Virology. 2014;462–463:343–350. doi: 10.1016/j.virol.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 51.Bruni R., Marcantonio C., Tritarelli E. An integrated approach identifies IFN-regulated microRNAs and targeted mRNAs modulated by different HCV replicon clones. BMC Genomics. 2011;12:485. doi: 10.1186/1471-2164-12-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng M., Si Y., Niu Y. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707–9718. doi: 10.1128/JVI.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami Y., Tanaka M., Toyoda H. Hepatic microRNA expression is associated with the response to interferon treatment of chronic hepatitis C. BMC Med Genomics. 2010;3:48. doi: 10.1186/1755-8794-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarasin-Filipowicz M., Krol J., Markiewicz I., Heim M.H., Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 55.Estrabaud E., Lapalus M., Broët P. Reduction of microRNA 122 expression in IFNL3 CT/TT carriers and during progression of fibrosis in patients with chronic hepatitis C. J Virol. 2014;88:6394–6402. doi: 10.1128/JVI.00016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Köberle V., Waidmann O., Kronenberger B. Serum microRNA-122 kinetics in patients with chronic hepatitis C virus infection during antiviral therapy. J Viral Hepat. 2013;20:530–535. doi: 10.1111/jvh.12075. [DOI] [PubMed] [Google Scholar]
- 57.Shirasaki T., Honda M., Shimakami T. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elhelw D.S., Mekky R.Y., El-Ekiaby N. Predictive prognostic role of miR-181a with discrepancy in the liver and serum of genotype 4 hepatitis C virus patients. Biomed Rep. 2014;2:843–848. doi: 10.3892/br.2014.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McFarland A.P., Horner S.M., Jarret A. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang M., Broering R., Trippler M. MicroRNA-155 controls Toll-like receptor 3- and hepatitis C virus-induced immune responses in the liver. J Viral Hepat. 2014;21:99–110. doi: 10.1111/jvh.12126. [DOI] [PubMed] [Google Scholar]
- 61.Bala S., Tilahun Y., Taha O. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsi E., Huang C.F., Dai C.Y. Peripheral blood mononuclear cells microRNA predicts treatment outcome of hepatitis C virus genotype 1 infection. Antiviral Res. 2014;105:135–142. doi: 10.1016/j.antiviral.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Choi Y., Dienes H.P., Krawczynski K. Kinetics of miR-122 expression in the liver during acute HCV infection. PLoS One. 2013;8:e76501. doi: 10.1371/journal.pone.0076501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fehr C., Conrad K.D., Niepmann M. Differential stimulation of hepatitis C virus RNA translation by microRNA-122 in different cell cycle phases. Cell Cycle. 2012;11:277–285. doi: 10.4161/cc.11.2.18699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou W., Bukong T.N., Kodys K., Szabo G. Alcohol facilitates HCV RNA replication via up-regulation of miR-122 expression and inhibition of cyclin G1 in human hepatoma cells. Alcohol Clin Exp Res. 2013;37:599–608. doi: 10.1111/acer.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bukong T.N., Hou W., Kodys K., Szabo G. Ethanol facilitates hepatitis C virus replication via up-regulation of GW182 and heat shock protein 90 in human hepatoma cells. Hepatology. 2013;57:70–80. doi: 10.1002/hep.26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sendi H., Mehrab-Mohseni M., Foureau D.M. miR-122 decreases HCV entry into hepatocytes through binding to the 3′ UTR of OCLN mRNA. Liver Int. 2014 doi: 10.1111/liv.12698. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 68.Hou W., Tian Q., Zheng J., Bonkovsky H.L. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494–1504. doi: 10.1002/hep.23401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jabłonowska E., Wójcik K., Szymańska B., Omulecka A., Cwiklińska H., Piekarska A. Hepatic HMOX1 expression positively correlates with Bach-1 and miR-122 in patients with HCV mono and HIV/HCV coinfection. PLoS One. 2014;9:e95564. doi: 10.1371/journal.pone.0095564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng X., Li Y., Walters K.A. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steuerwald N.M., Parsons J.C., Bennett K., Bates T.C., Bonkovsky H.L. Parallel microRNA and mRNA expression profiling of (genotype 1b) human hepatoma cells expressing hepatitis C virus. Liver Int. 2010;30:1490–1504. doi: 10.1111/j.1478-3231.2010.02321.x. [DOI] [PubMed] [Google Scholar]
- 72.Liu X., Wang T., Wakita T., Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 73.Schuppan D., Krebs A., Bauer M., Hahn E.G. Hepatitis C and liver fibrosis. Cell Death Differ. 2003;10(suppl 1):S59–S67. doi: 10.1038/sj.cdd.4401163. [DOI] [PubMed] [Google Scholar]
- 74.Marquez R.T., Bandyopadhyay S., Wendlandt E.B. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 75.Bandyopadhyay S., Friedman R.C., Marquez R.T. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis. 2011;203:1753–1762. doi: 10.1093/infdis/jir186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramachandran S., Ilias Basha H., Sarma N.J. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS One. 2013;8:e70744. doi: 10.1371/journal.pone.0070744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarma N.J., Tiriveedhi V., Subramanian V. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS One. 2012;7:e50826. doi: 10.1371/journal.pone.0050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarma N.J., Tiriveedhi V., Crippin J.S., Chapman W.C., Mohanakumar T. Hepatitis C virus-induced changes in microRNA 107 (miRNA-107) and miRNA-449a modulate CCL2 by targeting the interleukin-6 receptor complex in hepatitis. J Virol. 2014;88:3733–3743. doi: 10.1128/JVI.03060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goossens N., Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403–2412. doi: 10.1002/hep.26905. [DOI] [PubMed] [Google Scholar]
- 80.Singaravelu R., Chen R., Lyn R.K. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- 81.Boštjančič E., Bandelj E., Luzar B., Poljak M., Glavač D. Hepatic expression of miR-122, miR-126, miR-136 and miR-181a and their correlation to histopathological and clinical characteristics of patients with hepatitis C. J Viral Hepat. 2014 doi: 10.1111/jvh.12266. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 82.Raimondi S., Bruno S., Mondelli M.U., Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142–1154. doi: 10.1016/j.jhep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 83.Law P.T., Wong N. Emerging roles of microRNA in the intracellular signaling networks of hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26:437–449. doi: 10.1111/j.1440-1746.2010.06512.x. [DOI] [PubMed] [Google Scholar]
- 84.Huang S., He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giordano S., Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y., Wei W., Cheng N. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 87.Banaudha K., Kaliszewski M., Korolnek T. MicroRNA silencing of tumor suppressor DLC-1 promotes efficient hepatitis C virus replication in primary human hepatocytes. Hepatology. 2011;53:53–61. doi: 10.1002/hep.24016. [DOI] [PubMed] [Google Scholar]
- 88.Huang S., Xie Y., Yang P., Chen P., Zhang L. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by regulating Wnt1 in HepG2 cells. PLoS One. 2014;9:e81730. doi: 10.1371/journal.pone.0081730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ishida H., Tatsumi T., Hosui A. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011;412:92–97. doi: 10.1016/j.bbrc.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 90.Varnholt H., Drebber U., Schulze F. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 91.Ura S., Honda M., Yamashita T. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 92.Lemoinne S., Thabut D., Housset C. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. 2014;11:350–361. doi: 10.1038/nrgastro.2014.7. [DOI] [PubMed] [Google Scholar]
- 93.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 94.Shrivastava S., Petrone J., Steele R., Lauer G.M., Bisceglie A.M., Ray R.B. Upregulation of circulating miR-20a is correlated with hepatitis C virus mediated liver disease progression. Hepatology. 2013;58:863–871. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shwetha S., Gouthamchandra K., Chandra M., Ravishankar B., Khaja M.N., Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep. 2013;3:1555. doi: 10.1038/srep01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bihrer V., Friedrich-Rust M., Kronenberger B. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 97.Cermelli S., Ruggieri A., Marrero J.A., Ioannou G.N., Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Starkey Lewis P.J., Dear J., Platt V. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 99.Bala S., Petrasek J., Mundkur S. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Meer A.J., Farid W.R., Sonneveld M.J. Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122. J Viral Hepat. 2013;20:158–166. doi: 10.1111/jvh.12001. [DOI] [PubMed] [Google Scholar]
- 101.Bihrer V., Waidmann O., Friedrich-Rust M. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trebicka J., Anadol E., Elfimova N. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234–239. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 103.Abdalla M.A., Haj-Ahmad Y. Promising candidate urinary microrna biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis C virus egyptian patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]